Abstract
The interaction of Abs with their specific FcRs is of primary importance in host immune effector systems involved in infection and inflammation, and are the target for immune evasion by pathogens. FcγRIIa is a unique and the most widespread activating FcR in humans that through avid binding of immune complexes potently triggers inflammation. Polymorphisms of FcγRIIa (high responder/low responder [HR/LR]) are linked to susceptibility to infections, autoimmune diseases, and the efficacy of therapeutic Abs. In this article, we define the three-dimensional structure of the complex between the HR (arginine, R134) allele of FcγRIIa (FcγRIIa-HR) and the Fc region of a humanized IgG1 Ab, hu3S193. The structure suggests how the HR/LR polymorphism may influence FcγRIIa interactions with different IgG subclasses and glycoforms. In addition, mutagenesis defined the basis of the epitopes detected by FcR blocking mAbs specific for FcγRIIa (IV.3), FcγRIIb (X63-21), and a pan FcγRII Ab (8.7). The epitopes detected by these Abs are distinct, but all overlap with residues defined by crystallography to contact IgG. Finally, crystal structures of LR (histidine, H134) allele of FcγRIIa and FcγRIIa-HR reveal two distinct receptor dimers that may represent quaternary states on the cell surface. A model is presented whereby a dimer of FcγRIIa-HR binds Ag–Ab complexes in an arrangement that possibly occurs on the cell membrane as part of a larger signaling assembly.
The interaction between Ig complexes and FcRs induces potent and diverse immune responses. In normal immunity, these include inflammation, Ab-dependent killing of target cells, mast cell degranulation, phagocytosis, and regulation of Ag receptor activation of B cells. However, in pathological situations like autoimmunity, immune complex FcR-mediated activation of effector cells is a major pathway in the development of tissue injury and, indeed, the early events of disease pathogenesis (1, 2).
The FcγRII receptors (CD32 group of receptors) are key activating and inhibitory effectors of the IgG-mediated immune functions of leukocytes. FcγRIIa triggers both host protective and damaging proinflammatory activities, whereas FcγRIIb modulates signaling from the activating FcγRs, including FcγRIIa, and the B-cell Ag receptor complex. The evidence of many models of autoimmune disease and the association of receptor gene polymorphisms with human disease concur that the balanced integration of signals from activating receptors and inhibitory FcγRIIb is necessary for a normal host response to infection and resistance to autoimmune pathologies. Abs that specifically target these receptors to manipulate this balance of proinflammatory and inhibitory signals are of particular interest as therapeutics (3).
FcγRIIa is unique to higher primates and is the most widespread FcR. This low-affinity FcR is different from all other activating FcRs because its signaling ITAM is contained in the ligand-binding chain. Other activating FcRs and related multisubunit immunoreceptors, including the Ag receptors and NK receptors, signal via their noncovalent association with dimeric accessory molecules such as the common FcR-γ–chain dimer that contain ITAMs (4, 5). Animal and human studies indicate that FcγRIIa plays a major role in the development of destructive inflammation. Mice transgenic for human FcγRIIa exhibit extreme sensitivity to pathogenic Abs and also develop a spontaneous autoimmune disease with features of human rheumatoid arthritis including joint destruction (6) and are sensitive to thrombocytopenia-inducing Abs (7). The transgenic FcγRIIa mice are particularly susceptible to collagen-induced arthritis, which can be largely suppressed by small chemical inhibitors designed to bind FcγRIIa (8). In addition, in vitro studies of human plasmacytoid dendritic cells from systemic lupus erythematosus (SLE) patients show the targeting of DNA to TLR9 is dependent on Ab and FcγRIIa, as opposed to other FcRs (9).
Genetic polymorphisms of FcγRIIa have also been linked to susceptibility to several autoimmune diseases, notably SLE (10–13) and ulcerative colitis (14), as well as resistance to Gram-negative bacterial infection (15) and to the outcome of therapeutic Ab treatment in lymphoma (16). The most extensively studied polymorphism is the high-responder/low-responder (HR/LR) polymorphism, alleles of which code either arginine (HR) or histidine (LR) at position 134 [numbering is based on the experimentally determined N-terminal sequence (17); this polymorphic site is frequently referred to as position 131 in the amino acid sequence (18, 19)]. The functional differences between the HR (arginine, R134) allele of FcγRIIa (FcγRIIa-HR) and LR (histidine, H134) allele of FcγRIIa (FcγRIIa-LR) relate to different abilities to bind mouse IgG1 or human IgG2, respectively (19, 20). Indeed, the FcγRIIa-LR is the only receptor that binds human IgG2 (21), which interestingly is a major IgG class in autoimmunity (11) and in resistance to Gram-negative bacterial infection (15) and severe swine flu infection (22). A higher frequency of FcγRIIa-LR homozygous genotype was found to be associated with Plasmodium falciparum malaria resistance and consistently higher levels of anti-malarial IgG2 and IgG3 serum Ab (23).
Despite the extensive functional, genetic, and biochemical characterization of this receptor, the three-dimensional structural basis for the interaction of FcγRIIa with IgG and functional differences of the HR/LR polymorphism was unknown. Soluble FcγRIIa-LR glycoprotein can form a crystallographic dimer as described for orthorhombic P21212 crystals (24). Such a dimer of cellular receptors would place the two ITAM-containing cytoplasmic tails in close proximity, similar to other ITAM-activating molecules, in an arrangement presumably suitable for downstream intracellular signaling (25). It is generally accepted that, in activating leukocytes, the FcRs cluster or aggregate, a process that is driven by Ag–Ab complexes (26–28), yet the structural arrangement of receptors within these signaling assemblies is poorly understood.
In this article, the resolution of the crystal structure of FcγRIIa-LR is extended to 1.5 Å from orthorhombic P21212 crystals (24). In addition, we have determined the 2.3-Å resolution structure of glycosylated FcγRIIa-HR from orthorhombic C2221 crystals and show it forms a crystallographic dimer distinct from that of FcγRIIa-LR. We also report, to our knowledge, the first three-dimensional structure for the complex of FcγRIIa-HR and the Fc region of human IgG1 at a resolution of 3.8 Å. Interestingly, this FcγRIIa–Fc complex could itself form a dimer equivalent to the crystallographic dimer of FcγRIIa-HR (unlike FcγRIIa-LR) without interfering with ligand binding. Hence we predict signaling by FcγRIIa occurs through dimeric receptors with bound immune complexes (Ag–IgG), possibly as part of larger aggregates, which are important for intracellular signaling, cell activation, and ultimately inflammation.
Materials and Methods
Production and purification of recombinant human FcγRIIa and the IgG1 Fc
FcγRIIa-LR (H134) was prepared as previously described; a point mutation replacing S88 with F88 introduced during the production of soluble FcγRIIa-LR did not alter its ability to bind IgG or immune complexes (17). FcγRIIa-HR (R134) cDNA was produced by splice overlap extension PCR and expressed using a baculovirus system in SF21 insect cells using Life Technologies SF900 media (Invitrogen Australia Pty, Rowville, VIC, Australia) at 27°C for 72 h. The receptor was purified from supernatant by anion exchange over Q-Sepharose (GE Healthcare, Rydalmere, NSW, Australia), followed by affinity chromatography on heat-aggregated Ig (Novartis, Basel, Switzerland) coupled Sepharose, as previously described for FcγRIIa-LR (17). For crystallization, FcγRIIa-HR was prepared in 75 mM NaCl, 5 mM Tris-HCl pH 7.4 at between 5 and 10 mg/ml.
The Fc portion was released by plasmin proteolysis from a humanized IgG1, hu3S193, which was expressed in an NSO myeloma cell line (29). Purified recombinant hu3S193 at 5 mg/ml was dialyzed against 0.15 M NaCl, 50 mM Tris pH 8.0, and was digested with 1 U/ml plasmin (Roche, Basel, Switzerland) for 48 h at 21°C. The Fc was purified to homogeneity by separation from Fab by protein A-Sepharose, followed by removal of residual intact IgG by protein L-Sepharose (Pierce Protein Research Products, Thermo Fisher Scientific, Scoresby, VIC, Australia). Purified Fc was dialyzed against 75 mM NaCl, 5 mM Tris buffer pH 7.4 before crystallization.
Crystallization of FcγRIIa and complexes with the Fc of IgG1
Crystals of FcγRIIa-LR were produced in 5-µl hanging drops with the protein solution at 8 mg/ml in 0.2 M ammonium acetate, 0.1 M sodium citrate buffer pH 5.6, and 30% (w/v) polyethylene glycol (PEG) 4000. Crystals were nucleated at 37°C (typically 5–7 d for small crystals to appear), and were grown and maintained at 18°C. FcγRIIa-HR crystals were prepared and maintained at 18°C in 2 µl sitting droplets, with the protein at 4 mg/ml in 30% (w/v) PEG 4000 and 0.2 M ammonium sulfate.
Cocrystallization of IgG1-Fc was successful with both polymorphic variants of FcγRIIa, but only with wild-type FcγRIIa-HR were the crystals suitable for structure determination. However, both systems produced large, thin plates with growth habits of single crystals and crystalline clusters in 2 µl sitting and hanging droplets in 2–4 wk at 18°C. Typically, 1 µl protein solution (containing 5–8 mg/ml total protein with FcγRIIa and Fc mixed at a 1:1 molar ratio) was mixed with 1 µl of the reservoir solution that contained 0.2 M trisodium citrate or tripotassium citrate and 20% PEG 3350. A range of additives was tested with the best crystals of the complex grown from an initial protein concentration of 5.5 mg/ml in 5 mM Tris pH 7.4, 0.15 M NaCl, 2 mM MOPS, 0.2 M tripotassium citrate, and 20% PEG 3350. Supplies and reagents for crystallization were purchased from Hampton Research (Aliso Viejo, CA).
Data collection and structure determinations
For FcγRIIa-LR, a large (0.6 × 0.2 × 0.2 mm) orthorhombic P21212 crystal was briefly transferred into artificial mother liquor containing 20% (v/v) glycerol before flash-cooling to 100 K in a nitrogen cryostat (Cryojet; Oxford Instruments, Abingdon, U.K.). Diffraction data to 1.5-Å resolution were obtained at 100 K using synchrotron radiation on beamline 14-BMC (BioCARS) at the Advanced Photon Source and processed with the HKL package, version 1.97 (30). Initial crystallographic refinement involved rigid body fitting of domains 1 and 2 (residues 4–88 and 89–173, respectively) of LR-FcγRIIa using the coordinates of Protein Data Bank (PDB) entry 1FCG (24). Subsequent cycles of interactive model building and solvent fitting into σA-weighted 2Fo−Fc and Fo−Fc electron density maps were performed in TURBO-FRODO version 5.5 (BioGraphics, Marseille, France). Each cycle of model building was followed by simulated annealing, energy minimization, and individual temperature (B) factor refinement protocols against maximum likelihood target functions in the CNS version 1.0 program suite (31). Bulk solvent correction, anisotropic overall B factor refinements, and cross-validation by assigning 10% of intensities for calculation of an Rfree value were applied throughout the structure determination. Relevant values and statistics for evaluation of the X-ray diffraction data and the final atomic model are presented in Table I.
Table I.
Summary of data collection and crystallographic refinements
| FcγRIIa-LR | FcγRIIa-HR | FcγRIIa-HR–Fc | |
|---|---|---|---|
| Data collection | |||
| Space group | P21212 | C2221 | C2221 |
| Cell dimensions | |||
| a, b, c (Å) | 79.34, 100.41, 27.94 | 50.04, 77.94, 88.10 | 153.38, 255.55, 58.44 |
| Resolution (Å) | 20–1.5 (1.55–1.5)a | 30–2.3 (2.38–2.3) | 30–3.8 (3.94–3.8) |
| Rsym | 0.052 (0.32) | 0.10 (0.27) | 0.14 (0.35) |
| Mean I/σ(I)a | 21.8 (3.8) | 13.2 (4.9) | 10.8 (3.78) |
| Completeness (%) | 94.5 (85.5) | 97.2 (96.0) | 98.9 (99.7) |
| Redundancy | 6.1 (4.0) | 4.6 (4.3) | 5.0 (4.8) |
| Refinement | |||
| Resolution (Å) | 19.8–1.5 | 20.4–2.3 | 29.7–3.8 |
| No. of reflections | 34,590 | 7,554 | 11,110 |
| Rwork/Rfree | 0.20/0.23 | 0.21/0.28 | 0.25/0.42 |
| No. of atoms | |||
| Protein | 1,354 | 1,350 | 4,745 |
| Ligand | 14 | 0 | 376 |
| Water | 273 | 85 | 24 |
| B-factors | |||
| Protein | 22.7 | 20.7 | 34.5 |
| Water | 36.5 | 23.1 | 26.6 |
| RMSD bond lengths (Å) | 0.005 | 0.006 | 0.010 |
| RMSD bond angles (°) | 1.3 | 1.4 | 1.8 |
Values in parentheses are for the highest resolution shell.
RMSD, root-mean-square deviation.
Crystals of FcγRIIa-HR were extremely sensitive to glycerol, but crystals were relatively stable in artificial mother liquor containing 20% (v/v) PEG 400. Using this cryoprotectant, we used an orthorhombic C2221 crystal (0.2 × 0.1 × 0.1 mm) to collect X-ray diffraction data at 100 K on a MicroMax007/R-Axis IV2+ rotating anode generator system (Rigaku Americas, The Woodlands, TX), operated at 40 kV and 20 mA. Processing of X-ray diffraction data and crystallographic refinement to a resolution of 2.3 Å proceeded essentially as described earlier (Table I), but in this case, the coordinates of the high-resolution structure of FcγRIIa-LR were used as a starting template for molecular replacement. Two residues were replaced (F88 with S88 and H134 with R134) to correspond with the sequence of the wild-type and HR polymorphism of FcγRIIa, respectively.
Crystals of the FcγRIIa-HR–IgG1-Fc complex proved extremely sensitive to cryopreservation, with evident large increases in mosaic spread and uneven shrinkage of the unit cell such that different cryopreserved crystals were generally nonisomorphous. Moreover, the volumes of the crystals that were essentially thin plates (<10 µM in the smallest dimension) were not suitable for room temperature data collection. Our best crystals were flash-cooled in artificial mother liquor in the presence of 20% (v/v) glycerol and diffracted X-rays anisotropically to between 3.4 and 4.5 Å resolutions. Consequently, the structure determination extended to a resolution of 3.8 Å and was solved by molecular replacement using the coordinates for the unliganded form of FcγRIIa-HR (described in this article) and the Fc from PDB entry 1E4K (32). Crystals contained one FcγRIIa-HR–IgG1-Fc complex in the asymmetric unit. Crystallographic refinements were performed against a crystallographic residual target as described earlier, but only group and overall B factors were refined (Table I). The high Rfree and below average quality of the model geometry is due to the anisotropy and low resolution of the diffraction data, but in previous studies with glycosylated Fc molecules free and in complexes, similar high Rfree values and diffraction properties have been reported, even when data extend to higher resolutions (32). However, electron density maps were of sufficient quality for tracing the main chains for the FcγRIIa-HR monomer and the two H chains of the Fc of hu3S193 IgG1, and placement of 29 saccharide units at four N-linked glycosylation sites. Atomic coordinates and structure factors have been deposited in the PDB (PDB ID codes 3RY4, 3RY5, and 3RY6; http://www.pdb.org).
mAb and epitope mapping
The anti-FcγRIIa mAbs 8.7 (murine IgG1) (33) and IV.3 have been previously described (34). The anti-FcγRIIb mAb X63-21/7.2 was made by immunizing transgenic mice expressing the human FcγRIIa with mouse IIA1.6 lymphoma cells transfected with, and expressing, human FcγRIIb2. Mice were immunized on three occasions with 106 cells, boosted i.v. with 106 cells, and splenocytes fused with NS1 cells. Hybridoma cell culture supernatants were harvested from cloned X63-21/7.2 cells and IgG purified by affinity chromatography as described. Fab and F(ab′)2 fragments were generated by pepsin (Boehringer Mannheim, Mannheim, Germany) digestion at an enzyme/substrate ratio of 1:10 to 1:50 at 37°C for 1 h in 0.1 M sodium citrate pH 3.5 as described previously (33). Ab fragments were biotinylated with EZ-link biotin (Pierce Protein Research Products, Thermo Scientific). FcγRIIa-HR (R134) mutant constructs or with equivalent sequence elements exchanged between FcγRIIa and the IgE receptor, FcεRI, were produced as described previously (5, 35) and expressed by transient transfection of COS-7 or CHOP cells using Fugene-6 (Boehringer Mannheim) or Lipofectamine 2000 (Invitrogen Australia Pty). Transfected cells were reacted with unbiotinylated or biotinylated mAb fragments and further reacted with FITC-conjugated anti-mouse Ig or R-phycoerythrin–conjugated streptavidin as appropriate and analyzed by flow cytometry.
Results
Crystal structures of the FcγRIIa-HR and FcγRIIa-LR
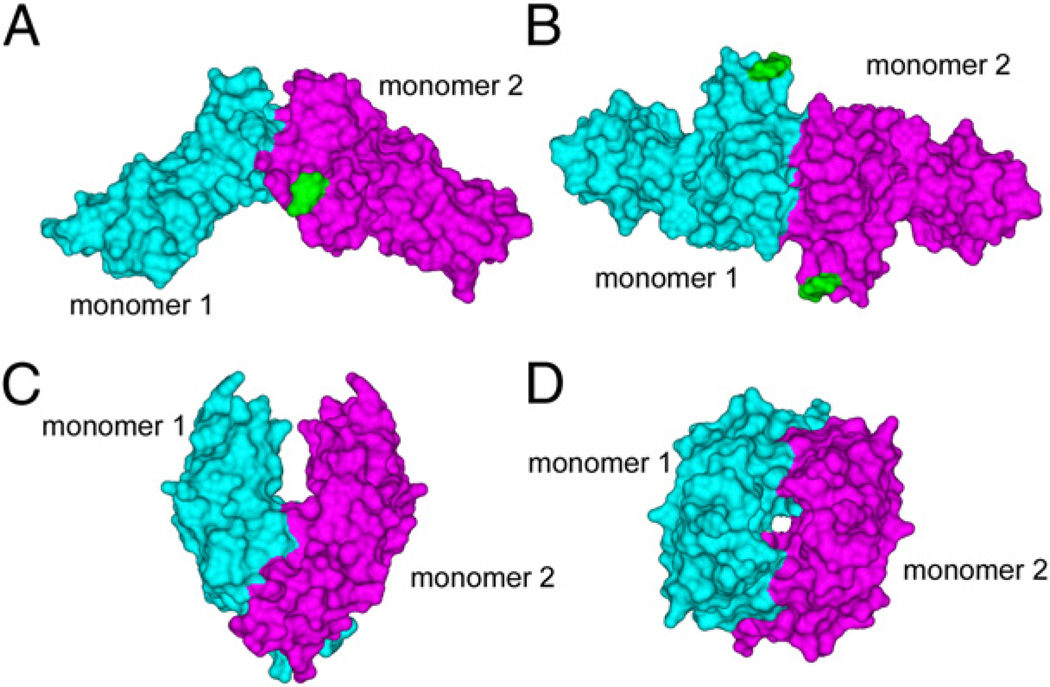
The two allelic variants of FcγRIIa were produced in insect cells and purified to homogeneity. Surface plasmon resonance established equilibrium dissociation constants (KD) for solid-phase human IgG1 of 0.5 ± 0.02 mM and 0.6 ± 0.03 mM for the HR and LR versions of FcγRIIa, respectively. This is consistent with affinities measured in previous studies of FcγRIIa (17) and for FcγRIIb (36), and is also consistent with the observed binding of IgG to whole cells (35). Thus, the relatively simple glycosylation pattern of the insect cells did not appear to significantly influence IgG binding activity of each of these forms. Higher resolution information was obtained for FcγRIIa-LR in this study (Fig. 1A, 1B), and the glycosylated HR form (Fig. 1C, 1D) was analyzed for the first time to our knowledge and in complex with the Fc of human IgG1 (Figs. 2, 3). The FcγRIIa-HR protein crystallized in the orthorhombic C2221 space group (Table I), which is distinct from the orthorhombic P21212 crystals of the LR form previously described (17, 24). Analysis of the crystal lattice to 2.3-Å resolution identified a dimer of FcγRIIa-HR that is markedly distinct from the LR dimer (Fig. 1) (24). The FcγRIIa-HR dimer interface consists of a total buried surface area of 1564 Å2 and involves regions of both extracellular Ig-like domains (D1 and D2) of the receptor monomers. Interactions in the FcγRIIa-HR dimer occur principally between the A′B strands and loop, as well as the E strand of D2 and the A′B and E strands of D1; notably, residues T26, R33, Q54, P55, S56, R58, E102, G103, T105, P142, and Q143 contribute to the receptor pairing. Importantly, the FcγRIIa-HR dimer interface is distinct from the FcγRIIa-LR interface that involves residues only of D2 of each monomer and does not overlap. As previously described, the FcγRIIa-LR interface involves residues from the C′ strands interacting with the F and G strands of neighboring monomers (24).
FIGURE 1.
Surface representations of the two distinct crystallographic dimers of FcγRIIa. A, Side view of the FcγRIIa-LR dimer with monomer 1 in cyan, monomer 2 in magenta, and the GlcNAc residue attached to N145 shown in green. B, End-on view of the FcγRIIa-LR dimer, looking down onto the surfaces involved in IgG binding. C, Side view of the FcγRIIa-HR dimer. D, End-on view of the FcγRIIa-HR dimer.
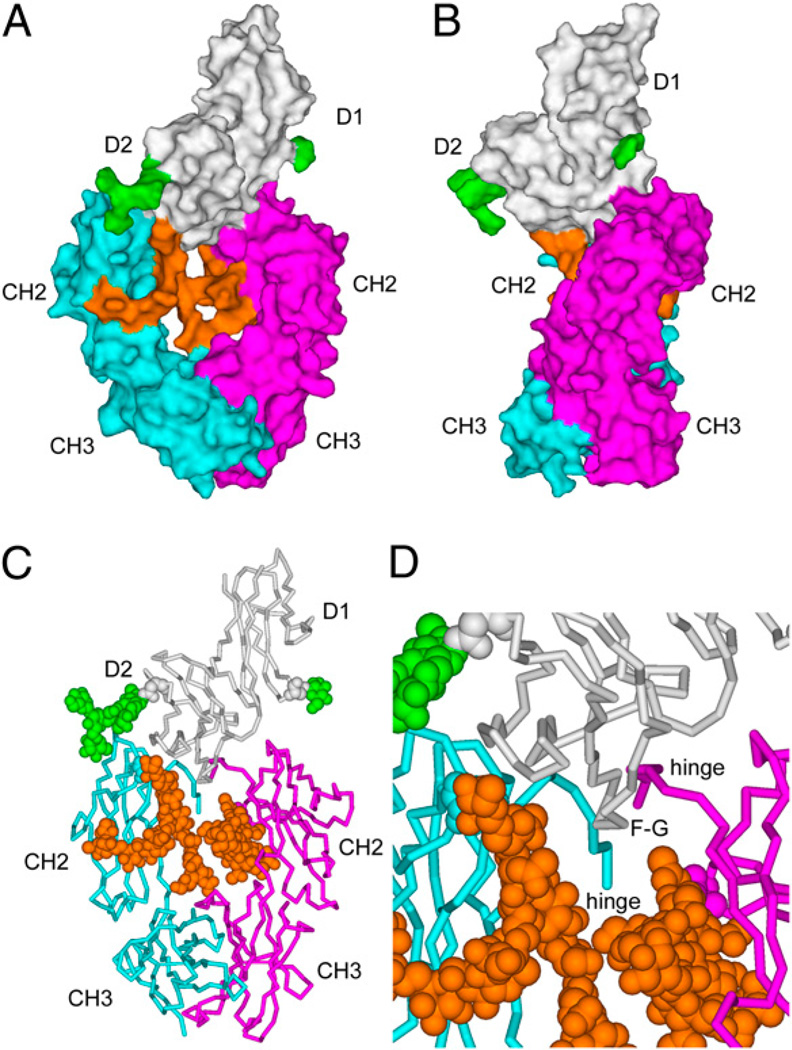
FIGURE 2.
Overview of the FcγRIIa-HR complex with Fc of human IgG1. A, Surface representation with FcγRIIa in white (N-linked oligosaccharides at positions N64 and N145 are in green) and the Fc in magenta (chain A) and cyan (chain B) for the two H chains. Oligosaccharides attached to N297 of the Fc are in orange. B, Surface view of the FcγRIIa–Fc complex rotated by 90° from its orientation in A. C, α-Carbon polypeptide trace of the FcγRIIa–Fc complex with the N-linked oligosaccharide chains displayed as space-filling (CPK) representations. D, Close-up of the FcγRIIa interaction with Fc. The extracellular domains 1 and 2 (D1 and D2) and the F–G loop of FcγRIIa are indicated, as are the constant domains, CH2 and CH3, of the H chains.
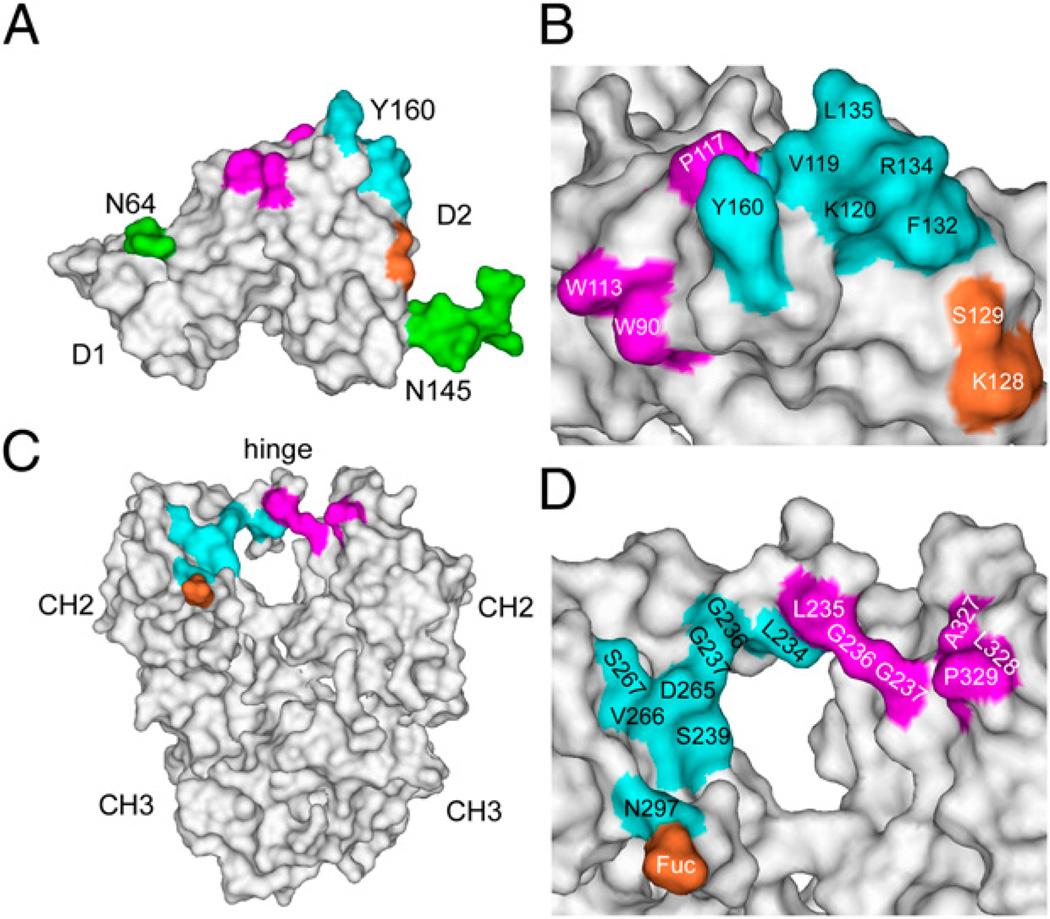
FIGURE 3.
Sites of interaction between FcγRIIa and human IgG1 revealed by the crystal structure of the FcγRIIa-HR complex with Fc. A, Binding site on FcR for its Fc ligand with residues that interact with Fc shaded on the solvent-accessible surface. The N-linked glycans at positions 64 and 145 are also included, but these do not participate directly in the interaction with Fc. B, Close-up of the FcR residues involved in the interaction with Fc. Note the site of interaction of the IgG fucose of N297 (S129 and K128) is shown in orange. C, Solvent-accessible view of the Fc ligand with the residues in contact with FcR shaded. Note the polymorphic R134 residue of FcγRIIa-HR binds in a shallow depression on the Fc formed by D265, V266, S267, and N297. D, Close-up of the Fc residues in contact with FcR. Numbering for the Fc residues is according to the Eu Index (62). The coloring scheme is the same as Fig. 2 with contacts shaded according to the Fc ligand.
The second major distinction between the HR and LR crystallographic forms is the relationship between the IgG binding surfaces and the dimer interfaces. Mutagenesis studies of FcγRIIa (24, 37) combined with homology modeling predict that the IgG binding surface involves residues from the A strands, the BC, C′E, and FG loops, which are not within the LR or HR dimer interfaces. Thus, for both dimers, the predicted IgG binding surfaces are accessible to solvent if the receptors are oriented in a hypothetical cell membrane (Fig. 1). However, in the LR dimer, the IgG binding surfaces of each monomer are “closely” apposed so that access of the IgG ligand for binding one receptor monomer is prevented by the neighboring monomer, whereas in the HR dimer, the relative position of the binding surfaces is more “open” and are predicted to allow access for binding of one or two IgG ligands.
Although both the LR and HR forms of FcγRIIa were glycosylated, we observed electron density corresponding to only a single N-acetylglucosamine (GlcNAc) moiety attached to N145 in the FcγRIIa-LR form of the receptor at a resolution of 1.5 Å (Fig. 1). The carbohydrates were disordered at N64 in both allelic forms used in these studies. However, we have observed carbohydrates at both N-linked sites in a heavily glycosylated sample of FcγRIIa-LR, but these crystals diffracted to a resolution of only 2.8 Å. In this glycoform, a GlcNAc[α1→6Fuc]β1→4GlcNAc trisaccharide was clearly visible at the N145 site, whereas a single GlcNAc was attached to N64 (P.A.R, W.F., and P.M.H., unpublished observations). We have attributed the decreased X-ray diffraction quality of crystals of the second FcγRIIa-LR glycoform to the oligosaccharide at position 145, which comes into contact with a symmetry-related molecule. Notably, in the two distinct dimers of FcγRIIa, the glycosylation sites are not involved in the interface region (Fig. 1). Thus, the carbohydrates may act to limit lateral interactions with other receptors in crystals and in their native context in membranes.
Overview of the FcγRIIa-HR complex with Fc of human IgG1
The 3.8-Å resolution crystal structure (Figs. 2, 3) was determined for FcγRIIa-HR bound to Fc of human IgG1. FcγRIIa bound its Fc ligand in an asymmetric fashion between the two CH2 domains and the lower hinge regions (Fig. 2), which is similar to FcγRIII (CD16) (32, 38), and consistent with mutagenesis of Fc and functional studies of IgG (39). The protruding loop connecting the F and G β-strands in D2 of FcγRIIa penetrates, with Y160 in the lead, between the lower hinge regions and the first β-strands of the two CH2 domains (Fig. 2C, 2D). As a result, the hinge regions were pushed from a central location and now lie almost perpendicular to the pseudo–2-fold axis of the Fc and are directed away from the FcγRIIa binding site.
Rather than a contiguous binding site, the binding surfaces of FcγRIIa that recognize Fc are composed of three separate regions (Fig. 3). First, a large, protruding ridge is formed by the tip of the F-G loop (Y160), the neighboring B-C loop (P117, V119, and K120), and the C′ β-strand (F132, R134, and L135; Fig. 3A, 3B). This ridge participates in the majority of binding interactions with the Fc ligand by fitting between the two lower hinge regions (Leu-Leu-Gly-Gly; positions 234–237) and binding to a large area (S239, D265, S267, V266, and N297) on one side of the Fc (chain B; Fig. 3C, 3D). Second, a small, yet important, binding site is formed by FcγRIIa residues W90 and W113 that trap P329 from Fc chain A (Fig. 3B). Such an arrangement has been observed in complexes of FcεRI and FcγRIII (32, 38, 40), and is almost certainly a feature of all conventional FcγRs. The Pro residue binding in this alcove on FcγRIIa is probably mimicked by a peptide that was selected by phage-display and blocks IgG binding (41). The third FcγRIIa binding region involves a small patch (S129 and K128), which contacts the Fc-associated carbohydrate of chain B (N297-glycan) and may influence recognition of various IgG glycoforms (see later).
Influence of carbohydrate on the FcγRIIa interaction with human IgG1
A considerable amount of carbohydrate was observed in the electron density maps calculated for the FcγRIIa-Fc complex (Fig. 2). Unlike the unliganded FcγRIIa-HR that had a notable absence of ordered carbohydrate (Fig. 1), the same receptor in complex with Fc displayed an ordered branched oligosaccharide chain at N145 and a clearly defined GlcNAc residue at N64 (Fig. 2). Both FcγRIIa N-linked carbohydrates are distant from the binding interaction with the Fc ligand.
Carbohydrates attached to N297 of the two H chains reside at the interface of the CH2 domains and are consequently ordered in most crystal structures of Fc and Fc–ligand complexes (32, 42, 43). In the hu3S193 Fc complex with FcγRIIa, we observed biantennary and fucosylated oligosaccharide on both H chains (Fig. 2). The oligosaccharide attached to H chain A was missing only the terminal disaccharide Neu5Acα2→6Gal of the Manα1→6 branch. In H chain B, a complete branched N-linked glycan, including the two terminal sialic acids, was observed. A bisecting GlcNAc residue was not observed for either Fc-associated glycan, which is not surprising because this moiety occurs in only 5–10% of normal human IgG.
Because of the asymmetric nature of the FcγRIIa–Fc interaction, only the carbohydrate associated with one H chain (B) is directly involved in the interaction through the proximity of the Fuc to S129 and K128 of the receptor (Figs. 2, 3). Interestingly, the F132 and R134 side chains of FcγRIIa-HR bind in a shallow depression on Fc (chain B), formed by D265, V266, S267, and N297, which is adjacent to the Fuc moiety. The binding of R134 near the Fuc moiety of the IgG-Fc may relate to the previously reported modest increase in affinity of FcγRIIa-HR for Fuc-deficient IgG1 (44). In addition, the structure provides a glimpse of the interactions with IgG involving the HR/LR (R134/H134) polymorphic site, which has been linked to disease susceptibility (45), and in H134/H134 individuals higher response rates to a therapeutic Ab, rituximab (16).
Structural comparison of FcγR interactions with different IgG subclasses
Amino acids involved in the FcγRIIa–Fc interaction were assessed for their role in determining the specificity of different FcRs and the variation in IgG subclass binding by FcγRIIa (Table II). The related FcγRIII and FcγRI that both bind IgG displayed low sequence identities (5/11 and 6/11, respectively) to the FcγRIIa binding-site residues, which may explain the unique specificity of FcγRIIa for various IgG subclasses. In addition, the related IgG inhibitory receptor, FcγRIIb, has R134 similar to the HR polymorphism, but differs by having S135 replacing L135 of FcγRIIa (Table II). It was not surprising that the IgE receptor (FcεRI) shared only 4/11 binding site residues used by FcγRIIa.
Table II.
Comparison of residues of the FcγRIIa–IgG1-Fc interaction with those of other human FcR and IgG subclasses
| Positiona | FcγRIIab | FcγRIII | FcγRI | FcεRI |
|---|---|---|---|---|
| Human FcRs | ||||
| 90 | W | W | W | W |
| 113 | W | W | W | W |
| 117 | P | A | I | D |
| 119 | V | H | V | V |
| 120 | K | K | N | K |
| 128 | K | K | K | F |
| 129 | S | G | A | A |
| 132 | F | Y | F | Y |
| 134 | H/R (R) | H | H | Y |
| 135 | L (S) | H | W | F |
| 160 | Y | S | K | Q |
| Eu Indexc | IgG1 | IgG2 | IgG3 | IgG4 |
| Human IgG subclasses | ||||
| 234 | L | V | L | F |
| 235 | L | A | L | L |
| 236 | G | – | G | G |
| 237 | G | G | G | G |
| 239 | S | S | S | S |
| 265 | D | D | D | D |
| 266 | V | V | V | V |
| 267 | S | S | S | S |
| 297 | N | N | N | N |
| 327 | A | G | A | G |
| 328 | L | L | L | L |
| 329 | P | P | P | P |
Position in amino acid sequence corresponding to the numbering scheme for crystal structures described in this article and reported previously (17, 24).
Residues in parentheses occur in FcγRIIb. The same numbering scheme has been used for FcγRIIb.
Numbering for IgG is the Eu Index (62).
–, deletion in the sequence.
The HR and LR alleles of FcγRIIa were initially named for their ability to bind mouse IgG1. However, the FcγRIIa-LR reacts efficiently with human IgG2, whereas the FcγRIIa-HR does not (21), and the relative reactivity of FcγRIIa toward human IgG subclasses is reported to be: IgG3 > IgG1/IgG2 > > IgG4 for LR-FcγRIIa (17) and IgG3/IgG1 > > IgG2 > IgG4 for FcγRIIa-HR (21). The near-equivalent binding of human IgG3 and human IgG1 is reflected in the identity of the Fc residues involved in the interaction with FcγRIIa-HR (Table II). It is likely that differences in affinity of FcγRIIa for the IgG subclasses are also contributed by different sequences and lengths of the hinge region. In IgG2, 8/12 (including a deletion at position 236 with respect to IgG1) contact residues were identical, whereas in IgG4, 10/12 residues were identical to IgG1. Interestingly, most of the differences in human IgG1 versus IgG2 are located near the R134/H134 polymorphic binding site, suggesting that cooperative conformational effects are responsible for the different binding capacities of the HR and LR receptor alleles toward the human IgG2 subclass. The LL to VA (234–235) substitutions and the deletion of G236 in the “lower hinge” region of IgG2 is where the F–G loop and Y160 residue of the receptor binds, which is immediately adjacent to the cavity where R134 is located. In contrast, the A to G (327) substitution in IgG2 is close to residue P329, which on the other side of the Fc is sandwiched between W90 and W113 of FcγRIIa-HR. Thus, the noted differences in IgG2 in the lower hinge may account for specificity of this subclass for the FcγRIIa-LR (also see later). It is noteworthy that like human IgG, the mouse IgG classes also vary in hinge sequence and length; in particular, mouse IgG1 originally used to distinguish between the HR and LR phenotypes contains a truncated lower hinge region, which is two residues shorter than in human IgG1 (L234 and L235 are absent), whereas 6/10 remaining FcγRIIa contact residues in mouse IgG1 are identical (S239, D265, S267, N297, A327, and P329). Thus, the sequence and length of the lower hinge appears to be important in dictating the strength of binding to the polymorphic variants of FcγRIIa.
The interaction of FcγRIIa and IgG is distinct from that of FcγRIII (CD16a) with IgG. The FcγRIII–IgG complex has been defined and predictions made for the interactions of human IgG1 with FcγRIIa-HR and FcγRI (32, 46). In FcγRIII, the binding site for Fc residue L235 of chain A involves residues T113, A114, and V155, and on this basis, the equivalent residues in FcγRIIa (K116, P117, and I158) were predicted to bind IgG (46). However, in the crystal structure of FcγRIIa-HR–Fc, whereas L235 on Fc binds close to the predicted binding site, only P117 of the receptor contacts the Fc ligand (Fig. 3B). Furthermore, L235 on the other H chain (B) in the FcγRIII–IgG complex is bound between FcγRIII residues H116 and H132 that correspond to FcγRIIa residues V119 and L135, which were predicted to bring L235 closer into hydrophobic contact with the receptor (46). In contrast, wedging of the FcγRIIa F-G loop into the lower hinge region where Y160 interacts with L234 (of H chain B) forces L235 of the Fc away (>12 Å) from the predicted V119/L135 hydrophobic binding pocket. Instead, V119 of FcγRIIa interacts with Fc residue G237 closer to the binding pocket for R134 of the HR allele, which as correctly predicted (46) does not have sufficient space (i.e., this residue adopts a bent or cramped conformation) in its binding site when compared with H134 of FcγRIIa-LR.
Structural mapping of epitopes of FcγRII blocking Abs
To define how three distinct anti-FcγRII Abs block IgG binding, we mapped their epitopes using a mutagenesis approach and correlated the findings to the crystal structures of FcγRIIa. The mAbs include IV.3 that specifically binds to FcγRIIa, X63-21/7.2 specifically recognizing FcγRIIb, and 8.7, which recognizes both FcγRIIa and FcγRIIb. All Abs inhibit immune complex IgG binding to the respective receptors.
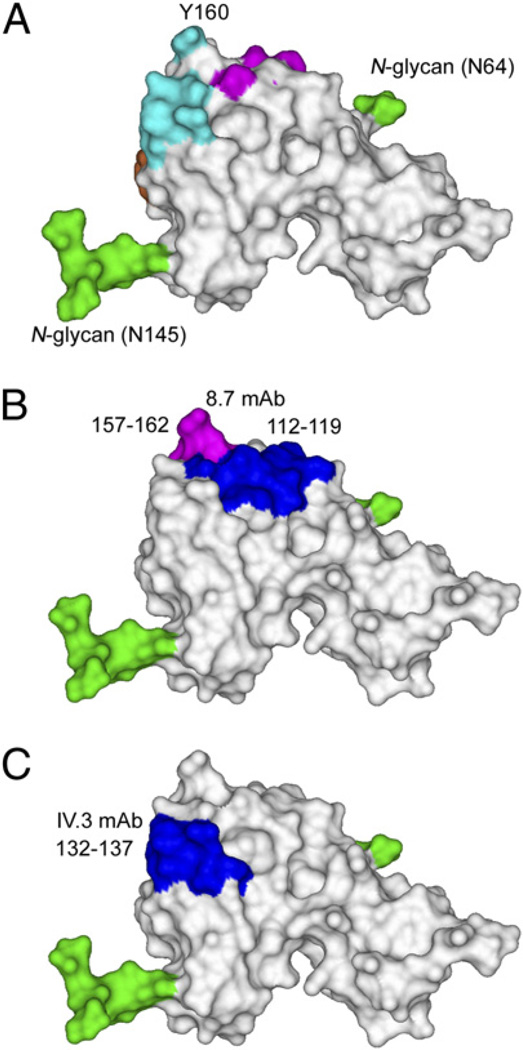
mAbs 8.7 and IV.3 block IgG binding and have been shown to bind to the second extracellular domain through the use of chimeric receptors containing sequence elements derived from FcγRIIa and FcεRI (35). This was evident from their binding to chimera εγ containing only the second domain of FcγRIIa (Supplemental Fig. 1B). Further analyses of these FcγRIIa/FcεRI receptor chimeras indicate that 8.7 recognizes a discontinuous epitope (Supplemental Figs. 1, 2). Ab 8.7 bound to only those chimeric receptors containing residues 112–119 together with 157–162 (chimeras 3 and 4) and failed to bind to any receptor lacking either one of these segments (chimeras 1 and 2; Supplemental Figs. 1, 2). Most importantly, 8.7 bound to chimera 4 wherein the only segments derived from FcγRIIa are residues 112–119 and 157–162. These two segments also encompass two IgG contact regions defined in the complex of FcγRIIa–IgG1-Fc (Fig. 3B).
Point mutations were then introduced into the FcγRIIa to further define the 8.7 epitope. First, residues L162 and F163 were mutated to introduce the equivalent N-linked glycosylation site of FcγRIIIa (47), N162 and V163, into FcγRIIa. Normal FcγRIIa bound aggregated IgG, as well as IV.3 and 8.7. However, the L162N mutant showed only weak binding of 8.7, whereas the binding of IV.3 was unaffected (Supplemental Fig. 3), indicating a role for this residue in the 8.7 epitope. Not surprisingly, the binding of aggregated IgG was diminished. The second component of the 8.7 epitope involves residues 112–119 and includes W113, which is pivotally involved in the interaction with IgG (Fig. 3B). The mutation of W90 and W113 completely abolished 8.7 binding, whereas the mutation of W90 alone had no effect, indicating that W113 is essential for the 8.7 epitope (Table III). Furthermore, both W113 and L162 are present in FcγRIIb to which 8.7 also binds.
Table III.
Summary of epitope mapping data using FcγRIIa mutants
| Residue Numbera | Binding Activity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Receptor Construct | 90 | 113 | 130 | 134–138 | 162 | 163 | 8.7 | IV.3 | X63-21 | HAGG |
| FcγRIIa-LR WT | W | W | Q | HLDPT | L | F | ++ | ++ | − | ++ |
| FcγRIIa-HR WT | ․ | ․ | ․ | R․ ․ ․ ․ | ․ | ․ | ++ | ++ | − | ND |
| FcγRIIb WT | ․ | ․ | K | RS ․ ․ N | ․ | ․ | ++ | − | ++ | ++ |
| FcγRIIa-LR W90A | A | ․ | ․ | ․ ․ ․ ․ ․ | ․ | ․ | ++ | ++ | ND | + |
| FcγRIIa-LR W90A,W113A | A | A | ․ | ․ ․ ․ ․ ․ | ․ | ․ | − | ++ | ND | ± |
| FcγRIIa-LR Q130K | ․ | ․ | K | ․ ․ ․ ․ ․ | ․ | ․ | ++ | ++ | ND | ++ |
| FcγRIIa-LR L135S | ․ | ․ | ․ | ․ S ․ ․ ․ | ․ | ․ | ++ | − | − | ++ |
| FcγRIIa-HR L135S | ․ | ․ | ․ | RS ․ ․ ․ | ․ | ․ | ++ | − | ++ | ++ |
| FcγRIIa-HR T138N | ․ | ․ | ․ | ․ ․ ․ ․ N | ․ | ․ | ++ | ++ | ND | ++ |
| FcγRIIa-HR L135S, T138N | ․ | ․ | ․ | ․ S ․ ․ N | ․ | ․ | ++ | − | ++ | ++ |
| FcγRIIa-HR L162N, F163V | ․ | ․ | ․ | ․ ․ ․ ․ ․ | N | V | ± | ++ | ND | + |
“․” indicates residues that are unaltered from the wild-type sequence of FcγRIIa-LR, FcγRIIa-HR, or FcγRIIb. Residue W113 is essential to the 8.7 epitope. Residues L135 and R134-S135 are essential to the IV.3 and X63-21/7.2 epitopes in FcγRIIa and FcγRIIb, respectively.
maximal binding equivalent to WT FcγRIIa;
intermediate binding;
weak detectable binding;
undetectable binding over background, as exemplified in Fig. 4 and Supplemental Fig. 3;
ND, no data.
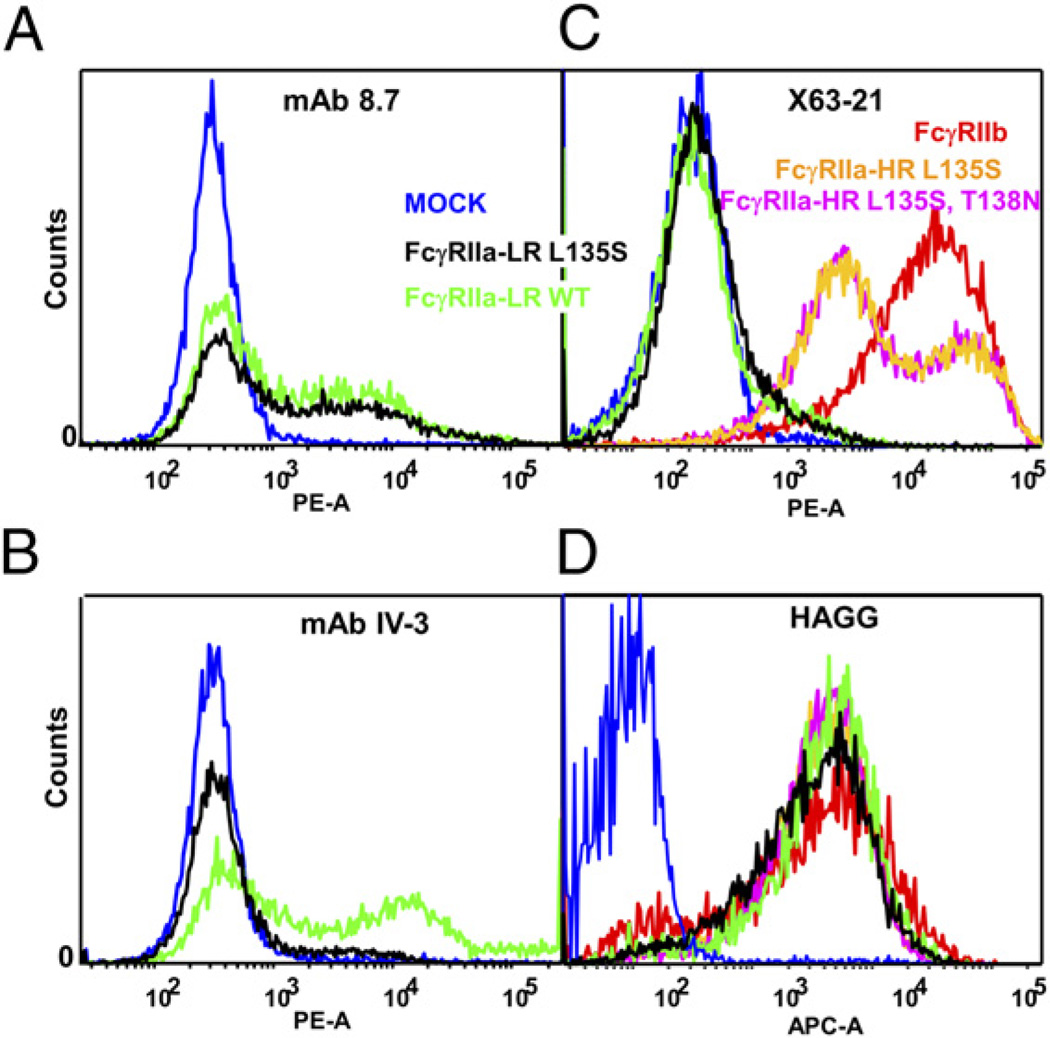
The IV.3 mAb specifically binds FcγRIIa, but not FcγRIIb, and its epitope was localized to residues 132–137. Replacement of this sequence in FcγRIIa by the equivalent six amino acids from FcεRI (chimera 6) destroys IV.3 binding, whereas all other chimeras containing this FcγRIIa sequence bound IV.3 (chimeras 1, 2, 3, 5; Table III, Supplemental Figs. 1, 2). This epitope contains the HR/LR polymorphism of FcγRIIa, but the fact that IV.3 binds to both FcγRIIa-HR and FcγRIIa-LR indicates either arginine or histidine at position 134 is compatible with IV.3 binding. However, in FcγRIIa and FcγRIIb, this sequence differs at residue 135 and it is also flanked by two additional sequence differences at residues 130 and 138. Mutation of Q130 or T138 in FcγRIIa to K130 or N138 of FcγRIIb had no detectable effect on IV.3 binding. However, replacement of L135 in both the FcγRIIa-HR and FcγRIIa-LR with S135 of FcγRIIb completely destroyed the IV.3 epitope and had no effect on 8.7 (Fig. 4, Table III), indicating L135 is an essential part of the IV.3 epitope and determines its selectivity for FcγRIIa.
FIGURE 4.
FACS analyses of mAb binding to mutants of FcγRII. Mock-transfected CHOP cells (blue line), FcγRIIa-LR WT (green line), or FcγRIIa-LR L135S mutant receptor (black line) were incubated with (A) biotinylated 8.7 F(ab′)2 or (B) biotinylated IV.3 Fab. C, FcγRIIb1 transfectants (red line) and mutant receptors FcγRIIa-HR L135S (orange line) and FcγRIIa-HR L135S T138N (magenta line) were reacted with the FcγRIIb-selective biotinylated F(ab′)2 of mAb X63-21/7.2. D, Ligand binding activity of the transfected cells was tested with heat-aggregated gamma globulin (HAGG) and allophycocyanin-conjugated anti-human IgG. C and D, Transfections were spiked with 1/40 EGFP-N1 (Clontech) and FL1+ cells gated to select only transfected cells for analysis.
The epitope detected by FcγRIIb-specific blocking mAb X63-21/7.2 was mapped to an area equivalent to that of the IV.3 epitope in FcγRIIa. In a reciprocal pattern to IV.3 binding, X63-21/7.2 fails to bind FcγRIIa or the FcγRIIa-LR L135S, but does stain FcγRIIa-HR L135S, T138N, and most importantly, FcγRIIa-HR L135S (Fig. 4), indicating both R134 and S135 are essential to the X63-21/7.2 epitope and S135 for its selectivity for FcγRIIb.
When the epitopes defined by the chimeric receptor and mutagenesis experiments were mapped onto the solvent-accessible surfaces of FcγRIIa, it was apparent that the epitopes recognized by 8.7 and IV.3 were likely to be nonoverlapping (Fig. 5). Importantly, both blocking Abs also bound to regions that were clearly used for interactions with IgG1-Fc as defined by the crystal structure of the FcγRIIa-HR–IgG1-Fc. Thus, these mAbs are clearly competitive inhibitors of binding of immune complexes to the FcγRII class of receptors.
FIGURE 5.
Structural location of epitopes recognized by FcγRIIa blocking mAb. A, FcγRIIa-HR with IgG1 binding residues mapped to the surface colored according to Fig. 3. B, Epitope regions defined to bind to 8.7. C, Epitope region defined to bind to IV.3. The views of FcγIIa are shown in the same orientation.
Model of the activation complex of FcγRIIa
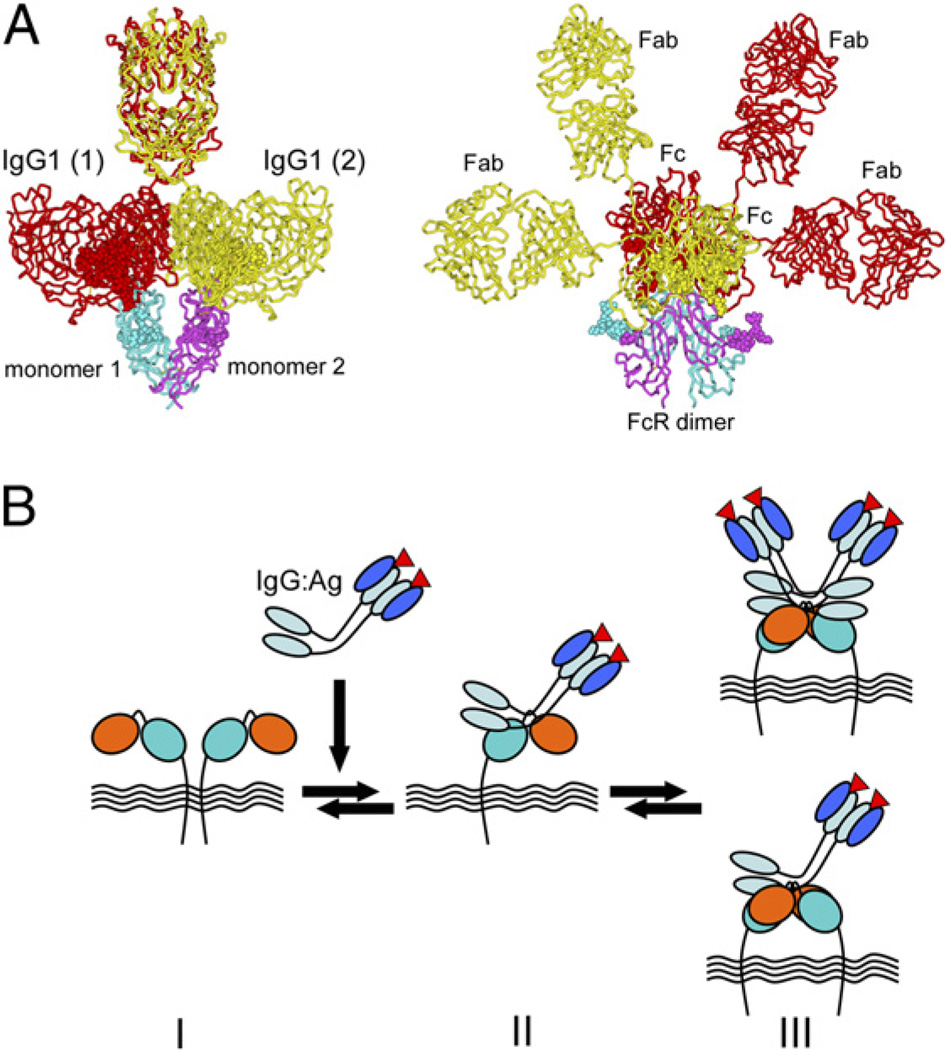
Immune complexes (Ag–IgG) drive the aggregation of FcγRIIa on the cell surface to initiate intracellular receptor phosphorylation and downstream signaling (25, 27, 48). However, the arrangement of Ag–IgG–FcγRIIa complexes within these aggregates has not been characterized. Furthermore, we previously demonstrated that a proportion of FcγRIIa is constitutively present as a dimer on the cell surface (25). Thus, the dimeric forms of FcγRIIa that we have observed in crystals of glycosylated receptor (Fig. 1) may represent the receptor presentation on the surface of cells. We attempted to overlay the coordinates for the FcγRIIa–Fc complex onto the two crystallographic FcγRIIa dimers. The dimer represented in crystals of FcγRIIa-LR was not capable of binding a single Fc without clashing with the neighboring receptor monomer. In contrast, the FcγRIIa-HR crystallographic dimer was capable of accommodating either one or two Fc ligands without any major steric clashes (Fig. 6). In this model, the hinge regions of the two trapped Fc ligands are nearly abutting, but are bent away from the proposed location of the FcγRIIa molecules and the membrane. As a result, the Fab arms, tethered by flexible hinge regions, are positioned a considerable distance away from the cell membrane and are readily available for binding Ag. This arrangement suggests that the elements in the hinges and CH2 domains of opposing receptor-bound Fc can interact (e.g., lower hinge residue E233 from one Fc can form a potential salt-bridge with K326 on the second Fc). This is entirely consistent with a comprehensive mutagenesis study where mutations in the lower hinge region including E233 reduced FcR binding to IgG1 (39). It is important to note that rather than the free IgG bound to FcγRIIa as in crystals and the model (Fig. 6), in vivo Ag–IgG binds to the receptors. However, the structures reported in this article may represent snapshots of the possible arrangements of receptors within the membrane and in signaling assemblies.
FIGURE 6.
Structural considerations for clustering and activation of FcγRIIa by immune complexes. A, Molecular model for the activation complex of FcγRIIa with two bound IgG ligands. The model was generated by rigid body superposition of the coordinates for the unliganded FcγRIIa-HR dimer with those of its complex with the Fc region of human IgG1, and the Fabs were derived from hu3S193 Fab (PDB code 1S3K). Two orthogonal views are shown of IgG bound to dimeric receptor. Sufficient space is available to accommodate the Fab arms that emerge as they are tethered to Fc by long, flexible hinge regions. B, Schematic view of FcR clustering based on the available crystallographic information. An inactive or resting FcγRIIa dimer (state I) dissociates for steric reasons with the addition of Ag–IgG (immune complexes) to produce an Ag–IgG–FcγRIIa complex (state II). Rearrangements of Ag–IgG–FcγRIIa complexes can form a dimeric activation complex (state III) composed of either two Ag–IgG bound to the FcR dimer (shown in A) or alternatively a single Ag–IgG interacting with receptor associated with an unliganded monomer. These component assemblies may cluster to form larger aggregates, similar to microdomains, and act as potent inducers of inflammation.
Discussion
The Fc region of IgG provides humoral immunity with a cell-based effector system through the binding of IgG immune complexes to their specific activating receptors: FcγRI (CD64), FcγRIII (CD16), and FcγRIIa (CD32) in humans. These interactions are critical for normal immunity, but in autoimmunity are capable, especially through FcγRIIa, of inducing powerful inflammatory responses with adverse consequences in diseases like rheumatoid arthritis and SLE (1).
In addition, the pivotal role of the Ab Fc in humoral immunity has directed the evolution of bacterial and viral virulence factors in their development of immune evasion mechanisms. Crystallographic studies of many immune system (host) and pathogen proteins in complex with Ab Fc regions have provided a wealth of information on the mechanisms of effector functions and immune evasion (32, 38, 40, 42, 49–52). These studies suggest that recognition strategies have converged toward the binding of proteins at one of two hot spots on the Fc portion of Ab. First, the conventional FcRs interact with the CH2 domains and adjacent lower hinge region, including FcγRIIa (this study), FcγRIII (32, 38), and FcεRI (40) as a complex with Cε2-Cε4 of IgE-Fc (53); this is also likely to be the case for FcγRI (46, 54). The second important Fc hot spot is located at the junction of the CH2 and CH3 domains, and is recognized by many molecules such as the MHC-related neonatal FcR (55). Numerous proteins produced by significant human pathogens also bind the CH2–CH3 domain junction such as protein A from Staphylococcus aureus (42); the equivalent site in IgA is the target for host and pathogen proteins (50, 52, 56). The CH2–CH3 domain junction has also been identified as a target for bacterial Ig-binding proteins across species (57). Furthermore, the CH2–CH3 domain junction is the primary site of rheumatoid factor recognition directed against IgG (49). In IgM, the CH3-CH4 domain junction is analogous to CH2–CH3 of IgG and appears to be a recognition site for both the Fcα/μ receptor (IgA and IgM receptor) (58) and some strains of P. falciparum erythrocyte membrane protein 1 (59).
Clinically important polymorphisms occur in FcγRIIa (R134/H134; HR/LR) and FcγRIII (V158/F158), which influence IgG binding and are known to be associated with susceptibility to Gram-negative infection (15), autoimmune diseases (10, 13, 45), and malaria (23). Furthermore, a structural understanding of the impact of the receptor polymorphisms on recognition of different IgG glycoforms or subclasses is of particular importance for the development of effective Ab-based therapies. For example, the homozygous H134/H134 (FcγRIIa) and V158/V158 (FcγRIII) genotypes were independently associated with higher response rates and freedom from disease progression in patients being treated for follicular lymphoma with a CD20-specific mAb, rituximab (16).
In this study, we observed the R134 of the FcγRIIa-HR binds in a shallow depression in the Fc close to the Fuc from the adjacent N297-linked carbohydrate on Fc. Removal of Fuc from human IgG1 has been shown to slightly improve the binding of the R134 (FcγRIIa-HR and FcγRIIb), but not the H134 (FcγRIIa-LR) form. The effect was even more pronounced in FcγRIII where binding to Fuc-deficient IgG1 was improved by up to 50-fold (F158 > V158) compared with native IgG1 (44). In the FcγRIII complex with Fc, the fucose does not participate in the interaction (32, 38). Furthermore, the carbohydrate on N162 of FcγRIII is needed for the high-affinity binding to Fuc-deficient IgG1, and this oligosaccharide (not present on FcγRIIa) is predicted to interact with the carbohydrate moieties of the Fc (60). Thus, different mechanisms may mediate the variation in binding of IgG glycoforms observed for FcγRII and FcγRIII. However, further studies are required to establish the link between IgG fucosylation and molecular interactions involving FcγRIIa polymorphisms.
The IgG binding regions on FcγRIIa are localized to two nonoverlapping sites on the receptor (Fig. 5A). The same regions on FcγRII that bind IgG were determined to be the predominant epitopes recognized by mAbs 8.7, IV.3, and X63-21/7.2, which establishes that these blocking Abs are direct inhibitors of immune complexes binding by the FcγRII class of receptors. The two FcγRIIa binding Abs, 8.7 (Fig. 5B) and IV.3 (Fig. 5C), bind to the nonoverlapping regions that correspond with the two distinct regions used by the receptor to bind IgG. The selectivity of mAb IV.3 for FcγRIIa (L135) and X63-21/7.2 for FcγRIIb (S135) is dependent on the difference at residue 135 between these two receptors. Although our studies have established the structural locations for the epitopes of these FcγRII blocking Abs, there remain significant challenges in developing intact IgG as potential therapeutics for immune complex-mediated disease. Importantly, we have determined the crystal structure of FcγRIIa in complex with IgG, which provides details of the target site for binding immune complexes by this human inflammatory receptor.
The Fc in the complex with FcγRIIa-HR was prepared from the hu3S193 IgG1 that has potent FcR-dependent, Ab-dependent cellular cytotoxicity against tumor cells bearing its target Ley Ag. We have previously determined the crystal structure of the hu3S193 Fab in complex with the Ley tetrasaccharide and shown that this compact epitope binds end-on into the Ag-binding cavity (29). Because the Ley epitopes are displayed at very high densities on the surface of tumor cells, avid crosslinking of Ley by hu3S193 IgG (61) would enhance clustering of FcRs (including FcγRIIa) on effector cells, thereby eliciting Ab-dependent cellular cytotoxicity. Similar mechanisms are certainly involved in FcR-mediated killing observed with other therapeutic Abs. In addition to the forces of Ag-driven clustering, it is possible that interactions between the FcRs themselves are involved in organization and activation of these cell surface receptors.
FcγRIIa, like all activating FcRs and indeed other multisubunit immunoreceptors including the Ag receptors, relies on ITAM-based signaling pathways to initiate signaling cascades. The ITAM signaling pathway typically relies on the noncovalent association of ligand-binding chains with covalently dimerized polypeptides such as the common FcR-γ–chain, the CD3 complex of TCR complex, or Igα/β associated with the B cell Ag receptor complex. However, in this respect, FcγRIIa is unique, because it does not associate with, nor require, separate ITAM-containing dimers for activation of cells. Receptor dimerization as anticipated in this study (Figs. 1, 6) would juxtapose the ITAM-containing cytoplasmic tails. We postulate that there is a dynamic interplay between quaternary states of FcγRIIa, which may be involved in immune complex activation leading to inflammation (Fig. 6). The structural information on FcγRIIa suggests that a minimum of three quaternary states may account for the available biochemical data on the interplay between Ag–IgG complexes and the cell surface-associated receptor. In such a model, FcγRIIa exists as a resting or inactive dimer (state I, Fig. 6) that requires dissociation before, or upon, binding an Ag–IgG complex (state II, Fig. 6). Lateral packing of receptors (as in crystals of the FcγRIIa-HR) with bound Ag–IgG complexes will bring two ITAMs together to form signaling competent complex (state III, Fig. 6), a building block for a larger cluster, which efficiently mediates cell activation. Depending on the presentation of Ag–Ab complexes, a variation on state III of one, not two, Ag–IgG complexes and a receptor dimer may also occur. In any event, for both cases, the liganded dimers contain cytoplasmic tails arranged analogously to the dimeric FcR-γ subunit of other immunoreceptors, and so could mediate intracellular signaling and inflammation.
In summary, we provide in this article a structural rationale for the interplay between glycosylated FcRγIIa monomers, their dimers, and binding of immune complexes on the cell surface that lead to ordered receptor clustering, signaling cascades, and inflammation. Collectively, the structural data suggest how the HR/LR polymorphism of FcγRIIa influences binding to different IgG subclasses and possibly glycoforms such as fucose-deficient IgG. Finally, the same sites that are involved in binding IgG were found to be targeted by blocking Abs specific for FcγRII.
Supplementary Material
Acknowledgments
This work was supported by project Grant 315625 from the National Health and Medical Research Council of Australia and the Victorian state government Operational Infrastructure Support Scheme. P.A.R. was supported by a National Health and Medical Research Council R. Douglas Wright Career Development award (Grant 365209). This work, including use of the BioCARS sector, was supported by the Australian Synchrotron Research Program, which is supported by the Common-wealth of Australia under the Major National Research Facilities Programme. Use of BioCARS Sector 14 was also supported by the National Institutes of Health, National Center for Research Resources (Grant RR07707). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research (Contract W-31-109-Eng-38).
Abbreviations used in this article
- FcγRIIa-HR
high-responder (arginine, R134) allele of FcγRIIa
- FcγRIIa-LR
low-responder (histidine, H134) allele of FcγRIIa
- GlcNAc
N-acetylglucosamine
- HR
high responder
- LR
low responder
- PDB
Protein Data Bank
- PEG
polyethylene glycol
- SLE
systemic lupus erythematosus
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hogarth PM. Fc receptors are major mediators of antibody based inflammation in autoimmunity. Curr. Opin. Immunol. 2002;14:798–802. doi: 10.1016/s0952-7915(02)00409-0. [DOI] [PubMed] [Google Scholar]
- 2.Takai T. Fc receptors and their role in immune regulation and autoimmunity. J. Clin. Immunol. 2005;25:1–18. doi: 10.1007/s10875-005-0353-8. [DOI] [PubMed] [Google Scholar]
- 3.Masuda A, Yoshida M, Shiomi H, Morita Y, Kutsumi H, Inokuchi H, Mizuno S, Nakamura A, Takai T, Blumberg RS, Azuma T. Role of Fc receptors as a therapeutic target. Inflamm. Allergy Drug Targets. 2009;8:80–86. doi: 10.2174/187152809787582525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinet JP. The gamma-zeta dimers of Fc receptors as connectors to signal transduction. Curr. Opin. Immunol. 1992;4:43–48. doi: 10.1016/0952-7915(92)90122-u. [DOI] [PubMed] [Google Scholar]
- 5.Wines BD, Trist HM, Ramsland PA, Hogarth PM. A common site of the Fc receptor gamma subunit interacts with the unrelated immuno-receptors FcalphaRI and FcepsilonRI. J. Biol. Chem. 2006;281:17108–17113. doi: 10.1074/jbc.M601640200. [DOI] [PubMed] [Google Scholar]
- 6.Tan Sardjono C, Mottram PL, Hogarth PM. The role of FcgammaRIIa as an inflammatory mediator in rheumatoid arthritis and systemic lupus erythematosus. Immunol. Cell Biol. 2003;81:374–381. doi: 10.1046/j.1440-1711.2003.01182.x. [DOI] [PubMed] [Google Scholar]
- 7.McKenzie SE, Taylor SM, Malladi P, Yuhan H, Cassel DL, Chien P, Schwartz E, Schreiber AD, Surrey S, Reilly MP. The role of the human Fc receptor Fc gamma RIIA in the immune clearance of platelets: a transgenic mouse model. J. Immunol. 1999;162:4311–4318. [PubMed] [Google Scholar]
- 8.Pietersz GA, Mottram PL, van de Velde NC, Sardjono CT, Esparon S, Ramsland PA, Moloney G, Baell JB, McCarthy TD, Matthews BR, et al. Inhibition of destructive autoimmune arthritis in FcgammaRIIa transgenic mice by small chemical entities. Immunol. Cell Biol. 2009;87:3–12. doi: 10.1038/icb.2008.82. [DOI] [PubMed] [Google Scholar]
- 9.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J. Clin. Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salmon JE, Millard S, Schachter LA, Arnett FC, Ginzler EM, Gourley MF, Ramsey-Goldman R, Peterson MG, Kimberly RP. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J. Clin. Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuniga R, Markowitz GS, Arkachaisri T, Imperatore EA, D’Agati VD, Salmon JE. Identification of IgG subclasses and C-reactive protein in lupus nephritis: the relationship between the composition of immune deposits and FCgamma receptor type IIA alleles. Arthritis Rheum. 2003;48:460–470. doi: 10.1002/art.10930. [DOI] [PubMed] [Google Scholar]
- 12.Loos BG, Leppers-Van de Straat FGJ, Van de Winkel JGJ, Van der Velden U. Fcgamma receptor polymorphisms in relation to periodontitis. J. Clin. Periodontol. 2003;30:595–602. doi: 10.1034/j.1600-051x.2003.00355.x. [DOI] [PubMed] [Google Scholar]
- 13.Dijstelbloem HM, Bijl M, Fijnheer R, Scheepers RH, Oost WW, Jansen MD, Sluiter WJ, Limburg PC, Derksen RH, van de Winkel JG, Kallenberg CG. Fcgamma receptor polymorphisms in systemic lupus erythematosus: association with disease and in vivo clearance of immune complexes. Arthritis Rheum. 2000;43:2793–2800. doi: 10.1002/1529-0131(200012)43:12<2793::AID-ANR20>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Asano K, Matsushita T, Umeno J, Hosono N, Takahashi A, Kawaguchi T, Matsumoto T, Matsui T, Kakuta Y, Kinouchi Y, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 2009;41:1325–1329. doi: 10.1038/ng.482. [DOI] [PubMed] [Google Scholar]
- 15.Sanders LA, Feldman RG, Voorhorst-Ogink MM, de Haas M, Rijkers GT, Capel PJ, Zegers BJ, van de Winkel JG. Human immunoglobulin G (IgG) Fc receptor IIA (CD32) polymorphism and IgG2-mediated bacterial phagocytosis by neutrophils. Infect. Immun. 1995;63:73–81. doi: 10.1128/iai.63.1.73-81.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 2003;21:3940–3947. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Powell MS, Barton PA, Emmanouilidis D, Wines BD, Neumann GM, Peitersz GA, Maxwell KF, Garrett TP, Hogarth PM. Biochemical analysis and crystallisation of Fc gamma RIIa, the low affinity receptor for IgG. Immunol. Lett. 1999;68:17–23. doi: 10.1016/s0165-2478(99)00025-5. [DOI] [PubMed] [Google Scholar]
- 18.Warmerdam PA, van de Winkel JG, Gosselin EJ, Capel PJ. Molecular basis for a polymorphism of human Fc gamma receptor II (CD32) J. Exp. Med. 1990;172:19–25. doi: 10.1084/jem.172.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tate BJ, Witort E, McKenzie IF, Hogarth PM. Expression of the high responder/non-responder human Fc gamma RII. Analysis by PCR and transfection into FcR-COS cells. Immunol. Cell Biol. 1992;70:79–87. doi: 10.1038/icb.1992.12. [DOI] [PubMed] [Google Scholar]
- 20.Tax WJ, Willems HW, Reekers PP, Capel PJ, Koene RA. Polymorphism in mitogenic effect of IgG1 monoclonal antibodies against T3 antigen on human T cells. Nature. 1983;304:445–447. doi: 10.1038/304445a0. [DOI] [PubMed] [Google Scholar]
- 21.Parren PW, Warmerdam PA, Boeije LC, Arts J, Westerdaal NA, Vlug A, Capel PJ, Aarden LA, van de Winkel JG. On the interaction of IgG subclasses with the low affinity Fc gamma RIIa (CD32) on human monocytes, neutrophils, and platelets. Analysis of a functional polymorphism to human IgG2. J. Clin. Invest. 1992;90:1537–1546. doi: 10.1172/JCI116022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon CL, Johnson PD, Permezel M, Holmes NE, Gutteridge G, McDonald CF, Eisen DP, Stewardson AJ, Edington J, Charles PG, et al. Association between severe pandemic 2009 influenza A (H1N1) virus infection and immunoglobulin G(2) subclass deficiency. Clin. Infect. Dis. 2010;50:672–678. doi: 10.1086/650462. [DOI] [PubMed] [Google Scholar]
- 23.Nasr A, Iriemenam NC, Giha HA, Balogun HA, Anders RF, Troye-Blomberg M, ElGhazali G, Berzins K. FcgammaRIIa (CD32) polymorphism and anti-malarial IgG subclass pattern among Fulani and sympatric ethnic groups living in eastern Sudan. Malar. J. 2009;8:43. doi: 10.1186/1475-2875-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxwell KF, Powell MS, Hulett MD, Barton PA, McKenzie IF, Garrett TP, Hogarth PM. Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa. Nat. Struct. Biol. 1999;6:437–442. doi: 10.1038/8241. [DOI] [PubMed] [Google Scholar]
- 25.Powell MS, Barnes NC, Bradford TM, Musgrave IF, Wines BD, Cambier JC, Hogarth PM. Alteration of the Fc gamma RIIa dimer interface affects receptor signaling but not ligand binding. J. Immunol. 2006;176:7489–7494. doi: 10.4049/jimmunol.176.12.7489. [DOI] [PubMed] [Google Scholar]
- 26.Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B. Investigation of early events in Fc epsilon RI-mediated signaling using a detailed mathematical model. J. Immunol. 2003;170:3769–3781. doi: 10.4049/jimmunol.170.7.3769. [DOI] [PubMed] [Google Scholar]
- 27.Metzger H. Transmembrane signaling: the joy of aggregation. J. Immunol. 1992;149:1477–1487. [PubMed] [Google Scholar]
- 28.Metzger H. Molecular versatility of antibodies. Immunol. Rev. 2002;185:186–205. doi: 10.1034/j.1600-065x.2002.18516.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramsland PA, Farrugia W, Bradford TM, Mark Hogarth P, Scott AM. Structural convergence of antibody binding of carbohydrate determinants in Lewis Y tumor antigens. J. Mol. Biol. 2004;340:809–818. doi: 10.1016/j.jmb.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Macromolecular Crystallography. Part A. Methods in Enzymology. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 33.Ierino FL, Hulett MD, McKenzie IF, Hogarth PM. Mapping epitopes of human Fc gamma RII (CDw32) with monoclonal antibodies and recombinant receptors. J. Immunol. 1993;150:1794–1803. [PubMed] [Google Scholar]
- 34.Looney RJ, Abraham GN, Anderson CL. Human monocytes and U937 cells bear two distinct Fc receptors for IgG. J. Immunol. 1986;136:1641–1647. [PubMed] [Google Scholar]
- 35.Hulett MD, McKenzie IF, Hogarth PM. Chimeric Fc receptors identify immunoglobulin-binding regions in human Fc gamma RII and Fc epsilon RI. Eur. J. Immunol. 1993;23:640–645. doi: 10.1002/eji.1830230310. [DOI] [PubMed] [Google Scholar]
- 36.Sondermann P, Jacob U. Human Fcgamma receptor IIb expressed in Escherichia coli reveals IgG binding capability. Biol. Chem. 1999;380:717–721. doi: 10.1515/BC.1999.090. [DOI] [PubMed] [Google Scholar]
- 37.Hulett MD, Witort E, Brinkworth RI, McKenzie IF, Hogarth PM. Multiple regions of human Fc gamma RII (CD32) contribute to the binding of IgG. J. Biol. Chem. 1995;270:21188–21194. doi: 10.1074/jbc.270.36.21188. [DOI] [PubMed] [Google Scholar]
- 38.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J. Biol. Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 39.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc γ RI, Fc γ RII, Fc γ RIII, and FcRn and design of IgG1 variants with improved binding to the Fc γ R. J. Biol. Chem. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- 40.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 41.Cendron AC, Wines BD, Brownlee RT, Ramsland PA, Pietersz GA, Hogarth PM. An FcgammaRIIa-binding peptide that mimics the interaction between FcgammaRIIa and IgG. Mol. Immunol. 2008;45:307–319. doi: 10.1016/j.molimm.2007.06.152. [DOI] [PubMed] [Google Scholar]
- 42.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 43.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 44.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SHA, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 45.Fossati G, Bucknall RC, Edwards SW. Fcgamma receptors in autoimmune diseases. Eur. J. Clin. Invest. 2001;31:821–831. doi: 10.1046/j.1365-2362.2001.00881.x. [DOI] [PubMed] [Google Scholar]
- 46.Sondermann P, Kaiser J, Jacob U. Molecular basis for immune complex recognition: a comparison of Fc-receptor structures. J. Mol. Biol. 2001;309:737–749. doi: 10.1006/jmbi.2001.4670. [DOI] [PubMed] [Google Scholar]
- 47.Drescher B, Witte T, Schmidt RE. Glycosylation of FcgammaRIII in N163 as mechanism of regulating receptor affinity. Immunology. 2003;110:335–340. doi: 10.1046/j.1365-2567.2003.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bournazos S, Hart SP, Chamberlain LH, Glennie MJ, Dransfield I. Association of FcgammaRIIa (CD32a) with lipid rafts regulates ligand binding activity. J. Immunol. 2009;182:8026–8036. doi: 10.4049/jimmunol.0900107. [DOI] [PubMed] [Google Scholar]
- 49.Corper AL, Sohi MK, Bonagura VR, Steinitz M, Jefferis R, Feinstein A, Beale D, Taussig MJ, Sutton BJ. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat. Struct. Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 50.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 51.Sprague ER, Wang C, Baker D, Bjorkman PJ. Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging. PLoS Biol. 2006;4:e148. doi: 10.1371/journal.pbio.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsland PA, Willoughby N, Trist HM, Farrugia W, Hogarth PM, Fraser JD, D.Wines B. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc. Natl. Acad. Sci. USA. 2007;104:15051–15056. doi: 10.1073/pnas.0706028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, Hunt J, Gould HJ, Beavil AJ, McDonnell JM, et al. Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI. Nat. Struct. Mol. Biol. 2011;18:571–576. doi: 10.1038/nsmb.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lund J, Winter G, Jones PT, Pound JD, Tanaka T, Walker MR, Artymiuk PJ, Arata Y, Burton DR, Jefferis R, Woof JM. Human Fc gamma RI and Fc gamma RII interact with distinct but overlapping sites on human IgG. J. Immunol. 1991;147:2657–2662. [PubMed] [Google Scholar]
- 55.Burmeister WP, Gastinel LN, Simister NE, Blum ML, Bjorkman PJ. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature. 1994;372:336–343. doi: 10.1038/372336a0. [DOI] [PubMed] [Google Scholar]
- 56.Pleass RJ, Areschoug T, Lindahl G, Woof JM. Streptococcal IgA-binding proteins bind in the Calpha 2-Calpha 3 interdomain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 2001;276:8197–8204. doi: 10.1074/jbc.M009396200. [DOI] [PubMed] [Google Scholar]
- 57.Lewis MJ, Meehan M, Owen P, Woof JM. A common theme in interaction of bacterial immunoglobulin-binding proteins with immunoglobulins illustrated in the equine system. J. Biol. Chem. 2008;283:17615–17623. doi: 10.1074/jbc.M709844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghumra A, Shi J, Mcintosh RS, Rasmussen IB, Braathen R, Johansen FE, Sandlie I, Mongini PK, Areschoug T, Lindahl G, et al. Structural requirements for the interaction of human IgM and IgA with the human Fcalpha/mu receptor. Eur. J. Immunol. 2009;39:1147–1156. doi: 10.1002/eji.200839184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghumra A, Semblat JP, McIntosh RS, Raza A, Rasmussen IB, Braathen R, Johansen FE, Sandlie I, Mongini PK, Rowe JA, Pleass RJ. Identification of residues in the Cmu4 domain of polymeric IgM essential for interaction with Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) J. Immunol. 2008;181:1988–2000. doi: 10.4049/jimmunol.181.3.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrara C, Stuart F, Sondermann P, Brünker P, Umaña P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J. Biol. Chem. 2006;281:5032–5036. doi: 10.1074/jbc.M510171200. [DOI] [PubMed] [Google Scholar]
- 61.Farrugia W, Scott AM, Ramsland PA. A possible role for metallic ions in the carbohydrate cluster recognition displayed by a Lewis Y specific antibody. PLoS ONE. 2009;4:e7777. doi: 10.1371/journal.pone.0007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. Public Health Service, U. S. Department of Health and Human Services. Bethesda, MD: National Institutes of Health; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.