Abstract
Enterovirus infections have been linked to type 1 diabetes in several studies. Enteroviruses also have tropism to pancreatic islets and can cause β-cell damage in experimental models. Viral persistence has been suspected to be an important pathogenetic factor. This study evaluates whether gut mucosa is a reservoir for enterovirus persistence in type 1 diabetic patients. Small-bowel mucosal biopsy samples from 39 type 1 diabetic patients, 41 control subjects, and 40 celiac disease patients were analyzed for the presence of enterovirus using in situ hybridization (ISH), RT-PCR, and immunohistochemistry. The presence of virus was compared with inflammatory markers such as infiltrating T cells, HLA-DR expression, and transglutaminase 2–targeted IgA deposits. Enterovirus RNA was found in diabetic patients more frequently than in control subjects and was associated with a clear inflammation response in the gut mucosa. Viral RNA was often detected in the absence of viral protein, suggesting defective replication of the virus. Patients remained virus positive in follow-up samples taken after 12 months’ observation. The results suggest that a large proportion of type 1 diabetic patients have prolonged/persistent enterovirus infection associated with an inflammation process in gut mucosa. This finding opens new opportunities for studying the viral etiology of type 1 diabetes.
Type 1 diabetes is one of the most common chronic diseases in developed countries, and its incidence has increased sharply since the second world war. It is caused by a selective destruction of insulin-producing β cells in the pancreas mediated by immunological mechanisms. Susceptibility to the disease is modulated by >40 different risk genes, with HLA genes contributing more than half of the genetic susceptibility. Environmental factors clearly influence the disease risk and contribute to the rapidly increasing incidence rates.
The connection between type 1 diabetes and enterovirus infections has been widely studied. Enteroviruses have been found in the blood and pancreas of type 1 diabetic patients in several studies, and they have also been associated with increased risk of type 1 diabetes in prospective studies (1–7). The recent discovery of the genetic polymorphisms of IFIH1 gene as diabetes risk-modulating factors has further strengthened this association, since these genes mediate the recognition of enteroviruses by the innate immune system (8,9). Diabetes-associated polymorphisms seem to be associated with a strong innate immune response, which may lead to enhanced inflammation response during virus infection (10). Other innate immune system genes (IRF7 network) also modulate the risk of T1D (11).
Frequent detection of the virus in diabetic patients has suggested a possible role of viral persistence. Enterovirus persistence has previously been described, e.g., in chronic cardiomyopathies (12) and in animal models (13–16), where enterovirus and other picornaviruses may persist for prolonged periods. These studies have suggested that enterovirus can persist as double-stranded RNA without clear protein synthesis (15,17). It is known from several viral diseases that persistent infection typically leads to a strong innate immune response, local inflammation, and immune-mediated tissue damage.
Celiac disease is characterized by small intestinal inflammation, mucosal damage, and increased mucosal permeability. The association between type 1 diabetes and celiac disease is well established. It is therefore intriguing to hypothesize whether enterovirus infection and enteropathy contribute to this association.
In spite of the recent progress in this research field, it is not known how enteroviruses could cause selective damage in insulin-producing β cells. Their primary replication site is the gut mucosa, from which the virus can spread to the blood (primary viremia) and infect internal organs. However, the possible presence of enteroviruses in human intestine has not been studied in detail. We recently published the first such study suggesting that enterovirus may be present in the small intestine in type 1 diabetic patients (18). The current study evaluates the hypothesis that gut mucosa is a reservoir for enterovirus persistence in type 1 diabetic patients, maintaining a continuous inflammatory state that can spread to the pancreas and play a crucial role in enterovirus-induced diabetes.
RESEARCH DESIGN AND METHODS
Study series.
The study series consisted of 120 individuals altogether who underwent gastroscopy because of various gastrointestinal complaints. Small-bowel mucosal biopsies were taken from distal duodenum for morphological analyses and for basic research. Samples were taken during the period 1995–2000 at the Department of Gastroenterology, Tampere University Hospital. Formalin-fixed samples were available from all 120 study subjects and frozen samples from 86 subjects.
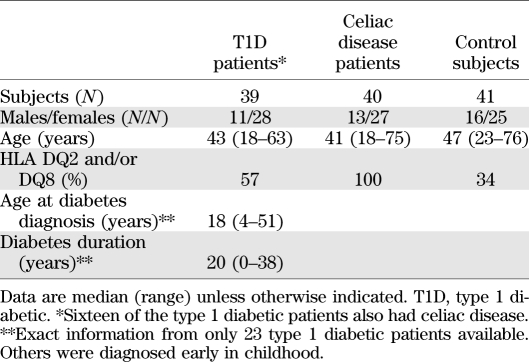
The study subjects comprised 39 type 1 diabetic patients, 40 patients with celiac disease who did not have type 1 diabetes, and 41 control subjects, including subjects who had neither of these diseases but nonspecific gastrointestinal symptoms (25 dyspepsia, 10 irritable bowel syndrome, 3 aphthous stomatitis, 1 ulcerative colitis, 1 collagenous colitis, and 1 eventually healthy). Furthermore, one control subject had psoriasis and one IgA nephropathy. All control subjects were nondiabetic and showed no signs of celiac disease (Table 1).
TABLE 1.
Summary of study subjects
The diagnosis of type 1 diabetes was based on World Health Organization criteria (19), and all the patients were on insulin treatment. Sixteen also had celiac disease, 10 thyroidal autoimmune disorder, 1 IgA nephropathy, 1 Addison disease, 1 alopecia, and 1 rheumatoid arthritis. The duration of diabetes varied from 0 to 38 years (median 20 years; exact information available from only 23 patients).
Celiac disease was diagnosed in study subjects by a positive endomysial antibody result and by small-bowel mucosal villous atrophy and crypt hyperplasia. Celiac disease had recently been diagnosed in all the patients, and all were on a normal gluten-containing diet at the time of the biopsy. One celiac disease patient had IgA nephropathy, and one had hyperthyroidism.
The study protocol was approved by the ethical committee of the Tampere University Hospital. All subjects gave informed consent.
In situ hybridization.
Primary screening of enterovirus was carried out on formalin-fixed and paraffin-embedded biopsy samples (5-μm sections) using an in situ hybridization (ISH) assay as previously described (4,18,20). This is based on a single enterovirus-specific probe targeting a highly conserved, group-common sequence in the 5′-noncoding region of the enteroviral genome. This assay covers most if not all known enterovirus types. All analyses were performed blind to the case-control status of the subjects. Positive hybridization signal was semiquantified by counting positive cells per microscopic field into the following categories: negative (no positive cells), weak positive (an average of 1–10 positive cells per microscopic field using 400-fold magnification), moderate positive (10–100 positive cells per field), and strong positive (>100 cells per field).
Immunohistochemical staining.
Formalin-fixed paraffin-embedded biopsy samples (5-μm sections) were stained with antienterovirus VP1 antibody (clone 5-D8/1, 1:300; DakoCytomation, Glostrup, Denmark) using Ventana BencMark LT (Ventana Medical Systems) and the ultraView Universal detection system as previously described (20). Known virus-positive tissue and cell culture samples were used to confirm the staining reliability of all separate staining batches. CD3+ intraepithelial lymphocytes (IELs) were stained with monoclonal antibody Leu-4 (Becton Dickinson, San Jose, CA), αβ+ IELs with monoclonal βF1 antibody (Endogen, Woburn, MA), and γδ+ IELs with T-cell receptor γ antibody (Endogen). Positive IELs were counted with a ×100 objective throughout the surface epithelium, and IEL reference values were set at 37 cells/mm for CD3+, 25 for αβ+, and 4.3 for γδ+ IELs as previously described (21). Mucosal HLA-DR expression was detected with monoclonal HLA-DR antibody (Becton Dickinson) and graded as previously described (22). IgA deposits were analyzed from unfixed frozen sections using double staining with fluorescein isothiocyanate–labeled rabbit antibody against human IgA (Dako, Glostrup, Denmark) and monoclonal mouse antibody against transglutaminase 2 (CUB7402; NeoMarkers, Fremont, CA) followed by rhodamine-conjugated anti-mouse IgG antibodies (Dako). It was earlier shown that the mucosal IgA deposits are specifically targeted against transglutaminase 2 (23,24). All evaluations were carried out blind to disease history or laboratory findings.
Enterovirus RT-PCR.
For RT-PCR, unfixed biopsy samples were stored frozen in optimal cutting temperature medium at −70°C. The biopsy samples were removed from the optimal cutting temperature medium and homogenized using a SilentCrusher S homogenizer (Heidolph, Schwabach, Germany). RNA was extracted with the RNeasy Mini Kit (Qiagen, Hilden, Germany). RT-PCR was performed using two independent methods: a previously described method amplifying a sequence common to all known enterovirus serotypes (25) and a real-time RT-PCR using the same primers and probes. A sample was considered enterovirus positive if it gave a positive signal in at least one of these assays.
Statistical analysis.
Statistical analyses were performed using SPSS 18.0 for Windows. Frequency comparison was performed with the Pearson χ2 and Fisher exact tests, and continuous variables were analyzed by independent sample t test.
RESULTS
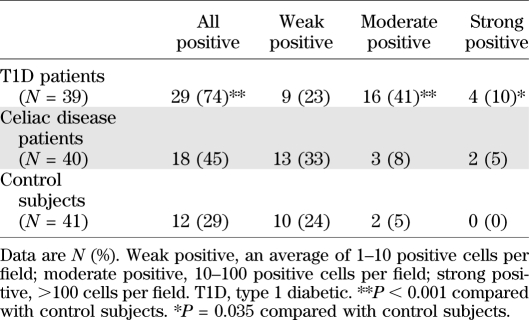
Small intestinal biopsy samples were first screened for the presence of enterovirus genome using ISH method. Seventy-four percent of the type 1 diabetic patients compared with 29% of the control subjects were found to be virus positive (P < 0.001) (Table 2 and Fig. 1). A higher frequency of clearly positive hybridization signals was seen in diabetic patients especially (51 vs. 5%; P < 0.001) (Table 2). Enterovirus RNA was mainly located in the epithelial cells of villi and crypts; however, occasional staining in lamina propria was also observed. A follow-up sample was available from three diabetic patients who were enterovirus positive in the initial sample (taken 1 year after the initial sample). All of these remained enterovirus positive. The detection of enteroviruses was not associated with age, sex, HLA DQ2 and/or DQ8 alleles, or duration of diabetes or celiac disease (data not shown).
TABLE 2.
Subjects positive for enterovirus RNA by ISH in small intestine biopsy samples
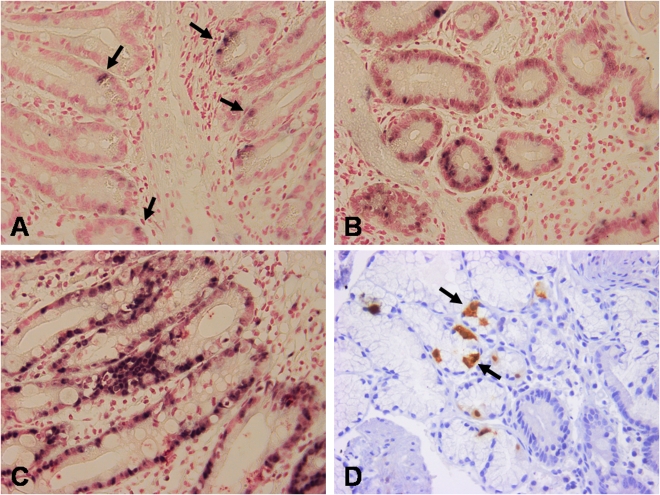
FIG. 1.
Detection of enterovirus RNA by ISH in intestinal biopsies of type 1 diabetic patients including weak-positive (A), moderate-positive (B), and strong-positive (C) cases. D: Immunohistochemical staining for enterovirus VP1 protein. Arrows point to cells positive for enterovirus RNA (A) and VP1 protein (D). (A high-quality digital representation of this figure is available in the online issue.)
Next, the presence of viral RNA was confirmed using RT-PCR. A freshly frozen biopsy sample was available from 26 of the diabetic patients, 34 celiac disease patients, and 31 control subjects, and RT-PCR found the virus in 19, 15, and 10% of them, respectively. The virus was detected more frequently in ISH-positive than in ISH-negative subjects (12 of 50 vs. 1 of 41; P < 0.01).
In the next phase, we analyzed whether the presence of viral RNA was associated with the production of viral VP1 protein (see Supplementary Table 5). The majority of subjects who were positive for virus RNA in ISH were negative for VP1 protein in immunohistochemistry. Overall, VP1 protein was found in 22, 20, and 22% of altogether 30 diabetic patients, 37 celiac disease patients, and 41 control subjects, respectively (Fig. 1). VP1 protein was mainly observed in the epithelial cells of the crypts. The proportion of VP1-negative subjects among all RNA-positive subjects tended to be higher among diabetic and celiac disease patients than in control subjects (78 and 86 vs. 66%).
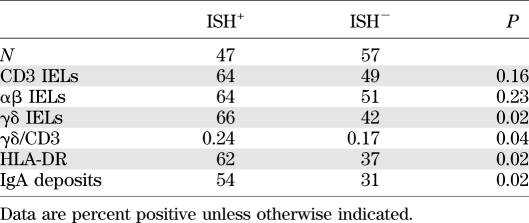
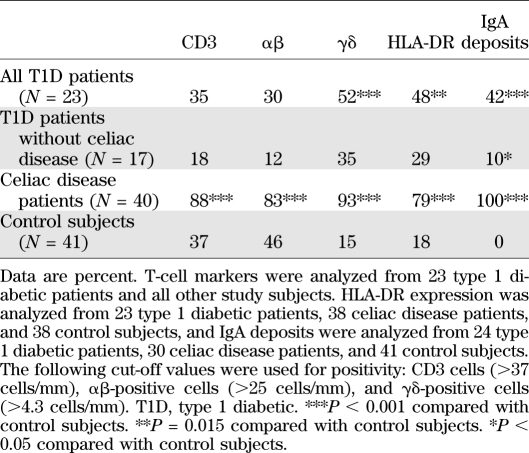
Finally, the presence of viral RNA (ISH) was correlated with inflammation markers in the intestinal mucosa. A total of 23 type 1 diabetic patients, 40 celiac disease patients, and 41 control subjects was analyzed for the presence of CD3+, αβ+, and γδ+ intraepithelial lymphocytes and HLA-DR+ cells. Viral RNA was associated with the infiltration of αβ+, γδ+, and HLA-DR+ cells (Table 3). The presence of inflammation markers was associated with enterovirus RNA positivity in the whole study cohort, and the trend was also seen in diabetic patients in γδ IELs (46 vs. 17%, respectively) and HLA-DR expression (46 vs. 0%), but because of the low number of diabetic patients positive for inflammation markers, the results did not reach statistical significance (data not shown). The mean γδ+-to-CD3+ cell ratio was also higher in virus-positive than in virus-negative subjects (0.24 vs. 0.17; P = 0.043). Besides T-cell–mediated inflammation, antibody-mediated inflammation (IgA deposits) was more frequent in virus-positive than in virus-negative subjects (54 vs. 31%; P = 0.023) (Table 3). As expected, the inflammatory markers were most frequent in celiac disease patients in general, but diabetic patients who did not have celiac disease also had a tendency of elevated γδ+ IELs, HLA-DR expression, and IgA deposits compared with control subjects (Table 4 and Supplementary Figs. 2 and 3). They also had a higher mean γδ+-to-CD3+ cell ratio (0.18 vs. 0.09; P = 0.028). However, the inflammation was clearly much weaker in the diabetes group than in the celiac disease group.
TABLE 3.
Inflammation markers in ISH+ and ISH− subjects
TABLE 4.
Subjects with increased density of CD3, αβ and γδ T cells, HLA-DR expression, and transglutaminase 2–targeted IgA deposits in the gut mucosa
DISCUSSION
The current study suggests that the intestinal mucosa of type 1 diabetic patients is frequently infected by enteroviruses. The findings fit well with viral persistence, a phenomenon commonly observed in picornavirus infections and that can lead to prolonged inflammation and tissue damage (26). This observation also confirms our earlier study showing a similar phenomenon in a smaller subset of diabetic patients and control subjects (18).
Several factors suggest that this observation may have clinical significance. First, the presence of enterovirus RNA was associated with increased inflammation activity in the gut mucosa, including both cell-mediated and antibody-mediated inflammation (accumulation of IELs and IgA deposits). Second, the virus was more frequently detected in type 1 diabetic patients than in the control subjects, suggesting that it may play a role in the disease process. Third, our findings are consistent with prolonged infection and viral persistence in gut mucosa: the virus was found in the follow-up sample taken after 1 year’s observation in all three diabetic patients who were initially virus positive and from whom such follow-up samples were available. Viral persistence was also supported by the fact that the viral genome (RNA) occurred frequently in the absence of viral proteins, and this was seen more often in the patient group than in the control group. Such an imbalance between viral RNA and protein expression has earlier been linked to enterovirus persistence, showing that enterovirus capsid protein synthesis is decreased in persistent infection and that virus replication occurs mainly on the RNA level without producing infective virus (15,17). An analogous finding in human endomyocardial tissue shows a presence of enteroviral RNA without production of viral VP1 protein in idiopathic dilated cardiomyopathy and chronic coronary disease (27). Altogether, the current study fits well with previous observations in orally infected mice showing that enterovirus can persist in intestinal mucosa (14,28). Accordingly, the virus detected in these patients may have persisted for a longer period. However, in light of the current study, it is not possible to conclude whether it infected these individuals before or after the diagnosis of type 1 diabetes. Studies have shown that a large proportion of type 1 diabetic patients are already positive for enterovirus in blood at the diagnosis of diabetes, and even earlier during the pathogenesis, close to the appearance of islet autoantibodies (6,7,29,30).
It is possible that individuals susceptible to type 1 diabetes are prone to enterovirus infections and their persistence. In theory, such susceptibility could be mediated by genetic factors conferring susceptibility to type 1 diabetes. It is known that some of these susceptibility genes, such as HLA and IFIH1 genes, influence immune responses against enteroviruses (9,31). However, there are no studies available on the possible role of these genes in the development of enterovirus persistence. In the current study, the detection of enterovirus RNA was not associated with HLA risk genes for type 1 diabetes, arguing against a major effect of HLA genes. In addition to genetic factors, hyperglycemia is known to predispose to infections. Therefore, we cannot exclude the possibility that diabetes itself could predispose to prolonged enterovirus infections. However, the detection of enterovirus was not associated with the duration of type 1 diabetes, which indirectly speaks against this possibility.
The intensity of staining and the number of positive cells in ISH varied between the study groups, with patients with type 1 diabetes showing significantly stronger positivity than celiac disease patients and control subjects. This suggests that the virus replicates at a higher level in diabetic subjects, producing more RNA molecules and infecting more cells. The very weak positives were relatively frequent also among disease control subjects, suggesting that a low-grade infection may have contributed to the nonspecific gastrointestinal symptoms in some of them.
Virus RNA was found using different techniques including ISH and RT-PCR assays. Even though they gave parallel results, it was evident that enterovirus positivity was more frequent in ISH than in RT-PCR. This could be explained by the fact that two separate biopsy samples were needed for these two methods, as well as by the methodological differences in the pretreatment of the samples and possible presence of PCR inhibitors, which impair the sensitivity of PCR (18).
In conclusion, we propose that persistent enterovirus infection in gut mucosa may play a role in the pathogenesis of type 1 diabetes. Gut mucosa may be an important virus reservoir from which the virus can spread to the pancreas, which is anatomically very close and has common lymphatic and vasculature networks. Also, the viral persistence in gut mucosa may maintain chronic inflammation milieu in this network, which can promote islet autoreactivity by bystander activation mechanism. For example, γδ+ T cells, which were clearly associated with the presence of enterovirus in intestinal mucosa, facilitate autoimmunity in enterovirus-induced myocarditis through inhibiting T regulatory cell activity (32).
ACKNOWLEDGMENTS
This study was financially supported by grants from the Juvenile Diabetes Research Foundation, the Sohlberg Foundation, the Sigrid Juselius Foundation, the Academy of Finland, and the Competitive Research Funding of the Tampere University Hospital. Additionally, part of the study was funded by the PEVNET project (European Seventh Framework Programme GA-261441-PEVNET).
H.H. is a minor (<5%) shareholder and a member of the board at Vactech Ltd, which develops vaccines against picornaviruses. No other potential conflicts of interest relevant to this article were reported.
M.O. designed the study, participated in the development of the methods, analyzed data, contributed to data interpretation, and helped prepare the manuscript. S.T. designed the study, contributed to data interpretation, and helped prepare the manuscript. S.O. and T.H. participated in the development of the methods and helped prepare the manuscript. P.C. analyzed data and helped prepare the manuscript. I.R. helped prepare the manuscript. M.M. collected samples, contributed to data interpretation, and helped prepare the manuscript. K.K. collected samples, analyzed data, contributed to data interpretation, and helped prepare the manuscript. H.H. designed the study, contributed to data interpretation, and helped prepare the manuscript. M.O. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Eeva Tolvanen and Maarit Patrikainen (Department of Virology, School of Medicine, University of Tampere) and Eini Eskola and Marja-Leena Koskinen (Department of Pathology, School of Medicine, University of Tampere) for technical assistance.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1157/-/DC1.
REFERENCES
- 1.Hyöty H. Enterovirus infections and type 1 diabetes. Ann Med 2002;34:138–147 [PubMed] [Google Scholar]
- 2.Ylipaasto P, Klingel K, Lindberg AM, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 2004;47:225–239 [DOI] [PubMed] [Google Scholar]
- 3.Dotta F, Censini S, van Halteren AG, et al. Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci USA 2007;104:5115–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oikarinen M, Tauriainen S, Honkanen T, et al. Analysis of pancreas tissue in a child positive for islet cell antibodies. Diabetologia 2008;51:1796–1802 [DOI] [PubMed] [Google Scholar]
- 5.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 2009;52:1143–1151 [DOI] [PubMed] [Google Scholar]
- 6.Tauriainen S, Oikarinen S, Oikarinen M, Hyöty H. Enteroviruses in the pathogenesis of type 1 diabetes. Semin Immunopathol 2011;33:45–55 [DOI] [PubMed] [Google Scholar]
- 7.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ 2011;342:d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem 2009;284:13348–13354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth DJ, Cooper JD, Bailey R, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 2006;38:617–619 [DOI] [PubMed] [Google Scholar]
- 10.Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol 2010;84:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinig M, Petretto E, Wallace C, et al. ; Cardiogenics Consortium A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature 2010;467:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Li Y, McClean DR, et al. Detection of enterovirus capsid protein VP1 in myocardium from cases of myocarditis or dilated cardiomyopathy by immunohistochemistry: further evidence of enterovirus persistence in myocytes. Med Microbiol Immunol (Berl) 2004;193:109–114 [DOI] [PubMed] [Google Scholar]
- 13.Drescher KM, Sosnowska D. Being a mouse in a man’s world: what TMEV has taught us about human disease. Front Biosci 2008;13:3775–3785 [DOI] [PubMed] [Google Scholar]
- 14.Harrath R, Bourlet T, Delézay O, et al. Coxsackievirus B3 replication and persistence in intestinal cells from mice infected orally and in the human CaCo-2 cell line. J Med Virol 2004;74:283–290 [DOI] [PubMed] [Google Scholar]
- 15.Tam PE, Messner RP. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J Virol 1999;73:10113–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feuer R, Ruller CM, An N, et al. Viral persistence and chronic immunopathology in the adult central nervous system following Coxsackievirus infection during the neonatal period. J Virol 2009;83:9356–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klingel K, Hohenadl C, Canu A, et al. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc Natl Acad Sci USA 1992;89:314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oikarinen M, Tauriainen S, Honkanen T, et al. Detection of enteroviruses in the intestine of type 1 diabetic patients. Clin Exp Immunol 2008;151:71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998;15:539–553 [DOI] [PubMed] [Google Scholar]
- 20.Oikarinen M, Tauriainen S, Penttilä P, et al. Evaluation of immunohistochemistry and in situ hybridization methods for the detection of enteroviruses using infected cell culture samples. J Clin Virol 2010;47:224–228 [DOI] [PubMed] [Google Scholar]
- 21.Järvinen TT, Kaukinen K, Laurila K, et al. Intraepithelial lymphocytes in celiac disease. Am J Gastroenterol 2003;98:1332–1337 [DOI] [PubMed] [Google Scholar]
- 22.Holm K, Savilahti E, Koskimies S, Lipsanen V, Mäki M. Immunohistochemical changes in the jejunum in first degree relatives of patients with coeliac disease and the coeliac disease marker DQ genes. HLA class II antigen expression, interleukin-2 receptor positive cells and dividing crypt cells. Gut 1994;35:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmi TT, Collin P, Korponay-Szabó IR, et al. Endomysial antibody-negative coeliac disease: clinical characteristics and intestinal autoantibody deposits. Gut 2006;55:1746–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korponay-Szabó IR, Halttunen T, Szalai Z, et al. In vivo targeting of intestinal and extraintestinal transglutaminase 2 by coeliac autoantibodies. Gut 2004;53:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lönnrot M, Sjöroos M, Salminen K, Maaronen M, Hyypiä T, Hyöty H. Diagnosis of enterovirus and rhinovirus infections by RT-PCR and time-resolved fluorometry with lanthanide chelate labeled probes. J Med Virol 1999;59:378–384 [PubMed] [Google Scholar]
- 26.Colbere-Garapin F, Pelletier I, Ouxilou L. Persistent infections by picornaviruses. In Molecular Biology of Picornaviruses. Semler BLWE, Ed. Washington, DC, ASM Press, 2002, p. 437–448 [Google Scholar]
- 27.Andréoletti L, Bourlet T, Moukassa D, et al. Enteroviruses can persist with or without active viral replication in cardiac tissue of patients with end-stage ischemic or dilated cardiomyopathy. J Infect Dis 2000;182:1222–1227 [DOI] [PubMed] [Google Scholar]
- 28.Jaïdane H, Gharbi J, Lobert PE, et al. Prolonged viral RNA detection in blood and lymphoid tissues from coxsackievirus B4 E2 orally-inoculated Swiss mice. Microbiol Immunol 2006;50:971–974 [DOI] [PubMed] [Google Scholar]
- 29.Stene LC, Oikarinen S, Hyöty H, et al. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY). Diabetes 2010;59:3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikarinen S, Martiskainen M, Tauriainen S, et al. Enterovirus RNA in blood is linked to the development of type 1 diabetes. Diabetes 2011;60:276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadeharju K, Knip M, Hiltunen M, Akerblom HK, Hyöty H. The HLA-DR phenotype modulates the humoral immune response to enterovirus antigens. Diabetologia 2003;46:1100–1105 [DOI] [PubMed] [Google Scholar]
- 32.Huber SA, Born W, O’Brien R. Dual functions of murine gammadelta cells in inflammation and autoimmunity in coxsackievirus B3-induced myocarditis: role of Vgamma1+ and Vgamma4+ cells. Microbes Infect 2005;7:537–543 [DOI] [PubMed] [Google Scholar]