Abstract
Background
RNA degradation is critical to the survival of all cells. With increasing evidence for pervasive transcription in cells, RNA degradation has gained recognition as a means of regulating gene expression. Yet, RNA degradation machinery has been studied extensively in only a few eukaryotic organisms, including Saccharomyces cerevisiae and humans. Giardia lamblia is a parasitic protist with unusual genomic traits: it is binucleated and tetraploid, has a very compact genome, displays a theme of genomic minimalism with cellular machinery commonly comprised of a reduced number of protein components, and has a remarkably large population of long, stable, noncoding, antisense RNAs.
Results
Here we use in silico approaches to investigate the major RNA degradation machinery in Giardia lamblia and compare it to a broad array of other parasitic protists. We have found key constituents of the deadenylation and decapping machinery and of the 5'-3' RNA degradation pathway. We have similarly found that all of the major 3'-5' RNA degradation pathways are present in Giardia, including both exosome-dependent and exosome-independent machinery. However, we observe significant loss of RNA degradation machinery genes that will result in important differences in the protein composition, and potentially functionality, of the various RNA degradation pathways. This is most apparent in the exosome, the central mediator of 3'-5' degradation, which apparently contains an altered core configuration in both Giardia and Plasmodium, with only four, instead of the canonical six, distinct subunits. Additionally the exosome in Giardia is missing both the Rrp6, Nab3, and Nrd1 proteins, known to be key regulators of noncoding transcript stability in other cells.
Conclusions
These findings suggest that although the full complement of the major RNA degradation mechanisms were present - and likely functional - early in eukaryotic evolution, the composition and function of the complexes is more variable than previously appreciated. We suggest that the missing components of the exosome complex provide an explanation for the stable abundance of sterile RNA species in Giardia.
Background
Cells control RNA levels through the regulation of both transcription and degradation. Organisms must degrade not only aberrantly folded or misprocessed RNAs, but also functional RNA transcripts that are no longer needed by the cell. In order to distinguish among and degrade only the appropriate RNA transcripts, cells have developed multiple RNA degradation processes and complexes. RNA degradation can occur by digestion inwards from the ends, using 5' to 3' and 3' to 5' exonucleases, or by digestion at internal sites using endonucleases. RNA degradation often, though not always, also involves deadenylation (for both 5' to 3' and 3' to 5' degradation) and decapping (for 5' to 3' degradation) of the transcripts. The range of RNA degradation machinery includes both nuclear and cytoplasmic components.
RNA degradation typically begins with the shortening of the long poly-A tail of mRNA transcripts. Although some RNA degradation pathways can apparently act on polyadenylated transcripts, e.g. nonstop decay (NSD), nonsense-mediated decay (NMD), and endonucleolytic cleavage (RNAi), the majority of 5' to 3' and 3' to 5' exonucleolytic activities require the prior removal of the poly-A tail. Two proteins, Ccr4p and/or Caf1p, have been shown to act as the catalytic core of the deadenylation machinery in all eukaryotic cells examined to date [1-3]. 5' to 3' degradation depends on the prior removal of the 5' cap structure followed by subsequent 5' to 3' exonucleolytic cleavage. In most cells this is performed by the Dcp1p/Dcp2p holoenzyme with the involvement of a wide and diverse array of additional protein machinery [4-6].
The exosome, a 3' to 5' exonuclease complex, is one of the important RNA degradation complexes and can be used as a means of classifying the different machinery into two groups: the exosome-dependent and the exosome-independent pathways. It is this convention that we use here to describe the RNA degradation machinery in Giardia lamblia and other parasitic protists. Exosomes have clear homologs in all three domains of life. Bacteria, archaea, and eukaryotes possess functionally analogous core 3' to 5' RNA degradation complexes in the bacterial polynucleotide phosphorylase (PNPase), the archaeal exosome, and the eukaryotic exosome, respectively. The similarity in the structure of all three mRNA degradation complexes is striking and suggests that the highly conserved structures are necessary for mRNA degradation and have been maintained throughout evolutionary history.
The bacterial PNPase exists as a homotrimer, in which each monomer possesses two tandem RNase PH domains in addition to single S1 and KH domains [7]. RNase PH domains have exonucleolytic activity, although only one of the two domains in each monomer is thought to be active, while the S1 and KH domains have RNA binding capacity [7]. The archaeal exosome ring is composed of repeating Rrp41/42 heterodimers arranged into a hexamer with three total copies of the stabilizing proteins Rrp4 and Csl4 acting as caps to the complex. The Rrp41 and Rrp42 subunits possess RNase PH domains, but Rrp41 is the only exonucleolytic component of the complex, again resulting in three active sites per complex [7,8]. Rrp4 and Csl4 both possess S1 domains and bind RNA.
In eukaryotes, the core exosome also exists as a ring structure made of a heterohexamer of proteins with RNase PH domains (the three Rrp 41-like proteins are Rrps41, 46 and Mtr3, and the three Rrp 42-like proteins are Rrps42, 43, and 45) with a trio of additional RNA-binding proteins which contain S1 domains (Rrps 4, 40, and Csl4) that broadly act as the entry to the pore of the exosome and in eukaryotic exosomes further act to stabilize the hexameric ring [9,10]. It is believed that, through gene duplication either early in the eukaryotic lineage or prior to the divergence of eukaryotes, rrp41 gave rise to both rrp46 and mtr3, while rrp42 gave rise to rrp43 and rrp45 [7]. The ring and stabilizing proteins are commonly associated with Rrp6 and Rrp44, both of which possess nucleolytic activity [9,11]. The core proteins display homology to archaeal exosome and bacterial PNPase proteins, whereas Rrp6 and Rrp44 display homology to bacterial RNases [12-14]. In some eukaryotes, the RNase PH domain of Rrp41 provides exonucleolytic activity, whereas in other species the activity is dependent upon Rrp6 and Rrp44 [9,11,15].
Although the exosome is an important complex involved in RNA degradation in eukaryotes, additional complexes also play a role in RNA degradation either through exosome-dependent or exosome-independent processes. Exosome-dependent complexes act mainly by preparing RNA substrates for degradation by the exosome, whereas exosome-independent complexes possess nucleolytic activity of their own. These additional complexes impart specificity to its function so that RNA is not degraded prematurely, and only a subset of RNA is targeted at any one time.
The exosome-dependent machinery includes the TRAMP complex, Pumilio (Puf) proteins, Nonsense mediated decay (NMD), Nonstop decay (NSD), and No-go decay (NGD) complexes. In the nucleus, the exosome can be found to be associated with the TRAMP complex, which aids in the degradation, maturation, and removal of secondary structures of RNA molecules through the post-transcriptional addition of a poly-A tail by TRAMP proteins Trf4/5 or via the helicase activity of TRAMP protein Mtr4, respectively [16,17]. In the cytoplasm a subset of Puf proteins bind mRNAs via sequence-specific elements in the 3' untranslated regions (UTRs) and recruit the deadenylation machinery [18,19]. Also in the cytoplasm, the NMD, NSD, and NGD pathways act as mRNA quality control and are activated in response to mRNAs containing premature termination codons (PTCs), no stop codon, or secondary structures such as stem loops, respectively. The NMD complex may possess endonucleolytic activity, but requires the exosome for complete degradation of RNAs.
The exosome-independent complexes are the RNAi machinery and the Ccr4-Not complex. RNAi acts to silence gene expression through endonucleolytic degradation of targeted mRNA transcripts or translation inhibition. RNAi machinery has been identified in a variety of eukaryotic organisms, from single-celled organisms to metazoans but is not ubiquitously present. Two components of the Ccr4-Not complex, which is conserved from S. cerevisiae to humans [20], have roles in mRNA deadenylation; Ccr4 and Caf1 deadenylate mRNA transcripts, although optimal degradation for many transcripts still requires the exosome [21].
The parasitic protists regulate gene expression through many different mechanisms, both transcriptional and post-transcriptional. Yet, while we understand much about transcriptional regulation, the study of mRNA degradation machinery in the parasitic protists is still in its early stages. This is perhaps especially surprising because RNA degradation is likely to play an unusually prominent role in organisms that exhibit diminished regulation of gene expression at the transcriptional level, as is known to be the case for several parasitic protists. For example, Trypanosoma brucei transcribes its genes polycistronically, implying that mRNA processing and degradation are its primary means of regulating gene expression [22-24]. And Giardia lamblia transcribes an abundance of full-length sterile antisense transcripts that are capped and polyadenylated [25], suggesting a role for mRNA degradation to eliminate these aberrant transcripts.
In this paper, we discuss our efforts to identify the mRNA degradation machinery in Giardia lamblia using in silico approaches. We additionally included several other parasitic protists (Entamoeba histolytica, Trichomonas vaginalis, Trypansoma brucei, and Plasmodium falciparum) in our analyses of the core and peripheral exosome components for comparison, building on the work of previous researchers in this field [1,23,24,26-33]. We focused especially on Giardia given its evolutionary divergence [34,35], severely reduced repertoire of transcriptional machinery [34,36], and unusual patterns of gene expression [37,38]. We identified an extensive collection of genes coding for proteins with significant sequence similarity to proteins that participate in RNA degradation pathways in other eukaryotes. Pathways such as the RNAi [39-41] and nonsense mediated decay pathways [28,42] previously have been identified in Giardia. However, these comparisons also revealed that a substantial number of protein constituents of mRNA degradation complexes in other eukaryotes are either absent or sufficiently divergent to thwart detection by similarity searches in these parasitic protists. We use this new knowledge to consider which protein components may comprise the most reduced core exosome structure in eukaryotes and to postulate explanations for observed patterns of mRNA transcripts in Giardia.
Results
Preparing the transcripts for degradation
Deadenylation machinery
Deadenylation, the removal of the 3' polyA tail, is typically performed by Ccr4p and Caf1p of the Ccr4-Not complex found in most eukaryotes studied to date, while the Pan2p/Pan3p complex plays a secondary role [1,3]. The Ccr4-Not complex is a multi-subunit complex with roles in transcription, mRNA and protein degradation, and cell division [20]. Members of the complex are the Not proteins 1-5, Ccr4, and the Caf proteins, Caf1 (also known as Pop2), Caf40, and Caf130. The Not1 protein acts as a scaffold to which other members of the complex attach and is necessary for S. cerevisiae viability [20]. Pop2p acts a scaffold for Ccr4p and also may have additional roles in decapping of transcripts in S. cerevisiae [2].
Giardia possesses only a subset of the genes associated with the Ccr4-Not complex that may be sufficient for its functionality in the parasite (Additional Files 1 and 2). We identified four candidate Not genes, as well as candidate genes for Caf1, confirming a previous identification of Caf1 in Giardia [1]. The classification of three of the Not genes as Not1, Not2, and Not4 was possible based on the presence of defined domains. The protein identified as most similar to Not1p has only a partial Not1 domain, while Not2p and Not4p possess complete domains. The fourth protein could possibly be either Not3p or Not5p based on sequence similarity search results, although the better scores and E-values obtained against Not3 proteins suggests it is more likely Not3p. Although we were unable to detect homologs of Ccr4, Caf40, Caf130 or CNot10 in the Giardia genome, the presence of the Not proteins and Caf1p should be sufficient to ensure a functional deadenylation complex.
The Pan2-Pan3 complex is thought to play a role in either initiating or subsequently trimming the poly-A tails in support of Ccr4-Caf1-Not complex activity. The complex functions through association of Pan3 with Pabp1 to recognize and associate with the poly-A tail of transcripts. We were able to detect genes with sequence similarity to Pan2 and Pabp1 in Giardia but could not detect a Pan3 homolog, raising questions about whether this complex would be present and functional in the parasite (Additional Files 1 and 2). A mammalian deadenylase, PARN, was also missing from Giardia. Of the other parasitic protists examined to date for deadenylation machinery, Trypanosoma brucei has been shown to contain both the Ccr4 -Not complex and the Pan2-Pan3 complex [1,26], while Plasmodium falciparum contained a majority of the required components of the Ccr4-Not complex but was also missing the Pan2-Pan3 complex [27].
Decapping machinery
All parasitic protists examined to date contain traditional 5' 7-methylguanosine caps, although a biochemically modified cap structure is found on the spliced leader RNA in Trypanosoma. In Giardia the presence of 5' 7-methylguanosine caps have been identified on both coding and sterile transcripts [25]. The removal of this cap is required for 5'-3' exonuclease processing. Decapping in other cells requires the decapping holoenzyme, comprised of Dcp1p and the catalytic subunit Dcp2p. A wide array of other machinery has been shown to interact with and mediate the activity of Dcp in vitro and in vivo, including Lsm (like-Sm) proteins, the Upf proteins, the Edc proteins, Dhh1p and Pat1p; while more recent research has suggested the presence of additional members of the decapping family, such as DcpS and Headless, and an astonishing diversity of proteins associated with mRNA storage and decay (e.g. TTP/Brf1 and 2; AUF1; HuR; KSRP; CUG-BP) in specific lineages, making it clear that the process of mRNA decay is both precise and organism-specific [43,44]. These proteins and the decapped mRNA often assemble into degradation complexes referred to as P-bodies (processing bodies).
Here we identify a Dhh1p and a divergent Giardia Dcp2-like protein (Additional Files 1 and 2). Like many eukaryotic helicases, Dhh1p protein possesses a DEXDc superfamily domain and when reciprocally BLAST against S. cerevisiae, the top identified gene is the Dhh1. The S. cerevisae Dcp2 protein has both dcp2 and nudix domains. The dcp2 domain aids in mRNA cap removal while the nudix hydrolase domain catalyzes catabolism of nucleotide diphosphates linked to other molecules. The identified Giardia protein contains a nudix hydrolase domain but is missing the dcp2 domain. However, when used as a query in BLASTp, significant hits identify it as a likely mRNA decapping enzyme and a possible Dcp2 homolog. We were unable to identify the Edc proteins, Pat1p, or any of the organism-specific alternative decapping machinery and P-body components, such as DcpS, Headless, TTP/Brf1and2. AUF1, etc. We think it likely therefore that the evolutionary divergence of Giardia has dictated either a unique array of P-body proteins specific to the Diplomond lineage or perhaps P-bodies are simply absent from Giardia.
Another set of proteins which have roles in decapping and mRNA degradation are the Lsm (like-Sm) proteins. Members of the Sm/Lsm protein family are conserved in bacteria and archaea [45]. This family of proteins plays roles in RNA processing, splicing and mRNA decapping [46,47] and some of the proteins colocalize with the mRNA decapping machinery [48]. The Lsm complex involved in mRNA degradation is made of Lsm proteins 1-7 and they form a heteroheptameric ring. In total we were able to identify 14 proteins with Sm-like domains using the Interpro domain accessory function in the Giardia genome database (Additional File 2). Of the 14 proteins, 12 had Conserved Domain Database (CDD) recognizable Lsm domains, yet only four of these proteins possess notable sequence similarity to Lsm proteins in S. cerevisiae. The remaining 8 proteins are likely members of the Sm protein family which have roles in pre-mRNA splicing [46]. The results we see here are consistent with what is seen in the other complexes which we observed, in that some machinery is present and recognizable while other components cannot be identified. The homologous complex in bacteria, known as the Hfq complex is a homohexamer [45], therefore possibilities exist that Lsm complex may still form in Giardia despite only identifying four Lsm proteins.
5' to 3' Degradation
Once mRNA transcripts are decapped, they are accessible to the 5' to 3' degradation machinery. The proteins responsible for 5' to 3' degradation of eukaryotic mRNAs are the Xrn proteins [6,49]. S. cerevisiae possesses two Xrn proteins: Rat1p (nuclear) and Xrn1p (cytoplasmic). The nuclear form is responsible for nuclear processing of RNAs while Xrn1p is responsible for degradation of cytoplasmic mRNA transcripts. The XRN domain is located in the N-terminus of the protein and provides 5' to 3' exonucleolytic activity. Although S. cerevisiae has two homologs, T. brucei possesses four Xrnp homologs [33]. All four are expressed in T. brucei, although only two of the four are needed for growth. In our search, we were able to identify two proteins with sequence similarity to Xrnp in Giardia (GL50803_24133 and GL50803_113365) (Additional Files 1 and 2). The former is more similar to S. cerevisiae Rat1p whereas the latter has more similarity with Xrn1p. The presence of these two exonucleases supports the likely presence of a functional 5' to 3' degradation pathway.
3' to 5' Degradation
Identification of exosome components
Eleven main proteins comprise the eukaryotic exosome: six RNase PH proteins (Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p, and Mtr3p), three stabilizing proteins (Rrp4p, Rrp40p and Csl4p), and two peripheral proteins (Rrp6p and Rrp44p). Rrp6p and Rrp44p are not always found in the exosome complex together within the same organism. For example, S. cerevisiae Rrp6p and Rrp44p are found only with the nuclear exosome and in both nuclear and cytoplasmic exosomes, respectively [13,14], while human Rrp6, and Rrp44 proteins (hDIS3 and hDIS3L) are localized to the nucleolus, the nucleus and cytoplasm, and primarily the cytoplasm respectively [50]. Human DIS3 and DIS3L display a lower affinity for the exosome than the S. cerevisiae homolog. In S. cerevisiae, H. sapiens, and A. thaliana, we found all eleven of the previously identified exosome genes, validating our similarity search protocol. However, in our investigation of the parasitic protists, we were unable to identify some specific exosome components. We first discuss our detailed characterization of the putative exosome proteins in Giardia, followed by our characterization of the exosome protein components in the other parasitic protists.
In the search for exosome components in Giardia, we had differential success depending on whether we used the PFAM or the CDD definitions. Using multiple approaches we identified only four of the expected six RNase PH domain proteins (Additional Files 1 and 2). GL50803_1890, GL50803_5632, and GL50803_40007 all contain RNase PH domains; GL50803_9847 does not have a true RNase PH domain but possesses the CDD multi-domain COG2123 that is associated with RNase PH domains (Additional File 1). It is unclear what arrangement of the four RNase PH domain proteins comprises the hexameric exosome in Giardia. Continuing this theme of genomic diminution, we were able to identify genes for only two of the three associated proteins, rrp4 and rrp40 but not csl4. We were also able to identify a gene for only one of the two exosome peripheral proteins: rrp44, identifiable by its RNase II-like domain (RNB), but not rrp6, or any other DNA Q domain-containing proteins. In addition to the putative Rrp44p homolog, we also were able to identify a second protein (GL50803_9912) containing an RNase II like domain in Giardia, but it displayed a sufficiently low similarity with Rrp44p that we cannot confirm a possible identity. The schematics in Additional File 1 provide a visual representation of the identified proteins in Giardia and their similarity to S. cerevisiae RNA degradation proteins.
In the other parasitic protists, T. brucei, P. falciparum, T. vaginalis, and E. histolytica, the identified protein repertoires for exosomes are more complete but often still partial (Table 1). T. brucei and E. histolytica had six proteins containing RNase PH domains, the correct number to construct a heterohexameric exosome. We note that our ability to find all six RNase PH domain proteins in T. brucei by similarity searches represents an improvement compared to previous studies [22,24] and indicates a high level of sensitivity for divergent protein sequences in our study. T. vaginalis had seven identifiable proteins containing RNase PH domains; this number more than accounts for enough proteins to form the hexameric exosome ring and reflects a common theme of gene family expansion in T. vaginalis [51]. However, P. falciparum, like Giardia, has only four proteins with recognizable RNase PH domains. Previous work from the DeRisi group was able to identify only three RNase PH proteins in P. falciparum. That work was published in 2007, where genome annotation was less complete than when we performed our search. They performed BLASTP and reciprocal BLASTP to identify the putative degradation proteins in Plasmodium [27].
Table 1.
Exosome components in the parasitic protists
| Core and peripheral eukaryotic exosome components | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. cerevisae | H. sapiens | A. thaliana | T. brucei (TREU) | P. falciparum | T. vaginalis |
E. histolyica (HM-1_IMSS) |
G. lamblia (WB) | Canonical Archaeal exosome | |
| Ring Components | |||||||||
| rrp41 | YGR195W | *NP_061910 | *AT3G61620 | Tb927.10.7450 | *NP_342241 | ||||
| rrp42 | YDL111C | NP_055819 | Tb927.1.2580 | NP_342240 | |||||
| rrp43 | YCR035C | NP_852480 | Tb11.01.8320 | ** | |||||
| rrp45 | YDR280W | NP_001029366 NP_005024 | AT4G27490 AT3G46210 AT3G07750 AT3G12990 AT3G60500 AT1G60080 | Tb927.6.670 | PF14_0256 PFB0415c MAL13P1.204 PF13_0340 | TVAG_250040 TVAG_441560 TVAG_287740 TVAG_027130 TVAG_453750 TVAG_189430 TVAG_220800 | EHI_040320 EHI_166910 EHI_126330 EHI_086520 EHI_188080 EHI_000580 | GL50803_ 1890 GL50803_5632 GL50803_40007 GL50803_ 9847 | ** |
| rrp46 | YGR095C | NP_064543.3 | Tb927.2.2180 | ** | |||||
| mtr3 | YGR158C | NP_478126 | Tb11.01.2820 | ** | |||||
| Stabilizers | |||||||||
| rrp4 | YHR069C | NP_055100 | *AT1G03360 | *Tb927.7.4670 | PFD0515w | TVAG_246740 | EHI_163510 | GL50803_33022 | NP_342242 |
| rrp40 | YOL142W | NP_057126 | AT2G25355 AT4G32175 | Tb09.160.5160 | MAL13P1.36 | TVAG_380110 | EHI_004770 | GL50803_17091 | ** |
| csl4 | YNL232W | NP_057130 | AT5G38890 | Tb927.5.1200 | MAL7P1.104 | TVAG_110240 TVAG_121320 | U | U | NP_341842 |
| Peripheral | |||||||||
| rrp6 | *YOR001W | *NP_001001998 NP_002676 | AT5G35910 AT2G32415 AT1G54440 |
Tb927.4.1630 | MAL13P1.311 PF14_0473 | TVAG_053630 TVAG_197890 TVAG_283650 | EHI_021400 EHI_064630 | U | ** |
| rrp44 | *YOL021C | NP_001121698 NP_055768 | AT2G17510 AT1G77680 AT5G02250 |
Tb11.02.5380 Tb11.01.0260 | MAL13P1.289 | TVAG_311220 | EHI_160720 | GL50803_112718 | ** |
The chart displays a selection of organisms and the components of their exosomes divided into the ring, stabilizing and peripheral components. Representative metazoan eukaryotes, S. cerevisiae, H. sapiens, and A. thaliana, are identified on the left most side of the chart, while several species of single-celled eukaryotic parasites are listed in the middle and right side of the chart. The far right column displays the typical proteins composing an archaeal exosome. Components of the exosome whose activity has been confirmed experimentally are marked with asterisks. Gene annotations are from Saccharmoyces Genome Database (S. cerevisiae), National Center for Biotechnology Information (H. sapiens sapiens and canonical archaeal exosome S. solfataricus), The Arabidopsis Information Resource (A. thaliana), and Eukaryotic Pathogen Database (T. brucei, P. falciparum, T. vaginalis, E. histolytica, and G. lamblia). Boxes are coded as follows; normal font/identified, bold font/unable to identify the exact identity, **/non-existant, and U/not identified.
Additionally, T. brucei, P. falciparum, and T. vaginalis possess all three exosome stabilizing proteins, whereas E. histolytica like Giardia, has only rrp4 and rrp40, but lacks a csl4 homolog (Table 1). Unlike Giardia all four of these parasitic protists contained putative homologs for both rrp6 and rrp44.
Exonucleolytic potential of RNase PH and Rrp44 homologous proteins
To define the exonucleolytic potential of the degenerate Giardia exosome, we first sought to classify the RNase PH proteins as either Rrp41-like or Rrp42-like. The archaeal exosome is composed of three Rrp41-42 heterodimers, and the exonucleolytic activity is maintained solely in the Rrp41p subunit [13,52]. In eukaryotic species the exosome is composed of three Rrp41-like and three Rrp42-like proteins, although the exonucleoytic activity of the Rrp41-like proteins is sometimes lost due to mutation of the catalytic or the phosphate binding sites [8]. Since the eukaryotic exosome likely evolved from the archaeal exosome, we expected to be able to identify at least one Rrp41-like and Rrp42-like protein in each of the parasitic protists. With the goal of identifying whether the RNase PH proteins were Rrp41-like or Rrp42-like in their origin, we constructed protein phylogenies.
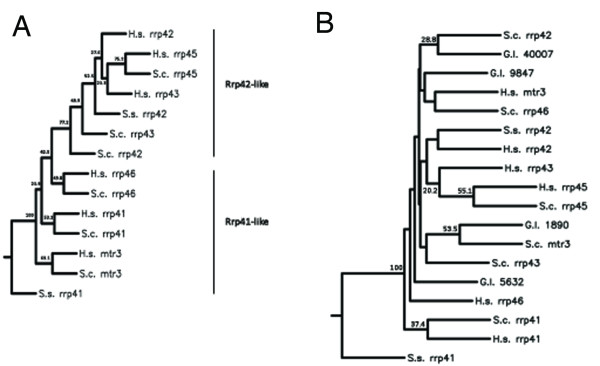
Our first step was construction of a protein phylogeny using Rrp41p-like and Rrp42p-like amino acid sequences from S. cerevisiae, H. sapiens, and S. solfataricus RNase PH protein amino acid sequences. Protein sequences were aligned in MUSCLE and trimmed using G-blocks as stated in the methods to limit our analysis to the more highly conserved regions of the proteins. This resulted in protein fragments of 110-115 amino acids from proteins of ~240 and ~300 amino acids for Rrp41-like and Rrp42-like proteins, respectively, for use in construction of the phylogeny. The retained amino acid sequences contained the RNase PH domain, with approximately ten amino acids flanking each end. The Phylip 3.69 package was run iteratively to generate 100 trees for estimation of bootstrap values. We observed distinct stratification of Rrp41-like and Rrp42-like proteins; however, node values showed only moderate support for placement into each Rrp subtype (Figure 1A), indicating high sequence divergence of these proteins.
Figure 1.
Rnase PH protein phylogenies reveal an absence of two subunits in Giardia. Gene trees were made to diagram the relatedness of the RNase PH protein amino acid sequences in S. serevisisae, H. sapiens, S. solfataricus, and G. lamblia. Node values shown are out of 100 and nodes with no labels had values below 20. (A) The protein parsimony tree displays relatedness between RNase PH proteins of S. cerevisiae, H. sapiens, and S. solfataricus. The tree is separated into Rrp41-like and Rrp42-like proteins with node values providing moderate support for the inferred clades. (B) Addition of putative Giardia RNase PH proteins results in disrupted clades and reduced bootstrap values.
However, with the addition of Giardia sequences, this divergence was profoundly exacerbated, and we observed considerable movement of sequences from previously strong clades that drastically altered the tree topology, eroding the stratification of the Rrp41-like and Rrp42-like proteins (Figure 1B). This phenomenon was observed even with the addition of single Giardia sequences (data not shown). Our inability to group these RNase PH proteins was not specific to Giardia, as similar difficulties were encountered with the use of RNase PH proteins from the other parasitic protists. Although frustrated in our attempt to classify the RNase PH domain proteins, our inability to produce informative gene trees about protein identity argues that high levels of sequence divergence are tolerated in the proteins that comprise the exosome ring.
As an alternative approach to classification of Giardia's RNase PH domain-containing proteins, we sought to determine which of the proteins possess the necessary amino acid residues for phosphate binding and catalytic activity. The amino acid alignment of the four genes containing the Giardia's RNase PH domains shows that while all possess the necessary residues for activity of the catalytic site, only GL50803_5632 has even one of the two amino acids required for phosphate binding (not shown). This technique, while not helping to classify the Giardia RNase PH proteins, does suggest that all RNase PH domain-containing genes in Giardia are nucleolytically inactive.
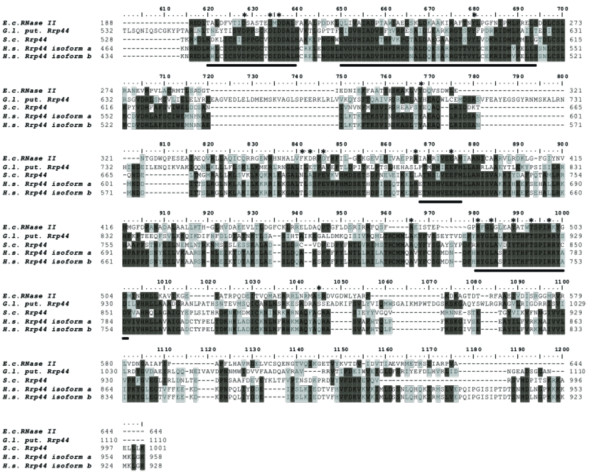
In contrast, examination of the putative Giardia Rrp44 homolog revealed that this enzyme is most likely an active enzyme with exonucleolytic activity. When compared with Escherichia coli RNase II, S. cerevisae and H. sapiens Rrp44 proteins, the putative Giardia homolog shares a high percentage of catalytic residues present in the enzymatic pocket [12,53], although it is missing the classical PIN domain characteristic of some classes of nucleases [53] (Figure 2). The E. coli RNase II has 17 identified residues that are found in the catalytic site of the enzyme that are important for enzyme catalysis [12,53]. Human Rrp44p homologs possess 13 of the 17 residues found within the catalytic site [53], while Giardia and S. cerevisiae each possess 12 of the 17 amino acids. In one position in both S. cerevisiae and Giardia Rrp44 proteins, there is an amino acid substitution where a negatively charged amino acid is replaced with another negatively charged amino acid; S. cerevisiae has an aspartate in place of glutamate at D363 while Giardia has an aspartate in place of glutamate at D360. The substitution is not likely to result in alterations in bond forming. In addition, the putative Giardia Rrp44 homolog possesses all four of the domains believed to play a role in the RNA binding activity of the enzyme. These RNA binding domains are highly conserved in species as divergent as H. sapiens and Giardia. Based on the presence of these highly conserved domains, and the number of residues identified as part of the catalytic sites, we believe that it is likely that the putative Giardia Rrp44p homolog possesses exonucleolytic activity and thus the exosome in conjunction with putative Rrp44p in Giardia is also likely active. The absence of an Rrp6p homolog means that Rrp44p is potentially the only nucleolytically active enzyme in the Giardia exosome complex.
Figure 2.
Identification of Rrp44 protein catalytic potential in Giardia. Alignment of E. coli RNase II with Rrp44 amino acid sequences from G. lamblia (G.l.), S. cerevisiae (S.c.), and H. sapiens (H.s.) show levels of high conservation. Important regions required for RNA binding are underlined. Dark gray represents amino acid identity while the light gray represents amino acid similarity. The conserved amino acids in the catalytic pocket of E. coli RNase II are noted by an asterisk above the sequence.
Exosome-dependent complexes and proteins
TRAMP complex
The TRAMP complex is a nuclear-localized, exosome-associated protein complex, responsible for polyadenylation of RNA molecules that will be targeted for further processing or degradation [16,17,54]. The RNA binding proteins Air1p/2p are responsible for identification and binding of the targeted RNA transcripts. The poly-A polymerase-like proteins, Trf4p/5p are responsible for the addition of the poly-A tails to all RNA species that are incorrectly folded or aberrantly produced. Lastly Mtr4p, an RNA helicase, is thought to remove the secondary structure of folded transcripts. The Trf5p/Air1p/Mtr4p complex localizes mainly to the nucleolus, while the Trf4p/Air2p/Mtr4p complex is found throughout the remaining nucleus in S. cerevisiae [16].
We identified genes with sequence and domain similarity to Trf4/5 and Mtr4 in Giardia (Additional Files 1 and 2). The Giardia Trf4p/5p proteins contain the canonical nucleotidyltransferase domains, suggesting that they are functionally capable of adding poly-A tails to RNAs. However, we identified only one gene with a partial Air1 domain in the Giardia genome. S. cerevisiae proteins Air1p and Air2p both contain an Air1 domain, and deletion of either Air1p or 2p does not inhibit the processing of some snoRNAs and rRNAs, while deletion of both proteins leads to a dramatic increase in unprocessed snoRNAs [17]. Therefore, it is possible that both Giardia TRAMP complexes function normally in the presence of a single Air1 domain-containing protein.
Puf proteins
Members of the Puf family of proteins are identified in a variety of eukaryotes with a variable number of genes in each species [55-57]; different members of the Puf protein family are either nuclear or cytoplasmic in their localization. Although in some cell types, Puf proteins act to stabilize mRNA transcripts and increase levels of translation, more typically Puf proteins reduce mRNA expression either through inhibition of translation or through mediation of mRNA decay. For example, Puf5 in yeast binds to Pop2 (Caf1) to recruit the Ccr4p deadenylase to the 3' UTRs of mRNAs in the cytoplasm [18,19]. Given the unusually short 3' UTRs of Giardia mRNAs, it was not apparent that the Puf proteins would be relevant in the parasite, but we were able to identify a full repertoire of five genes containing Puf repeats in Giardia. Higher eukaryotes generally have fewer members of the family [56-58]. Puf proteins normally possess eight Puf repeats in the C-terminus [56], although studies in S. cerevisiae suggest that six Puf repeats are sufficient for RNA binding [59]. Of the five identified proteins in Giardia, one contains eight Puf repeats, three contain between five and seven repeats in the C-terminal half of the protein, and one may be a pseudogene with only a partial N-terminal RNA binding domain with only three repeats (Additional Files 1 and 2). Amino acid sequence comparison with S. cerevisiae Puf protein sequences suggests that the putative Giardia Puf proteins may be homologs of S. cerevisae Puf proteins that primarily bind ribosomal RNAs and mRNAs encoding nuclear localized proteins. This designation was obtained using Giardia Puf proteins as query for BLASTP in the Saccharomyces genome database.
Nonsense-mediated decay
Nonsense-mediated decay (NMD) is responsible for the translation-coupled degradation of mRNA transcripts containing premature termination codons (PTCs) [60,61], Yet, while PTCs are defined as any stop codon upstream of an exon junction complex, NMD is functional in single exon transcripts. Thus, while stop codons upstream of exon junction complexes may trigger the NMD mechanism, they are not the only stimulus. An alternative explanation is that the distance between the stop codon and the poly-A binding protein (PABP) is what triggers NMD, such that increasing the distance between the stop codon and the PABP increases the chance of NMD activation.
The NMD complex has been previously shown to be functional in Giardia, although the precise means by which transcripts lacking introns in Giardia are targeted for degradation by the pathway has yet to be identified [28,42]. These studies by Sun and colleagues identified 7 of 14 NMD associated factors by sequence similarity. The putative homologs of the following NMD genes in Giardia were identified: upf1, eRF1 and eRF3, sMG1, hrp1, xrn1 and xrn2, and ski7 (Additional File 2) [28].
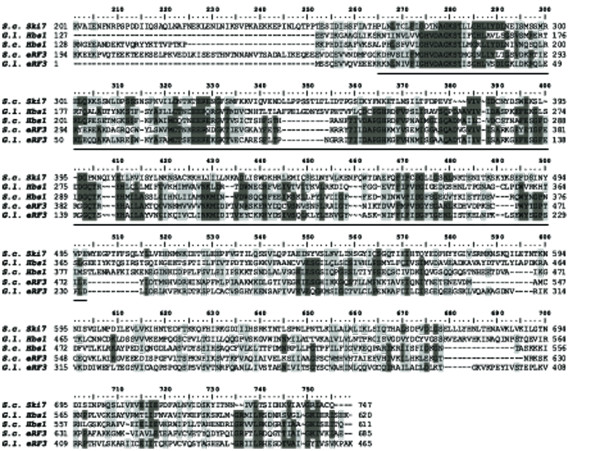
eRF1p and eRF3p are eukaryotic release factors that function in normal translation termination to remove the ribosome from the mRNA [62]. The proteins Ef1αp, eRF3p, Hbs1p, and Ski7p belong to the EF1 family, and all possess elongation factor GTP binding domains (Additional File 1). We believe that the proposed Ski7p homolog was incorrectly identified by Sun and colleagues and is actually the Giardia Hbs1p homolog. Amino acid alignments indicate that the Giardia Hbs1 protein sequence is approximately twice as similar to the S. cerevisiae eRF3 protein sequence as it is to the S. cerevisiae Ski7 protein sequence (Figure 3). Additionally, Hbs1 proteins possess two translation factor domains, both of which are absent from Ski7 (Additional File 1) and are present in the proposed Giardia Ski7p protein. With the alignment data, domain identification, and with the knowledge that ski7 has only previously been found in some Saccharomyces species [62], we believe that the gene previously identified at ski7 is actually hbs1. However, the relationship between the members of the protein family make it likely that Ski7p and other EF1αp homologs may be interchangeable in their functionality in different eukaryotes, so that even without Ski7p, mRNA quality control pathways such as NMD and NSD may still remain functional [62].
Figure 3.
Identification of EF1 family proteins in Giardia. Alignment of members of the EF1 family from G. lamblia (G.l.) and S. cerevisiae (S.c.). Important regions required for RNA binding are underlined. Dark gray represents amino acid identity while the light gray represents amino acid similarity. The GTPase domains common to members of the family are underlined. The Hbs1 and eRF3 homologs are on average twice as similar to each other than they are to Ski7.
Non-stop decay
Acting as a counterpoint to the NMD mechanism of RNA degradation, and sharing some of the same protein machinery, non-stop decay (NSD) is again translation-dependent but is alternatively activated when the ribosome fails to encounter a stop codon during translation of an mRNA transcript [63,64]. In S. cerevisiae, the ribosome continues reading into the poly-A tail and stalls at the end of the transcript, followed by recruitment of Ski7p - and possibly other proteins - to the ribosome. Given that Ski7p has only been identified in a subset of Saccharomyces species, and a NSD-like mechanism has been functionally identified in HeLa cells with the demonstration that an mRNA transcript lacking a stop codon is degraded more quickly than the same transcript with a stop codon [64], it seems likely that machinery beyond Ski7 must be involved, although the HeLa cell machinery responsible for nonstop transcript degradation has not been identified. In Giardia we identified proteins with sequence and domain similarity to eRF3p and Hbs1p along with two identical Ef1α homologs. Since these proteins all share functional domains with Ski7p (Additional Files 1 and 2), they may possibly be able to trigger a NSD like mechanism in the absence of Ski7p [62,64].
No-go decay
Like NMD and NSD, No-go decay (NGD) is also translation-dependent, but it is triggered when the ribosome stalls upon encountering secondary structure in an mRNA transcript. Stalling of the ribosome due to other mRNA sequence-related factors can also induce NGD [65]. The stalling of the ribosome causes the recruitment of Dom34p and Hbs1p; Dom34p is a homolog of eRF1p, while Hbs1p is a homolog of eRF3p; Endonucleolytic cleavage ensues and the two halves of the mRNA transcript are degraded. Giardia possesses proteins with sequence and domain similarity to both Dom34p and Hbs1p (Additional Files 1 and 2).
RNA interference machinery
The RNAi machinery consists of Dicer and Argonaute proteins. Dicer possesses single Paz and RNase III domains [66-68]. The guide strand of the dsRNA is loaded into an Argonaute homolog that forms the RNA-induced silencing complex (RISC) [66,67]. Argonaute homologs contain one Piwi and one Paz domain each [68]; the Piwi domain binds the RNA at its 3' end, while the PAZ domain possesses a nuclease-like activity that cleaves or inhibits translation of the bound target mRNA transcript. In some organisms, the dsRNA signal can be amplified through the use of an RNA dependent RNA polymerase (RdRP), which uses the Dicer product as a primer for subsequent dsRNA synthesis.
RNAi has been documented in a broad range of eukaryotes such as T. brucei, Drosophila melanogaster, H. sapiens, and Schizosaccharomyces pombe with roles in gene silencing and heterochromatin formation [5,47]. However, RNAi machinery is absent from other eukaryotes such as P. falciparum and S. cerevisiae [68,69], and in Toxoplasma gondii, the RNAi machinery has resisted functional characterization [70]. Giardia's RNAi machinery has been previously well-characterized and consists of an RdRP, Dicer and Argonaute homologs [39,68,71] (Additional File 2). Knockdown of Giardia RdRP or Dicer results in altered gene expression, especially concerning the variant surface proteins (VSP) on the plasma membrane of trophozoites, while Argonaute is an essential gene [39]. The observation of different effects in Giardia depending on which gene in the RNAi pathway is knocked down, leads us to believe that some proteins in the pathway have roles in the cell beyond that of just RNAi.
Discussion
The importance of RNA degradation is evident from the multitude of pathways and mechanisms present in eukaryotic cells. RNA transcripts must be degraded to remove both non-functional transcripts and transcripts that are no longer needed for translation. Indeed, given the recent discovery of the ubiquitous nature of transcription along eukaryotic genomes [72], there is a rising appreciation for the role of RNA degradation in the regulation of gene expression. The exosome serves as a basic machinery of RNA degradation in eukaryotic cells, but the complex does not act alone. Additional complexes such as TRAMP in the nucleus and NSD, NMD, and NGD in the cytoplasm assist the exosome with degradation of mRNA transcripts. Furthermore, complexes such as the RNAi pathway degrade dsRNA into shorter transcripts that are used for gene regulation and are not dependent on the exosome for their function. The diversity of these complexes allows complete degradation of all RNA species found in the cell.
In this study we used a bioinformatics approach to investigate the exosome of the parasitic protists with a special focus on the RNA degradation machinery of Giardia lamblia. Although our analyses described here specified only the Giardia Genome isolate, we found that Giardia isolates P15 and GS possessed the same exosome components as the Genome strain (data not shown). We were able to identify several putative components of Giardia's exosome, including four RNase PH proteins for the ring of the exosome; Rrp4 and Rrp40, as stabilizing proteins for the ring structure; and lastly an Rrp44 homolog that we predict possesses the requisite nucleolytic activity. There are no trends in the expression patterns of proteins from the different complexes, when looking at normal growth and development, and also stress responses of Giardia.
One of the most striking results of our research is the detection of only four RNase PH proteins. Our findings expand the repertoire of putative RNase PH proteins beyond the two (A8BNT9/GL50803_1890 and C6LWS9/GL50803_5632) identified through a bioinformatics analysis in a recent publication from Clayton and colleagues [73]. The other paper did not define their search parameters in detail, and we suspect that the difference in our findings is the consequence of more relaxed search parameters in the searches described in this paper. We stress, however, that all four putative RNase PH proteins passed a reciprocal BLASTp search test and contained either strong consensus PH domains, or, in the case of GL50803_9847, the more generic, but related COG2123 domain. Likewise, we identified four putative RNase PH proteins in Plasmodium falciparum, one more than DeRisi and colleagues identified [27], although the fourth one has subsequently been identified in online databases. Given the extreme sequence divergence that often characterizes proteins in the parasitic protists compared to other eukaryotes, it is possible that an in silico approach is inadequate to identify the full protein family. Indeed, this proved to be the case in earlier studies in T. brucei, P. falciparum, and C. reinhardti [24,27,74]. However, our search algorithm was capable of detecting all six RNase PH domain proteins in T. brucei, suggesting an enhanced sensitivity relative to previous efforts. Thus, our inability to identify all exosome components in Giardia and Plasmodium that were previously identified in S. cerevisiae and H. sapiens is likely either the result of true gene absence or sequence divergence of the unidentified proteins. The Giardia genome sequencing project has 11× coverage of the genome with an estimated 96% of the genome sequenced [36], while the P. falciparum sequencing project has a minimum of 9× coverage for its chromosomes [75], further reducing the likelihood that genes could be missed because they are not yet part of the genome database
If Giardia and Plasmodium are indeed limited to four RNase PH proteins, this would mark a departure from the typical eukaryotic exosome structure and suggests that these proteins must be present in multiple copies within the exosome ring in order for Giardia and Plasmodium to have functional hexameric exosomes. We believe it unlikely that Giardia's and P. falciparum's exosomes differ this dramatically from all other known exosome structures in eukaryotes and archaea and from bacterial PNPase by having fewer than six RNase PH subunits building the exosome ring structure. The hexameric structure has been conserved in all domains of life and is still the most likely structure for Giardia and Plasmodium. Studies involving C. reinhardtii demonstrate that absence of mtr3 exosome component may be tolerated in some eukaryotes [74], providing some precedent to the scenario presented here. Therefore, one central finding from our research is that the core structure of the eukaryotic exosome may not be universal, and the exosome may be able to be composed of different combinations of Rrp41-like and Rrp42-like proteins, with potential for different stoichiometric ratios of the subunits. Unfortunately, the extreme sequence divergence of the parasitic protist RNase PH domain proteins interferes with our ability to accurately classify them, and we can therefore not be more specific in our description of the construction of the Giardia and Plasmodium altered exosomes.
Additionally, our inability to identify a csl4 homolog is initially striking since the archaeal exosome possesses a csl4 homolog, and S. cerevisiae is nonviable in csl4 deletion mutants [9]. However, T. brucei conditional mutants survive, with slowed growth rates, in the absence of the Csl4, although exosome functionality was never determined in the T. brucei Csl4 conditional mutants [24]. Tolerance of deleted genes is likely different in every organism but because Rrp4, Rrp40 and Csl4 all possess S1-like domains, it seems likely that Rrp4 or Rrp40 may be able to act as a replacement for Csl4.
Perhaps most surprisingly, while we were able to identify a likely Rrp44 homolog, we did not identify an Rrp6, or indeed any RNase D-like, homolog. The absence of Rrp6 appears to be specific only to Giardia since it was found in the other parasitic protists we examined. Similar studies of C. reinhardtii were also unable to identify an Rrp6 homolog, but they were able to detect several putative open reading frames with homology to RNase D domains [74] that likely serve as divergent substitutes. In S. cerevisiae, Rrp6 is nuclear localized and acts to degrade unstructured RNAs. In S. cerevisiae, deletion of Rrp6 is tolerated, but cells display steady state increases in RNA transcripts such as antisense and cryptic unstable transcripts (CUTs) [76,77]. These transcripts are normally produced but fail to accumulate to significant levels because they are degraded by Rrp6 [76].
The absence of an Rrp6 homolog in Giardia is intriguing in light of the abundance of stable noncoding antisense transcripts in the parasite [37,38,78]. The Nab3 and Nrd1 proteins were also unidentified. These proteins play roles in maintaining undetectable levels of CUTs in S. cerevisiae [76,77]. CUTs and Giardia antisense transcripts are similar because both can be produced from cryptic promoter sequences in the genome or from bidirectional transcription at defined promoters [38,76,77,79]. However, Giardia's antisense transcripts are quite different from CUTs. CUTs are capped at their 5' ends and are approximately 300 nucleotides or shorter in length [77]. Whereas Giardia antisense transcripts are also capped at their 5'end, they can be thousands of nucleotides long and are polyadenyated [25,37]. Perhaps, most importantly, Giardia's antisense transcripts are apparently stable, long-lived transcripts [37,78] and therefore, while cryptic, cannot accurately be termed CUTs. We suggest that the absence of Giardia's Rrp6 and other proteins such as Nrd1 and Nab3 [80,81] may play a role in the relative stability of these antisense transcripts, an observation for which we have previously been unable to provide a molecular explanation.
If the exosome is independently incapable of recognizing and degrading sterile transcripts in Giardia, we need to look to the other exosome-dependent and exosome-independent machineries to serve this role. In particular, we focus our attention on the cytoplasmic RNA degradation machinery since recent evidence has shown that antisense transcripts are exported from the nucleus into the cytoplasm in Giardia (Teodorovic and Elmendorf, in prep.). The non-exosome RNA quality control machinery is also ancient, with homologs in archaea: an EF1α homolog (aEF1A), an eRF1 homolog (aRF1), and an eRF1 family member (aDom34) [62,82,83]. Eukaryotes possess an expanded EF1 family consisting of EF1α, eRF3, Hbs1, and in some Saccharomyces species, Ski7. Interpreting the function of this machinery is complicated, however, since most of the protein components have dual roles in translation and in translation-dependent RNA quality control pathways. Thus, although Giardia possesses a full complement of these proteins, other than Ski7, we can't yet conclude that NSD and/or NGD are functional within the parasite.
Conclusion
The key finding from this study is the identification of RNA degradation pathways in a group of highly divergent eukaryotes, the parasitic protists. These protists display an unusual reluctance to regulate gene expression at the transcriptional level, instead often engaging in promiscuous transcription and reliance on post-transcriptional regulation of gene expression. A better understanding of the RNA degradation machinery in these parasitic protists provides us with the opportunity to answer intriguing basic molecular biology questions about the range of structural variation that is permissible in a functional eukaryotic exosome. Our research has revealed the surprising finding that the 'canonical' eukaryotic exosome composition of six different RNasePH domain-containing proteins (three Rrp41-like and three Rrp42-like subunits) is not universal, and both Giardia and Plasmodium each contain only four identifiable RNasePH domain-containing proteins, while Trichomonas contains a record seventh protein. While a theme of 'genomic minimalism' has been identified previously in Giardia, our findings presented here emphasize the extreme nature of this mechanistic diminution.
Our research also helps us to understand the unusual status of ncRNAs in Giardia. Our laboratory has previously published on the abundance and atypical stability of long ncRNA transcripts in the parasite. While we have now understood their origins for several years, we have previously been unable to explain their stability. Our detailed examination of the exosome in Giardia reveals the absence of Rrp6 and Nab3-Nrd1, and that difference may at last provide an explanation for this phenomenon.
Methods
Searching for degradation machinery of the RNA exosome
Sequence similarity and protein domain searches were performed to identify potential RNA degradation machinery components of the exosome in Giardia lamblia (WB), Entamoeba histolytica (HM-1:IMSS), and Trichomonas vaginalis (G3). Parallel searches were performed on organisms with previously identified exosome machinery (e.g. Trypanosoma brucei (TREU), P. falciparum (3D7), Arabidopsis thaliana, and Homo sapiens) to validate the efficacy of our search protocol.
We used S. cerevisiae RNA degradation machinery protein sequences as queries in our searches because many of the yeast homologs have been verified functionally. When putative homologs could not be identified using S. cerevisiae sequences, we additionally used human and T. brucei RNA degradation machinery protein sequences as queries in our searches. We performed initial searches using the BLASTP algorithm with default search parameters against deprecated and accepted open reading frames (ORFs) in the parasite genomes available in the Eukaryotic Pathogen Database Resource (EupathDB.org). The default parameters were set according to Washington University BLASTP default parameters (cpus = 2, topcomboN = 1, V = 100, B = 20, hspmax = 1000000, gi E = 1e-3, wordmask = seg, hspsepQmax = 4000, span1). For genes that could not be identified using BLASTP alone, we used the Interpro domain accessory function in the Giardia genome database (GiardiaDB.org) to search the genome with PFAM domains or Conserved Domains that define the RNA degradation machinery components. The domains we searched for include the RNase PH (PF01138), AIR1 (COG5082), NOT1 (PF0454), NOT2_3_5 (PF04153), NOT3 (PF04065), Exo_endo_phosph (PF03372), S1 (PF00575), KH (PF00013), Hit-like (PF01230), DCPS (PF05652), DCP1 (PF06058), RCD (PF04078), and the DNAQ_like_exo_superfamily domains.
Parameters for acceptance or refusal of queried results
When using BLASTP, acceptance of queried protein output was based on E-value (≤ 0.05) and the presence of conserved RNA-binding or exonucleolytic protein domains found in other eukaryotic homologs. For proteins that were not conclusively identified in the initial search, all 'hits', regardless of E-value, were examined for the presence of the conserved RNA-binding or exonucleolytic protein domains when compared to the query sequence. This flexible search protocol is often necessary in organisms as divergent as the parasitic protists examined here. Parasite sequences at this stage were finally validated in a reciprocal search against the S. cerevisiae genome at the Saccharomyces Genome Database (yeastgenome.org). Parasite genes that identified the initial S. cerevisiae search sequence as the top hit were accepted as correct. The proteins in the resulting list were categorized and assigned to protein complexes.
Amino acid alignments
All amino acid alignments were made using MUSCLE 3.8.31 [84] (parameters: gapopen -12.0, gapextend -1.0, center at 0.0). The RNase PH protein alignments were performed using a BLOSUM45 matrix, while all other alignments were made using default parameters (PAM 200 scoring matrix). G-Blocks was used to exclude portions of the RNase PH amino acid sequences with limited sequence similarity beyond the RNase PH domain [85,86]. G-Blocks settings were always set to the default parameters except for maximum number of contiguous non-conserved positions (10), minimum block length (4), allowed gap positions (with half), and use of similarity matrices (yes). All alignments are displayed using Bioedit Sequence Alignment Editor graphic display [87].
Protein Phylogenies
Amino acid alignments of the RNase PH proteins from MUSCLE were used in the Phylip-3.69 package for the production of gene trees. All programs mentioned below are part of the Phylip-3.69 package [88]. Seqboot was used to bootstrap the data set 100 times with block size set at 1 (regular boostrap). Next, Protpars was used to infer relatedness of the sequences to one another from analysis of the 100 data sets produced from Seqboot. Solfolobus solfataricus Rrp41 was set as the outgroup. Lastly the freeware program Consense was used to produce the final gene tree. The consensus type used was extended majority rule. Extended majority rule allows groups of sequences that appear in more than 50% of trees to be included in the final tree and other sequences which fall below this level are then added until all sequences are accounted for.
Authors' contributions
CW conceived of the study, performed the described bioinformatics and phylogenetic analyses, and drafted the manuscript. HE participated in its design and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Comparison of domain profiles in RNA degradation machinery between Saccharomyces cerevisiae and Giardia lamblia. This file is in PDF format and contains schematic representations of protein components of the RNA degradation machinery complexes. Functional domains and protein lengths are indicated in these schematics to allow a fuller understanding of the repertoire of RNA degradation machinery pathways in Giardia lamblia.
Classification of the Putative RNA Degradation Machinery Protein Components in Giardia lamblia. This file is in Excel format and contains a table of the complete list of identified RNA degradation complexes and their protein constituents in Giardia lamblia.
Contributor Information
Christopher W Williams, Email: cww7@georgetown.edu.
Heidi G Elmendorf, Email: hge@georgetown.edu.
Acknowledgements
We would like to thank D. Hagen, M. Poelchau, C. Childers, C. Elsik, and M. Hamilton for their discussions and insight about gene tree production especially. The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. This work was supported by the National Institutes of Health [F31AI078714 to C.W.].
References
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Research. 2008;36(10):3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Chiang YC, Lee DJ, Yao G, Zhang CX, Denis CL. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Research. 2007;35(9):3002–3015. doi: 10.1093/nar/gkm196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen JJ, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104(3):377–386. doi: 10.1016/S0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Degradation Of Messenger-Rna In Eukaryotes. Cell. 1995;81(2):179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Sachs AB. Messenger-Rna Degradation In Eukaryotes. Cell. 1993;74(3):413–421. doi: 10.1016/0092-8674(93)80043-E. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. mRNA stability in eukaryotes. Current Opinion In Genetics & Development. 2000;10(2):193–198. doi: 10.1016/S0959-437X(00)00063-0. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12(7):575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. Structural basis of 3 ' end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Molecular Cell. 2005;20(3):473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Liu QS, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127(6):1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD. Structural Components And Architectures Of Rna Exosomes. Rna Exosome. pp. 9–28. [PMC free article] [PubMed]
- Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. Journal Of Biological Chemistry. 2000;275(42):33158–33166. doi: 10.1074/jbc.M005493200. [DOI] [PubMed] [Google Scholar]
- Barbas A, Matos RG, Amblar M, Lopez-Vinas E, Gomez-Puertas P, Arraiano CM. Determination of Key Residues for Catalysis and RNA Cleavage Specificity ONE MUTATION TURNS RNase II INTO A "SUPER-ENZYME". Journal Of Biological Chemistry. 2009;284(31):20486–20498. doi: 10.1074/jbc.M109.020693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. Journal Of Cell Science. 2009;122(10):1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Raijmakers R, Pruijn GJM. Cell and molecular biology of the exosome: How to make or break an RNA. International Review Of Cytology - A Survey Of Cell Biology, Vol 251. 2006;251:159. doi: 10.1016/S0074-7696(06)51005-8. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Dutko JA, Mian IS, Belostotsky DA. Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3 '- > 5 ' exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Research. 2002;30(3):695–700. doi: 10.1093/nar/30.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Wang XY. Nuclear RNA surveillance: no sign of substrates tailing off. Critical Reviews In Biochemistry And Molecular Biology. 2009;44(1):16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyade nylation complex. Cell. 2005;121(5):713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nature Structural & Molecular Biology. 2006;13(6):533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley interdisciplinary reviews RNA. 2011;2(4):471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Chen C-YA, Shyu A-B. Mechanisms of deadenylation-dependent decay. Wiley Interdisciplinary Reviews: RNA. 2011;2(2):167. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orso I, De Gaudenzi JG, Frasch ACC. RNA-binding proteins and mRNA turnover in trypanosomes. Trends Parasitol. 2003;19(4):151–155. doi: 10.1016/S1471-4922(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Clayton C, Schwede A, Stewart M, Robles A, Benz C, Po J, Wurst M, Quieroz R, Archer S. Control of mRNA degradation in trypanosomes. Biochemical Society Transactions. 2008;36:520–521. doi: 10.1042/BST0360520. [DOI] [PubMed] [Google Scholar]
- Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20(14):3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Altura MA, Witmer M, Singer SM, Elmendorf HG, Shuman S. Yeast-like mRNA capping apparatus in Giardia lamblia. Journal of Biological Chemistry. 2005;280(13):12077–12086. doi: 10.1074/jbc.M412063200. [DOI] [PubMed] [Google Scholar]
- Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C. The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Research. 2009;37(16):5511–5528. doi: 10.1093/nar/gkp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock JL, Fischer KF, DeRisi JL. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biology. 2007;8(7) doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Su LH, Sun CH. Incomplete nonsense-mediated mRNA decay in Giardia lamblia. International Journal For Parasitology. 2008;38(11):1305–1317. doi: 10.1016/j.ijpara.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Yi-Hsiu Chen L-HS, Yu-Chang Huang, Yi-Ting Wang, Yu-Yun Kao, Chin-Hung Sun. UPF1, a Conserved Nonsense-Mediated mRNA Decay Factor, Regulates Cyst Wall Protein Transcripts in Giardia lamblia. PLoS ONE. 2008;3(10) doi: 10.1371/journal.pone.0003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile S, Cristodero M, Clayton C, Estevez AM. The subcellular localisation of trypanosome RRP6 and its association with the exosome. Molecular And Biochemical Parasitology. 2007;151(1):52–58. doi: 10.1016/j.molbiopara.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Haile S, Estevez AM, Clayton C. A role for the exosome in the in vivo degradation of unstable mRNAs. Rna-A Publication Of The Rna Society. 2003;9(12):1491–1501. doi: 10.1261/rna.5940703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. Journal Of Biological Chemistry. 2003;278(37):34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estevez AM, Clayton C. Roles of a Trypanosoma brucei 5 '- > 3 ' exoribonuclease homolog in mRNA degradation. Rna-A Publication Of The Rna Society. 2006;12(12):2171–2186. doi: 10.1261/rna.291506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best AA, Morrison HG, McArthur AG, Sogin ML, Olsen GJ. Evolution of eukaryotic transcription: Insights from the genome of Giardia lamblia. Genome Research. 2004;14(8):1537–1547. doi: 10.1101/gr.2256604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA. Phylogenetic Meaning of the Kingdom Concept - an Unusual Ribosomal-Rna from Giardia-Lamblia. Science. 1989;243(4887):75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ. et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Singer SM, Nash TE. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Research. 2001;29(22):4674–4683. doi: 10.1093/nar/29.22.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodorovic S, Walls CD, Elmendorf HG. Bidirectional transcription is an inherent feature of Giardia lamblia promoters and contributes to an abundance of sterile antisense transcripts throughout the genome. Nucleic Acids Research. 2007;35(8):2544–2553. doi: 10.1093/nar/gkm105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456(7223):750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Li F, Zhou K, Cande WZ, Doudna JA. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harbor Symposia On Quantitative Biology. 2006;71:73–80. doi: 10.1101/sqb.2006.71.042. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou KH, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, Su L-H, Huang Y-C, Wang Y-T, Kao Y-Y, Sun C-H. UPF1, a Conserved Nonsense-Mediated mRNA Decay Factor, Regulates Cyst Wall Protein Transcripts in Giardia lamblia. PLoS ONE. 2008;3(10):e3609. doi: 10.1371/journal.pone.0003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Current Opinion In Cell Biology. 2009;21(3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The Control of mRNA Decapping and P-Body Formation. Molecular Cell. 2008;32(5):605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. 2005;12(12):1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- He WH, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Current Opinion In Cell Biology. 2000;12(3):346–350. doi: 10.1016/S0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. Rna-A Publication Of The Rna Society. 2002;8(12):1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nature Reviews Molecular Cell Biology. pp. 467–478. [DOI] [PMC free article] [PubMed]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP. et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. Embo Journal. 2010;29(14):2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UCM, Besteiro S. et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315(5809):207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends In Biochemical Sciences. 2008;33(10):501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Molecular Cell. 2007;27(2):324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, D'Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The Nuclear Poly(A)-Binding Protein Interacts with the Exosome to Promote Synthesis of Noncoding Small Nucleolar RNAs. Molecular Cell. 2010;37(1):34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Hoek M, Zanders T, Cross GAM. Trypanosoma brucei expression-site-associated-gene-8 protein interacts with a Pumilio family protein. Mol Biochem Parasitol. 2002;120(2):269–283. doi: 10.1016/S0166-6851(02)00009-9. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3 ' UTR regulation as a way of life. Trends Genet. 2002;18(3):150–157. doi: 10.1016/S0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Muller K, Matuschewski K, Silvie O. The Puf-Family RNA-Binding Protein Puf2 Controls Sporozoite Conversion to Liver Stages in the Malaria Parasite. Plos One. 2011;6(5) doi: 10.1371/journal.pone.0019860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Carrington M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends In Parasitology. 2011;27(1):23–30. doi: 10.1016/j.pt.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2(3):342–354. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S, Wen JK. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16(2):107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- Wen J, Brogna S. Nonsense-mediated mRNA decay. Biochem Soc Trans. 2008;36:514–516. doi: 10.1042/BST0360514. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8 doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295(5563):2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295(5563):2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440(7083):561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Doudna JA. Structural insights into RNA interference. Current Opinion In Structural Biology. 2010;20(1):90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes & Development. 2005;19(5):517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cellular Microbiology. 2004;6(6):509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Harrison BR, Yazgan O, Krebs JE. Life without RNAi: noncoding RNAs and their functions in Saccharomyces cerevisiae. Biochemistry And Cell Biology-Biochimie Et Biologie Cellulaire. 2009;87(5):767–779. doi: 10.1139/O09-043. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6(6):509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Chen XW, Collins LJ, Biggs PJ, Penny D. High Throughput Genome-Wide Survey of Small RNAs from the Parasitic Protists Giardia intestinalis and Trichomonas vaginalis. Genome Biology And Evolution. 2009;1:165–175. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky D. Exosome complex and pervasive transcription in eukaryotic genomes. Curr Opin Cell Biol. 2009;21(3):352–358. doi: 10.1016/j.ceb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Clayton C, Estevez A. The Exosomes Of Trypanosomes And Other Protists. Rna Exosome. pp. 39–49. [DOI] [PubMed]
- Zimmer SL, Fei ZJ, Stern DB. Genome-based analysis of Chlamydomonas reinhardtii exoribonucleases and poly(A) polymerases predicts unexpected organellar and exosomal features. Genetics. 2008;179(1):125–136. doi: 10.1534/genetics.107.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in s. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu ZY, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457(7232):1038–U1038. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Birkeland SR, Preheim SP, Davids BJ, Cipriano MJ, Palm D, Reiner DS, Svard SG, Gillin FD, McArthur AG. Transcriptome analyses of the Giardia lamblia life cycle. Mol Biochem Parasitol. 2010;174(1):62–65. doi: 10.1016/j.molbiopara.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The nuclear RNA surveillance machinery: The ncRNAs and genome structure in budding link between yeast? Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms. 2008;1779(4):239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-Binding proteins Nrd1 and Nab3. Molecular Cell. 2006;23(6):841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the Nrd1-Nab3 pathway in genome surveillance. Molecular Cell. 2006;23(6):853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1 alpha complex. Proceedings Of The National Academy Of Sciences Of The United States Of America. pp. 17575–17579. [DOI] [PMC free article] [PubMed]
- Saito K, Kobayashi K, Wada M, Kikuno I, Takusagawa A, Mochizuki M, Uchiumi T, Ishitani R, Nureki O, Ito K. Omnipotent role of archaeal elongation factor 1 alpha (EF1 alpha) in translational elongation and termination, and quality control of protein synthesis. Proceedings Of The National Academy Of Sciences Of The United States Of America. pp. 19242–19247. [DOI] [PMC free article] [PubMed]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989. pp. 164–166.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of domain profiles in RNA degradation machinery between Saccharomyces cerevisiae and Giardia lamblia. This file is in PDF format and contains schematic representations of protein components of the RNA degradation machinery complexes. Functional domains and protein lengths are indicated in these schematics to allow a fuller understanding of the repertoire of RNA degradation machinery pathways in Giardia lamblia.
Classification of the Putative RNA Degradation Machinery Protein Components in Giardia lamblia. This file is in Excel format and contains a table of the complete list of identified RNA degradation complexes and their protein constituents in Giardia lamblia.