Abstract
Several insect lineages have developed diverse strategies to sequester toxic pyrrolizidine alkaloids from food-plants for their own defense. Here, we show that in two highly divergent insect taxa, the hemimetabolous grasshoppers and the holometabolous butterflies, an almost identical strategy evolved independently for safe accumulation of pyrrolizidine alkaloids. This strategy involves a pyrrolizidine alkaloid N-oxygenase that transfers the pyrrolizidine alkaloids to their respective N-oxide, enabling the insects to avoid high concentrations of toxic pyrrolizidine alkaloids in the hemolymph. We have identified a pyrrolizidine alkaloid N-oxygenase, which is a flavin-dependent monooxygenase, of the grasshopper Zonocerus variegatus. After heterologous expression in E. coli, this enzyme shows high specificity for pyrrolizidine alkaloids of various structural types and for the tropane alkaloid atropine as substrates, a property that has been described previously for a pyrrolizidine alkaloid N-oxygenase of the arctiid moth Grammia geneura. Phylogenetic analyses of insect flavin-dependent monooxygenase sequences suggest that independent gene duplication events preceded the establishment of this specific enzyme in the lineages of the grasshoppers and of arctiid moths. Two further flavin-dependent monooxygenase sequences have been identified from Z. variegatus sharing amino acid identities of approximately 78% to the pyrrolizidine alkaloid N-oxygenase. After heterologous expression, both enzymes are also able to catalyze the N-oxygenation of pyrrolizidine alkaloids, albeit with a 400-fold lower specific activity. With respect to the high sequence identity between the three Z. variegatus sequences this ability to N-oxygenize pyrrolizidine alkaloids is interpreted as a relict of a former bifunctional ancestor gene of which one of the gene copies optimized this activity for the specific adaptation to pyrrolizidine alkaloid containing food plants.
Introduction
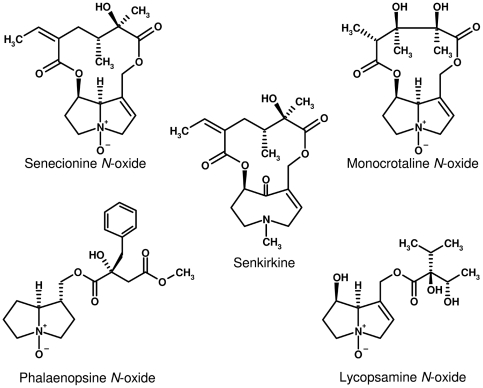
Chemical defense against herbivory is essential for plants to be able to survive in their natural habitat. During evolution, many insect herbivores have developed counterstrategies to cope with these toxic compounds. In some cases, they have even acquired these chemicals for their own benefit. One of the best studied examples of plant toxins sequestered by adapted insects are the pyrrolizidine alkaloids (PAs) that are found in certain lineages scattered within the angiosperms [1]. PAs occur in plants usually in their polar non-toxic N-oxide form (Figure 1). After ingestion by a vertebrate or insect herbivore, the N-oxides are easily reduced to the protoxic free base, the substrate for cytochrome P450-mediated bioactivation [2], [3].
Figure 1. Structures of characteristic pyrrolizidine alkaloids.
Structures are given in the N-oxide form with the exception of the otonecine derivative, senkirkine.
Strategies for PA sequestration in adapted insects have evolved in various insect lineages under the identical selection pressure to avoid higher concentrations of PAs in the form of their free base in the hemolymph (for recent reviews see [1], [3], [4], [5]). Leaf beetles of the genus Platyphora (Chrysomelidae, Coleoptera) have developed a strategy to transfer the free base from the hemolymph into defense secretions with such an efficiency that all other tissues outside the secretory glands are almost devoid of PAs [6]. In the related leaf beetle genus Oreina (Chrysomelidae, Coleoptera), the reduction of ingested PAs is suppressed. Instead, the N-oxides are directly absorbed and accumulated in their hemolymph and defense glands [7], [8]. A third strategy to handle sequestered PAs is the stabilization of the PAs by enzyme-catalyzed N-oxidation of the alkaloids within the insect. This mechanism is realized in larvae of the tiger moth family (Arctiidae, Lepidoptera) [9]–[13], in several Longitarsus flea beetle species [14], [15], and in the grasshopper genus Zonocerus [13], [16]. Recently, we have been able to show that the enzyme responsible for the N-oxygenation of PAs in arctiids belongs to the family of flavin-dependent monooxygenases (FMOs) [12], [13] and was recruited by gene duplication early in this lineage during the adaptation to PA-containing plants [17]. To date, these PA N-oxygenases are the only functional characterized FMOs of insects.
FMOs are well characterized in vertebrates, where they are involved in the detoxification of nucleophilic nitrogen- and sulfur-containing xenobiotics [18]. Although insects have to cope with a wide variety of xenobiotics, none of these so-called microsomal multisubstrate FMOs has been detected in insects, suggesting that the xenobiotic metabolism is guaranteed by cytochrome P450 monooxygenases (CYPs). Despite the different mechanism by which these two classes of monooxygenases operate, both convert lipophilic compounds into more hydrophilic metabolites that are readily excreted and have reduced bioactivity [3], [18]. Both, FMOs and CYPs have been shown to catalyze the N-oxygenation of PAs in vertebrates [19], [20].
Here, we report that PA N-oxygenation as one strategy the for insect adaptation to PA-containing food plants evolved independently in the grasshopper Z. variegatus and the arctiid moths. We show that convergent evolution resulted in two almost identical systems. This identity refers to the findings that, in both cases, an FMO has been recruited, and that the respective enzymes show almost identical substrate specificity.
Materials and Methods
Collection and rearing of Zonocerus
Larvae and adults of Zonocerus variegatus were collected in the vicinity of Accra, Ghana, and reared in the laboratory under a light/dark regime of 16/8 h at 20 to 25°C. Egg pots were incubated in moist sand at a constant temperature of 30°C. Nymphs and adults were fed with leaves of Rubus fruticosus unless otherwise stated. Leaves of R. fruticosus are regarded as non-toxic as they do not contain PAs.
RNA isolation and cDNA synthesis
Adult insects were dissected, and fat body tissue was frozen in liquid nitrogen before total RNA was isolated by using TRIZOL® reagent (Invitrogen); 2 µg RNA was used for cDNA synthesis with Superscript III reverse transcriptase (Invitrogen) and an oligo(dT)17 primer (P1, 0.1 µM; Table S1) at 55°C in a total volume of 20 µl.
Identification and expression of FMO-like cDNA sequences
Degenerate primers P2 and P3 (for primer sequences see Table S1) were designed based on an alignment of FMO sequences of the lepidopteran species Arctia caja, A. villica, Tyria jacobaeae, and Grammia geneura (Syn.: G. incorrupta) in combination with two FMO-like sequences identified in each of the genomes of Anopheles gambiae and Drosophila melanogaster. In a semi-nested PCR approach, 3 µl of Z. variegatus cDNA was amplified with AccuTaq LA polymerase (Sigma) and primer pair P2/P1 by using a touch-down protocol with decreasing annealing temperatures from 60°C to 52°C (−0.5°C per cycle for 16 cycles and 52°C constant for 20 further cycles). The reaction was diluted 1∶100, and 3 µl was used as template for the second PCR with primer pair P2/P3 and a decreasing annealing temperature from 55°C to 47°C in a total volume of 25 µl. The resulting fragment of 473 bp was subcloned by using the pGEM-T Easy vector (Promega). Sequencing revealed similarity to the FMO-encoding sequences of insects. Gene-specific primers were designed and used for 3′- and 5′-rapid amplification of cDNA ends (RACE, P4–P7) as described previously [21]. 3′-RACE resulted in the identification of two sequences covering the 3′-cDNA end and sharing 86% sequence identity. One of these sequences overlapped with the fragment identified with the degenerate primers and with the 5′-RACE fragment and was assembled to the full-length sequence (ZvFMOa). According to the second 3′-end cDNA, gene-specific primers (P8–P10) were designed that were used for identification by 5′-RACE. The resulting second full-length sequence was named ZvFMOc. For the amplification of the full open reading frames (ORFs) of ZvFMOa and ZvFMOc, primer pairs P11/P12 and P13/P14 were used, respectively, at an annealing temperature of 55°C with Platinum pfx DNA polymerase (Invitrogen), which possesses proof-reading activity. The resulting fragments were digested with NdeI/XhoI and cloned into an NdeI/XhoI-linearized pET22b vector for heterologous expression with a C-terminal hexahistidine tag in E. coli BL21(DE3). Restriction of the fragment resulting from amplification with primer pair P13/P14 (ZvFMOc) indicated that the PCR product was not homogeneous, as a fragment was detected that contained an internal, non-predicted restriction site for XhoI. Cloning of this fragment into the pGEM-T easy vector, screening with XhoI, and sequencing resulted in the identification of another FMO-encoding sequence, termed ZvPNO. For this sequence, the 3′- and 5′-cDNA ends were identified by RACE with the primers P15–P19. The full ORF of ZvPNO was amplified with primer pair P20/P21 at 55°C with Platinum pfx DNA polymerase, digested with BsaI and NotI, and cloned into an NcoI/NotI-linearized pET28a vector for expression with a C-terminal hexahistidine tag in E. coli BL21(DE3). Expression of the recombinant proteins ZvFMOa, ZvFMOc, and ZvPNO was induced with 0.1 mM isopropyl β-D-thiogalactoside at 30°C, before the proteins were purified by metal chelate affinity chromatography by using Ni2+-nitrilotriacetic acid-agarose (Qiagen). For determination of the native M r, the purified enzyme was applied to a Superdex200HighLoad column (GE Healthcare) using 20 mM glycine/NaOH buffer pH 9.0 containing 200 mM NaCl. As reference proteins thyroglobulin (669 kDa), ferritin (440 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa) have been used.
Sequence analysis
Comparison of amino acid sequences was performed with the Bestfit software of the Wisconsin Sequence Analysis Package (version10, Genetics Computer Group, Madison, WI). An alignment of amino acid sequences (Figure S1) was generated with ClustalX [22] and used to estimate phylogenies with the PHYLIP program package [23]. The maximum likelihood tree was calculated with PROML by using the Jones-Taylor-Thornton model for amino acid changes [24]. Bootstrap values were estimated with the programs SEQBOOT and CONSENSE. The bootstrap estimates are the result of 1000 replicates. The sequences reported in this paper have been submitted to the EMBL Nucleotide Sequence Database with the accession numbers FR696371 (ZvFMOa), FR696372 (ZvFMOc), and FR696373 (ZvPNO).
Assay for PA N-oxygenase activity
Enzyme activity was assayed in a 100 mM glycine/NaOH buffer pH 9 containing 0.2 mM NADPH at 37°C photometrically or in a radioassay as described previously [12]. All substrates were assayed at a concentration of 0.2 mM by using the photometric assay or the radioassay (0.023 µCi/assay), except for retronecine, supinidine, and isoretronecanol, which were tested as 3H-labeled substrates, and phalaenopsine as a 14C-labeled substrate in the radioassay (0.023 and 0.045 µCi/assay, respectively). For enzyme kinetics, initial rates were determined by varying the concentration of alkaloid (1–40 µM for senecionine, 4–60 µM for monocrotaline and atropine, and 0.1–1.5 mM for heliotrine and phalaenopsine) while maintaining the concentration of NADPH at 200 µM. The identity of senecionine N-oxide, seneciphylline N-oxide, monocrotaline N-oxide, phalaenopsine N-oxide, and atropine N-oxide was confirmed by liquid chromatography mass spectrometry (LC-MS). For LC-MS analysis, samples were diluted by using 50% acetonitrile containing 1% formic acid. Solutions were infused directly into a 3200 QTrap MS instrument (Applied Biosystems/MDS Sciex). The mass spectrometer was equipped with an electrospray ionization interface (ESI, Turbo V) and was operated in positive-ion mode and enhanced product ion (EPI) scan mode. Ionization and EPI conditions were optimized individually for each compound in its non-oxidated form, and the obtained settings were used to analyze extracts of the corresponding enzymatic assays. Nitrogen was used as a curtain and auxiliary gas.
Results
Identification and Heterologous Expression of PA N-Oxygenase of Z. variegatus
In contrast to the arctiid moths in which the PA N-oxygenase is a soluble protein in the hemolymph, the alkaloid N-oxygenating activity in the grasshopper Z. variegatus is associated with the fat body [13]. Separate incubation of a particulate fraction and the supernatant obtained from the insects fat body with 14C-seneciphylline showed that N-oxygenation activity is detectable in the soluble fraction, but not in the pellet. These results indicate that PNO is expressed as a soluble protein, excluding the involvement of a membrane-bound CYP. Therefore, we hypothesized that the enzyme might belong to the FMOs in this species. Using cDNA preparations of fat body tissue of Z. variegatus, we applied a polymerase chain reaction (PCR)-based approach, in order to identify FMO-encoding cDNA sequences. Degenerate primers were designed according to an alignment of FMO sequences of Drosophila melanogaster and Anopheles gambiae present in the database and of four arctiid species identified recently in our group, viz., Arctia caja, A. villica, Tyria jacobaeae, and Grammia geneura [17]. Three cDNA sequences were identified, ZvFMOa, ZvFMOc, and ZvPNO, with open reading frames of 1242 bp, 1245 bp, and 1242 bp in length, encoding proteins with a subunit size of 47,726 Da, 47,939 Da, and 47,793 Da, respectively. The isoelectric point of the three proteins is predicted to be 6.6, 6.2, and 6.0, respectively. The sequence motifs most characteristic for FMO, viz., the nucleotide-binding sites (Rossman folds, consensus GxGxxG) for binding of FAD as a prosthetic group and of the NADPH cofactor, respectively, and the FMO-identifying sequence (consensus FxGxxxHxxxY/F) are highly conserved in sequence position in comparison with the FMOs of other eukaryotes [25]–[28] (Figure S1). For none of the three sequences an N-terminal signal peptide was predicted as it was shown to be present in the SNO of Tyria jacobaeae and the FMOs of other arctiid species [12], [17]. No transmembrane helices were detected as they are present in mammalian FMOs close to the C-terminus for membrane attachment [29]. The FMO-like sequences of Z. variegatus share amino acid identities of 77% to 78% between each other and identities of 35% to 40% to the sequences encoding the senecionine N-oxygenase of T. jacobaeae (Lepidoptera), to the FMOs of Drosophila melanogaster (Diptera), and to the human FMO1 (Table 1).
Table 1. Degree of identity between amino acid sequences encoding FMOs of Z. variegatus, other insects, and the FMO1 of human.
| (%) | ZvFMOa | ZvFMOc | ZvPNO | TjSNO | DmFMO3006 | DmFMO3174 | HsFMO1 |
| ZvFMOa | 100 | 77.0 | 77.8 | 35.2 | 36.4 | 36.3 | 39.8 |
| ZvFMOc | 77.0 | 35.3 | 38.2 | 36.6 | 38.8 | ||
| ZvPNO | 36.3 | 38.2 | 37.1 | 37.7 |
For comparison, the full coding regions were used. TjSNO, Senecionine N-oxygenase of Tyria jacobaeae; DmFMO3006 and DmFMO3174, FMO sequences of unknown function present in the genome of Drosophila melanogaster; HsFMO1, FMO1 of human.
Biochemical Characterization of PA N-Oxygenase of Z. variegatus
After the expression and purification by metal chelate affinity chromatography, ZvFMOa, ZvFMOc, and ZvPNO showed, in assays for PA N-oxygenation, specific activities of 123 pkat/mg, 146 pkat/mg, and 59 nkat/mg, respectively (100 mM glycine/NaOH buffer pH 9.0 with 0.2 mM senecionine as substrate). The identity of the reaction product as senecionine N-oxide was confirmed by LC-MS (liquid chromatography with mass spectrometry). Therefore, ZvPNO was designated as PA N-oxygenase (PNO). Recombinant ZvPNO shows a pH optimum of 9.0 in potassium phosphate buffer (10 mM), Tris/HCl buffer (10 mM), and in glycine/NaOH buffer (100 mM), respectively (Figure S2). Using the potassium phosphate-based buffer system (pH9.0) in the standard assay, linear rates of product formation were maximal at a temperature of 42°C. Using a temperature range of 20–42°C, the activation energy was estimated to be 70.7 kJ/mol. The molecular mass of the heterologously expressed protein has been estimated by gel filtration chromatography to be 91,200 Da, indicating that the active enzyme is a homodimer.
Substrate Specificity of Recombinant FMO of Z. variegatus
Z. variegatus is a generalist feeding on a wide array of food plants and encountering a huge diversity of plant allelochemicals [30]. Therefore, in testing the substrate specificity of recombinant ZvFMOa, ZvFMOc, and ZvPNO, we used, in addition to various structures of PAs, other alkaloids and compounds known to be accepted as substrates of mammalian and yeast FMO (Table 2). With regard to ZvPNO, PA structures of the five main structural types are N-oxygenized. The otonecin derivative senkirkine, which carries a methyl group at the ring-bound nitrogen, is not a substrate (Figure 1). Atropine, a tropane alkaloid common to certain solaneceous plants, is converted to its N-oxide form with almost the same activity as the best substrates tested, e.g., senecionine and seneciphylline. Of the substrates described for mammalian FMOs and for the FMO from Saccharomyces cerevisiae, cysteamine is accepted by the PNO of Zonocerus, whereas L-cysteine, dimethylaniline, hydroxylamine, or glutathione are not substrates. ZvFMOa and ZvFMOc show low but unequivocal activity with most of the substrates that show activity with ZvPNO.
Table 2. Substrate specificity of recombinant flavin-dependent monooxygenases of Z. variegatus.
| Substrate | Specific activity (nkat/mg) | |||
| GgPNO | ZvPNO | ZvFMOa | ZvFMOc | |
| Pyrrolizidine alkaloids | ||||
| Senecionine type | ||||
| Senecionine | 55.0 | 32.0 | 0.1 | 0.1 |
| Seneciphylline | 53.2 | 29.4 | 0.6 | 0.1 |
| Senecivernine | - | 24.0 | 1.3 | 0.6 |
| Retrorsine | - | 18.6 | 1.5 | 0.5 |
| Senkirkine | n.d. | n.d. | n.d. | n.d. |
| Monocrotaline type | ||||
| Monocrotaline | 90.2 | 23.4 | 1.5 | 0.9 |
| Axillarine | 62.2 | 18.6 | n.d. | n.d. |
| Axillaridine | - | 11.8 | n.d. | n.d. |
| Lycopsamine type | ||||
| Heliotrine | 44.2 | 19.5 | n.d. | n.d. |
| Rinderine | 38.8 | 11.5 | 1.3 | 0.4 |
| Indicine | - | 4.8 | n.d. | n.d. |
| Triangularine type | ||||
| Sarracine | - | 16.0 | 0.4 | 0.3 |
| Phalaenopsine type | ||||
| Phalaenopsine | 17.1 | 9.6 | n.d. | n.d. |
| Necine bases | ||||
| Retronecine | n.d. | 10.2 | n.d. | 0.1 |
| Heliotridine | - | n.d. | n.d. | n.d. |
| Supinidine | n.d. | n.d. | n.d. | n.d. |
| Other alkaloids | ||||
| Ephedrine | - | n.d. | n.d. | n.d. |
| Nicotine | n.d. | 2.3 | 0.3 | 0.4 |
| Atropine | 18.9 | 29.1 | 0.3 | n.d. |
| Other substrates | ||||
| Dimethylaniline | n.d. | n.d. | n.d. | n.d. |
| Cysteamine | n.d. | 7.0 | 0.5 | 0.1 |
| L-Cysteine | n.d. | n.d. | n.d. | n.d. |
| Hydroxylamine | - | n.d. | n.d. | n.d. |
| Glutathione | 37.0 | n.d. | n.d. | n.d. |
n.d., not detectable; -, not tested.
PA N-oxygenase (ZvPNO) and two related FMOs of Z. variegatus (ZvFMOa and ZvFMOc) have been assayed in comparison to the previously characterized recombinant PA N-oxygenase (GgPNO) of G. geneura (Arctiidae, Lepidoptera) [17].
For ZvPNO, enzyme kinetics have been established by using selected substrates representing the major classes of PAs and of atropine. The apparent K m values of 1.1 µM (senecionine), 11.9 µM (monocrotaline), 263.9 (heliotrine), 837.6 µM (phalaenopsine), and 9.8 µM (atropine) show that the PNO has a higher affinity to the macrocyclic diesters senecionine and monocrotaline than to the monoesters heliotrine and phalaenopsine. The affinity to the tropane alkaloid atropine is in the same range as that for the macrocyclic diester type. The values for the catalytic efficiency (k cat/K m) support the interpretation that senecionine is the most efficient substrate for the PNO followed by monocrotaline and atropine (Table 3).
Table 3. Enzyme kinetics of recombinant ZvPNO.
| K m (µM) | V max (nkat/mg) | k cat (1/s) | k cat/K m (s−1 mol−1 l) | |
| senecionine | 1.08±0.07 | 20.73±0.24 | 0.99 | 916 666 |
| monocrotaline | 11.93±0.76 | 19.99±0.40 | 0.96 | 80 469 |
| heliotrine | 263.89±10.06 | 2.49±0.03 | 0.12 | 454 |
| phalaenopsine | 837.61±77.37 | 2.66±0.11 | 0.13 | 155 |
| atropine | 9.83±1.04 | 7.68±0.23 | 0,37 | 37 640 |
For calculations a molecular mass of ZvPNO of 47793 g/mol was used.
Comparative Sequence Analysis of Insect FMOs
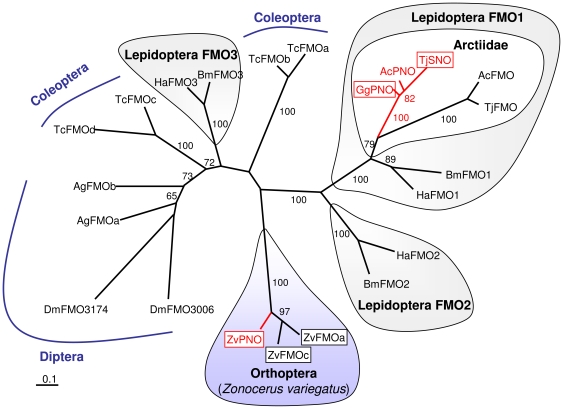
The amino acid sequences of the three FMOs of Z. variegatus and of various FMOs of insects have been used for phylogenetic analysis (Figure 2). The sequences form well-supported individual clusters with regard to the sequences of the Diptera (D. melanogaster and A. gambiae) and of Z. variegatus, two clusters for the sequences of Tribolium castaneum, and three separate clusters for lepidopteran sequences, recently termed FMO1 to FMO3 [17]. Within the Lepidoptera, gene duplication events have been shown to be responsible for the separation of the related FMO1 and FMO2 cluster and, within the Arctiids, for two separated clusters, of which one encodes the PA N-oxygenases [17]. The tree topology clearly supports two independent origins of PA N-oxygenizing enzymes in insects, one early in the arctiid lineage and one in grasshoppers.
Figure 2. Unrooted maximum-likelihood tree of amino acid sequences derived from cDNA encoding FMOs of various insect species.
Framed sequences were heterologously expressed and functionally analyzed. The other sequences should be regarded as putative FMO-coding cDNA. Branch lengths are proportional to the number of amino acid substitutions per site (scale: 0.1 substitutions per site). Bootstrap proportions resulted from 1000 replicates and are given for values >50. Ac, Arctia caja; Ag, Anopheles gambiae; Bm, Bombyx mori; Dm, Drosophila melanogaster; Gg, Grammia geneura; Ha, Helicoverpa armigera; Tc, Tribolium castaneum; Tj, Tyria jacobaeae; Zv, Zonocerus variegatus. Accession numbers for all sequences are listed in Figure S1.
Discussion
Our studies show that, in Arctiids and in the grasshopper Z. variegatus, FMOs were recruited independently as N-oxygenases for the safe handling of plant-derived PAs in these insects. Therefore, PA N-oxygenation in insects seems to be a unique feature of FMOs. Of note, CYPs in insects form a far larger gene family than FMOs (the genomes of D. melanogaster and B. mori encode 83 and 86 putative CYPs, respectively, in contrast to only 2 and 3 putative FMOs, respectively [12], [17], [31], [32]) and are well-known for their importance in the metabolism of natural and artificial xenobiotics, including insecticides [33]. Specific and inducible CYP-encoding genes in Papilio butterflies (Papilionidae, Lepidoptera) for the detoxification of furanocoumarins represent only one of several fascinating examples of gene evolution during adaptation of insects to plant allelochemicals [34], [35]. The array of substrates of the respective enzymes has been shown to be correlated with the feeding habits of the insects, i.e., a broad substrate specificity in generalists that feed on a wide diversity of host plants containing a broad spectrum of allelochemicals, and a narrow substrate specificity in specialist insects that feed on only one or a few plant species [36]. The same has been shown for FMOs involved in PA N-oxygenation of a specialist and a generalist arctiid species [17]. Concretely, the respective enzyme of the specialist T. jacobaeae is highly specific for PAs of the toxic senecionine type, the predominant PA type of its almost exclusive food plant Jacobaea vulgaris (syn. Senecio jacobaea). In contrast, the N-oxygenase of the generalist G. geneura (Syn.: G. incorrupta) accepts a wider range of substrates, including the non-toxic 1,2-saturated PA phalaenopsine and the tropane alkaloid atropine [17]. Most interestingly, the substrate specificity of the PA N-oxygenase of G. geneura is almost identical to that and of the PNO of Z. variegatus that also accepts atropine as a substrate. We postulate that the almost identical substrate specificity of the PA N-oxygenases of the two unrelated insects is the result of convergent evolution under identical selection pressure of generalist feeding. The wide substrate specificity of the PA N-oxygenase of Z. variegatus correlates with the extremely polyphagous behavior of this grasshopper, which is ranked as one of the most important economically pests of agriculture in west and central Africa [37], [38].
Of note, the N-oxygenation of PAs in specialized insects is not a detoxification mechanism that converts xenobiotics into more polar metabolites for efficient excretion from the body. Instead, this enzymatic conversion allows the sequestration of these plant toxins in a deactivated, metabolically safe form [5]. The recruitment of an enzyme that catalyzes PA N-oxygenation can be regarded as a “key innovation” during the insect's adaptation to PA-containing food plants [17]. In arctiids, this enzyme was the prerequisite for the evolution of several further, highly specific adaptations, such as the positive feeding response triggered by traces of PAs sensed by specialized taste receptors, the ability to transfer the alkaloids from the larvae to the adult stage, and the insect's behavior that ensures optimal egg protection by PAs of both parents governed by PA-derived pheromones [5], [39].
The high degree of sequence identity of the PA N-oxygenase to two further FMOs identified from Z. variegatus, and the fact that the latter two enzymes are also able to catalyze the N-oxygenation of alkaloids, albeit with a much lower specific activity, indicate the close relationship of these sequences. The data suggest a duplication of an ancestor gene encoding an FMO of unknown function that already possessed the ability to N-oxygenize PAs, most probably as a side activity. One of the gene copies was recruited and optimized for N-oxygenation of plant-derived PAs. The finding that PAs are strong phagostimulants for Zonocerus [40] suggests a central role of PAs in the insect's ecology. Further research has to show whether this is also the case for the N-oxygenation of further plant-derived toxins, such as atropine or nicotine, that are accepted as a substrate by the PNO of Z. variegatus.
Supporting Information
Amino acid alignment of flavin-dependent monooxygenases of various insect species. Only the central part of the alignment that was used for the estimation of the phylogenies is shown, spanning the region from amino acid position 5 to 402 with respect to ZvPNO. The sequence motifs for binding of FAD and NADPH and the FMO-identifying sequence are boxed. The accession numbers of the three FMO sequences of Zonocerus variegatus and of all sequences taken from the databases are given at the end of the alignment.
(PDF)
PNO activity in three buffer systems with pH variation. Enzyme activities have been determined with senecionine as substrate at 37°C.
(PDF)
Sequences of primers used for the identification and cloning of cDNAs of flavin-dependent monooxygenases of Zonocerus variegatus . Recognition sites of restriction endonucleases used for cloning are underlined.
(DOC)
Acknowledgments
We thank S. Adom, P. Adjei, and U. S. Issah for the identification and collection of Zonocerus individuals at Ghana, M. Doose and B. Schemmerling for her excellent technical assistance, C. Theuring (Technical University Braunschweig) for providing us most of the substrates, M. Schilhabel (IKMB, Kiel University) for sequencing support, and T. Hartmann for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Hartmann T, Ober D. Defense by pyrrolizidine alkaloids: developed by plants and recruited by insects. In: Schaller A, editor. Induced Plant Resistance to Herbivory. Berlin: Springer Science+Business Media B.V; 2008. pp. 213–231. [Google Scholar]
- 2.Mattocks AR. Chemistry and Toxicology of Pyrrolizidine Alkaloids. London: Academic Press; 1986. [Google Scholar]
- 3.Langel D, Ober D. Evolutionary recruitment of a flavin-dependent monooxygenase for stabilization of sequestered pyrrolizidine alkaloids in arctiids. Phytochemistry. 2011 doi: 10.1016/j.phytochem.2010.12.014. In Press: DOI: 10.1016/j.phytochem.2010.1012.1014. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann T, Ober D. Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. In: Leeper FJ, Vederas JC, editors. Topics in Current Chemistry. Berlin, Heidelberg: Springer; 2000. pp. 207–244. [Google Scholar]
- 5.Hartmann T. Pyrrolizidine alkaloids: the successful adoption of a plant chemical defense. In: Conner WE, editor. Tiger Moths and Woolly Bears Behavior, Ecology, and Evolution of the Arctiidae. New York: Oxford University Press; 2009. pp. 55–81. [Google Scholar]
- 6.Hartmann T, Theuring C, Witte L, Pasteels JM. Sequestration, metabolism and partial synthesis of tertiary pyrrolizidine alkaloids by the neotropical leaf-beetle Platyphora boucardi. Insect Biochemistry and Molecular Biology. 2001;31:1041–1056. doi: 10.1016/s0965-1748(01)00052-2. [DOI] [PubMed] [Google Scholar]
- 7.Pasteels JM, Rowell-Rahier M, Randoux T, Braekman JC, Daloze D. Pyrrolizidine alkaloids of probable host-plant origin in the pronotal and elytral secretion of the leaf beetle Oreina cacaliae. Entomologia Experimentalis et Applicata. 1988;49:55–58. [Google Scholar]
- 8.Ehmke A, Rowell-Rahier M, Pasteels JM, Hartmann T. Sequestration of ingested 14C-senecionine N-oxide in the exocrine defensive secretions of chrysomelid beetles. Journal of Chemical Ecology. 1991;17:2367–2380. doi: 10.1007/BF00994588. [DOI] [PubMed] [Google Scholar]
- 9.von Nickisch-Rosenegk E, Wink M. Sequestration of pyrrolizidine alkaloids in several arctiid moths (Lepidoptera: Arctiidae). Journal of Chemical Ecology. 1993;19:1889–1903. doi: 10.1007/BF00983794. [DOI] [PubMed] [Google Scholar]
- 10.Rothschild M, Aplin RT, Cockrum PA, Edgar JA, Fairweather P, et al. Pyrrolizidine alkaloids in arctiid moths (Lep.) with a discussion on host plant relationships and the role of these secondary plant substances in the Arctiidae. Biological Journal of the Linnean Society. 1979;12:305–326. [Google Scholar]
- 11.Hartmann T, Biller A, Witte L, Ernst L, Boppré M. Transformation of plant pyrrolizidine alkaloids into novel insect alkaloids by arctiid moths (Lepidoptera). Biochemical Systematics and Ecology. 1990;18:549–554. [Google Scholar]
- 12.Naumann C, Hartmann T, Ober D. Evolutionary recruitment of a flavin-dependent monooxygenase for the detoxification of host plant-acquired pyrrolizidine alkaloids in the alkaloid-defended arctiid moth Tyria jacobaeae. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6085–6090. doi: 10.1073/pnas.082674499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindigkeit R, Biller A, Buch M, Schiebel HM, Boppré M, et al. The two faces of pyrrolizidine alkaloids: The role of the tertiary amine and its N-oxide in chemical defense of insects with acquired plant alkaloids. European Journal of Biochemistry. 1997;245:626–636. doi: 10.1111/j.1432-1033.1997.00626.x. [DOI] [PubMed] [Google Scholar]
- 14.Dobler S, Haberer W, Witte L, Hartmann T. Selective sequestration of pyrrolizidine alkaloids from diverse host plants by Longitarsus flea beetles. Journal of Chemical Ecology. 2000;26:1281–1298. [Google Scholar]
- 15.Narberhaus I, Theuring C, Hartmann T, Dobler S. Uptake and metabolism of pyrrolizidine alkaloids in Longitarsus flea beetles (Coleoptera: Chrysomelidae) adapted and non-adapted to alkaloid-containing host plants. Journal of Comparative Physiology B, Biochemical, Systemic, and Environmental Physiology. 2003;173:483–491. doi: 10.1007/s00360-003-0356-6. [DOI] [PubMed] [Google Scholar]
- 16.Bernays EA, Edgar JA, Rothschild M. Pyrrolizidine alkaloids sequestered and stored by the aposematic grasshopper, Zonocerus variegatus. Journal of Zoology. 1977;182:85–87. [Google Scholar]
- 17.Sehlmeyer S, Wang L, Langel D, Heckel DG, Mohagheghi H, et al. Flavin-dependent monooxygenases as a detoxification mechanism in insects: new insights from the arctiids (Lepidoptera). PloS One. 2010;5:e10435. doi: 10.1371/journal.pone.0010435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cashman JR. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochemical and Biophysical Research Communications. 2005;338:599–604. doi: 10.1016/j.bbrc.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Huan J-Y, Miranda CL, Buhler DR, Cheeke PR. Species differences in the hepatic microsomal enzyme metabolism of the pyrrolizidine alkaloids. Toxicology Letters. 1998;99:127–137. doi: 10.1016/s0378-4274(98)00152-0. [DOI] [PubMed] [Google Scholar]
- 20.Williams DE, Reed RL, Kedzierski B, Ziegler DM, Buhler DR. The role of flavin-containing monooxygenase in the N-oxidation of the pyrrolizidine alkaloid senecionine. Drug Metabolism and Disposition. 1989;17:380–386. [PubMed] [Google Scholar]
- 21.Ober D, Hartmann T. Deoxyhypusine synthase from tobacco: cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. Journal of Biological Chemistry. 1999;274:32040–32047. doi: 10.1074/jbc.274.45.32040. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenstein J. 2005. (2005) PHYLIP, Phylogeny Inference Package (Univ. of Washington, Seattle), Version 3.6.
- 24.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Cashman JR. Structural and catalytic properties of the mammalian flavin-containing monooxygenase. Chemical Research in Toxicology. 1995;8:165–181. doi: 10.1021/tx00044a001. [DOI] [PubMed] [Google Scholar]
- 26.Fraaije MW, Kamerbeek NM, van Berkel WJH, Janssen DB. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Letters. 2002;518:43–47. doi: 10.1016/s0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- 27.Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Mechanism of action of a flavin-containing monooxygenase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9832–9837. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alfieri A, Malito E, Orru R, Fraaije MW, Mattevi A. Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6572–6577. doi: 10.1073/pnas.0800859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips IR, Dolphin CT, Clair P, Hadley MR, Hutt AJ, et al. The molecular biology of the flavin-containing monooxygenases of man. Chemico-Biological Interactions. 1995;96:17–32. doi: 10.1016/0009-2797(94)03580-2. [DOI] [PubMed] [Google Scholar]
- 30.Chapman RF, Page WW, McCaffery AR. Bionomics of the variegated grasshopper (Zonocerus variegatus) in West and Central Africa. Annual Review of Entomology. 1986;31:479–505. [Google Scholar]
- 31.Tijet N, Helvig C, Feyereisen R. The cytochrome P450 gene superfamily in Drosophila melanogaster: Annotation, intron-exon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/s0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- 32.Ai J, Yu Q, Cheng T, Dai F, Zhang X, et al. Characterization of multiple CYP9A genes in the silkworm, Bombyx mori. Molecular Biology Reports. 2010;37:1657–1664. doi: 10.1007/s11033-009-9580-9. [DOI] [PubMed] [Google Scholar]
- 33.Feyereisen R. Insect Cytochrome P450. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Amsterdam: Elsevier; 2005. pp. 1–77. [Google Scholar]
- 34.Hung CF, Berenbaum MR, Schuler MA. Isolation and characterization of CYP6B4, a furanocoumarin-inducible cytochrome P450 from a polyphagous caterpillar (Lepidoptera: Papilionidae). Insect Biochemistry and Molecular Biology. 1997;27:377–385. doi: 10.1016/s0965-1748(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Schuler MA, Berenbaum MR. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: Specificity and substrate encounter rate. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 2):14593–14598. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernays EA, Chapman RF. Plant secondary compounds and grasshoppers: beyond plant defenses. Journal of Chemical Ecology. 2000;26:1773–1794. [Google Scholar]
- 38.Kekeunou S, Weise S, Messi J, Tamo M. Farmers' perception on the importance of variegated grasshopper (Zonocerus variegatus (L.)) in the agricultural production systems of the humid forest zone of Southern Cameroon. Journal of Ethnobiology and Ethnomedicine. 2006;2:17. doi: 10.1186/1746-4269-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisner T, Meinwald J. The chemistry of sexual selection. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:50–55. doi: 10.1073/pnas.92.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boppré M, Fischer OW. Zonocerus und Chromolaena in West Africa. A chemoecological approach towards pest management. In: Krall S, Wilps H, editors. New Trends in Locust Control. Eschborn: GTZ; 1994. pp. 107–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid alignment of flavin-dependent monooxygenases of various insect species. Only the central part of the alignment that was used for the estimation of the phylogenies is shown, spanning the region from amino acid position 5 to 402 with respect to ZvPNO. The sequence motifs for binding of FAD and NADPH and the FMO-identifying sequence are boxed. The accession numbers of the three FMO sequences of Zonocerus variegatus and of all sequences taken from the databases are given at the end of the alignment.
(PDF)
PNO activity in three buffer systems with pH variation. Enzyme activities have been determined with senecionine as substrate at 37°C.
(PDF)
Sequences of primers used for the identification and cloning of cDNAs of flavin-dependent monooxygenases of Zonocerus variegatus . Recognition sites of restriction endonucleases used for cloning are underlined.
(DOC)