Abstract
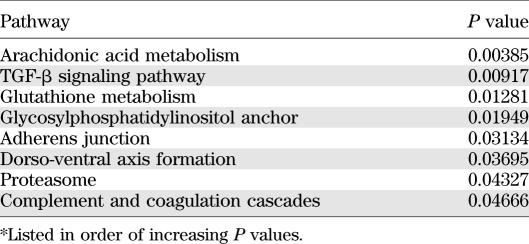
Clinical studies suggest metabolic memory to hyperglycemia. We tested whether diabetes leads to persistent systematic in vitro gene expression alterations in patients with type 1 diabetes (T1D) compared with their monozygotic, nondiabetic twins. Microarray gene expression was determined in skin fibroblasts (SFs) of five twin pairs cultured in high glucose (HG) for ∼6 weeks. The Exploratory Visual Analysis System tested group differences in gene expression levels within KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. An overabundance of differentially expressed genes was found in eight pathways: arachidonic acid metabolism (P = 0.003849), transforming growth factor-β signaling (P = 0.009167), glutathione metabolism (P = 0.01281), glycosylphosphatidylinositol anchor (P = 0.01949), adherens junction (P = 0.03134), dorsal-ventral axis formation (P = 0.03695), proteasome (P = 0.04327), and complement and coagulation cascade (P = 0.04666). Several genes involved in epigenetic mechanisms were also differentially expressed. All differentially expressed pathways and all the epigenetically relevant differentially expressed genes have previously been related to HG in vitro or to diabetes and its complications in animal and human studies. However, this is the first in vitro study demonstrating diabetes-relevant gene expression differences between T1D-discordant identical twins. These SF gene expression differences, persistent despite the HG in vitro conditions, likely reflect “metabolic memory”, and discordant identical twins thus represent an excellent model for studying diabetic epigenetic processes in humans.
There are indications from clinical studies that “metabolic memory”, i.e., epigenetic modifications that can sustain biologic responses to environmental perturbations, is operating in diabetes. Thus, the Diabetes Control and Complications Trial (DCCT) and the follow-up Epidemiology of Diabetes Interventions and Complications (EDIC) studies found that the benefits of reduced microvascular complications of intensified over standard glycemic control during the DCCT extended into the first several years of the EDIC study, even though the glycemic group differences were no longer present (1). This durable effect of glycemia was also postulated by us to explain some of the findings after successful pancreas transplantation. Thus, despite 5 years of insulin-free normoglycemia, established diabetic nephropathy lesions were unchanged from baseline (2). Only after 10 years of normoglycemia was reversal of lesions seen in the renal biopsies (3). This could have been due to metabolic memory causing persistence of the diabetic renal pathophysiology. Interestingly, the time frame of these pancreas transplant/diabetic nephropathy reversal studies is consistent with the duration of the durable benefit of intensified glycemic control on retinopathy complications in the EDIC follow-up studies (1).
There has been an explosion of new knowledge on the complex biology of epigenetic regulation of gene expression, including the interacting influences of DNA methylation and histone modifications and micro-RNA (miRNA) mechanisms (4). There are in vitro evidences of these processes in diabetes as originally suggested by Lorenzi and colleagues in 1990 (5), before many epigenetic concepts were developed. More recently, Villeneuve and Natarajan (6) reported histone modifications in genes relevant to diabetes and its complications in cells exposed to high glucose (HG). They also reported that the HG-induced increased expression of profibrotic genes such as collagen and connective tissue growth factor in cultured mesangial cells was associated with changes in key epigenetic histone lysine methylation marks at their promoters (7). HG also increased the expression of the nuclear factor-κB (NF-κB) p65 subunit, which was sustained after transfer of these human aortic endothelial cells to normal glucose (NG) (8). This was suggested to be through persistently increased H3K4me1 by the histone methyltransferase Set7 recruited to the p65 proximal promoter, and decreased histone 3 lysine 4 monomethylation (H3K9me) due to recruitment of the histone H3 demethylase LSD1 (lysine-specific demethylase 1). Thus, HG in vitro induces chromatin remodeling and epigenetic changes resulting, during subsequent incubation of cells in NG, in persistent increases in gene expression in pathways linked to the pathogenesis of diabetes complications. There is also growing interest in the role of miRNAs in the pathogenesis of diabetes complications (6). There is evidence for upregulation of miR-192, miR-216a, and miR-217 in association with increased collagen expression in mesangial cells treated with transforming growth factor-β (TGF-β) and in diabetic mice (6).
Lymphocytes from type 1 diabetic (T1D) patients showed upregulation of H3K9me2 in the promoter and coding regions of multiple genes (9), many of which were in autoimmune- and inflammation-related pathways (9). However, these studies did not determine the persistence of these in vitro phenomena, nor could they dissect differences consequent to the diabetic state from differences relating to the genetic propensity to develop diabetes (9). In order to address such questions, we have developed a human model for the study of epigenetic phenomena related to diabetes. Identical, or monozygotic, twins may have different phenotypes, despite being genetically identical. In fact, there are high discordance rates for common diseases between monozygotic twins, suggesting the involvement of environmental or epigenetic factors. Thus, identical twin studies can play an essential role in estimating phenotypic heritability, offering an opportunity to study epigenetic variations as dynamic quantitative traits (10). Here, we describe gene expression studies in cultured skin fibroblasts (SFs) from identical twins discordant for T1D.
We have previously demonstrated pathway differences in microarray gene expression studies in SFs grown in HG from T1D patients who were “fast” or “slow track” for the development of glomerular lesions and clinical manifestations of diabetic nephropathy, suggesting that these cells were genetically and/or epigenetically programmed to react differently under similar in vivo and identical in vitro conditions based on diabetic nephropathy risk. Our aim with the twins study was to determine whether such “programmed” behaviors in SFs from T1D patients studied in HG in vitro are at least in part epigenetic. Thus, if genetic, we would not expect to find systematic differences between identical twins, whereas if epigenetic, systematic differences would more likely be related to prior in vivo exposure to hyperglycemia in the T1D twin.
RESEARCH DESIGN AND METHODS
Subjects.
Participants were five identical twin pairs discordant for T1D. All T1D twins were ≤30 years old at diabetes onset and were on insulin within 6 months. Seven to eight highly variable DNA markers were tested on cheek cell swabs to confirm monozygosity with >99% certainty (Proactive Genetics, Martinez, GA). These studies were approved by the Committee for the Use of Human Subjects in Research of the University of Minnesota. All subjects were 10 years of age or older, had glycated hemoglobin (HbA1c) ≥7.0, and diabetes duration of at least 5 years. Informed consent (and assent in children) was obtained from all participants.
Clinical studies.
Blood pressure (BP) was measured with a Dinamap monitor. Hypertension was defined as BP ≥130/85 mmHg in adults or >95th percentile for age and sex norms in children (11) or by the use of antihypertensive drugs. HbA1c was measured with the Tosoh method (Tosoh Medics).
Skin biopsy and cell culture.
The methods for skin biopsy, SF culture, and RNA isolation have been previously detailed (12–15). Cells were grown, from acquisition, in HG (25 mmol/L). At confluency, these cells are long and spindle shaped and grow in multilayer. Cells are coded by the single technician performing the explants but not the gene expression studies. Those were done by a technician who was masked to the subjects’ identity. Forth- to sixth-passage SFs were seeded in 150-cm2 flasks at a density of 104 cells/cm2 and grown in Dulbecco's modified Eagle's medium (DMEM) in HG with 10% FCS and antibiotics for 24 h. After exposure to serum-deprived DMEM for 48 h, SFs were cultured for an additional 36 h in HG medium supplemented with 10% FCS.
RNA isolation.
Total RNA was isolated and its integrity evaluated using the RNA 6000 LabChip kit and Agilent 2100 bioanalyzer (Hewlett Packard, Palo Alto, CA).
Microarray: target preparation, hybridization, and data filtering.
Microarray expression was performed using the Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Santa Clara, CA), which analyzes the expression level of >47,000 transcripts and variants, including 38,500 well-characterized human genes, and is comprised of more than 54,000 probe sets. In brief, biotinylated cRNA are generated from samples of total RNA (15). The cRNA hybridization, all further processing steps, and data scanning and acquisition were performed at the Affymetrix/University of Minnesota Core Laboratory of the Biomedical Genomics Facility.
These microarray data were then preprocessed using Genedata Expressionist Pro 4.5 software (GDE; Genedata Inc., Waltham, MA), available at the Minnesota Supercomputing Institute. Quality analyses and data processing were run in the “Refiner” module using a variety of algorithms, including Affymetrix MAS 5, which normalizes and scales the data, assesses the signal to noise (S/N), and determines gene transcript “present” or “absent” status. Codes for a patient’s grouping were broken, and the GDE “Analyst” module was used for statistical testing by paired Student t test (P ≤ 0.05) for differential gene expression and data plotting. The selected gene expression level data were then used for pathway analysis (15).
Pathway analysis.
The primary objective for the use of this pathway analysis methodology was to determine whether differentially expressed genes were over-represented or enriched in particular biochemical pathways defined by the KEGG pathway database (available online from http://www.genome.jp/kegg/pathway.html). For each pathway we constructed a 2 × 2 contingency table with columns labeled “significant genes” (K) and “nonsignificant genes” (N-K) and rows labeled “present in pathway” (M) and “not present in pathway” (N-M) for a total of N genes. We then determined whether the significance of gene expression differences is independent of being present in a given pathway using a Fisher exact test that is based on the hypergeometric distribution. We also performed a 1,000-fold permutation test that randomly redistributes genes across pathways to generate a null distribution of significant gene counts within each pathway. We confirmed that both approaches to determining statistical significance gave nearly identical results. All enrichment analyses were conducted using the Exploratory Visual Analysis software at (http://endeavour.dartmouth.edu/eva/EVA.html) (15–17). A pathway was considered to have a statistically significant enrichment of differentially expressed genes at a type I error rate of α = 0.05. This level of significance was selected to maximize our power to identify pathways that are associated with in vivo exposure to hyperglycemia in the diabetic twins. To help guard against type I errors, we determined, as part of our interpretation, whether prior evidence exists from human, in vitro, or animal studies for the pathways’ relationship to diabetes and/or its complications. For those more concerned about type I than type II errors, it is possible to apply a more stringent significance cutoff and subsequent interpretation.
RESULTS
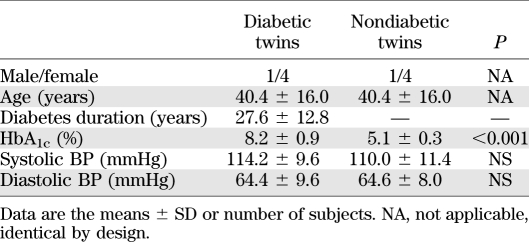
Clinical and demographic characteristics of these discordant identical twins are presented in Table 1.
TABLE 1.
Demographic and clinical characteristics of diabetic and nondiabetic twins
Pathway analyses.
There were eight pathways that had a significantly greater proportion of genes that were differentially expressed between the discordant twin pairs than expected by chance (Table 2).
TABLE 2.
Pathway analyses*
Other genes.
Interestingly, in paired analyses, we also found differences between these identical twins in the expression levels of genes regulating epigenetic processes. Thus, SET 7 (histone lysine methyltransferase) (P = 0.0281), H3K4 methyltransferase (MLL3) (P = 0.0021), and histone deacetylase 8 (P = 0.031) were downregulated whereas histone deacetylase 4 was upregulated (P = 0.0186) in the T1D twin.
DISCUSSION
There are at least 32 peer-reviewed publications from 12 separate laboratories supporting the in vitro study of SFs as a model to study diabetes and its complications, especially diabetic nephropathy. Skin biopsies are well accepted by research subjects and SFs grow well under appropriate culture conditions, retaining many of their original phenotypic characteristics. Using a culture system whereby cells from different individuals are exposed to identical culture conditions for several weeks and multiple passages likely reduces influences of acute metabolic disturbances may thus better reflect longer-term epigenetic influences of the diabetic state in these genetically identical T1D discordant twins.
We incubated the SFs in HG in order to seek proof for the concept that SF in vitro behaviors can reflect memory for the in vivo metabolic differences between the diabetic and nondiabetic identical twins. Thus, this study was designed to test whether long-term hyperglycemia changes cellular behavior in a persistent and systematic way, above and beyond the effects of several weeks of in vitro exposure of the nondiabetic twins’ cells to these HG levels. In this in vitro condition, systematic differences between the twin pairs would more likely reflect the long-term consequences of in vivo hyperglycemia rather than acute effects of HG on SF behaviors. Studies of these cells under NG conditions would also be of interest, perhaps revealing time lines and mechanisms for reversal of the in vivo effects.
Statistical hypothesis testing seeks to find a balance between maximizing the power (1-type II error) to detect a valid signal and minimizing the probability of finding a false-positive (type I error). This balance between type I and type II errors is often a source of philosophical debate between experimental biologists and statisticians. Statisticians tend to be more concerned about type I errors and thus establish very conservative significance criteria that control for false-positives in the context of multiple testing. Statistical methods such as Bonferroni that correct for all tests performed assume that the universal null hypothesis of no valid relationships is true. Experimental biologists, on the other hand, are often more concerned with type II errors and are unwilling to assume that the universal null hypothesis is true. This is because biological experiments almost always build on the prior work of others. Given a set of prior work, biologists can use their knowledge of the end point being studied and its related biological processes to help determine whether a set of statistical results are likely to provide evidence in support of a given hypothesis. Here we carried out statistical analyses to determine whether differentially expressed genes are enriched in biological pathways defined by the KEGG database. We established a liberal statistical significance criteria of α = 0.05 that was uncorrected for the number of pathways that we examined. We chose not to correct for the number of pathways considered because the existing biological knowledge about diabetes and its complications suggests that the universal null hypothesis is unlikely to be true. Further, we chose to maximize our power to detect pathways by using a more liberal significance threshold. Using this approach, we found a total of eight pathways that were enriched for differentially expressed genes. The statistical concern is that a liberal threshold would yield a number of false-positives. We chose to address this concern through biological interpretation using prior knowledge from published studies. We review these results below. Suffice it to say that with more conservative statistical approaches, the present studies could be entirely unrevealing, whereas potentially important observations emerged with the more liberal approaches used here. Particularly in very slowly progressing chronic conditions such as diabetes complications, subtle cellular perturbations sustained over decades are more likely to occur than more dramatic pathway disturbances. If so, new analytic strategies may be needed to better understand these processes.
Identical twins discordant for diabetes could represent an ideal human model for the study of epigenetic processes related to the diabetic state. Although systematic differences between these twin pairs could also represent differences relating to the triggering factors leading to diabetes, this is unlikely given that such factors probably vary among T1D patients and may be unlikely to be influencing cell behavior so many years after T1D onset. Moreover, one might predict such long-lasting influences of T1D triggers to be in inflammatory pathways, but, in one study, monocyte gene expression profiles did not differ among T1D-discordant identical twins, whereas both the diabetic and nondiabetic twins differed from controls in such pathways (18). In a study of Vα24JαT-cell clones activated by anti-CD3, differences between discordant identical twin pairs were found in a large number of genes in multiple pathways, not restricted to inflammatory processes (19). Importantly, the gene expression differences found in the current study were in pathways that are known to be affected when normal cells are placed in HG, and in pathways that are, in most instances, highly relevant to diabetes complications.
For example, the cyclooxygenase direction of the arachidonic acid pathway can initiate the inflammatory response through conversion of arachidonic acid into an unstable endoperoxide intermediate that is converted to prostacyclin and thromboxane A2, resulting in activation of NF-κB and inducible nitric oxide synthase, culminating in mitochondrial stress, which has been linked to diabetes complications (20). The 12-lipoxygenase direction of the arachidonic acid pathway mediates some actions of angiotensin II, including increasing extracellular matrix synthesis in mesangial cells (21). 12-Lipoxygenase also has a role in p38-mitogen activated protein (MAP) kinase and α5 (IV) collagen synthesis in diabetic rats and cultured mouse podocytes (22), linking this pathway to diabetic nephropathy. This link was strengthened by the demonstration that the 12-lipoxygenase genotype is associated with albuminuria in T2D patients with poor glycemic control (23).
The TGF-β pathway has long been considered to play a pivotal role in diabetes complications (24,25). Thus, as reviewed elsewhere (24,25), studies in cell culture systems, animal models, and humans support the hypothesis that increased TGF-β production mediates hypertrophic and fibrotic manifestations of diabetic nephropathy. This appears to result from the convergence of oxidative stress, nonenzymatic glycation, and hemodynamic processes on the upregulation of TGF-β signaling and, consequently, increases in extracellular matrix molecule production by glomerular, tubular, and interstitial cells (24,25).
Glutathione metabolism, representing one of the most important antioxidant pathways, has been linked to diabetes complications (26–29). Ceriello et al. (28) reported inadequate antioxidant enzyme responses to HG, including that of glutathione peroxidase in SFs of T1D patients with diabetic nephropathy, whereas these responses were normal in T1D patients without nephropathy. We found no statistically significant evidence of heritability in SF glutathione peroxidase gene expression in T1D sibling pairs who were concordant for diabetic nephropathy lesions (14), which is consistent with epigenetic explanations for the SF behavior differences noted by Ceriello et al. (28).
The complement pathway has also been linked to diabetes complications. The eyes of T2D patients showed deposition of the C5b-9 terminal product of complement activation (the membrane attack complex) in luminal endothelium (30) and choriocapillaris (31). These findings were confirmed in streptozotocin-diabetic rats and in T2D patients’ eyes, and were associated with decreased levels of glycosylphosphatidylinositol-anchored complement inhibitors (30), thus linking with yet another differentially expressed pathway among the discordant identical twins. Membrane attack complex deposition was also found in endoneurial microvessels of patients with diabetic neuropathy (32), and in glomeruli, tubular basement membranes, and vessel walls in advanced diabetic nephropathy (33). Mannose binding lectin (MBL) can directly activate the complement system. High levels of MBL early in the course of T1D independently predicted the later development of overt nephropathy. Moreover, high MBL expression genotypes were more common in T1D patients with overt nephropathy (34–36).
Increased endothelial permeability has been associated with both diabetic nephropathy and retinopathy (37,38). Endothelial permeability can be modulated by alterations in the adherens junction pathway proteins. Vascular endothelial cadherin (VE-cadherin) associates with vascular endothelial growth factor (VEGF) receptor 2, markedly reducing VEGF's induction of phospholipase Cγ and MAP kinase (39). HG induces F-actin reorganization and associated adherens junction proteins in cultured rat heart endothelial cells (40). Increased retinal vascular permeability in diabetic rats was associated with decreased VE-cadherin expression (41). Interestingly, VE-cadherin is also a critical endothelial regulator of TGF-β signaling through recruitment of components of the TGF-β receptor complex, including TGF-β receptor and endoglin (42). We found that both latent TGF binding protein-1, necessary for local TGF-β activity (12), and endoglin (43) gene expression were decreased in SFs of T1D patients with very slow versus rapid development of diabetic nephropathy and versus normal controls, which is consistent with protective mechanisms.
As already mentioned, oxidative stress mechanisms are likely very important in the pathogenesis of diabetic nephropathy (14,26–29). Based on an earlier report suggesting that inhibition of the proteasome protects oxidative stress–induced endothelial dysfunction (44), Luo et al. (45) tested this idea in a diabetic rat model. Nontoxic proteasome inhibition reduced proteinuria and diabetic nephropathy lesions, upregulated Nrf2, and improved the diminished renal antioxidant enzyme gene expression in diabetic rats (45).
Interestingly, there were also systematic differences in expression in genes important in epigenetic processes. For example, the histone methyltransferase Set7 was increased in expression in the nondiabetic twin SFs exposed to HG in vitro. As mentioned above, transient exposure of aortic endothelial cells and normal mice to HG led to persistent aortic endothelial cell epigenetic changes in the promoter of the NF-κB p65 subunit. Apparently, transient HG increased H3K4me1 in the promoter region of p65 by Set7, causing a sustained increase in p65 gene expression and, consequently, in potentially damaging NF-κB-responsive genes (8). We also found increased expression of the H3K4 methyltransferase (MLL3) gene in SFs of nondiabetic twins exposed to HG. Studies showed that the TGF-β1–induced increases in the expression of α1 (I) collagen, connective tissue growth factor, and plasminogen activator inhibitor 1 in cultured rat mesangial cells correlated positively with increased H3K4me1, H3K4me2, and H3K4me3 at their promoters (7). The histone deacetylase 8 gene, also upregulated in nondiabetic twins exposed to HG, has been shown to be activated in the retina and its capillary cells in diabetic rats, and this persists despite the subsequent induction of good glycemic control (46). Bovine retinal endothelial cells exposed to HG for 4 days showed similar responses (46). Finally, we also found downregulation of histone deacetylase 4 in the nondiabetic twins. Exposure of rat cardiac myocytes to HG resulted in a reduction in the proportion of acetylated histone-4 associating with the insulin-like growth factor 1 receptor (IGF-1R) promoter. HG also resulted in decreased IGF-1R mRNA and IGF-1R protein levels, and this was linked to the HG-induced increase in apoptosis in these cells (47).
In summary, these studies show that, despite 6 weeks and multiple passages in identical in vitro HG conditions, SFs of the T1D twins have gene expression differences from nondiabetic identical twins in pathways that have, in most instances, been previously implicated in diabetes complications. Although there are currently no statistical procedures that allow us to calculate the probability of identifying such pathway clustering, as opposed to random unassociated pathways, intuitively, the likelihood of this happening by chance would seem to be extremely low. Similarly, the probability that the differences in expression levels of genes related to epigenetic processes would all be for molecules previously reported to be persistently altered by HG in different cell lines and in some animal models would also appear to be very small. Moreover, given that the nondiabetic twin cells were in HG for weeks and that persistent epigenetic alterations can be induced in cells within 16 h (8), differences between these discordant twins may have been blunted. It is, therefore, possible that a more complete picture of the prolonged impact of in vivo hyperglycemia would emerge from in vitro discordant identical twin studies that are also carried out in an NG environment. It should also be noted that many studies from different laboratories have demonstrated multiple in vitro behavior differences between SFs of T1D patients with and without diabetic nephropathy. Since, regularly, T1D cohorts with diabetic nephropathy have, on average, worse glycemia, and given the strong evidence for familial concordance for diabetic nephropathy risk (48–54), it will be important to design studies that can dissect the genetic versus the epigenetic processes in the pathogenesis of diabetes complications.
ACKNOWLEDGMENTS
This work was supported by a research grant from the Juvenile Diabetes Research Foundation (JDRF) and by funds from the Pennock Professorship. M.L.C. is a recipient of a JDRF Career Development Award.
No potential conflicts of interest relevant to this article were reported.
M.L.C. and M.M. researched data and wrote the manuscript. Y.K. and N.K. reviewed and edited the manuscript and contributed to discussion. J.H.M. and J.C.M. analyzed data, reviewed and edited the manuscript, and contributed to discussion. S.S.R. reviewed and edited the manuscript. M.L.C. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Cathy Bagne (University of Minnesota) for her clinical coordinator efforts, Paul Walker (University of Minnesota) for his technical work, and Patricia L. Erickson (University of Minnesota) for her assistance in manuscript preparation.
REFERENCES
- 1.White NH, Sun W, Cleary PA, et al. Prolonged effect of intensive therapy on the risk of retinopathy complications in patients with type 1 diabetes mellitus: 10 years after the Diabetes Control and Complications Trial. Arch Ophthalmol 2008;126:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fioretto P, Mauer SM, Bilous RW, Goetz FC, Sutherland DER, Steffes MW. Effects of pancreas transplantation on glomerular structure in insulin-dependent diabetic patients with their own kidneys. Lancet 1993;342:1193–1196 [DOI] [PubMed] [Google Scholar]
- 3.Fioretto P, Steffes MW, Sutherland DER, Goetz FC, Mauer M. Reversal of lesions of diabetic nephropathy after pancreas transplantation. N Engl J Med 1998;339:69–75 [DOI] [PubMed] [Google Scholar]
- 4.Chernov AV, Baranovskaya S, Golubkov VS, et al. Microarray-based transcriptional and epigenetic profiling of matrix metalloproteinases, collagens, and related genes in cancer. J Biol Chem 2010;285:19647–19659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy S, Sala R, Cagliero E, Lorenzi M. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA 1990;87:404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol 2010;299:F14–F25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 2010;21:2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao F, Smith DD, Zhang L, Min A, Feng W, Natarajan R. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 2008;57:3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet 2011;27:116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaelber DC, Pickett F. Simple table to identify children and adolescents needing further evaluation of blood pressure. Pediatrics 2009;123:e972–e974 [DOI] [PubMed] [Google Scholar]
- 12.Huang C, Kim Y, Caramori MLA, et al. Cellular basis of diabetic nephropathy II: the TGF β-system and diabetic nephropathy lesions in type 1 diabetes. Diabetes 2002;51:3577–3581 [DOI] [PubMed] [Google Scholar]
- 13.Huang C, Kim Y, Caramori ML, et al. Cellular basis of diabetic nephropathy: III. In vitro GLUT1 mRNA expression and risk of diabetic nephropathy in type 1 diabetic patients. Diabetologia 2004;47:1789–1794 [DOI] [PubMed] [Google Scholar]
- 14.Caramori ML, Kim Y, Fioretto P, et al. Cellular basis of diabetic nephropathy: IV. Antioxidant enzyme mRNA expression levels in skin fibroblasts of type 1 diabetic sibling pairs. Nephrol Dial Transplant 2006;21:3122–3126 [DOI] [PubMed] [Google Scholar]
- 15.Huang C, Kim Y, Caramori ML, et al. Diabetic nephropathy is associated with gene expression levels of oxidative phosphorylation and related pathways. Diabetes 2006;55:1826–1831 [DOI] [PubMed] [Google Scholar]
- 16.Reif DM, Dudek SM, Shaffer CM, Wang J, Moore JH. Exploratory visual analysis of pharmacogenomic results. Pac Symp Biocomput 2005;10:296–307 [PubMed] [Google Scholar]
- 17.Reif DM, Moore JH. Visual analysis of statistical results from microarray studies of human breast cancer. Oncol Rep 2006;15(Spec no):1043–1047 [DOI] [PubMed] [Google Scholar]
- 18.Beyan H, Drexhage RC, van der Heul Nieuwenhuijsen L, et al. Monocyte gene-expression profiles associated with childhood-onset type 1 diabetes and disease risk: a study of identical twins. Diabetes 2010;59:1751–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson SB, Kent SC, Horton HF, et al. Multiple differences in gene expression in regulatory Valpha 24Jalpha Q T cells from identical twins discordant for type I diabetes. Proc Natl Acad Sci USA 2000;97:7411–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayilavarapu S, Kantarci A, Fredman G, et al. Diabetes-induced oxidative stress is mediated by Ca2+-independent phospholipase A2 in neutrophils. J Immunol 2010;184:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy MA, Adler SG, Kim Y-S, et al. Interaction of MAPK and 12-lipoxygenase pathways in growth and matrix protein expression in rat mesangial cells. Am J Physiol Renal Physiol 2002;283:F985–F994 [DOI] [PubMed] [Google Scholar]
- 22.Kang S-W, Natarajan R, Shahed A, et al. Role of 12-lipoxygenase in the stimulation of p38 mitogen-activated protein kinase and collagen α5(IV) in experimental diabetic nephropathy and in glucose-stimulated podocytes. J Am Soc Nephrol 2003;14:3178–3187 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Freedman BI, Burdon KP, et al. Association of arachidonate 12-lipoxygenase genotype variation and glycemic control with albuminuria in type 2 diabetes. Am J Kidney Dis 2008;52:242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziyadeh FN. Different roles for TGF-β and VEGF in the pathogenesis of the cardinal features of diabetic nephropathy. Diabetes Res Clin Pract 2008;82(Suppl. 1):S38–S41 [DOI] [PubMed] [Google Scholar]
- 25.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-β as a key mediator. Diabetes 1995;44:1139–1146 [DOI] [PubMed] [Google Scholar]
- 26.Dave GS, Kalia K. Hyperglycemia induced oxidative stress in type-1 and type-2 diabetic patients with and without nephropathy. Cell Mol Biol (Noisy-le-grand) 2007;53:68–78 [PubMed] [Google Scholar]
- 27.Yildirim Z, Uçgun NI, Kiliç N, Gürsel E, Sepici-Dinçel A. Antioxidant enzymes and diabetic retinopathy. Ann N Y Acad Sci 2007;1100:199–206 [DOI] [PubMed] [Google Scholar]
- 28.Ceriello A, Morocutti A, Mercuri F, et al. Defective intracellular antioxidant enzyme production in type 1 diabetic patients with nephropathy. Diabetes 2000;49:2170–2177 [DOI] [PubMed] [Google Scholar]
- 29.Hodgkinson AD, Bartlett T, Oates PJ, Millward BA, Demaine AG. The response of antioxidant genes to hyperglycemia is abnormal in patients with type 1 diabetes and diabetic nephropathy. Diabetes 2003;52:846–851 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Gerhardinger C, Lorenzi M. Early complement activation and decreased levels of glycosylphosphatidylinositol-anchored complement inhibitors in human and experimental diabetic retinopathy. Diabetes 2002;51:3499–3504 [DOI] [PubMed] [Google Scholar]
- 31.Gerl VB, Bohl J, Pitz S, Stoffelns B, Pfeiffer N, Bhakdi S. Extensive deposits of complement C3d and C5b-9 in the choriocapillaris of eyes of patients with diabetic retinopathy. Invest Ophthalmol Vis Sci 2002;43:1104–1108 [PubMed] [Google Scholar]
- 32.Rosoklija GB, Dwork AJ, Younger DS, Karlikaya G, Latov N, Hays AP. Local activation of the complement system in endoneurial microvessels of diabetic neuropathy. Acta Neuropathol 2000;99:55–62 [DOI] [PubMed] [Google Scholar]
- 33.Falk RJ, Dalmasso AP, Kim Y, et al. Neoantigen of the polymerized ninth component of complement. Characterization of a monoclonal antibody and immunohistochemical localization in renal disease. J Clin Invest 1983;72:560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovind P, Hansen TK, Tarnow L, et al. Mannose-binding lectin as a predictor of microalbuminuria in type 1 diabetes: an inception cohort study. Diabetes 2005;54:1523–1527 [DOI] [PubMed] [Google Scholar]
- 35.Hansen TK, Tarnow L, Thiel S, et al. Association between mannose-binding lectin and vascular complications in type 1 diabetes. Diabetes 2004;53:1570–1576 [DOI] [PubMed] [Google Scholar]
- 36.Østergaard J, Hansen TK, Thiel S, Flyvbjerg A. Complement activation and diabetic vascular complications. Clin Chim Acta 2005;361:10–19 [DOI] [PubMed] [Google Scholar]
- 37.Nørgaard K, Jensen T, Feldt-Rasmussen B. Transcapillary escape rate of albumin in hypertensive patients with type 1 (insulin-dependent) diabetes mellitus. Diabetologia 1993;36:57–61 [DOI] [PubMed] [Google Scholar]
- 38.Chagnac A, Korzets A, Zevin D, et al. Effect of enalapril on the microvascular albumin leakage in patients with diabetic microangiopathy and normal or mildly elevated blood pressure. Clin Nephrol 1994;41:144–149 [PubMed] [Google Scholar]
- 39.Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE-cadherin in the control of vascular permeability. J Cell Sci 2008;121:2115–2122 [DOI] [PubMed] [Google Scholar]
- 40.Lee H-Z, Yeh F-T, Wu C-H. The effect of elevated extracellular glucose on adherens junction proteins in cultured rat heart endothelial cells. Life Sci 2004;74:2085–2096 [DOI] [PubMed] [Google Scholar]
- 41.Davidson MK, Russ PK, Glick GG, Hoffman LH, Chang MS, Haselton FR. Reduced expression of the adherens junction protein cadherin-5 in a diabetic retina. Am J Ophthalmol 2000;129:267–269 [DOI] [PubMed] [Google Scholar]
- 42.Rudini N, Felici A, Giampietro C, et al. VE-cadherin is a critical endothelial regulator of TGF-β signalling. EMBO J 2008;27:993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez-Muñoz P, Mauer M, Kim Y, et al. Cellular basis of diabetic nephropathy: V. Endoglin expression levels and diabetic nephropathy risk in patients with type 1 diabetes. J Diabetes Complications 2010;24:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenz M, Wilck N, Meiners S, et al. Proteasome inhibition prevents experimentally induced endothelial dysfunction. Life Sci 2009;84:929–934 [DOI] [PubMed] [Google Scholar]
- 45.Luo ZF, Qi W, Feng B, et al. Prevention of diabetic nephropathy in rats through enhanced renal antioxidative capacity by inhibition of the proteasome. Life Sci 2011;88:512–520 [DOI] [PubMed] [Google Scholar]
- 46.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem 2010;110:1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu XY, Geng YJ, Liang JL, et al. High levels of glucose induce apoptosis in cardiomyocyte via epigenetic regulation of the insulin-like growth factor receptor. Exp Cell Res 2010;316:2903–2909 [DOI] [PubMed] [Google Scholar]
- 48.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 1989;320:1161–1165 [DOI] [PubMed] [Google Scholar]
- 49.Borch-Johnsen K, Nørgaard K, Hommel E, et al. Is diabetic nephropathy an inherited complication? Kidney Int 1992;41:719–722 [DOI] [PubMed] [Google Scholar]
- 50.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996;39:940–945 [DOI] [PubMed] [Google Scholar]
- 51.Freedman BI, Tuttle AB, Spray BJ. Familial predisposition to nephropathy in African-Americans with non-insulin-dependent diabetes mellitus. Am J Kidney Dis 1995;25:710–713 [DOI] [PubMed] [Google Scholar]
- 52.Faronato PP, Maioli M, Tonolo G, et al. ; The Italian NIDDM Nephropathy Study Group Clustering of albumin excretion rate abnormalities in Caucasian patients with NIDDM. Diabetologia 1997;40:816–823 [DOI] [PubMed] [Google Scholar]
- 53.Canani LH, Gerchman F, Gross JL. Familial clustering of diabetic nephropathy in Brazilian type 2 diabetic patients. Diabetes 1999;48:909–913 [DOI] [PubMed] [Google Scholar]
- 54.Fava S, Azzopardi J, Hattersley AT, Watkins PJ. Increased prevalence of proteinuria in diabetic sibs of proteinuric type 2 diabetic subjects. Am J Kidney Dis 2000;35:708–712 [DOI] [PubMed] [Google Scholar]