Abstract
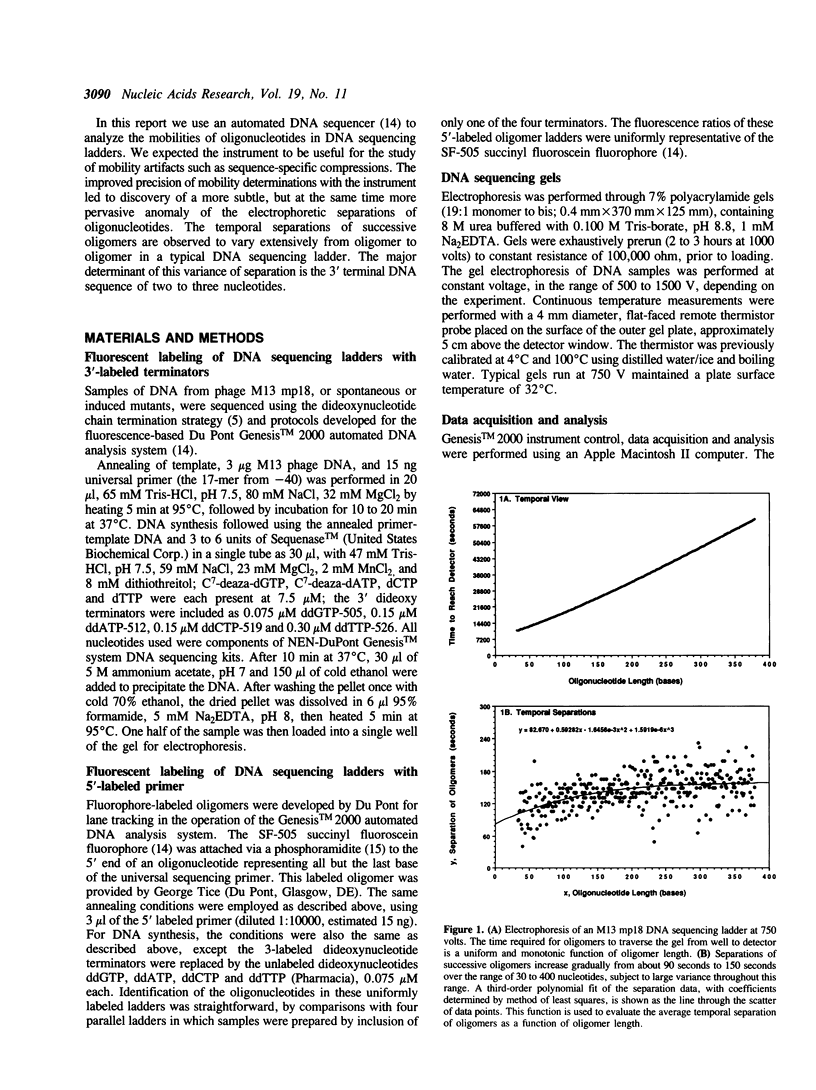
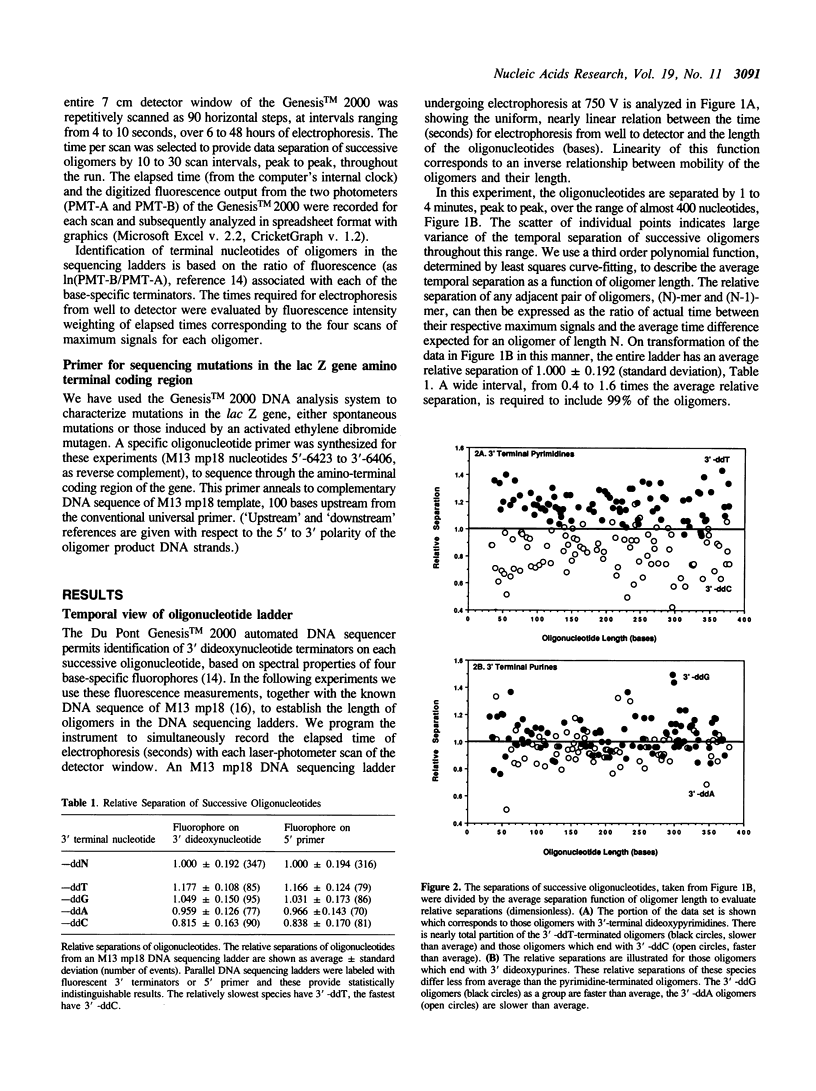
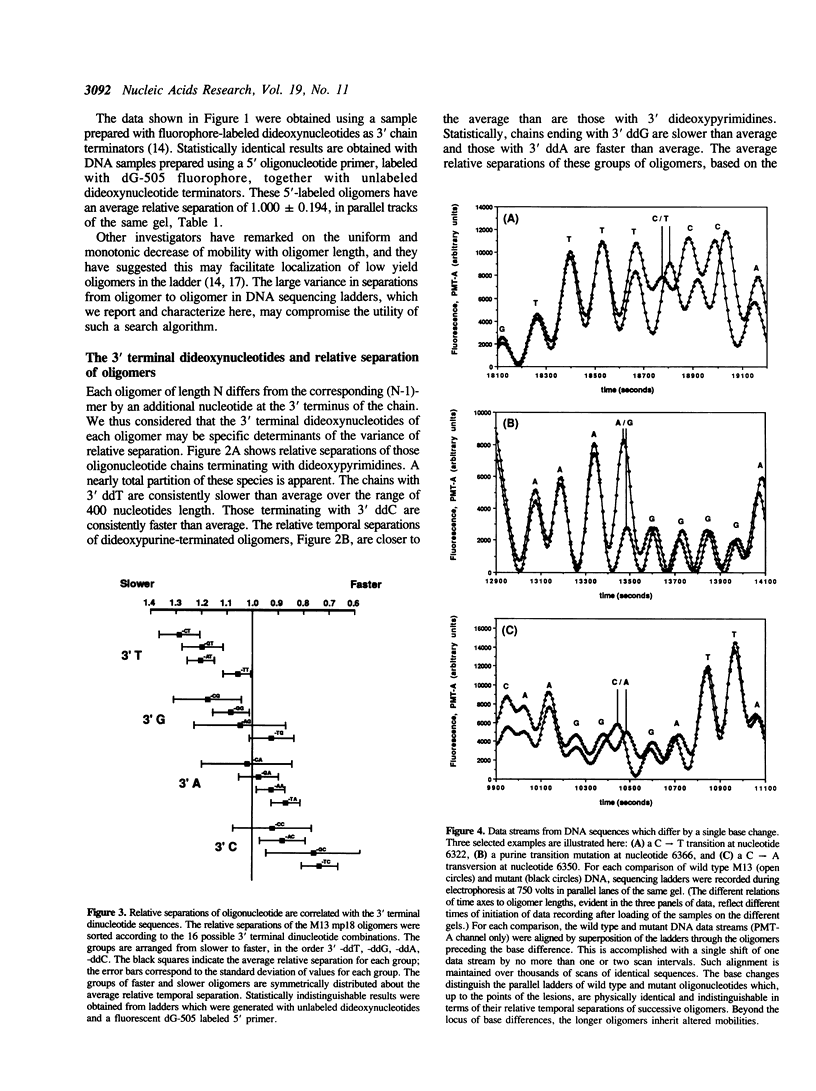
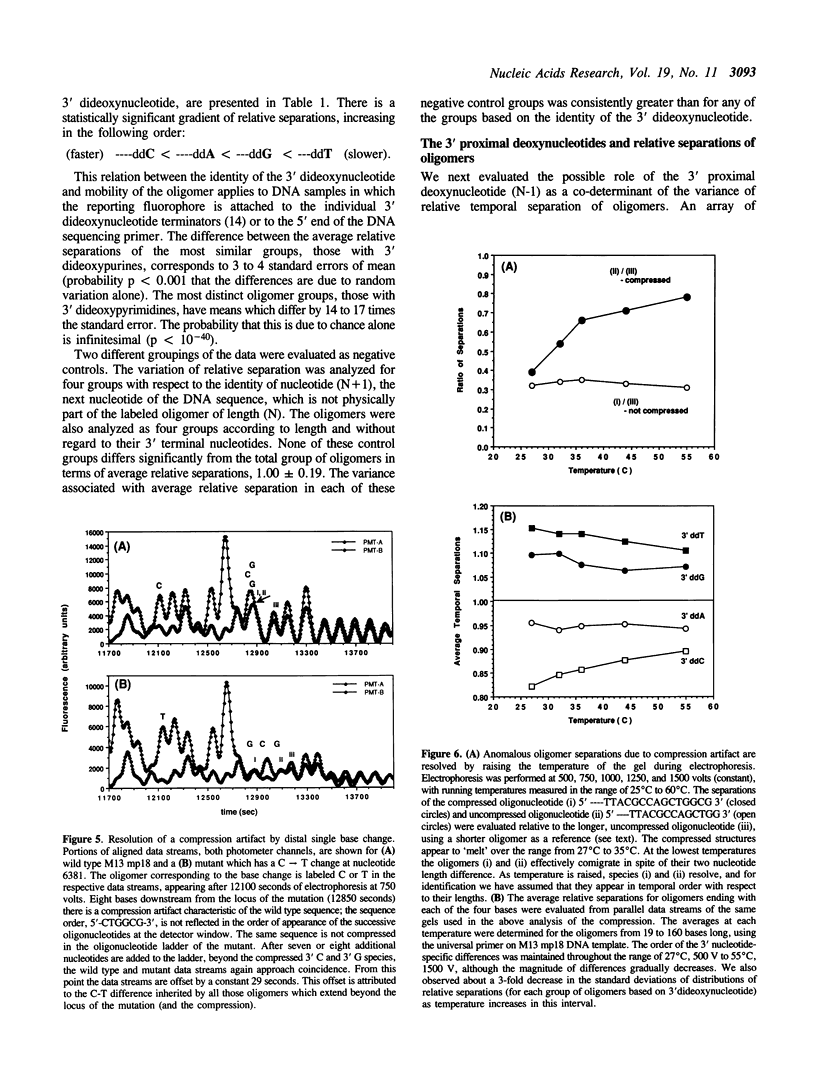
Electrophoretic separation of oligonucleotides in denaturing polyacrylamide gels is primarily a function of length-dependent mobility. The 3' terminal nucleotide sequence of the oligonucleotide is a significant, secondary determinant of mobility and separation. Oligomers with 3'-ddT migrate more slowly than expected on the basis of length alone, and thus are better separated from the preceding, shorter oligomers in the sequencing ladder. Oligomers with 3'-ddC are relatively faster than expected, and are therefore less separated. At the 3' penultimate position, -dC- increases and -dT- reduces separation. Purines at the 3' terminal or penultimate positions of oligonucleotides affect separation less than the pyrimidines. These results suggest specific interactions among neighboring nucleotides with important effects on the conformation of oligonucleotides during electrophoresis. These interactions are compared to compression artifacts, which represent more extreme anomalies of length-dependent separation of oligonucleotides. Knowledge of base-specific effects on electrophoretic behavior of DNA oligomers supplements the usual information available for determination of sequences; additionally it provides an avenue to thermodynamic and hydrodynamic investigations of DNA structure.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansorge W., Sproat B., Stegemann J., Schwager C., Zenke M. Automated DNA sequencing: ultrasensitive detection of fluorescent bands during electrophoresis. Nucleic Acids Res. 1987 Jun 11;15(11):4593–4602. doi: 10.1093/nar/15.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S., Pohl F. M. DNA sequencing with direct blotting electrophoresis. EMBO J. 1984 Dec 1;3(12):2905–2909. doi: 10.1002/j.1460-2075.1984.tb02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren B. W., Simon M. I., Lai E. The basis of high resolution separation of small DNAs by asymmetric-voltage field inversion electrophoresis and its application to DNA sequencing gels. Nucleic Acids Res. 1990 Mar 25;18(6):1481–1487. doi: 10.1093/nar/18.6.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerman N., Fawcett J. K., Cameran A. Molecular structure of a deoxyribose-dinucleotide, sodium thymidylyl-(5' yields to 3')-thymidylate-(5') hydrate (pTpT), and a possible structural model for polythymidylate. J Mol Biol. 1976 Nov 15;107(4):601–621. doi: 10.1016/s0022-2836(76)80086-1. [DOI] [PubMed] [Google Scholar]
- Deutsch J. M. Theoretical studies of DNA during gel electrophoresis. Science. 1988 May 13;240(4854):922–924. doi: 10.1126/science.3363374. [DOI] [PubMed] [Google Scholar]
- Edmondson S. P., Gray D. M. Analysis of the electrophoretic properties of double-stranded DNA and RNA in agarose gels at a finite voltage gradient. Biopolymers. 1984 Dec;23(12):2725–2742. doi: 10.1002/bip.360231204. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Holzwarth G., McKee C. B., Steiger S., Crater G. Transient orientation of linear DNA molecules during pulsed-field gel electrophoresis. Nucleic Acids Res. 1987 Dec 10;15(23):10031–10044. doi: 10.1093/nar/15.23.10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S., Frisch H. L. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 1982 May;21(5):995–997. doi: 10.1002/bip.360210511. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J. Mobility of DNA in gel electrophoresis. Biopolymers. 1982 Nov;21(11):2315–2316. doi: 10.1002/bip.360211116. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noolandi J, Rousseau J, Slater GW, Turmel C, Lalande M. Self-trapping and anomalous dispersion of DNA in electrophoresis. Phys Rev Lett. 1987 Jun 8;58(23):2428–2431. doi: 10.1103/PhysRevLett.58.2428. [DOI] [PubMed] [Google Scholar]
- Olvera de la Cruz M, Deutsch JM, Edwards SF. Electrophoresis in strong fields. Phys Rev A Gen Phys. 1986 Mar;33(3):2047–2055. doi: 10.1103/physreva.33.2047. [DOI] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Sanders J. Z., Kaiser R. J., Hughes P., Dodd C., Connell C. R., Heiner C., Kent S. B., Hood L. E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986 Jun 12;321(6071):674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Aldridge P. K., Callis J. B. Observation of individual DNA molecules undergoing gel electrophoresis. Science. 1989 Jan 13;243(4888):203–206. doi: 10.1126/science.2911733. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Wu R. Sequence analysis of short DNA fragments. Methods Enzymol. 1980;65(1):620–638. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


