Abstract
Fluxes of greenhouse gases to the atmosphere are heavily influenced by microbiological activity. Microbial enzymes involved in the production and consumption of greenhouse gases often contain metal cofactors. While extensive research has examined the influence of Fe bioavailability on microbial CO2 cycling, fewer studies have explored metal requirements for microbial production and consumption of the second- and third-most abundant greenhouse gases, methane (CH4), and nitrous oxide (N2O). Here we review the current state of biochemical, physiological, and environmental research on transition metal requirements for microbial CH4 and N2O cycling. Methanogenic archaea require large amounts of Fe, Ni, and Co (and some Mo/W and Zn). Low bioavailability of Fe, Ni, and Co limits methanogenesis in pure and mixed cultures and environmental studies. Anaerobic methane oxidation by anaerobic methanotrophic archaea (ANME) likely occurs via reverse methanogenesis since ANME possess most of the enzymes in the methanogenic pathway. Aerobic CH4 oxidation uses Cu or Fe for the first step depending on Cu availability, and additional Fe, Cu, and Mo for later steps. N2O production via classical anaerobic denitrification is primarily Fe-based, whereas aerobic pathways (nitrifier denitrification and archaeal ammonia oxidation) require Cu in addition to, or possibly in place of, Fe. Genes encoding the Cu-containing N2O reductase, the only known enzyme capable of microbial N2O conversion to N2, have only been found in classical denitrifiers. Accumulation of N2O due to low Cu has been observed in pure cultures and a lake ecosystem, but not in marine systems. Future research is needed on metalloenzymes involved in the production of N2O by enrichment cultures of ammonia oxidizing archaea, biological mechanisms for scavenging scarce metals, and possible links between metal bioavailability and greenhouse gas fluxes in anaerobic environments where metals may be limiting due to sulfide-metal scavenging.
Keywords: methane, nitrous oxide, microbes, metals, enzymes
Introduction
With increasing concern about the future impacts of global climate change, a detailed understanding of the sources and sinks of greenhouse gases is essential to mitigating their environmental impact. Although CO2 is the most abundant greenhouse gas, many other climatically important gases exist (Montzka et al., 2011). The second- and third-most abundant naturally produced greenhouse gases are methane (currently ~1.8 ppm; Heimann, 2011) and nitrous oxide (currently ~322 ppb; Montzka et al., 2011) and are ~25× and 300× more efficient at absorbing infrared radiation than CO2, respectively. Furthermore, CH4 is oxidized to CO2 in the atmosphere, contributing to rising CO2 levels. N2O has a very long residence time in the atmosphere (120 years) and reacts with atomic O to form nitric oxide (NO), which is involved in ozone destruction (Montzka et al., 2011).
Both humans and microbes play important, and often interwoven, roles in the production of CH4 and N2O. Humans have bred and expanded the habitats of ruminants that contain methanogenic archaea in their guts, cultivated rice paddies, and built wastewater and sewage treatment plants where methanogens proliferate. Humans have also extensively applied inorganic N as fertilizer to agricultural soils, leading to increased microbial N2O emissions in soils, wetlands, and coastal hypoxic zones (Schlesinger, 2009). Understanding the controls and regulation of microbial greenhouse gas emissions is therefore fundamental to quantifying and managing both natural and anthropogenic fluxes of these gases.
This review will focus on two potent greenhouse gases, CH4 and N2O, both of which are produced as natural by-products of microbial energy-generating metabolisms. CH4 is the final product of the anaerobic degradation of organic matter, whereas N2O formation results from incomplete conversion of nitrate or nitrite to N2. At the heart of the pathways that generate these gases are enzymes that catalyze redox reactions. Many of these enzymes contain transition metals as cofactors for electron transport or as catalytic centers at active sites. Important transition metals in the pathway of CH4 production (methanogenesis) and anaerobic methane oxidation include Fe, Ni, Co, Mo/W, and Zn, whereas aerobic (and intra-aerobic) methanotrophy and N2O production require Fe-, Cu-, and Mo-containing proteins. Only one protein, the Cu-rich nitrous oxidase reductase, is known to reduce N2O to N2. Physiological studies of pure cultures have shown that optimal metal concentrations for microbial metabolism are orders of magnitude higher than in situ concentrations in most aquatic environments. These findings lead to the question: are some microbes perennially metal-limited in nature? If so, does metal availability exert influence on the flux of greenhouse gases? If not, what mechanisms do microbes use in natural environments to acquire trace metals?
Previous environmental studies of metal requirements for microbes have largely focused on those microbes directly involved in CO2 cycling, principally phytoplankton that consume CO2 during photosynthesis. Driving these studies was the “Fe hypothesis” by John Martin positing that CO2-consuming marine phytoplankton could be fertilized by Fe (Martin and Fitzwater, 1988). Far less attention has been aimed at metal requirements for microbes involved in the cycling of non-CO2 greenhouse gases, although many of these organisms also live in ecosystems with very low metal bioavailability. A notable exception is the enormous wealth of literature generated by the wastewater scientific community about metal (particularly Fe, Ni, and Co) controls on methanogenesis in anaerobic digesters (see Demirel and Scherer, 2011).
The purpose of this article is to review the current state of literature on the metalloenzymes and trace metal physiology of organisms involved in CH4 and N2O processing. In each section, we discuss environmental studies if they exist and compare metal concentrations for optimal growth of pure cultures to measured values of trace metals in natural environments. In the final sections of the article, we compare the metal requirements of microbes involved in the CH4 and N2O cycles and discuss future research directions. Readers are referred to previous reviews (Rogers and Whitman, 1991; Conrad, 1996) for other aspects of microbial controls on greenhouse gas cycling.
Methanogenesis
Methanogenesis is the microbial process whereby CO2, acetate, or methyl-compounds are converted to methane in order to generate ATP through the build-up of a sodium ion or proton gradient. All methanogens are in the Euryarchaeota phylum (Whitman et al., 2006) and account for 75–80% of the annual global production of CH4 (IPCC, 2007). Approximately two-thirds of biologically produced CH4 is generated by aceticlastic methanogens that oxidize the carbonyl group of acetate to CO2 and reduce the methyl group to CH4 (Rogers and Whitman, 1991; Ferry, 2010b). The remaining one-third of biogenic methane is produced by reduction of CO2 with electrons from H2 or formate and the conversion of methyl groups from compounds such as methanol, methylamines, and dimethylsulfide. Therefore, methanogenesis is divided into three pathways: hydrogenotrophic (i.e., CO2 reduction), aceticlastic (acetate disproportionation), and methylotrophic (C1 utilization). The three pathways differ in the enzymes used to generate methyl-tetrahydro(methano/sarcina)pterin (CH3-H4(M/S)PT), the intermediate common to all pathways, but converge in the last two steps used to generate CH4 (Ferry, 2010b).
In the first section we review the metal inventory of metalloenzymes in each of the three pathways. This is not intended as a definitive review as many of these enzymes are still understudied and more complete reviews of methanogenic enzymes are available (Thauer, 1998; Thauer et al., 2008, 2010). Instead, our goal is to estimate the transition metal stoichiometry of the three methanogenic pathways, illustrated in Figure 1. In the next section we discuss physiological studies of the metal requirements for methanogenesis and relate these findings back to the metal content of each pathway. Pure culture, anaerobic digester, and environmental studies are reviewed. Lastly, a summary of previous studies examining metal requirements for methanogenesis is provided, and topics warranting further research are discussed. This general format is also used in the sections on methanotrophy (see Methanotrophy) and nitrous oxide cycling (see Nitrous Oxide Production and Consumption).
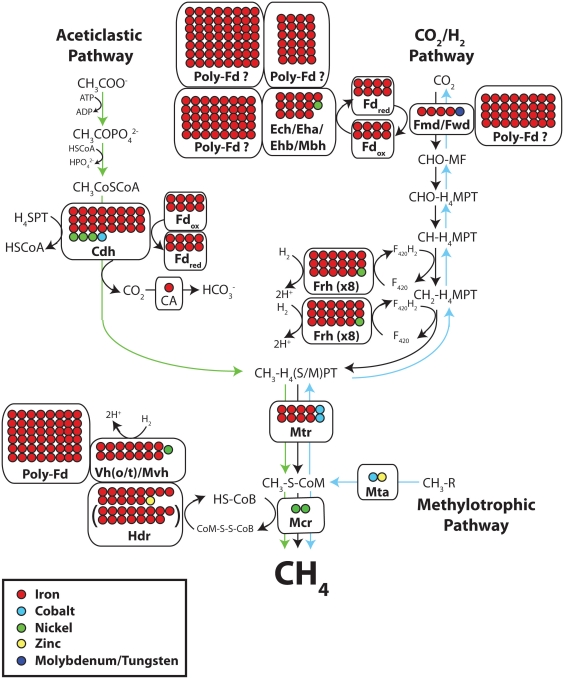
Figure 1.
Metal content of metalloenzymes in the three pathways of methanogenesis: H2/CO2 (black arrows), aceticlastic (green arrows), and methylotrophic (blue arrows). Each circle represents one metal atom. Parentheses show varying metal content of a given enzyme. Question marks mean that enzyme may not be present in all methanogens (6×[Fe4S4] and 10×[Fe4S4]polyferredoxins are encoded in the eha operon and a 14×[Fe4S4]polyferredoxin is encoded in the ehb operon). Abbreviations: CA, carbonic anhydrase; Cdh, carbon monoxide dehydrogenase/acetyl-CoA synthase; Ech/Eha/Ehb/Mbh, energy-converting hydrogenase; Fd, ferredoxin; Fmd/Fwd, Mo/W formylmethanofuran dehydrogenase; Frh, F420-reducing hydrogenase; Hdr, heterodisulfide reductase; Hmd, Ni-free Fe hydrogenase; Mcr, methyl coenzyme M reductase; Mta, methanol-coenzyme M methyltransferase; Mtr, CH3-H4M(S)PT-coenzyme M methyltransferase; Vh(o/t)/Mvh, Ni–Fe hydrogenase. For simplicity, only metalloenzymes are labeled. See the Section “Metalloenzymes in Methanogenesis” for more details, abbreviations for intermediates and enzyme substitutions that occur in specific conditions and species.
Metalloenzymes in methanogenesis
Methanogenesis is one of the most metal-rich enzymatic pathways in biology (Zerkle et al., 2005). Depending on the pathway, exact metal requirements may differ, but the general trends remain the same: Fe is the most abundant metal, followed by Ni and Co, and smaller amounts of Mo (and/or W) and Zn. Fe is primarily present as Fe–S clusters used for electron transport and/or catalysis. Ni is either bound to Fe–S clusters or in the center of a porphyrin unique to methanogens, cofactor F430. Co(balt) is present in cobamides involved in methyl group transfer. Zn occurs as a single structural atom in several enzymes. Mo or W is bound to a pterin cofactor to form “molybdopterin” or “tungstopterin,” which catalyze two-electron redox reactions. Other alkali metals and metalloids, such as Na and Se, are essential for methanogenesis, but the discussion here is limited to the transition metals. So far, no Cu-dependent methanogenesis enzymes have been identified, which is striking given the huge importance of Cu in aerobic methanotrophy (see the Section “Aerobic Methanotrophy”). It is plausible that scarce Cu in anaerobic early-earth marine ecosystems selected for use of other metals (i.e., Fe, Ni, and Co) in primitive methanogens whereas Cu was more bioavailable to aerobic methanotrophs after the rise of atmospheric oxygen (Dupont et al., 2006, 2010; David and Alm, 2010).
Most methanogen orders – Class I (Methanobacteriales, Methanococcales, and Methanopyrales) and Class II (Methanomicrobiales) – obtain energy by reducing CO2 to CH4 using electrons from H2 or in some cases formate (Anderson et al., 2009). These methanogens do not contain cytochrome proteins, in which Fe is bound to heme. Only one order of methanogens, the Methanosarcinales (Class III), contain cytochromes and are capable of metabolizing a wider range of substrates for methanogenesis, including methanol, methylamines, methyl sulfides, acetate, and/or CO2 with H2. Members of the Methanosarcinales play a key ecological role because the aceticlastic pathway is estimated to account for two-thirds of biologically sourced methane (Rogers and Whitman, 1991).
The Fe requirement for methanogenesis is vast: almost every metalloenzyme involved in the pathway contains multiple Fe2S2, Fe3S4, or Fe4S4 clusters. The first enzyme in the CO2 reduction pathway, formylmethanofuran dehydrogenase (abbreviated Fmd for the Mo form and Fwd for the W form) can bind up to nine Fe4S4 clusters: one cluster in the Mo/W-pterin binding subunit and up to eight additional clusters in polyferredoxin (Vorholt et al., 1996; Figure 1). It is likely that Fwd can bind additional Fe4S4 clusters (Ferry, 1999). Ferredoxins that transfer electrons from H2 to other methanogenesis enzymes require additional Fe in the form of two Fe4S4 clusters (Daas et al., 1994).
All hydrogenases involved in methanogenesis are Ni–Fe enzymes that oxidize H2 and reduce ferredoxin, coenzyme F420, and other electron carriers (Thauer et al., 2010). The four different types of Ni–Fe hydrogenases involved in methanogenesis all contain abundant Fe. The energy-converting membrane-associated hydrogenase [abbreviated Ech, Eha, Ehb, or Mbh depending on the class of methanogen (Anderson et al., 2009)] contains three Fe4S4 clusters and a Ni–Fe active site (Thauer et al., 2010), and can contain additional polyferredoxin subunits with 6, 10, or 14 Fe4S4 clusters (Tersteegen and Hedderich, 1999; Figure 1).
Another Ni–Fe hydrogenase is used to reduce coenzyme F420. Coenzyme F420, a flavin derivative that plays a critical role in two intermediate electron-transfer steps in methanogenesis, is reduced with H2 by the cytoplasmic hydrogenase Frh (Alex et al., 1990). The Frh enzyme complex contains a Ni–Fe active site and four Fe4S4 clusters (Figure 1), and forms large aggregates, multiplying its metal requirements approximately eightfold (Fox et al., 1987). Under conditions of Ni limitation, some methanogens without cytochromes substitute Frh for a Ni-free Fe hydrogenase (Hmd) to decrease Ni requirements (Afting et al., 1998). The Ni-free Fe hydrogenase has a significantly lower Fe requirement, using only two Fe atoms per complex (Zirngibl et al., 1992; Shima et al., 2008; Thauer et al., 2010). When formate is used as an electron source instead of H2, Frh is replaced by formate dehydrogenase (Fdh), which contains one Mo/W-pterin cofactor, two Zn atoms and between 21 and 24 Fe atoms (Schauer and Ferry, 1986).
Co(balt)-containing methyltransferases are essential for methanogenesis. All methanogens utilize the energy-conserving CH3-H4M(S)PT-coenzyme M methyltransferase (Mtr) to transfer the methyl group from CH3-H4M(S)PT to HS-CoM. The Mtr enzyme complex contains two cobamide cofactors (with one Co each) and eight Fe atoms (Gartner et al., 1993). A wide range of other Co(balt)-containing methyltransferases are required for methyl coenzyme M formation in methylotrophic methanogens, some of which contain additional metals (Thauer, 1998). For instance, the methanol-coenzyme M methyltransferase enzyme (Mta) expressed by methanol-utilizing methanogens contains one Zn in addition to one cobalamin Co (Hagemeier et al., 2006).
Heterodisulfide reductase (Hdr) is an Fe–S and Zn-containing enzyme that catalyzes the reduction of heterodisulfide (CoM-S-S-CoB) to form HS-CoM and HS-CoB in the second-to-last step in methanogenesis. In methanogens without cytochromes, Hdr has three subunits (HdrABC) whereas methanogens with cytochromes have a two-subunit protein (HdrDE; Hedderich et al., 2005; Thauer et al., 2008). HdrABC contains seven Fe4S4 clusters and one structural Zn atom (Hamann et al., 2007) and forms a tight complex with the Ni–Fe hydrogenase Mvh, which contains one Ni–Fe active site, one Fe2S2 cluster, two Fe4S4 clusters, one Fe3S4 cluster, and polyferredoxin with 12 Fe4S4 clusters (Reeve et al., 1989; Figure 1). In Methanosarcinales, HdrDE is reduced by methanophenazine, a lipid-soluble electron, and proton carrier with very low redox potential. HdrDE contains three Fe4S4 clusters, one heme b cofactor, and one structural Zn atom (Heiden et al., 1994; Künkel et al., 1997; Simianu et al., 1998). Methanophenazine is in turn reduced by the Ni–Fe hydrogenase Vht/Vho, which contains one Ni–Fe center, two b-type hemes, one Fe3S4 cluster, and two Fe4S4 clusters (Deppenmeier et al., 1992; Thauer et al., 2010). In the marine strain Methanosarcina acetivorans, Vht/Vho is replaced by Rnf, which contains six Fe4S4 clusters and no Ni, and uses reduced ferredoxin instead of H2 (Li et al., 2006; Ferry, 2010a).
Methyl coenzyme M reductase (Mcr), common to all methanogenic pathways, catalyzes the final step in methanogenesis: the reduction of CH3-S-CoM to CH4 and the formation of CoM–CoB heterodisulfide with electrons derived from HS-CoB. The crystal structure of Mcr has been solved and contains two coenzyme F430 Ni tetrapyrroles (Ermler et al., 1997). Previous studies have shown that the F430 content of cells is dependent on cellular Ni content (Diekert et al., 1980, 1981; Lin et al., 1989). Methanogens actively transport Ni and Co using ATP-dependent uptake systems in order to fulfill enzymatic requirements (Jarrell and Sprott, 1982; Baudet et al., 1988; Rodionov et al., 2006; Zhang et al., 2009).
When grown on acetate, methanogens use two metalloenzymes for the conversion of the methyl group from acetate to CH3-H4SPT, which differ from those used in CO2 reduction and methylotrophic pathways. The most metal-rich aceticlastic enzyme is CO dehydrogenase/acetyl-CoA synthase (Cdh), which cleaves the methyl group off of acetyl-CoA and transfers it to CH3-H4SPT. The Cdh complex contains one Fe4S4 cluster bridged to an Ni–Ni site (Funk et al., 2004), four Fe4S4 clusters and a NiFe4S4 cluster (Gong et al., 2008), and reduces a 2×[Fe4S4]ferredoxin (Terlesky and Ferry, 1988; Ferry, 2010b). The CO2 by-product of Cdh is converted to bicarbonate by an Fe-containing carbonic anhydrase (CA) instead of the Zn-form, which is commonly in CAs from other species (Macauley et al., 2009). Overall, the metal content of enzymes involved in aceticlastic methanogenesis is similar to the H2/CO2 pathway (Figure 1). The Fe and Mo/W requirements are likely lower in the aceticlastic pathway because the Fe-rich polyferredoxins involved in the CO2 reduction route are not present, nor is Fmd/Fwd.
Physiological studies of trace metal requirements for methanogenesis
Previous physiological studies of metals as micronutrients for methanogenesis have been compromised by significant metal contamination in growth vessels and sampling equipment. The requirements for gas-tight material to maintain anaerobic conditions has favored the use of glass culturing bottles with butyl rubber stoppers and stainless steel needles for sampling, both of which are notoriously dirty with respect to metals. In fact, Ni requirements for methanogens were overlooked in early studies due to the high Ni content (~10%) of stainless steel syringe needles. Dissolution of Ni in stainless steel needles by H2S in the media supplied ample Ni for growth of methanogens on media not supplemented with Ni salts (S. Zinder, personal comm, 2011; Diekert et al., 1981; Whitman et al., 1982). The need for anaerobic conditions excludes the use of acid-washed plastics, which have extremely low metal contamination and are commonly used for trace metal limitation experiments with aerobes, but are permeable to O2 diffusion. To address these concerns, researchers have acid-washed glassware with sulfuric acid, treated butyl rubber stoppers, and polypropylene pipet tips with NaHCO3/EDTA and covered cannula needles with Teflon tubing (Whitman et al., 1982; Sowers and Ferry, 1985). Such methods were used in some, though not all, of the studies reviewed in the Section “Pure Cultures.” To our knowledge, Teflon-lined glass bottles have not been used to minimize metal contamination in previous anaerobic studies, but may be a good option for future research.
The experiments discussed in Section “Mixed Cultures and Anaerobic Bioreactors” on mixed cultures and anaerobic bioreactors were conducted, for the most part, not in defined media but rather in sludge or wastewater treatment effluent. Studies of metal requirements for pure cultures and anaerobic digesters typically report only total added metal, although these reactors usually have higher background metal contents than pure cultures. However, the ubiquitous presence of sulfide, carbonate, and phosphate in methanogenic cultures leads to metal precipitation, so dissolved metal concentrations generally decline throughout experiments (Callander and Barford, 1983a,b). Indeed, precipitation–dissolution kinetics of metal sulfides may play a key role in bioavailability of metals in anaerobic digesters (Gonzalez-Gil et al., 1999; Jansen et al., 2007). However, increased rates of methanogenesis after addition of a particular metal are clear evidence of environmental (albeit human-contributed) metal limitation as compared to axenic cultures.
Pure cultures
Based on the metal content of the enzymes involved in methanogenesis, one would predict that methanogens require very high concentrations of Fe, relatively high levels of Ni and Co, trace Zn and Mo/W, and negligible amounts of Cu and Mn. This prediction does not take into account complications if enzymes have differing Km values for their respective substrates, which could result in shifts in enzyme ratios, nor does it consider metal requirements for metabolic pathways besides methanogenesis. Nevertheless, it roughly agrees with the cellular metal contents of 11 previously analyzed methanogens, which contained 700–2800 ppm Fe, 17–180 ppm Ni, 23–1810 ppm Zn, 10–120 ppm Co, 10–70 ppm Mo, <10–160 ppm Cu, and 2–27 ppm Mn, in terms of dry weight (Diekert et al., 1981; Whitman et al., 1982; Scherer et al., 1983). These data suggest that the metal requirements for methanogenesis have a strong influence on whole cell metal contents.
Early studies on Methanobacterium thermoautotrophicum (renamed Methanothermobacter thermautotrophicus) grown on H2 and CO2 showed that low concentrations of Fe, Ni, Co, and Mo in growth media could limit growth (Taylor and Pirt, 1977; Schönheit et al., 1979). For the formation of 1 g of biomass (dry weight), 10 μmol of Fe, 150 nmol of Ni, 20 nmol of Co, and 20 nmol of Mo were required, a stoichiometry of 500 Fe: 7.5 Ni: 1 Co: 1 Mo (Schönheit et al., 1979). Optimal growth was observed at 300–500 μM Fe (Patel et al., 1978) and 1 μM Ni, while addition of Cu, Zn, and Mn to the media did not stimulate growth (Schönheit et al., 1979; Table 1). Interestingly, optimal growth of another H2/CO2-utilizing methanogen, Methanococcus voltae, occurred at much lower Fe and Ni concentrations (10 and 0.2 μM, respectively), despite the more rigorous cleaning protocols used in this study (Whitman et al., 1982).
Table 1.
Optimal dissolved metal concentrations (calculated from total amounts of salt added) in defined growth media for pure methanogenic cultures grown on H2/CO2, formate, acetate, methanol, or trimethylamine.
| Metal | Substrate | Concentration (μM) | Species | Reference |
|---|---|---|---|---|
| Fe | H2/CO2 | 300–500 | Methanospirillum hungatei, Methanobacterium bryantii strain MOH | Patel et al. (1978) |
| H2/CO2 | >15 | Methanococcus voltae | Whitman et al. (1982) | |
| Acetate | 100 | Methanothrix soehngenii VNBF1 | Fathepure (1987) | |
| Methanol | 50 | Methanosarcina barkeri | Lin et al. (1990) | |
| Trimethylamine | 5 | Methanococcoides methylutens | Sowers and Ferry (1985) | |
| Ni | H2/CO2 | 1 | Methanobacterium thermoautotrophicum2 | Schönheit et al. (1979) |
| H2/CO2 | 0.2 | Methanococcus voltae | Whitman et al. (1982) | |
| Acetate | 2 | Methanothrix soehngenii VNBF1 | Fathepure (1987) | |
| Methanol | 0.1 | Methanosarcina barkeri | Scherer and Sahm (1981) | |
| Trimethylamine | 0.25 | Methanococcoides methylutens | Sowers and Ferry (1985) | |
| Co | Acetate | 2 | Methanothrix soehngenii VNBF1 | Fathepure (1987) |
| Methanol | 1 | Methanosarcina barkeri | Scherer and Sahm (1981) | |
| Trimethylamine | 0.1 | Methanococcoides methylutens | Sowers and Ferry (1985) | |
| Mo | Acetate | 2 | Methanothrix soehngenii VNBF1 | Fathepure (1987) |
| Methanol (with NH4+) | 0.5 | Methanosarcina barkeri | Scherer and Sahm (1981), Scherer (1988) | |
| Methanol (N2-fixing) | 5 | Methanosarcina barkeri | Scherer (1988) | |
| W | H2/CO2 | 1 | Methanocorpusculum parvum | Zellner and Winter (1987) |
| Formate | 0.5 | Methanocorpusculum parvum | Zellner and Winter (1987) |
1Renamed Methanosaeta concilii; 2Renamed Methanothermobacter thermautotrophicus.
Adapted from Takashima and Speece (1990).
How do metal requirements change when methanogens are grown on substrates other than H2/CO2? Optimal growth of Methanothrix soehngenii VNBF (renamed Methanosaeta concilii) grown on acetate was observed at 100 μM Fe, 2 μM Ni, 2 μM Co, and 2 μM Mo (Fathepure, 1987; Table 1). Optimal growth of methanol-utilizing Methanosarcina barkeri was observed at lower concentrations: 50 μM Fe, 0.1 μM Ni, 1 μM Co and 0.5 μM Mo, and addition of Cu and Mn had no effect on growth (Scherer and Sahm, 1981; Lin et al., 1990). Optimal growth of trimethylamine-grown Methanococcoides methylutens was lower still: 5 μM Fe, 0.25 μM Ni, and 0.1 μM Co, with no stimulation by addition of Mo, Zn, Mn, W, and Cu (Sowers and Ferry, 1985). These studies suggest that metal requirements are higher on acetate than methyl-compounds, although the opposite – particularly for Co – has been found in mixed culture anaerobic digester studies (see the Section “Mixed Cultures and Anaerobic Bioreactors”).
In some methanogens, W-containing formylmethanofuran dehydrogenases (Fwd) and W-containing formate dehydrogenases (W-Fdh) are expressed in place of Mo-containing forms of the enzymes. W was first shown to stimulate growth of Methanococcus vannielii on formate (Jones and Stadtman, 1977). A subsequent study revealed that optimal growth on H2/CO2 required more W (1 μM) than growth on formate (0.5 μM) and that more W was incorporated into cells during growth on H2/CO2 than on formate (Zellner and Winter, 1987), suggesting that Fwd has a higher W requirement than W-Fdh.
In addition to metal cofactors in enzymes directly involved in methanogenesis, additional metals are required for other metabolic pathways in methanogens, such as nitrogen fixation. In a study of metal requirements for two strains of M. barkeri, Scherer (1988) found that optimal Mo media concentrations for diazotrophic growth were 10× greater (5 μM Mo) than NH4-based growth (0.5 μM Mo). One of the two strains (strain 227) could grow just as well with vanadium (V) instead of Mo in the media, suggesting that it contained a V-nitrogenase (Scherer, 1988) as later confirmed by Chien et al. (2000). However, differing results have been published regarding the extent to which V stimulates nitrogen fixation in this strain (Lobo and Zinder, 1988). Other methanogens, such as Methanococcus maripaludis, lack the V-nitrogenase and have an absolute requirement for Mo for diazotrophy (Kessler et al., 1997).
Mixed cultures and anaerobic bioreactors
The potential of metals to increase the efficiency of methanogenesis in industrial settings has led to a steady stream of studies on metal requirements for optimal functioning of anaerobic waste (sewage, agricultural, and food-processing) treatment bioreactors since the late 1970s (Demirel and Scherer, 2011). The production of methane from small organic molecules such as acetate and methanol in waste treatment reactors is a beneficial by-product because it can be captured and used as a renewable fuel source.
Several studies suggest that the addition of Fe, Ni, and Co can increase the rate of aceticlastic methanogenesis in anaerobic digesters. An early study of aceticlastic methanogenesis in mixed cultures originally obtained from sewage sludge and maintained in a defined medium showed that the optimal soluble Fe concentrations for methanogenesis were 0.2–2 mM (lower than the total 5–10 mM FeCl2 added due to precipitation as ferrous carbonate), whereas background soluble Fe was ~25 μM (Hoban and van den Berg, 1979). The authors suggested that mixed methanogenic cultures might have higher Fe requirements than pure cultures. Addition of Ni and Co stimulated aceticlastic methanogenesis and increased the thickness of biofilms in anaerobic food waste digesters (Murray and van den Berg, 1981), municipal sludge digesters (Speece et al., 1983), and synthetic wastewaters (Kida et al., 2001). Addition of Mo (50 nM) had a smaller effect (Murray and van den Berg, 1981). In another study, individual addition of Fe, Ni, Co, Mn, Zn, Mo, and Cu to a wastewater treatment sludge did not produce any significant increase in the conversion of acetate to methane (Florencio et al., 1993). It is possible that the sludge in the latter study contained sufficient metal carry-over that these metals were not limiting for aceticlastic methanogenesis. Metal speciation almost certainly influences bioavailability in these bioreactors, where up to 95% of dissolved Ni and Co is present in strongly bound forms (Jansen et al., 2005). It is currently unknown whether methanogens can directly take up soluble metal-sulfide complexes or metal-organic complexes (Jansen et al., 2007). Additionally, temperature may exert influence on the trace metal requirements; higher trace metal requirements have been found for thermophilic than mesophilic mixed reactors, despite smaller biomass yield (Takashima et al., 2011).
Limitation of methanogenesis by low Co(balt) bioavailability has been identified in numerous studies of anaerobic mixed cultures grown on methanol. A study of methylotrophic methanogens in wastewater treatment sludge showed a pronounced increase in methanogenic activity when Co was added, and a significant decline when it was absent from the media (Florencio et al., 1993). Follow-up studies have confirmed the high Co(balt) requirements for methanol-grown anaerobic bioreactors (Zandvoort et al., 2006). The same trend was observed for Ni, although to a lesser extent, but minimal effect was found for other metals (Fe, Zn, Mo, Cu, and Zn). Optimal Ni and Co concentrations for methanogenesis was ~2 μM (Florencio et al., 1993, 1994; Zandvoort et al., 2002). The increased need for Co for methanol-based growth vs. other substrates is likely due to additional Co(balt)-containing methyltransferases required for growth on methyl-compounds. This concurs with previous findings that the highest amount of corrinoids are produced in methanogens grown on methanol compared to other substrates, with about 3× as much corrinoids in methanol-grown cell vs. acetate-grown cells (Krzycki and Zeikus, 1980) and that cobalamin production increases with media Co concentration up to ~50 μM (Lin et al., 1989).
Environmental studies
Very few studies have investigated the effects of metal additions on natural methanogenic ecosystems. To our knowledge, the only published experiment was performed in five North American peatlands (Basiliko and Yavitt, 2001). Ambient dissolved metal concentrations in mineral-poor peatlands were low (2–20 nM Ni, 30 nM Co, and <2 μM Fe) due to the high metal-binding capacity of peat. In mineral-rich peatlands, concentrations were 40–60 nM Ni, 2–5 μM Co, and 20–30 μM Fe. The authors found that combined addition of Fe (12 μM), Ni (12 μM), and Co (6 μM) to mineral-poor surface samples resulted in an enhancement of CH4 production, whereas no effect or inhibition was observed at mineral-rich peatlands. The effect of individual metals was not tested. No effect was observed for CO2 production in mineral-poor peatlands, suggesting that heterotrophic microbes are not as metal-limited as methanogens. Addition of the chelator citrate, but not EDTA, led to increased CH4 production in the peat with the lowest metal concentrations, suggesting either that methanogens can take up metal–citrate complexes or that they can compete with citrate for metal-binding.
Other anoxic ecosystems have similar trace metal concentrations to peat bogs, suggesting that metal limitation of methanogenesis may be widespread in natural environments. The concentration range of Ni, Co, and Mo in five representative anoxic waters was consistently below the optima for growth of pure cultures (Tables 1 and 2). Optimal methanogenic growth occurs at 0.2–2 μM Ni and in natural anoxic waters Ni ranges from 0.002 to 0.07 μM. Optimal media concentrations of Co are 0.1–2 μM and in natural environments Co is generally <0.02 μM. Optimal Mo concentrations are 0.5–5 μM; ambient Mo is 0.01–0.2 μMin anoxic environments. Zn is also generally very low (<0.04 μM) in anoxic waters, and may contribute to trace metal limitation of methanogenesis, although pure culture studies are not available to determine optimal Zn concentrations. The opposite is true for W: few data are available on in situ W concentrations, but in most ecosystems W is below the growth optima of 0.5–1 μM. Optimal Fe concentrations are present in anoxic sediment pore waters (such as the Santa Monica Basin) where Fe reaches 200 μM, but other anoxic waters have significantly lower Fe (Table 2). Therefore, it is quite reasonable to expect that bioavailability of trace metals may be important limiting growth factors in natural methanogenic environments.
Table 2.
Compiled dissolved trace metal concentrations in representative anoxic waters.
| Sample site | Fe | Ni | Co | Cu | Zn | Mo | Reference |
|---|---|---|---|---|---|---|---|
| Peat bog waters | 0.4–40 | 0.002–0.07 | 0.002–4 | 0.02–2 | Basiliko and Yavitt (2001), Bragazza (2006) | ||
| Santa Monica basin sediment pore waters (>5 cm depth) | 100–200 | 0.01 | 0.01–0.02 | <0.005 | 0.2 | Shaw et al. (1990) | |
| Black Sea sulfidic water column (>200 m) | 0.01–0.3 | 0.01 | <0.004 | <0.002 | <0.004 | <0.01 | Haraldsson and Westerlund (1988), Emerson and Huested (1991) |
| Framvaren Fjord water column (>20 m) | 0.03–2 | <0.07 | 0.001–0.01 | <0.001 | 0.002–0.01 | <0.04 | Haraldsson and Westerlund (1988), Emerson and Huested (1991) |
| Baltic Sea water column (>150 m) | 0.1–2 | 0.01 | <0.002 | <0.007 | 0.01–0.04 | Kremling (1983) |
All concentrations are in units of micromolar (μM).
Summary and areas for future research
Based on the metal content of enzymes in methanogenesis, one would predict (1) roughly equal Fe, Ni, and Zn requirements for CO2 reduction, aceticlastic, and methylotrophic pathways; (2) higher Mo/W requirements for CO2 reduction and methylotrophic pathways than aceticlastic methanogenesis; and (3) higher Co(balt) requirement for the methylotrophic pathway than CO2 reduction and aceticlastic pathways. Previous studies are difficult to compare because few of them simultaneously vary methanogenic pathways using the same experimental conditions and/or organism. Every laboratory likely had different levels of trace metal contamination and metal-sulfide precipitation, and thus the given optimal metal concentrations are all estimates to varying degrees. Nonetheless, a few trends are clear: (1) Fe concentrations required for optimal growth in media are much higher than all other metals (in the 10−5 to 10−4 M range); (2) optimal Ni, Co, Mo, and W concentrations are in the 10−7 to 10−6 M range, and higher Mo concentrations (10−5 M) are needed for diazotrophic growth; (3) addition of Cu and Mn is not needed for growth. Few studies have investigated Zn requirements for methanogenesis, although Zn is present in at least two enzymes in the pathway.
Studies of anaerobic digesters and peatlands suggest that Fe, Ni, and Co – and possibly other metals – may limit methanogenesis in the natural ecosystems. The high Co(balt) requirement for methylotrophic methanogenesis in anaerobic digesters is of particular interest. To our knowledge, only one study has looked into the possibility of metal limitation of methanogenesis in a natural setting (Basiliko and Yavitt, 2001). More environmental studies are essential to better understand the nature of metal bioavailability and/or limitation in the ecosystems that contribute 75–80% of the world’s annual CH4 sources, but preliminary comparison of optimal concentrations for growth of pure cultures and in situ levels shows that trace metal (particularly Ni and Co) limitation of methanogenesis may be widespread in anoxic settings.
Methanotrophy
Methanotrophy is the microbial process whereby CH4 is oxidized as an energy and carbon source. Aerobic methanotrophy is estimated to consume ~35% (0.6 Gt) of the total global CH4 produced per year, whereas anaerobic methanotrophy is estimated to consume ~18% (0.3 Gt; Thauer, 2011). Aerobic methanotrophy is performed by certain bacteria that combine molecular oxygen with CH4 to produce methanol (CH3OH). Methanol is then oxidized to formaldehyde (CH2O), which is incorporated into organic compounds using either the serine or ribulose monophosphate pathway (Semrau et al., 2010). Sulfate-dependent anaerobic methane oxidation (AOM) occurs primarily in anoxic marine sediments and is performed by archaea that have not been isolated in pure culture, but are most closely related to Methanosarcinales and Methanomicrobiales. These archaea are believed to oxidize methane anaerobically through a reverse methanogenesis pathway (Scheller et al., 2010; Thauer, 2011). Thus, phylogenetically unrelated aerobic and anaerobic methanotrophs accomplish the same task using very different enzymes (Chistoserdova et al., 2005) and are therefore predicted to have different metal requirements. Cu requirements for aerobic methanotrophy have been the subject of a great deal of research, whereas metal requirements for AOM are just beginning to be explored, but are expected to be similar to metal requirements for methanogenesis.
Aerobic methanotrophy
Aerobic methanotrophs (abbreviated “MOB” for “methane-oxidizing bacteria”) are commonly found at oxic/anoxic interfaces of environments where CH4 is produced. Methanotroph genera, beginning with “Methylo-,” are found in the Gammaproteobacteria and Alphaproteobacteria (Hanson and Hanson, 1996; Semrau et al., 2010). Recently, MOB have also been discovered in the Verrucomicrobia (Op den Camp et al., 2009). Furthermore, the newly discovered freshwater phylum NC10 bacterium Methylomirabilis oxyfera performs intra-aerobic methane oxidation using pMMO and internally produced O2 from NO (Ettwig et al., 2010; Wu et al., 2011). In addition to being important in greenhouse gas processing, some MOB play important roles in pollutant degradation and bioremediation (Semrau, 2011).
Metalloenzymes in aerobic methanotrophy
Classical aerobic methanotrophy
The metal requirements for MOB are largely regulated by copper (Cu) availability. When Cu is abundant, MOB express a membrane-bound Cu (and possibly Fe and Zn)-containing enzyme (particulate methane monooxygenase, pMMO) that catalyzes the first step of CH4 oxidation. When Cu is lacking, some MOB can synthesize a soluble protein for methane oxidation that contains Fe instead of Cu (soluble methane monooxygenase, sMMO). Subsequent enzymatic steps in aerobic methanotrophy rely primarily on Fe, with lesser amounts of Ca, Mo, and possibly Zn (Figure 2). Below is a general overview of the metal content of enzymes in methanotrophy. For a more extensive discussion of the biochemistry of aerobic methanotrophy, the reader is referred to other reviews (Hanson and Hanson, 1996; Hakemian and Rosenzweig, 2007; Trotsenko and Murrell, 2008; Semrau et al., 2010; Chistoserdova, 2011).
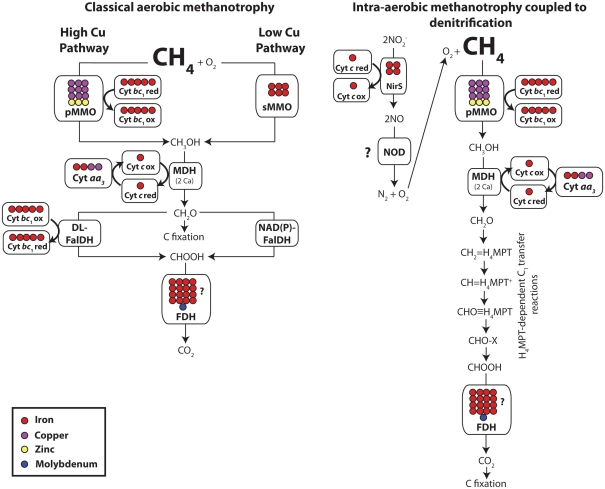
Figure 2.
Metal content of the enzymes involved in aerobic methanotrophy at high or low Cu, and intra-aerobic methane oxidation coupled with denitrification. Each circle represents one metal atom. Abbreviations: Cyt, cytochrome; DL-FalDH, dye-linked formaldehyde dehydrogenase; FDH, formate dehydrogenase; MDH, methanol dehydrogenase; pMMO, Cu-particulate methane monooxygenase; sMMO, soluble methane monooxygenase; NirS, Fe–nitrite reductase; NOD, NO dismutase (putative enzyme). Putative intra-aerobic methane oxidation coupled with denitrification pathway is adapted from Wu et al. (2011). For simplicity, only metalloenzymes are labeled. Question marks mean that the metal content of the enzyme is not well known, and for the case of NOD, that the enzyme is unknown. See the Section “Metalloenzymes in Aerobic Methanotrophy” for more details.
The first step of aerobic methanotrophy, the oxidation of CH4 to CH3OH, is catalyzed by methane monooxygenase (MMO). All aerobic methanotrophs (with the exception of facultative methanotrophic species in the genus Methylocella) are capable of expressing pMMO (Hanson and Hanson, 1996). The metal content of pMMO has been a matter of debate for many years, namely regarding whether the active site coordinates Fe or Cu for function (Bollinger, 2010). Crystal structures and experiments have provided strong evidence that the active site is a two Cu center in the pMMO-B subunit (Lieberman and Rosenzweig, 2005; Balasubramanian and Rosenzweig, 2007; Hakemian et al., 2008; Balasubramanian et al., 2010). An additional mono-nuclear Cu atom is present in one of crystal structures (from Methylococcus capsulatus Bath) but not in Methylosinus trichosporium OB3b and is not required for CH4 oxidation (Balasubramanian et al., 2010). Crystal structures predict coordination of another metal ion (Zn in M. capsulatus Bath and Cu in M. trichosporium OB3b), positioned between the pMMO-B and pMMO-C subunits (Lieberman and Rosenzweig, 2005; Hakemian et al., 2008). Since pMMO is a heterotrimer, it therefore contains nine Cu atoms (and possibly three Zn atoms; Figure 2). Recent Mössbauer evidence suggests that Fe could also be present in pMMO, as supported by previous protein preparations that contained Fe (Martinho et al., 2007; Semrau et al., 2010). The electron donor to pMMO is a cytochrome bc1 complex, containing three heme groups and one Fe2S2 cluster (Zahn and DiSpirito, 1996).
Several strains of aerobic methanotrophs are also capable of expressing a cytoplasmic soluble MMO (sMMO), which contains a dimeric hydroxylase with two Fe ions in a di-Fe active site cluster and an additional Fe2S2 cluster in a reductase subunit (Lipscomb, 1994; Merkx et al., 2001). Expression of particulate vs. soluble MMO is regulated by Cu availability (Murrell et al., 2000). Work with pure cultures has shown that a “Cu switch” occurs at 0.85–1 μmol/g dry weight: at lower Cu:biomass ratios, sMMO is expressed, and at higher Cu:biomass ratios, pMMO is expressed (Stanley et al., 1983; Hanson and Hanson, 1996; Nielsen et al., 1997). In contrast to pMMO, the electron donor to sMMO is NADH (Lipscomb, 1994; Merkx et al., 2001).
After CH4 is oxidized to CH3OH by either pMMO or sMMO, the periplasmic pyrroloquinoline quinone-dependent enzyme methanol dehydrogenase (abbreviated MDH or Mxa) oxidizes CH3OH to formaldehyde (CHOH) using two cytochrome c proteins (cL and cH) as electron acceptors (Hanson and Hanson, 1996). These cytochromes are then oxidized by cytochrome aa3, containing two hemes and two Cu atoms (Figure 2). MDH/Mxa does not contain any transition metal cofactors, but does possess 2 mol of Ca per mole of tetramer. Approximately 50% of the CHOH produced by MDH/Mxa is assimilated into biomass, and the other half is further oxidized to formate (CHOOH) and then to CO2 to generate reducing equivalents (Hanson and Hanson, 1996; Trotsenko and Murrell, 2008; Chistoserdova, 2011). This step is mediated by either a membrane-bound cytochrome-linked formaldehyde dehydrogenase (DL-FalDH) when cells are grown under high Cu conditions or a soluble NAD(P)+-linked enzyme (N-FalDH) when cells are grown under low Cu conditions (Zahn et al., 2001). Neither protein contains metal cofactors, but DL-FalDH does require cytochrome bc1 as an electron acceptor (Zahn et al., 2001). The final step in aerobic methane oxidation is formate oxidation to CO2, catalyzed by NAD+–formate dehydrogenase (Fdh). The only characterized Fdh protein in MOB (from M. trichosporium OB3b) contains four FexSx clusters and one molybdopterin cofactor (Jollie and Lipscomb, 1991; Figure 2). Multiple Fdh enzymes are present in M. capsulatus Bath genome (Ward et al., 2004) and it is possible that one or more of them contains W in place of Mo, similar to Fdh proteins from methylotrophs (Laukel et al., 2003).
Intra-aerobic methane oxidation coupled to denitrification
The genome of the denitrifying bacterium M. oxyfera, coupled to proteomic, transcriptomic, and stable isotope labeling experiments, have revealed that this organism also contains pMMO but uses a different pathway for methane oxidation than other MOB. Instead of obtaining O2 from the environment, M. oxyfera generates its own supply of O2 internally from NO. Therefore, this metabolism has been called intra-aerobic methane oxidation coupled to denitrification (Ettwig et al., 2010; Wu et al., 2011). In the proposed pathway, an Fe-based cytochrome cd1 nitrite reductase [cd1NIR or NirS; see the Section “Nitrate/Nitrite Reduction to N2O/N2 Pathway (Denitrification)”], likely containing four Fe atoms (Fülöp et al., 1995), is used to reduce to NO (Ettwig et al., 2010; Wu et al., 2011). In contrast to classical denitrification, where NO is reduced to N2O and then N2, M. oxyfera disproportionates two molecules of NO to O2 and N2 (Figure 2). The NO dismutase enzyme that performs this reaction is currently unknown. The O2 generated by this uncharacterized NO dismutase is then used to oxidize CH4 to CH3OH using pMMO, followed by oxidation of CH3OH to CH2O by MDH/Mxa (see the Section “Classical Aerobic Methanotrophy”; Ettwig et al., 2010; Wu et al., 2011). Instead of using FalDH for oxidation of CH2O to CHOOH, M. oxyfera contains several genes encoding enzymes responsible for tetrahydromethanopterin (H4MPT)-dependent C1 transfer reactions (Ettwig et al., 2010; Wu et al., 2011). The enzymes involved in these reactions are not known to contain metals (Vorholt, 2002). In the last step, CHOOH is oxidized to CO2 by formate dehydrogenase [Fdh; see the Section “Nitrate/Nitrite Reduction to N2O/N2 Pathway (Denitrification)”]. Carbon assimilation in M. oxyfera likely proceeds via CO2 fixation with the Calvin–Benson–Bassham cycle instead of the serine and ribulose monophosphate (RuMP) pathways used by other MOB (Ettwig et al., 2010; Wu et al., 2011). This pathway remains to be confirmed biochemically, but likely requires abundant Cu for pMMO.
Physiological studies of metal requirements for aerobic methanotrophy
Pure cultures
Studies of the physiological importance of metals for MOB have focused largely on Cu. Addition of Cu to the growth media in strains encoding both the particulate and the soluble forms of MMO fundamentally changes methanotroph physiology by triggering the “Cu switch,” turning on the expression of pMMO (Stanley et al., 1983; Choi et al., 2003). Further increases in Cu concentration can lead to up to 55-fold higher pMMO expression, with optimal pMMO activity occurring at 60 μM Cu for M. capsulatus Bath (Choi et al., 2003; Semrau et al., 2010). Cellular Cu contents in laboratory cultures of aerobic methanotrophs amended with high Cu in the growth media can be extremely high, up to ~250 nmol Cu/mg protein (Choi et al., 2003).
In order to maintain Cu homeostasis and protect against the potentially toxic effects of high levels of Cu, some aerobic methanotrophs express the Cu-chelating “chalkophore” molecule methanobactin (Kim et al., 2004; Balasubramanian and Rosenzweig, 2008). Methanobactin is a 1217-Da molecule that binds one atom of Cu(I) with high affinity (Kim et al., 2004; Choi et al., 2006) and accumulates in the growth media when Cu is <0.7 μM (DiSpirito et al., 1998). Cu-loaded methanobactin is actively transported into methanotrophic cells when Cu concentrations rise between 0.7 and 1 μM (Semrau et al., 2010; Balasubramanian et al., 2011). Furthermore, it has recently been shown that M. capsulatus Bath contains numerous proteins on its surface that respond to changing Cu concentrations in growth medium, including abundant Fe-containing cytochrome c peroxidases and multi-heme c-type cytochromes (Karlsen et al., 2011). It is perhaps interesting to note that abundant extracellular iron is evident on other methanotrophs in the genera Crenothrix and Clonothrix (Kolk, 1938; Vigliotta et al., 2007), although separate pathways and functional roles may be at play in these cases.
Environmental studies
Environmental studies on Cu availability to MOB are lacking, which is striking considering that Cu has been known to be involved in methanotrophy for many years. Knapp et al. (2007) suggest that the reason that correlations between environmental dissolved Cu (which in oxic settings is <1 μM; Table 3) and methanotrophic activity have not been noted is because some MOB can access mineral- and organic-bound Cu using small Cu-binding peptides, most notably methanobactin. In the absence of dissolved Cu, M. trichosporium OB3b acquired sufficient Cu from Cu-containing minerals to express pMMO, but only when methanobactin was present (Knapp et al., 2007). CH4 oxidation rates were highest at ~100 ppm mineral Cu (Kulczycki et al., 2011). Low Cu minerals were dissolved more quickly by methanobactin than high Cu minerals (Kulczycki et al., 2007). Similarly, increasing concentrations of surface sites on minerals lead to decreased Cu bioavailability to M. trichosporium OB3b in common soil minerals (Morton et al., 2000). Therefore, particulate Cu content and sediment or soil mineral geochemistry may display a more robust correlation with methanotrophic activity than dissolved Cu levels in aquatic systems or pore waters (Knapp et al., 2007; Fru, 2011), although this remains to be tested in natural systems.
Table 3.
Compiled dissolved trace metal concentrations in representative oxic waters.
| Sample site | Fe | Ni | Co | Cu | Zn | Mo | Reference |
|---|---|---|---|---|---|---|---|
| Seawater | <0.001 | 0.002–0.01 | <0.001 | <0.001–0.005 | <0.001–0.009 | 0.1 | Bruland (1980), Collier (1985), Johnson et al. (1997) |
| Soil pore water | 1–20 | 0.1–0.5 | 0.1–2 | Kinniburgh and Miles (1983) | |||
| Oxic lake water | 0.01–10 | <0.001–0.03 | 0.01–0.8 | 0.02–0.6 | 0.004–0.3 | Wetzel (2001) |
All concentrations are in units of micromolar (μM).
Anaerobic methanotrophy
The process of anaerobic methane oxidation (AOM) has been known for over 30 years based on geochemical data (Martens and Berner, 1977), but the identity of the microorganisms carrying out this metabolism remained elusive until the early 2000s due to the extreme difficulty of culturing AOM microbes. Combined molecular and stable isotope studies revealed that archaeal groups (anaerobic methanotrophic archaea, abbreviated “ANME”) were responsible for AOM activity in CH4 seep sediments through syntrophic associations with sulfate-reducing bacteria (Hinrichs et al., 1999; Boetius et al., 2000; Orphan et al., 2001, 2002). Described ANME lineages currently belong to three distinct phylogenetic groups, with members of the ANME-1 forming a novel clade between the Methanomicrobiales and Methanosarcinales orders, and groups ANME-2 and ANME-3 belonging to the Methanosarcinales. Together, these methanotrophic lineages are believed to be responsible for the oxidation of ~80% of the methane naturally seeping upward from marine sediments, or ~7–25% of total global methane (Reeburgh, 2007). Although ANME have not been cultured, metagenomic, and environmental proteomic studies have provided insight into their metabolic strategies for AOM (Hallam et al., 2003, 2004; Krüger et al., 2003; Meyerdiercks et al., 2005; Pernthaler et al., 2008).
Metalloenzymes in anaerobic methanotrophy
Evidence is accumulating that ANME anaerobically oxidize methane by running the methanogenesis pathway in reverse (Scheller et al., 2010), as was predicted based on the close phylogenetic relationship of ANME to methanogens in the orders Methanomicrobiales and Methanosarcinales (Knittel and Boetius, 2009). The discovery of gene homologs to mcrA in ANME provided the first clues that these microbes were using reverse methanogenesis to oxidize CH4 (Hallam et al., 2003). High concentrations of the Ni-containing cofactor F430 and a slightly modified variant of F430 (Mayr et al., 2008) were found in microbial mats performing AOM in the Black Sea (Krüger et al., 2003), adding further support for the model. Immunolocalization and biochemical studies have provided clear evidence that Mcr catalyzes the first step of AOM in ANME (Heller et al., 2008; Scheller et al., 2010) and metagenomic studies of ANME communities identified the presence of genes encoding almost all of the other methanogenic enzymes with the exception of the Ni–Fe hydrogenases Eha/Ehb and two of the three subunits of the Ni–Fe hydrogenase Mvh (Hallam et al., 2004; Meyerdiercks et al., 2005; Thauer, 2011). Therefore, it is reasonable to predict that metal requirements for the enzymes involved in AOM are similar to those used for methanogenesis, involving high levels of Fe, Ni, Co, and additional Mo/W and Zn (see the Section “Metalloenzymes in Methanogenesis”1), but possibly with slightly lower Fe and Ni requirements due to the absence of two of the Ni–Fe hydrogenases involved in methanogenesis. The finding that ANME can also fix N2 (Dekas et al., 2009) suggests that they may have additional Fe and Mo/V requirements. ANME occur in anoxic and sulfidic environments where Ni, Co, Mo, and possibly Fe likely occur at levels lower than their growth optima (Table 2), assuming that their metal requirements are similar to those of methanogens. For a full review of the biochemistry of AOM, readers are referred to Thauer and Shima (2008).
Summary
Extensive research has been focused on the importance of Cu for particulate methane monooxygenase, resulting in a detailed understanding of physiological and biochemical aspects of Cu in pure cultures of MOB. However, few studies have attempted to relate Cu bioavailability, particularly in mineral form, to rates of aerobic CH4 oxidation in the environment. Furthermore, few studies have investigated additional metal requirements, particularly Fe, but also small amounts of Mo and Zn, for aerobic and intra-aerobic methanotrophy, although clearly Fe in particular is important for these metabolisms (Figure 2) and cell surface Cu acquisition mechanisms (Karlsen et al., 2011). In contrast to aerobic methanotrophy, essentially nothing is known about the metal requirements for AOM besides the need for large amounts of Ni in cofactor F430 (Krüger et al., 2003). Presumably, metal requirements for reverse methanogenesis by ANME are similar to those of the canonical seven-step methanogenic pathway, but this hypothesis remains to be tested.
Nitrous Oxide Production and Consumption
A diverse range of organisms using several different pathways are known to produce N2O. Whereas CH4 production is only known to occur within the archaea, N2O production is more widespread. Pathways for biological N2O production include dissimilatory nitrate/nitrite reduction (e.g., denitrification), nitrifier denitrification (Wrage et al., 2001), hydroxylamine oxidation, and NOx detoxification (also known as the “nitrosative stress” pathway; Stein, 2011). N2O is also produced by MOB (Sutka et al., 2006; Campbell et al., 2011) and enrichment cultures of ammonia oxidizing archaea (AOA; Jung et al., 2011; Santoro et al., 2011). Bacterial denitrification and nitrifier denitrification together account for ~70% of the global N2O flux (IPCC, 2007), although recent data suggest that AOA may also be an important source of N2O in marine and terrestrial systems (Jung et al., 2011; Santoro et al., 2011). For an in-depth review of the methods used to identify N2O-producing bacteria by their key enzymes, readers are referred to Stein (2011).
Metalloenzymes involved in N2O production primarily contain Fe and/or Cu, with additional Mo needed for the dissimilatory nitrate reduction pathway to N2O and/or N2 and possibly additional Zn in nitrifier denitrification. A schematic of the metal content of proteins involved in N2O production and consumption is shown in Figure 3. Below we review biochemical and physiological studies of the metal requirements for N2O production through the standard and nitrifier denitrification pathways, and touch on new discoveries of metalloenzymes putatively involved in archaeal ammonia oxidation. Metalloenzymes involved in the nitrosative stress pathway are mentioned briefly, but are outside the scope of this review.
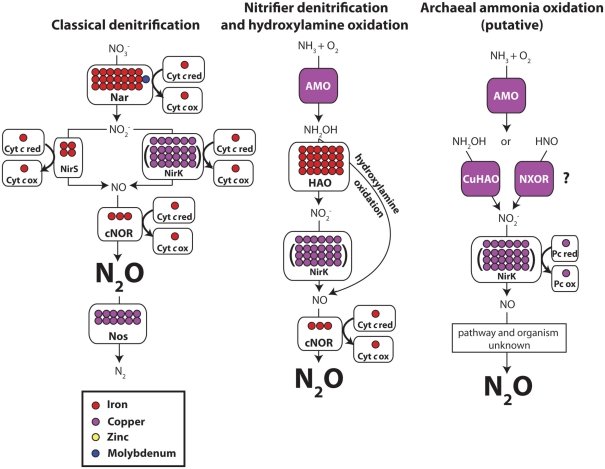
Figure 3.
Metal content of three pathways of N2O production: classical denitrification, bacterial nitrifier denitrification, and archaeal ammonia oxidation. Each circle represents one metal atom. Parentheses show varying metal content of a given enzyme. Abbreviations: AMO, ammonia monooxygenase; HAO, hydroxylamine oxidoreductase; Nar, dissimilatory nitrate reductase; NirS, Fe–nitrite reductase; NirS, Cu–nitrite reductase; cNOR, nitric oxide reductase; Nos, nitrous oxide reductase; NXOR, nitroxyl oxidoreductase (putative enzyme). Putative archaeal ammonia oxidation pathway is adapted from Walker et al. (2010). For simplicity, only metalloenzymes are labeled. Question marks depict unknown or unconfirmed enzymes. Enzymes colored a solid purple indicate that an unknown amount of Cu is present. See the Section “Metalloenzymes Involved in the Production and Consumption of Nitrous Oxide” for more details.
Metalloenzymes involved in the production and consumption of nitrous oxide
Nitrate/nitrite reduction to N2O/N2 pathway (denitrification)
Nitrous oxide is a gaseous intermediate in the process of denitrification, the anaerobic microbial respiratory process whereby is reduced to N2 in the sequence: The process is leaky and therefore N2O is almost always found in trace abundances in environments where denitrification occurs (Rogers and Whitman, 1991). Since is typically present at higher abundances than in natural environments, denitrification typically starts with dissimilatory nitrate reduction. The most well-studied dissimilatory nitrate reductase is the membrane-bound NarGHI complex, which contains two b-type cytochromes (each binding one Fe atom), one Fe3S4 cluster, four Fe4S4 clusters, and a molybdopterin (Mo) active site (Bertero et al., 2003; Jormakka et al., 2004; Figure 3). It is possible for dissimilatory nitrate reduction to occur with Fe or V in place of Mo, although such alternative nitrate reductases have so far only been found in metal-reducing bacteria (Antipov et al., 1998, 2005).
The next step in denitrification, nitrite reduction to nitric oxide (NO), is catalyzed by either a cytochrome cd1 (four Fe)-containing nitrite reductase (abbreviated cd1NIR or NirS; Fülöp et al., 1995) or a Cu-containing nitrite reductase (abbreviated CuNIR or NirK) that possesses either 6 or 24 Cu atoms depending on its subunit content (Godden et al., 1991; Nojiri et al., 2007; Figure 3).
NO reduction to N2O occurs almost instantaneously after its formation due to the toxicity of NO. In denitrifying bacteria, NO reductases (NORs) are members of the heme/copper cytochrome oxidase enzyme superfamily that substitute Fe in place of Cu at their active site (Hendriks et al., 2000). Other enzymes in this family cannot efficiently reduce NO, suggesting that the replacement of Cu with Fe is important for the enzyme mechanism. The most common type of NOR in denitrifying bacteria is NorBC (or cNOR), a cytochrome bc complex that contains Fe in the form of two hemes and one non-heme Fe and uses cytochrome c as an electron donor (Hendriks et al., 2000; Zumft, 2005; Figure 3). The nitrosative stress pathway uses a different NO reductase called qNOR, which contains only the NorB subunit and accepts electrons from quinols (Hendriks et al., 2000; Zumft, 2005). Two additional Fe-containing NOR enzymes, the flavohemoglobin Hmp and the flavorubredoxin NorVW, are important for relieving nitrosative stress under anoxic conditions by NO reduction to N2O (Poole and Hughes, 2000; Gardner et al., 2003). Fungal NORs are members of the cytochrome P-460 family and also contain only Fe (Hendriks et al., 2000).
After its production, N2O can either escape to the atmosphere or be further reduced to N2 by nitrous oxide reductase (Nos). Nos is a Cu-rich enzyme, binding 12 atoms of Cu per homodimer (Brown et al., 2000; Figure 3). Low pH and/or trace amounts of O2 tend to increase the ratio of N2O:N2 released because Nos is inhibited at low pH and is more sensitive to O2 than other reductases in denitrification (Knowles, 1982).
Bacterial ammonia oxidation to N2O (nitrifier denitrification and hydroxylamine oxidation)
Autotrophic ammonia oxidizing Bacteria (AOB; Shaw et al., 2006) and MOB (Yoshinari, 1985; Ren et al., 2000) are capable of aerobic production of N2O during the process of ammonia oxidation N2O is produced from two offshoots of the nitrification pathway: nitrifier denitrification (Ritchie and Nicholas, 1972; Poth and Focht, 1985; Wrage et al., 2001; Shaw et al., 2006) and hydroxylamine oxidation (Cantera and Stein, 2007b; Stein, 2011). In nitrifier denitrification, is reduced to NO, and then N2O or N2, instead of being oxidized to In hydroxylamine oxidation, NH2OH is oxidized to NO, which is then reduced to N2O. Little is known about the controls on N2O:N2 ratios for both of these processes.
In the first step of nitrifier denitrification and hydroxylamine oxidation, ammonia (NH3) is oxidized to hydroxylamine (NH2OH), catalyzed by the enzyme ammonia monooxygenase (AMO). Although the crystal structure of AMO has not been solved, AMO shares high similarity to pMMO of aerobic methanotrophs (Holmes et al., 1995; Arp and Stein, 2003 and references therein), suggesting that it also contains three Cu atoms per monomer and possibly additional Zn and Fe (see the Section “Metalloenzymes in Aerobic Methanotrophy”; Figure 3). A Cu requirement for AMO is supported by experiments showing that Cu addition activates AMO (Ensign et al., 1993).
The next step of nitrifier denitrification, NH2OH oxidation to is catalyzed by the enzyme hydroxylamine oxidoreductase (HAO), which contains 24 Fe atoms in c-type cytochromes (Igarashi et al., 1997; Figure 3). Orthologs of the copper-containing NirK enzymes are typically used for the reduction of to NO (Casciotti and Ward, 2001; Cantera and Stein, 2007a,b). NO, a toxic intermediate, is then reduced to N2O by an unknown enzyme, likely either NorBC (Beaumont et al., 2004; Casciotti and Ward, 2005), the tetraheme cytochrome c554 (Upadhyay et al., 2006), or NorS (Stein, 2011). In hydroxylamine oxidation, NH2OH is oxidized by HAO to NO instead of (Figure 3). Reduction of NO to N2O is then catalyzed by cytochrome c554 or NorS as with nitrifier denitrification (Stein, 2011).
Although N2 has been shown to be produced by the nitrifier denitrification pathway (Poth, 1986), nos genes have not been found in the genomes of bacterial nitrifiers (Stein et al., 2007) so it is unclear how N2 is formed during nitrifier denitrification.
Archaeal ammonia oxidation
Archaeal nitrification was first reported in 2005 (Könneke et al., 2005) and archaeal enrichment cultures have recently been shown to produce N2O (Jung et al., 2011; Santoro et al., 2011). For a review of AOA, see Hatzenpichler (in preparation). The genomes of AOA Cenarchaeum symbiosum and Nitrosopumilus maritimus suggest that AOA have higher Cu requirements than AOB (Hallam et al., 2006; Walker et al., 2010). C. symbiosum and N. maritimus contain amo genes with conserved motifs that bind the metal centers in M. capsulatus Bath, suggesting that archaeal AMO also contains abundant Cu (Hallam et al., 2006; Walker et al., 2010). Both AOA genomes lack genes to encode a bacterial-type HAO complex. AOA may use a Cu-based electron-transfer system instead of the Fe-based system used by AOB, as implied by the abundance of genes encoding copper-containing proteins, including numerous multi-copper oxidases and small Cu-binding plastocyanin-like domains (Hallam et al., 2006; Walker et al., 2010). The N. maritimus genome contains nirK genes (Walker et al., 2010) and AOA nirK homologs have been found to be widespread in environmental samples (Bartossek et al., 2010). Walker et al. (2010) propose a model for the AOA nitrification pathway whereby the products of AMO (either NH2OH, or alternatively the highly reactive intermediate nitroxyl, HNO) are oxidized to nitrous acid (HNO2) by a Cu-containing protein, either a Cu-HAO or a multi-copper oxidase acting as a nitroxyl oxidoreductase (NXOR; Figure 3). Following these reactions, is reduced to NO by NirK, with plastocyanin acting as an electron donor. Future studies are needed to test whether pure cultures of AOA can produce N2O or whether an N intermediate such as NO is produced by AOA and reduced to N2O by bacterial partners. If AOA can indeed produce N2O as supported by isotopic studies (Santoro et al., 2011), future biochemical studies are required to determine what pathway is used, and whether they can consume it.
Physiological studies
As reviewed in the Section “Metalloenzymes Involved in the Production and Consumption of Nitrous Oxide,” the metal content of enzymes involved in N2O production and consumption is composed primarily of Fe and Cu (Figure 3). The influence of Cu on N2O processing by denitrifying bacteria has been the subject of a number of pure culture and environmental investigations. The influence of Fe availability on growth of AOB and denitrifiers is less studied, and very little is known about the importance of Fe for N2O processing in these microbes. Research is also lacking on the influence of Fe and Cu availability on N2O processing during nitrifier denitrification and in AOA enrichment cultures. These pathways are of particular interest due to the likelihood of high Cu requirements for their first step (AMO), whereas N2O can be produced (but not consumed) without Cu by denitrifiers.
Pure cultures
The importance of Cu for the consumption of N2O by denitrifying bacteria has been clearly demonstrated by laboratory studies (Iwasaki and Terai, 1982; Matsubara et al., 1982; Granger and Ward, 2003; Twining et al., 2007). These studies showed that N2O accumulates when denitrifying bacteria are grown without added Cu, whereas complete denitrification to N2 occurs when Cu is present above a certain threshold. In early studies with Pseudomonas perfectomarinus and two Alcaligenes strains, background Cu contamination in growth medium was not quantified and high levels of Cu were added in the replete condition (1–6 μM; Iwasaki and Terai, 1982; Matsubara et al., 1982). In follow-up studies with Paracoccus denitrificans and Pseudomonas denitrificans, trace metal-clean growth protocols were utilized and total dissolved Cu background contamination was measured to be 0.3–0.5 nM (Granger and Ward, 2003; Twining et al., 2007). At less than 1 nM total dissolved Cu, N2O accumulated; above 1 nM, it was consumed (Granger and Ward, 2003; Twining et al., 2007). Enzyme activity of nitrous oxide reductase (Nos) was below the detection limit in denitrifiers grown at 0.3 nM total dissolved Cu, suggesting that the reason for build-up of N2O was Cu limitation of Nos (Granger and Ward, 2003). Supplementation of 10 nM Cu to Cu-limited cultures caused increased growth rate and complete consumption of N2O (Granger and Ward, 2003). These laboratory studies led to the hypothesis that low Cu concentrations in natural aquatic ecosystems may promote N2O accumulation (see the Section “Environmental Studies”).
Metal requirements for AOB have not been studied as extensively as those for denitrifiers, but preliminary studies support a high importance of Fe for nitrification. An early study of nitrifying bacterial cultures showed that addition of Fe alone produced a 56% increase in production whereas addition of Cu alone produced a 17% increase (Lees and Meiklejohn, 1948). The few subsequent studies of metal requirements for AOB have focused on Fe requirements (Meiklejohn, 1953; Wei et al., 2006). Both AMO and HAO activity were reduced in Nitrosomonas europaea cultures grown at 0.2 vs. 10 μM Fe (Wei et al., 2006). Lowered HAO activity is expected due to the high heme content of HAO in AOB cells (Whittaker et al., 2000). AMO activity dependence on Fe suggests that Fe is a component of AMO or that Fe is present in the electron donor to AMO. The Fe content of AMO is difficult to assess due to the lack of in vitro protein preparations. It is also possible that AOB scavenge/co-opt Fe-loaded siderophores produced by other bacteria; if supplied with exogenous siderophores, N. europaea expresses genes for siderophore transporters and ferric transporters when Fe is limiting (Wei et al., 2006; Stein et al., 2007). Lastly, the fact that AOB cells are bright red in color further supports the major requirement for Fe in their metabolism.
Environmental studies
Several recent field studies have suggested that Cu limitation prohibits N2O consumption. Cu addition stimulated N2O consumption in Linsley Pond, a temperate lake ecosystem with 3 nM Cu (Twining et al., 2007). Similarly, the ratio of N2O/N2 decreased as water-extractable Cu concentration increased in freshwater sediments (Jacinthe and Tedesco, 2009). While scarce Cu can limit denitrification, excess Cu (above 100 mg kg−1 sediment) in polluted ecosystems can inhibit the pathway (Sakadevan et al., 1999; Magalhães et al., 2011). On the other hand, denitrification in three major marine oxygen minimum zones does not appear to be Cu-limited: addition of Cu had minimal effect on denitrification rates in seawater incubations with background Cu concentrations of 1–3 nM (Ward et al., 2008). Therefore, it is likely that marine denitrifiers have evolved mechanisms to satisfy their Cu requirements by taking up Cu that is strongly bound to natural ligands, which can bind 99% of the total Cu in temperate lakes (Twining et al., 2007) and in the ocean (Moffett and Dupont, 2007).
Metal bioavailability varies considerably between freshwater and seawater oxic environments. In general, dissolved Fe and Cu are present at much lower concentrations in seawater than in lakes and soil pore waters (Table 3). Therefore, N2O production by marine ammonia oxidizers may be Fe or Cu-limited and/or they may have evolved strategies to scavenge these metals.
Summary
In contrast to CH4, N2O can be produced by diverse microorganisms via numerous pathways containing varying amounts of Fe and Cu, small amounts of Mo and possibly Zn. To date, only one enzyme has been found to convert N2O to N2: the Cu-containing Nos enzyme found in denitrifiers. The absence of nos genes from the genomes of AOA and AOB (Hallam et al., 2006; Stein et al., 2007; Walker et al., 2010) suggests either that N2O is their final gaseous product or that they have alternative nitrous oxide reductases. The ability of some AOB to produce N2 (Poth, 1986) lends support to the presence of alternative reductases. A search for such alternative enzymes deserves attention, as ammonia oxidizers are important players in the global N2O budget. Research on the biochemistry of N2O production in AOA enrichment cultures is essential to understand the exciting findings of recent studies suggesting that large amounts of Cu may be involved in AOA metabolism.
Comparisons, Conclusion, and Future Research
Trace metals present in microbial enzymes involved in CH4 and N2O cycling include Fe, Co, Ni, Cu, Zn, Mo, and W. While Fe-containing enzymes are present in almost all pathways we reviewed (with the possible exception of archaeal ammonia oxidation), other metals differ by metabolism. For instance, Ni, Co, and W are present in methanogenic enzymes, but not in enzymes involved in aerobic methanotrophy and N2O cycling. In contrast, Cu is not used in methanogenesis, but is very important for aerobic methanotrophy and N2O cycling. Small amounts of Mo and Zn are common in most pathways. If ongoing biochemical studies confirm that anaerobic methanotrophs use the reverse methanogenesis pathway to oxidize methane, they likely require the same suite of metals as methanogens: Fe, Co, Ni, Zn, and Mo/W.
How do metal requirements for CH4 and N2O cycling compare with other ecologically important microbes? The metal stoichiometry of 15 marine eukaryotic phytoplankton showed an average trace metal stoichiometry of Fe1Mn0.53Zn0.08Cu0.05Co0.03Mo0.005 (Ho et al., 2003; Barton et al., 2007). High Mn requirements are likely reflective of the oxygen-generating Mn-containing photosystem II complex. The elemental stoichiometry of CH4 and N2O processing microbes is not nearly as well-constrained as that of eukaryotic phytoplankton, but preliminary studies of methanogens suggest that, after Fe, Ni is an extremely important metal for methanogenesis and anaerobic methanotrophy. Very little is known about the metal stoichiometry of aerobic and intra-aerobic methanotrophs and N2O-producing microbes, although it is predicted that their Cu content is much higher than both methanogens and phytoplankton, especially in the case of archaeal ammonia oxidizers. Metallomics – the comprehensive analysis of the entirety of metal-containing species within a cell (Szpunar, 2005) – may reveal still more unexpected metals or natural products excreted by microbes to alter metal bioavailability. For example, a recent metallomics study of the hyperthermophilic sulfur-reducing Archeon, Pyrococcus furiosus, revealed 158 unassigned metalloprotein peaks, 75 of which contained metals that the organism was not known to assimilate, such as Pb and U (Cvetkovic et al., 2010). Furthermore, it is likely that the metal requirements for each of the pathways reviewed here are amplified by additional metalloenzymes involved in complex metal cofactor biosynthesis, such is the case for hydrogenases and nitrogenases (Rubio and Ludden, 2008; Shepard et al., 2011).
More studies are needed to address gaps in our knowledge of microbial trace metal physiology. Ideally these studies will make use of well-defined media and trace metal-clean conditions, such as Teflon-lined glass bottles for anaerobic studies. Understudied topics include Zn requirements for methanogenesis, trace metal content of ANME enzymes (other than the well-studied Mcr), the influence of Cu on N2O production by archaeal enrichment cultures, the biochemistry of Cu-enzymes in archaeal ammonia oxidizers, and strategies for metal-binding ligand production (i.e., methanobactin) in greenhouse gas cycling microbes. The search for alternative N2O-consuming enzymes in ammonia oxidizers deserves particular attention, as suggested by the ability of some bacterial nitrifiers lacking the Cu-containing nitrous oxide reductase enzyme to convert N2O to N2.
In order to better connect the findings from biochemical and physiological studies of pure cultures with the larger picture of global greenhouse gas cycling and environmental metal bioavailability, more in situ studies are necessary. Preliminary studies suggest that rates of methanogenesis in peatlands may be controlled by Fe, Ni, and Co availability, since addition of all three metals stimulated methanogenesis in mineral-poor peats. It is currently unknown whether one metal was the primary limiting growth element or whether multiple metals were co-limiting (Saito et al., 2008). If co-limitation was occurring, the addition of all three elements would result in higher rates of methanogenesis than any metal added solely. On the other hand, other systems and pathways may be dominantly regulated by the bioavailability of only one scarce metal; this seems to be the case for Co(balt) limitation of methylotrophic methanogenesis in anaerobic digesters, Cu limitation of N2O consumption by freshwater denitrifying bacteria and possibly Cu limitation of ammonia oxidizing archaea as suggested by numerous genes encoding Cu-containing proteins in AOA genomes.
Lastly, relationships between metal bioavailability and fluxes of greenhouse gases remain largely unexplored, particularly in anaerobic ecosystems. High sulfide environments likely pose particular challenges for microbial metal acquisition due to abiotic sulfide scavenging and precipitation of metal sulfides. Of the bioessential metals involved in greenhouse gas cycling, Mo, Fe, and Cu are most susceptible to sulfide scavenging, followed by Co, Ni, and Zn (Morse and Luther, 1999). Microbial metal limitation due to sulfide drawdown may have driven major evolutionary episodes and geochemical transitions in Earth history (Anbar and Knoll, 2002; Scott et al., 2008; Dupont et al., 2010). Several such scenarios have been proposed for metal limitation of microbial greenhouse gas cycling: a decline in seawater Ni due to cooling of the Earth’s mantle in the late Archean may have limited methanogenesis and contributed to the Great Oxidation Event 2.4 billion years ago (Konhauser et al., 2009) while low Cu due to sulfide scavenging in the Proterozoic ocean could have led to global warming through the build-up of N2O (Buick, 2007). The question of how metal bioavailability influences greenhouse gas cycling in ancient and modern ecosystems is just beginning to be investigated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge funding from the National Aeronautics and Space Administration (NASA) Astrobiology Postdoctoral Fellowship (to Jennifer B. Glass), the NASA Astrobiology Institute (NNA04CC06A), and the U.S. Department of Energy’s Office of Biological and Environmental Research Early Career Award (to Victoria J. Orphan). Thanks to Roland Hatzenpichler, Patricia Tavormina, Shawn McGlynn, Anne Dekas, Joshua Steele, Hiroyuki Imachi, Lisa Stein and Aubrey Zerkle for helpful comments on drafts of this manuscript.
References
- Afting C., Hochheimer A., Thauer R. (1998). Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch. Microbiol. 169, 206–210 10.1007/s002030050562 [DOI] [PubMed] [Google Scholar]
- Alex L. A., Reeve J. N., Orme-Johnson W. H., Walsh C. T. (1990). Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum delta H. Biochemistry 29, 7237–7244 10.1021/bi00483a011 [DOI] [PubMed] [Google Scholar]
- Anbar A. D., Knoll A. H. (2002). Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297, 1137–1142 10.1126/science.1069651 [DOI] [PubMed] [Google Scholar]
- Anderson I., Ulrich L. E., Lupa B., Susanti D., Porat I., Hooper S. D., Lykidis A., Sieprawska-Lupa M., Dharmarajan L., Goltsman E., Lapidus A., Saunders E., Han C., Land M., Lucas S., Mukhopadhyay B., Whitman W. B., Woese C., Bristow J., Kyrpides N. (2009). Genomic characterization of Methanomicrobiales reveals three classes of methanogens. PLoS ONE 4, e5797. 10.1371/journal.pone.0005797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov A. N., Lyalikova N. N., Khijniak T. V., L’vov N. P. (1998). Molybdenum-free nitrate reductases from vanadate-reducing bacteria. FEBS Lett. 441, 257–260 10.1016/S0014-5793(98)01510-5 [DOI] [PubMed] [Google Scholar]
- Antipov A. N., Morozkina E. V., Sorokin D. Y., Golubeva L. I., Zvyagilskaya R. A., L’vov N. P. (2005). Characterization of molybdenum-free nitrate reductase from haloalkalophilic bacterium Halomonas sp strain AGJ 1-3. Biochemistry 70, 799–803 [DOI] [PubMed] [Google Scholar]
- Arp D. J., Stein L. Y. (2003). Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38, 471–495 10.1080/10409230390267446 [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Kenney G. E., Rosenzweig A. C. (2011). Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 286, 37313–37319 10.1074/jbc.M111.249326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Rosenzweig A. C. (2007). Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc. Chem. Res. 40, 573–580 10.1021/ar700004s [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Rosenzweig A. C. (2008). Copper methanobactin: a molecule whose time has come. Curr. Opin. Chem. Biol. 12, 245–249 10.1016/j.cbpa.2008.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Smith S. M., Rawat S., Yatsunyk L. A., Stemmler T. L., Rosenzweig A. C. (2010). Oxidation of methane by a biological dicopper centre. Nature 465, 115–119 10.1038/nature08992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L. L., Goulhen F., Bruschi M., Woodards N. A., Plunkett R. M., Rietmeijer F. J. M. (2007). The bacterial metallome: composition and stability with specific reference to the anaerobic bacterium Desulfovibrio desulfuricans. Biometals 20, 291–302 10.1007/s10534-006-9059-2 [DOI] [PubMed] [Google Scholar]
- Bartossek R., Nicol G. W., Lanzen A., Klenk H. P., Schleper C. (2010). Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ. Microbiol. 12, 1075–1088 10.1111/j.1462-2920.2010.02153.x [DOI] [PubMed] [Google Scholar]
- Basiliko N., Yavitt J. B. (2001). Influence of Ni, Co, Fe, and Na additions on methane production in Sphagnum-dominated Northern American peatlands. Biogeochemistry 52, 133–153 10.1023/A:1006461803585 [DOI] [Google Scholar]
- Baudet C., Sprott G. D., Patel G. B. (1988). Adsorption and uptake of nickel in Methanothrix concilii. Arch. Microbiol. 150, 338–342 10.1007/BF00408304 [DOI] [Google Scholar]
- Beaumont H. J. E., Van Schooten B., Lens S. I., Westerhoff H. V., Van Spanning R. J. M. (2004). Nitrosomonas europaea expresses a nitric oxide reductase during nitrification. J. Bacteriol. 186, 4417–4421 10.1128/JB.186.13.4417-4421.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero M. G., Rothery R. A., Palak M., Hou C., Lim D., Blasco F., Weiner J. H., Strynadka N. C. J. (2003). Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10, 681–687 10.1038/nsb969 [DOI] [PubMed] [Google Scholar]
- Boetius A., Ravenschlag K., Schubert C. J., Rickert D., Widdel F., Gieseke A., Amann R., Jørgensen B. B., Witte U., Pfannkuche O. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626 10.1038/35036572 [DOI] [PubMed] [Google Scholar]
- Bollinger J. M. (2010). Biochemistry: getting the metal right. Nature 465, 40–41 10.1038/465040a [DOI] [PubMed] [Google Scholar]
- Bragazza L. (2006). Heavy metals in bog waters: an alternative way to assess atmospheric precipitation quality? Glob. Planet. Change 53, 290–298 10.1016/j.gloplacha.2006.03.011 [DOI] [Google Scholar]
- Brown K., Tegoni M., Prudencio M., Pereira A. S., Besson S., Moura J. J., Moura I., Cambillau C. (2000). A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 7, 191–195 10.1038/73288 [DOI] [PubMed] [Google Scholar]
- Bruland K. W. (1980). Oceanographic distributions of cadmium, zinc, nickel, and copper in the North Pacific. Earth Planet. Sci. Lett. 47, 176–198 10.1016/0012-821X(80)90035-7 [DOI] [Google Scholar]
- Buick R. (2007). Did the Proterozoic Canfield Ocean cause a laughing gas greenhouse? Geobiology 5, 97–100 10.1111/j.1472-4669.2007.00110.x [DOI] [Google Scholar]
- Callander I., Barford J. (1983a). Precipitation, chelation, and the availability of metals as nutrients in anaerobic digestion. I. Methodology. Biotechnol. Bioeng. 25, 1947–1957 10.1002/bit.260250806 [DOI] [PubMed] [Google Scholar]
- Callander I., Barford J. (1983b). Precipitation, chelation, and the availability of metals as nutrients in anaerobic digestion. II. Applications. Biotechnol. Bioeng. 25, 1959–1972 10.1002/bit.260250806 [DOI] [PubMed] [Google Scholar]
- Campbell M. A., Nyerges G., Kozlowski J. A., Poret-Peterson A. T., Stein L. Y., Klotz M. G. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322, 82–89 10.1111/j.1574-6968.2011.02340.x [DOI] [PubMed] [Google Scholar]
- Cantera J. J. L., Stein L. Y. (2007a). Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9, 765–776 10.1111/j.1462-2920.2006.01198.x [DOI] [PubMed] [Google Scholar]
- Cantera J. J. L., Stein L. Y. (2007b). Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch. Microbiol. 188, 349–354 10.1007/s00203-007-0255-4 [DOI] [PubMed] [Google Scholar]
- Casciotti K. L., Ward B. B. (2001). Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67, 2213–2221 10.1128/AEM.67.5.2213-2221.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciotti K. L., Ward B. B. (2005). Phylogenetic analysis of nitric oxide reductase gene homologues from aerobic ammonia oxidizing bacteria. FEMS Microbiol. Ecol. 52, 197–205 10.1016/j.femsec.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Chien Y. T., Auerbuch V., Brabban A. D., Zinder S. H. (2000). Analysis of genes encoding an alternative nitrogenase in the archaeon Methanosarcina barkeri 227. J. Bacteriol. 182, 3247–3253 10.1128/JB.182.11.3247-3253.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ. Microbiol. 13, 2603–2622 10.1111/j.1462-2920.2011.02464.x [DOI] [PubMed] [Google Scholar]
- Chistoserdova L., Vorholt J. A., Lidstrom M. E. (2005). A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol. 6, 208–213 10.1186/gb-2005-6-2-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D. W., Corbin J., Do Y. S., Semrau J. D., Antholine W. E., Hargrove M. S., Pohl N. L., Boyd E. S., Geesey G., Hartsel S. C., Shafe P. H., McEllistrem M. T., Kisting C. J., Campbell D., Rao V., de la Mora A. M., Dispirito A. A. (2006). Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry 45, 1442–1453 10.1021/bi0614939 [DOI] [PubMed] [Google Scholar]
- Choi D.-W., Kunz R. C., Boyd E. S., Semrau J. D., Antholine W. E., Han J.-I., Zahn J. A., Boyd J. M., De La Mora A. M., DiSpirito A. A. (2003). The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH: quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 185, 5755–5764 10.1128/JB.185.19.5755-5764.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier R. W. (1985). Molybdenum in the northeast Pacific Ocean. Limnol. Oceanogr. 30, 1351–1354 10.4319/lo.1985.30.6.1351 [DOI] [Google Scholar]
- Conrad R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Mol. Biol. Rev. 60, 609–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole Ii F. L., Jenney F. E., Jr., Lancaster W. A., Praissman J. L., Shanmukh S., Vaccaro B. J., Trauger S. A., Kalisiak E., Apon J. V., Siuzdak G., Yannone S. M., Tainer J. A., Adams M. W. W. (2010). Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782 10.1038/nature09265 [DOI] [PubMed] [Google Scholar]
- Daas P. J. H., Hagen W. R., Keltjens J. T., Vogels G. D. (1994). Characterization and determination of the redox properties of the 2[4Fe-4S] ferredoxin from Methanosarcina barkeri strain MS. FEBS Lett. 356, 342–344 10.1016/0014-5793(94)01313-6 [DOI] [PubMed] [Google Scholar]
- David L. A., Alm E. J. (2010). Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469, 93–96 10.1038/nature09649 [DOI] [PubMed] [Google Scholar]
- Dekas A., Poretsky R. S., Orphan V. J. (2009). Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426 10.1126/science.1178223 [DOI] [PubMed] [Google Scholar]
- Demirel B., Scherer P. (2011). Trace element requirements of agricultural biogas digesters during biological conversion of renewable biomass to methane. Biomass Bioenergy 35, 992–998 10.1016/j.biombioe.2010.12.022 [DOI] [Google Scholar]
- Deppenmeier U., Blaut M., Schmidt B., Gottschalk G. (1992). Purification and properties of a F420-nonreactive, membrane-bound hydrogenase from Methanosarcina strain Gö1. Arch. Microbiol. 157, 505–511 [DOI] [PubMed] [Google Scholar]
- Diekert G., Konheiser U., Piechulla K., Thauer R. K. (1981). Nickel requirement and factor F430 content of methanogenic bacteria. J. Bacteriol. 148, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert G., Weber B., Thauer R. K. (1980). Nickel dependence of factor F430 content in Methanobacterium thermoautotrophicum. Arch. Microbiol. 127, 273–277 10.1007/BF00427204 [DOI] [PubMed] [Google Scholar]
- DiSpirito A. A., Zahn J. A., Graham D. W., Kim H. J., Larive C. K., Derrick T. S., Cox C. D., Taylor A. (1998). Copper-binding compounds from Methylosinus trichosporium OB3b. J. Bacteriol. 180, 3606–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C. L., Butcher A., Ruben R. E., Bourne P. E., Caetano-Anollés G. (2010). History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U.S.A. 107, 10567–10572 10.1073/pnas.1009565107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C. L., Yang S., Palenik B., Bourne P. E. (2006). Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl. Acad. Sci. U.S.A. 103, 17822. 10.1073/pnas.0605798103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. R., Huested S. S. (1991). Ocean anoxia and the concentrations of molybdenum and vanadium in seawater. Mar. Chem. 34, 177–196 10.1016/0304-4203(91)90002-E [DOI] [Google Scholar]
- Ensign S. A., Hyman M. R., Arp D. J. (1993). In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J. Bacteriol. 175, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermler U., Grabarse W., Shima S., Goubeaud M., Thauer R. K. (1997). Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278, 1457–1462 10.1126/science.278.5342.1457 [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Butler M. K., Le Paslier D., Pelletier E., Mangenot S., Kuypers M. M. M., Schreiber F., Dutilh B. E., Zedelius J., De Beer D., Gloerich J., Wessels H. J., van Alen T., Luesken F., Wu M. L., van de Pas-Schoonen K. T., Op den Camp H. J., Janssen-Megens E. M., Francoijs K. J., Stunnenberg H., Weissenbach J., Jetten M. S., Strous M (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548 10.1038/nature08883 [DOI] [PubMed] [Google Scholar]
- Fathepure B. Z. (1987). Factors affecting the methanogenic activity of Methanothrix soehngenii VNBF. Appl. Environ. Microbiol. 53, 2978–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry J. G. (1999). Enzymology of one carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 23, 13–38 10.1111/j.1574-6976.1999.tb00390.x [DOI] [PubMed] [Google Scholar]
- Ferry J. G. (2010a). The chemical biology of methanogenesis. Planet. Space Sci. 58, 1775–1783 10.1016/j.pss.2010.08.014 [DOI] [Google Scholar]
- Ferry J. G. (2010b). How to make a living by exhaling methane. Annu. Rev. Microbiol. 64, 453–473 10.1146/annurev.micro.112408.134051 [DOI] [PubMed] [Google Scholar]
- Florencio L., Field J. A., Lettinga G. (1994). Importance of cobalt for individual trophic groups in an anaerobic methanol-degrading consortium. Appl. Environ. Microbiol. 60, 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florencio L., Jenicek P., Field J. A., Lettinga G. (1993). Effect of cobalt on the anaerobic degradation of methanol. J. Ferment. Bioeng. 75, 368–374 10.1016/0922-338X(93)90136-V [DOI] [Google Scholar]
- Fox J. A., Livingston D. J., Orme-Johnson W. H., Walsh C. T. (1987). 8-Hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum: 1. Purification and characterization. Biochemistry 26, 4219–4227 10.1021/bi00388a007 [DOI] [PubMed] [Google Scholar]
- Fru E. C. (2011). Copper biogeochemistry: a cornerstone in aerobic methanotrophic bacterial ecology and activity? Geomicrobiol. J. 28, 601–614 10.1080/01490451.2011.581325 [DOI] [Google Scholar]
- Fülöp V., Moir J. W. B., Ferguson S. J., Hajdu J. (1995). The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell 81, 369–377 10.1016/0092-8674(95)90390-9 [DOI] [PubMed] [Google Scholar]
- Funk T., Gu W., Friedrich S., Wang H., Gencic S., Grahame D. A., Cramer S. P. (2004). Chemically distinct Ni sites in the A-cluster in subunit of the acetyl-CoA decarbonylase/synthase complex from Methanosarcina thermophila: Ni L-edge absorption and X-ray magnetic circular dichroism analyses. J. Am. Chem. Soc. 126, 88–95 10.1021/ja0366033 [DOI] [PubMed] [Google Scholar]
- Gardner A. M., Gessner C. R., Gardner P. R. (2003). Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. J. Biol. Chem. 278, 10081–10086 10.1074/jbc.M208366200 [DOI] [PubMed] [Google Scholar]
- Gartner P., Ecker A., Fischer R., Linder D., Fuchs G., Thauer R. K. (1993). Purification and properties of N5 methyltetrahydromethanopterin: coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Eur. J. Biochem. 213, 537–545 10.1111/j.1432-1033.1993.tb17792.x [DOI] [PubMed] [Google Scholar]
- Godden J. W., Turley S., Teller D. C., Adman E. T., Liu M. Y., Payne W. J., Legall J. (1991). The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science 253, 438–442 10.1126/science.1862344 [DOI] [PubMed] [Google Scholar]
- Gong W., Hao B., Wei Z., Ferguson D. J., Tallant T., Krzycki J. A., Chan M. K. (2008). Structure of the 2 2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarbonylase/synthase complex. Proc. Natl. Acad. Sci. U.S.A. 105, 9558–9563 10.1073/pnas.0709416105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil G., Kleerebezem R., Lettinga G. (1999). Effects of nickel and cobalt on kinetics of methanol conversion by methanogenic sludge as assessed by on-line CH4 monitoring. Appl. Environ. Microbiol. 65, 1789–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger J., Ward B. B. (2003). Accumulation of nitrogen oxides in copper-limited cultures of denitrifying bacteria. Limnol. Oceanogr. 48, 313–318 10.4319/lo.2003.48.1.0313 [DOI] [Google Scholar]
- Hagemeier C. H., Kruer M., Thauer R. K., Warkentin E., Ermler U. (2006). Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc. Natl. Acad. Sci. U.S.A. 103, 18917–18922 10.1073/pnas.0603650103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakemian A. S., Kondapalli K. C., Telser J., Hoffman B. M., Stemmler T. L., Rosenzweig A. C. (2008). The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 47, 6793–6801 10.1021/bi800598h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakemian A. S., Rosenzweig A. C. (2007). The biochemistry of methane oxidation. Annu. Rev. Biochem. 76, 223–241 10.1146/annurev.biochem.76.061505.175355 [DOI] [PubMed] [Google Scholar]
- Hallam S. J., Girguis P. R., Preston C. M., Richardson P. M., Delong E. F. (2003). Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69, 5483–5491 10.1128/AEM.69.9.5483-5491.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. J., Mincer T. J., Schleper C., Preston C. M., Roberts K., Richardson P. M., DeLong E. F. (2006). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4, e95. 10.1371/journal.pbio.0040095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. J., Putman N., Preston C. M., Detter J. C., Rokhsar D., Richardson P. M., DeLong E. F. (2004). Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462 10.1126/science.1100025 [DOI] [PubMed] [Google Scholar]
- Hamann N., Mander G. J., Shokes J. E., Scott R. A., Bennati M., Hedderich R. (2007). A cysteine-rich CCG domain contains a novel [4Fe-4S] cluster binding motif as deduced from studies with subunit B of heterodisulfide reductase from Methanothermobacter marburgensis. Biochemistry 46, 12875–12885 10.1021/bi700679u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R. S., Hanson T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsson C., Westerlund S. (1988). Trace metals in the water columns of the Black Sea and Framvaren Fjord. Mar. Chem. 23, 417–424 10.1016/0304-4203(88)90108-9 [DOI] [Google Scholar]
- Hedderich R., Hamann N., Bennati M. (2005). Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol. Chem. 386, 961–970 10.1515/BC.2005.112 [DOI] [PubMed] [Google Scholar]
- Heiden S., Hedderich R., Setzke E., Thauer R. K. (1994). Purification of a two subunit cytochrome b containing heterodisulfide reductase from methanol grown Methanosarcina barkeri. Eur. J. Biochem. 221, 855–861 10.1111/j.1432-1033.1994.tb18800.x [DOI] [PubMed] [Google Scholar]
- Heimann M. (2011). Atmospheric science: enigma of the recent methane budget. Nature 476, 157–158 10.1038/476157a [DOI] [PubMed] [Google Scholar]
- Heller C., Hoppert M., Reitner J. (2008). Immunological localization of coenzyme M reductase in anaerobic methane-oxidizing archaea of ANME 1 and ANME 2 type. Geomicrobiol. J. 25, 149–156 10.1080/01490450802006884 [DOI] [Google Scholar]
- Hendriks J., Oubrie A., Castresana J., Urbani A., Gemeinhardt S., Saraste M. (2000). Nitric oxide reductases in bacteria. Biochim. Biophys. Acta 1459, 266–273 10.1016/S0005-2728(00)00161-4 [DOI] [PubMed] [Google Scholar]
- Hinrichs K.-U., Hayes J. M., Sylva S. P., Brewer P. G., DeLong E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805 10.1038/19751 [DOI] [PubMed] [Google Scholar]
- Ho T. Y., Quigg A., Finkel Z. V., Milligan A. J., Wyman K., Falkowski P. G., Morel F. M. M. (2003). The elemental composition of some marine phytoplankton. J. Phycol. 39, 1145–1159 10.1111/j.0022-3646.2003.03-090.x [DOI] [Google Scholar]
- Hoban D., van den Berg L. (1979). Effect of iron on conversion of acetic acid to methane during methanogenic fermentations. J. Appl. Microbiol. 47, 153–159 10.1111/j.1365-2672.1979.tb01179.x [DOI] [PubMed] [Google Scholar]
- Holmes A. J., Costello A., Lidstrom M. E., Murrell J. C. (1995). Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208 10.1111/j.1574-6968.1995.tb07834.x [DOI] [PubMed] [Google Scholar]
- Igarashi N., Moriyama H., Fujiwara T., Fukumori Y., Tanaka N. (1997). The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea. Nat. Struc. Biol. 4, 276–284 10.1038/nsb0497-276 [DOI] [PubMed] [Google Scholar]
- IPCC. (2007). The Physical Science Basis: Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. New York: Cambridge University Press [Google Scholar]
- Iwasaki H., Terai H. (1982). N2 and N2O produced during growth of denitrifying bacteria in copper-depleted and -supplemented media. J. Gen. Appl. Microbiol. 28, 189–193 10.2323/jgam.28.189 [DOI] [Google Scholar]
- Jacinthe P. A., Tedesco L. P. (2009). Impact of elevated copper on the rate and gaseous products of denitrification in freshwater sediments. J. Environ. Qual. 38, 1183–1192 10.2134/jeq2007.0666 [DOI] [PubMed] [Google Scholar]
- Jansen S., Gonzalez-Gil G., van Leeuwen H. P. (2007). The impact of Co and Ni speciation on methanogenesis in sulfidic media – biouptake versus metal dissolution. Enzyme Microb. Technol. 40, 823–830 10.1016/j.enzmictec.2006.06.019 [DOI] [Google Scholar]
- Jansen S., Steffen F., Threels W. F., van Leeuwen H. P. (2005). Speciation of Co (II) and Ni (II) in anaerobic bioreactors measured by competitive ligand exchange-adsorptive stripping voltammetry. Environ. Sci. Technol. 39, 9493–9499 10.1021/es050492l [DOI] [PubMed] [Google Scholar]
- Jarrell K., Sprott G. (1982). Nickel transport in Methanobacterium bryantii. J. Bacteriol. 151, 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. S., Gordon R. M., Coale K. H. (1997). What controls dissolved iron concentrations in the world ocean? Mar. Chem. 57, 137–161 10.1016/S0304-4203(97)00043-1 [DOI] [Google Scholar]
- Jollie D., Lipscomb J. D. (1991). Formate dehydrogenase from Methylosinus trichosporium OB3b. Purification and spectroscopic characterization of the cofactors. J. Biol. Chem. 266, 21853–21863 [PubMed] [Google Scholar]
- Jones J., Stadtman T. C. (1977). Methanococcus vannielii: culture and effects of selenium and tungsten on growth. J. Bacteriol. 130, 1404–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M., Richardson D., Byrne B., Iwata S. (2004). Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 12, 95–104 10.1016/j.str.2003.11.020 [DOI] [PubMed] [Google Scholar]
- Jung M.-Y., Park S.-J., Min D., Kim J.-S., Rijpstra W. I. C., Sinninghe Damsté J. S., Kim G.-J., Madsen E. L., Rhee S.-K. (2011). Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I. 1a from an agricultural soil. Appl. Environ. Microbiol. 77, 8635–8647 10.1128/AEM.05787-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen O. A., Larsen O., Jensen H. B. (2011). The copper responding surfaceome of Methylococccus capsulatus Bath. FEMS Microbiol. Lett. 323, 97–104 10.1111/j.1574-6968.2011.02365.x [DOI] [PubMed] [Google Scholar]
- Kessler P. S., Mclarnan J., Leigh J. A. (1997). Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J. Bacteriol. 179, 541–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida K., Shigematsu T., Kijima J., Numaguchi M., Mochinaga Y., Abe N., Morimura S. (2001). Influence of Ni2+ and Co2+ on methanogenic activity and the amounts of coenzymes involved in methanogenesis. J. Biosci. Bioeng. 91, 590–595 10.1016/S1389-1723(01)80179-1 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Graham D. W., DiSpirito A. A., Alterman M. A., Galeva N., Larive C. K., Asunskis D., Sherwood P. M. A. (2004). Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305, 1612–1615 10.1126/science.1100860 [DOI] [PubMed] [Google Scholar]
- Kinniburgh D. G., Miles D. L. (1983). Extraction and chemical analysis of interstitial water from soils and rocks. Environ. Sci. Technol. 17, 362–368 10.1021/es00112a011 [DOI] [Google Scholar]
- Knapp C. W., Fowle D. A., Kulczycki E., Roberts J. A., Graham D. W. (2007). Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc. Natl. Acad. Sci. U.S.A. 104, 12040–12045 10.1073/pnas.0702879104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel K., Boetius A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 10.1146/annurev.micro.61.080706.093130 [DOI] [PubMed] [Google Scholar]
- Knowles R. (1982). Denitrification. Microbiol. Mol. Biol. Rev. 46, 43–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk L. A. (1938). A comparison of the filamentous iron organisms, Clonothrix fusca Roze and Crenothrix polyspora Cohn. Am. J. Bot. 25, 11–17 10.2307/2436625 [DOI] [Google Scholar]
- Konhauser K. O., Pecoits E., Lalonde S. V., Papineau D., Nisbet E. G., Barley M. E., Arndt N. T., Zahnle K., Kamber B. S. (2009). Oceanic nickel depletion and a methanogen famine before the Great Oxidation Event. Nature 458, 750–753 10.1038/nature07858 [DOI] [PubMed] [Google Scholar]
- Könneke M., Bernhard A. E., De La Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Kremling K. (1983). The behavior of Zn, Cd, Cu, Ni, Co, Fe, and Mn in anoxic Baltic waters. Mar. Chem. 13, 87–108 10.1016/0304-4203(83)90019-1 [DOI] [Google Scholar]
- Krüger M., Meyerdierks A., Glöckner F. O., Amann R., Widdel F., Kube M., Reinhardt R., Kahnt R., Böcher R., Thauer R. K., Shima S. (2003). A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–881 10.1038/nature02207 [DOI] [PubMed] [Google Scholar]
- Krzycki J., Zeikus J. (1980). Quantification of corrinoids in methanogenic bacteria. Curr. Microbiol. 3, 243–245 10.1007/BF02602456 [DOI] [PubMed] [Google Scholar]
- Kulczycki E., Fowle D. A., Kenward P. A., Leslie K., Graham D. W., Roberts J. A. (2011). Stimulation of methanotroph activity by Cu-substituted borosilicate glass. Geomicrobiol. J. 28, 1–10 10.1080/01490451003614971 [DOI] [Google Scholar]
- Kulczycki E., Fowle D. A., Knapp C., Graham D. W., Roberts J. A. (2007). Methanobactin-promoted dissolution of Cu-substituted boro-silicate glass. Geobiology 5, 251–263 10.1111/j.1472-4669.2007.00102.x [DOI] [Google Scholar]
- Künkel A., Vaupel M., Heim S., Thauer R. K., Hedderich R. (1997). Heterodisulfide reductase from methanol grown cells of Methanosarcina barkeri is not a flavoenzyme. Eur. J. Biochem. 244, 226–234 10.1111/j.1432-1033.1997.00226.x [DOI] [PubMed] [Google Scholar]
- Laukel M., Chistoserdova L., Lidstrom M. E., Vorholt J. A. (2003). The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270, 325–333 10.1046/j.1432-1033.2003.03391.x [DOI] [PubMed] [Google Scholar]
- Lees H., Meiklejohn J. (1948). Trace elements and nitrification. Nature 161, 398–399 10.1038/161398a0 [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Lessner D. J., Karger B. L., Ferry J. G. (2006). Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188, 702–710 10.1128/JB.00933-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R. L., Rosenzweig A. C. (2005). Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434, 177–182 10.1038/nature03311 [DOI] [PubMed] [Google Scholar]
- Lin D., Kakizono T., Nishio N., Nagai S. (1990). Enhanced cytochrome formation and stimulated methanogenesis rate by the increased ferrous concentrations in Methanosarcina barkeri culture. FEMS Microbiol. Lett. 68, 89–92 10.1111/j.1574-6968.1990.tb04128.x [DOI] [Google Scholar]
- Lin D. G., Nishio N., Mazumder T. K., Nagai S. (1989). Influence of Co2+, Ni2+ and Fe2+ on the production of tetrapyrroles by Methanosarcina barkeri. Appl. Microbiol. Biotechnol. 30, 196–200 10.1007/BF00264011 [DOI] [Google Scholar]
- Lipscomb J. D. (1994). Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 48, 371–399 10.1146/annurev.mi.48.100194.002103 [DOI] [PubMed] [Google Scholar]
- Lobo A. L., Zinder S. H. (1988). Diazotrophy and nitrogenase activity in the archaebacterium Methanosarcina barkeri 227. Appl. Environ. Microbiol. 54, 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley S. R., Zimmerman S. A., Apolinario E. E., Evilia C., Hou Y.-M., Ferry J. G., Sowers K. R. (2009). The archetype gamma-class carbonic anhydrase (Cam) contains iron when synthesized in vivo. Biochemistry 48, 817–819 10.1021/bi802246s [DOI] [PubMed] [Google Scholar]
- Magalhães C. M., Machado A., Matos P., Bordalo A. A. (2011). Impact of copper on the diversity, abundance and transcription of nitrite and nitrous oxide reductase genes in an urban European estuary. FEMS Microbiol. Ecol. 77, 274–284 10.1111/j.1574-6941.2011.01107.x [DOI] [PubMed] [Google Scholar]
- Martens C. S., Berner R. A. (1977). Interstitial water chemistry of anoxic Long Island Sound sediments. 1. Dissolved gases. Limnol. Oceanogr. 22, 10–25 10.4319/lo.1977.22.1.0010 [DOI] [Google Scholar]
- Martin J. H., Fitzwater S. E. (1988). Iron deficiency limits phytoplankton growth in the north-east Pacific subarctic. Nature 331, 341–343 10.1038/331660a0 [DOI] [Google Scholar]
- Martinho M., Choi D. W., DiSpirito A. A., Antholine W. E., Semrau J. D., Münck E. (2007). Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J. Am. Chem. Soc. 129, 15783–15785 10.1021/ja077682b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T., Frunzke K., Zumft W. (1982). Modulation by copper of the products of nitrite respiration in Pseudomonas perfectomarinus. J. Bacteriol. 149, 816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr S., Latkoczy C., Krüger M., Günther D., Shima S., Thauer R. K., Widdel F., Jaun B. (2008). Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J. Am. Chem. Soc. 130, 10758–10767 10.1021/ja802929z [DOI] [PubMed] [Google Scholar]
- Meiklejohn J. (1953). Iron and the nitrifying bacteria. J. Gen. Microbiol. 8, 58–65 [DOI] [PubMed] [Google Scholar]
- Merkx M., Kopp D. A., Sazinsky M. H., Blazyk J. L., Müller J., Lippard S. J. (2001). Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew. Chem. Int. Ed. Engl. 40, 2782–2807 [DOI] [PubMed] [Google Scholar]
- Meyerdiercks A., Kube M., Lombardot T., Knittel K., Bauer M., Glöckner F. O., Reinhardt R., Amann R. (2005). Insights into the genomes of archaea mediating the anaerobic oxidation of methane. Environ. Microbiol. 7, 1937–1951 10.1111/j.1462-2920.2005.00844.x [DOI] [PubMed] [Google Scholar]
- Moffett J. W., Dupont C. (2007). Cu complexation by organic ligands in the sub-arctic NW Pacific and Bering Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 54, 586–595 10.1016/j.dsr.2006.12.013 [DOI] [Google Scholar]
- Montzka S., Dlugokencky E., Butler J. (2011). Non-CO2 greenhouse gases and climate change. Nature 476, 43–50 10.1038/nature10322 [DOI] [PubMed] [Google Scholar]
- Morse J. W., Luther G. W. (1999). Chemical influences on trace metal-sulfide interactions in anoxic sediments. Geochim. Cosmochim. Acta 63, 3373–3378 10.1016/S0016-7037(99)00258-6 [DOI] [Google Scholar]
- Morton J. D., Hayes K. F., Semrau J. D. (2000). Bioavailability of chelated and soil-adsorbed copper to Methylosinus trichosporium OB3b. Environ. Sci. Technol. 34, 4917–4922 10.1021/es001330m [DOI] [Google Scholar]
- Murray W. D., van den Berg L. (1981). Effects of nickel, cobalt, and molybdenum on performance of methanogenic fixed-film reactors. Appl. Environ. Microbiol. 42, 502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell J. C., McDonald I. R., Gilbert B. (2000). Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8, 221–225 10.1016/S0966-842X(00)01739-X [DOI] [PubMed] [Google Scholar]
- Nielsen A. K., Gerdes K., Murrell J. C. (1997). Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 25, 399–409 10.1046/j.1365-2958.1997.4801846.x [DOI] [PubMed] [Google Scholar]
- Nojiri M., Xie Y., Inoue T., Matsumura H., Kataoka K., Deligeer, Yamaguchi K., Kai Y., Suzuki S. (2007). Structure and function of a hexameric copper-containing nitrite reductase. Proc. Natl. Acad. Sci. U.S.A. 104, 4315–4320 10.1073/pnas.0609195104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp H. J. M., Islam T., Stott M. B., Harhangi H. R., Hynes A., Schouten S., Jetten M. S. M., Birkeland N.-K., Pol A., Dunfield P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- Orphan V. J., House C. H., Hinrichs K.-U., McKeegan K. D., DeLong E. F. (2001). Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487 10.1126/science.1061338 [DOI] [PubMed] [Google Scholar]
- Orphan V. J., House C. H., Hinrichs K.-U., McKeegan K. D., DeLong E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U.S.A. 99, 7663–7668 10.1073/pnas.072210299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G., Khan A., Roth L. (1978). Optimum levels of sulphate and iron for the cultivation of pure cultures of methanogens in synthetic media. J. Appl. Microbiol. 45, 347–356 10.1111/j.1365-2672.1978.tb04235.x [DOI] [PubMed] [Google Scholar]
- Pernthaler A., Dekas A. E., Brown C. T., Goffredi S. K., Embaye T., Orphan V. J. (2008). Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 7052–7057 10.1073/pnas.0711303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R. K., Hughes M. N. (2000). New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775–783 10.1046/j.1365-2958.2000.01889.x [DOI] [PubMed] [Google Scholar]
- Poth M. (1986). Dinitrogen production from nitrite by a Nitrosomonas isolate. Appl. Environ. Microbiol. 52, 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth M., Focht D. D. (1985). 15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49, 1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeburgh W. S. (2007). Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513 10.1021/cr050362v [DOI] [PubMed] [Google Scholar]
- Reeve J. N., Beckler G. S., Cram D. S., Hamilton P. T., Brown J. W., Krzycki J. A., Kolodziej A. F., Alex L., Orme-Johnson W. H., Walsh C. T. (1989). A hydrogenase-linked gene in Methanobacterium thermoautotrophicum strain delta H encodes a polyferredoxin. Proc. Natl. Acad. Sci. U.S.A. 86, 3031–3035 10.1073/pnas.86.9.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Roy R., Knowles R. (2000). Production and consumption of nitric oxide by three methanotrophic bacteria. Appl. Environ. Microbiol. 66, 3891–3897 10.1128/AEM.66.4.1311-1320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie G. A., Nicholas D. J. (1972). Identification of the sources of nitrous oxide produced by oxidative and reductive processes in Nitrosomonas europaea. Biochem. J. 126, 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Hebbeln P., Gelfand M. S., Eitinger T. (2006). Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188, 317–327 10.1128/JB.188.1.317-327.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. E., Whitman W. B. (1991). Microbial Production and Consumption of Greenhouse Gases: Methane, Nitrogen Oxides, and Halomethanes. Washington: American Society for Microbiology [Google Scholar]
- Rubio L. M., Ludden P. W. (2008). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111 10.1146/annurev.micro.62.081307.162737 [DOI] [PubMed] [Google Scholar]
- Saito M. A., Goepfert T. J., Ritt J. T. (2008). Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability. Limnol. Oceanogr. 53, 276–290 10.4319/lo.2008.53.1.0266 [DOI] [Google Scholar]
- Sakadevan K., Zheng H., Bavor H. J. (1999). Impact of heavy metals on denitrification in surface wetland sediments receiving wastewater. Water Sci. Technol. 40, 349–355 10.1016/S0273-1223(99)00471-0 [DOI] [Google Scholar]
- Santoro A. E., Buchwald C., McIlvin M. R., Casciotti K. L. (2011). Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333, 1282–1285 10.1126/science.1208239 [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. (1986). Composition of the coenzyme F420-dependent formate dehydrogenase from Methanobacterium formicicum. J. Bacteriol. 165, 405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller S., Goenrich M., Boecher R., Thauer R. K., Jaun B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608 10.1038/nature09015 [DOI] [PubMed] [Google Scholar]
- Scherer P. (1988). Vanadium and molybdenum requirement for the fixation of molecular nitrogen by two Methanosarcina strains. Arch. Microbiol. 151, 44–48 10.1007/BF00444667 [DOI] [Google Scholar]
- Scherer P., Lippert H., Wolff G. (1983). Composition of the major elements and trace elements of 10 methanogenic bacteria determined by inductively coupled plasma emission spectrometry. Biol. Trace Elem. Res. 5, 149–163 10.1007/BF02916619 [DOI] [PubMed] [Google Scholar]
- Scherer P., Sahm H. (1981). Effect of trace elements and vitamins on the growth of Methanosarcina barkeri. Acta Biotechnol. 1, 57–65 10.1002/abio.370010108 [DOI] [Google Scholar]
- Schlesinger W. H. (2009). On the fate of anthropogenic nitrogen. Proc. Natl. Acad. Sci. U.S.A. 106, 203–208 10.1073/pnas.0810193105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönheit P., Moll J., Thauer R. K. (1979). Nickel, cobalt, and molybdenum requirement for growth of Methanobacterium thermoautotrophicum. Arch. Microbiol. 123, 105–107 10.1007/BF00403508 [DOI] [PubMed] [Google Scholar]
- Scott C., Lyons T. W., Bekker A., Shen Y., Poulton S. W., Chu X., Anbar A. D. (2008). Tracing the stepwise oxygenation of the Proterozoic ocean. Nature 452, 456–459 10.1038/nature06811 [DOI] [PubMed] [Google Scholar]
- Semrau J. D. (2011). Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front. Microbiol. 2:209. 10.3389/fmicb.2011.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau J. D., DiSpirito A. A., Yoon S. (2010). Methanotrophs and copper. FEMS Microbiol. Rev. 34, 496–531 [DOI] [PubMed] [Google Scholar]
- Shaw L. J., Nicol G. W., Smith Z., Fear J., Prosser J. I., Baggs E. M. (2006). Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8, 214–222 10.1111/j.1462-2920.2005.00882.x [DOI] [PubMed] [Google Scholar]
- Shaw T. J., Gieskes J. M., Jahnke R. A. (1990). Early diagenesis in differing depositional environments: the response of transition metals in pore water. Geochim. Cosmochim. Acta 54, 1233–1246 10.1016/0016-7037(90)90149-F [DOI] [Google Scholar]
- Shepard E. M., Boyd E. S., Broderick J. B., Peters J. W. (2011). Biosynthesis of complex iron-sulfur enzymes. Curr. Opin. Chem. Biol. 15, 319–327 10.1016/j.cbpa.2011.02.012 [DOI] [PubMed] [Google Scholar]
- Shima S., Pilak O., Vogt S., Schick M., Stagni M. S., Meyer-Klaucke W., Warkentin E., Thauer R. K., Ermler U. (2008). The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site. Science 321, 572–575 10.1126/science.1158978 [DOI] [PubMed] [Google Scholar]
- Simianu M., Murakami E., Brewer J. M., Ragsdale S. W. (1998). Purification and properties of the heme-and iron-sulfur-containing heterodisulfide reductase from Methanosarcina thermophila. Biochemistry 37, 10027–10039 10.1021/bi9726483 [DOI] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. (1985). Trace metal and vitamin requirements of Methanococcoides methylutens grown with trimethylamine. Arch. Microbiol. 142, 148–151 10.1007/BF00447058 [DOI] [Google Scholar]
- Speece R. E., Parkin G. F., Gallagher D. (1983). Nickel stimulation of anaerobic digestion. Water Res. 17, 677–683 10.1016/0043-1354(83)90237-3 [DOI] [Google Scholar]
- Stanley S. H., Prior S. D., Leak D., Dalton H. (1983). Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol. Lett. 5, 487–492 10.1007/BF00132233 [DOI] [Google Scholar]
- Stein L. Y. (2011). Surveying N2O-producing pathways in bacteria. Meth. Enzymol. 486, 131–152 10.1016/B978-0-12-381294-0.00006-7 [DOI] [PubMed] [Google Scholar]
- Stein L. Y., Arp D. J., Berube P. M., Chain P. S., Hauser L., Jetten M. S., Klotz M. G., Larimer F. W., Norton J. M., Op den Camp H. J. M., Shin M., Wei X. (2007). Whole genome analysis of the ammonia oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ. Microbiol. 9, 2993–3007 10.1111/j.1462-2920.2007.01409.x [DOI] [PubMed] [Google Scholar]
- Sutka R. L., Ostrom N. E., Ostrom P. H., Breznak J. A., Gandhi H., Pitt A. J., Li F. (2006). Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 72, 638–644 10.1128/AEM.72.1.638-644.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpunar J. (2005). Advances in analytical methodology for bioinorganic speciation analysis: metallomics, metalloproteomics and heteroatom- tagged proteomics and metabolomics. Analyst 130, 442–465 10.1039/b418265k [DOI] [PubMed] [Google Scholar]
- Takashima M., Shimada K., Speece R. E. (2011). Minimum requirements for trace metals (iron, nickel, cobalt, and zinc) in thermophilic and mesophilic methane fermentation from glucose. Water Environ. Res. 83, 339–346 10.2175/106143010X12780288628895 [DOI] [PubMed] [Google Scholar]
- Takashima M., Speece R. E. (1990). Mineral requirements for methane fermentation. Crit. Rev. Environ. Control 19, 465–479 10.1080/10643389009388378 [DOI] [Google Scholar]
- Taylor G. T., Pirt S. J. (1977). Nutrition and factors limiting the growth of a methanogenic bacterium (Methanobacterium thermoautotrophicum). Arch. Microbiol. 113, 17–22 10.1007/BF00428574 [DOI] [PubMed] [Google Scholar]
- Terlesky K., Ferry J. (1988). Purification and characterization of a ferredoxin from acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263, 4080–4082 [PubMed] [Google Scholar]
- Tersteegen A., Hedderich R. (1999). Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Eur. J. Biochem. 264, 930–943 10.1046/j.1432-1327.1999.00692.x [DOI] [PubMed] [Google Scholar]
- Thauer R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144, 2377–2406 10.1099/00221287-144-9-2377 [DOI] [PubMed] [Google Scholar]
- Thauer R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299 10.1016/j.mib.2011.03.003 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A.-K., Goenrich M., Schick M., Hiromoto T., Shima S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536 10.1146/annurev.biochem.030508.152103 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A.-K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Shima S. (2008). Methane as fuel for anaerobic microorganisms. Ann. N. Y. Acad. Sci. 1125, 158–170 10.1196/annals.1419.000 [DOI] [PubMed] [Google Scholar]
- Trotsenko Y. A., Murrell J. C. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229 10.1016/S0065-2164(07)00005-6 [DOI] [PubMed] [Google Scholar]
- Twining B. S., Mylon S. E., Benoit G. (2007). Potential role of copper availability in nitrous oxide accumulation in a temperate lake. Limnol. Oceanogr. 52, 1354–1366 10.4319/lo.2007.52.4.1354 [DOI] [Google Scholar]
- Upadhyay A. K., Hooper A. B., Hendrich M. P. (2006). NO reductase activity of the tetraheme cytochrome c554 of Nitrosomonas europaea. J. Am. Chem. Soc. 128, 4330–4337 10.1021/ja055183+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigliotta G., Nutricati E., Carata E., Tredici S. M., De Stefano M., Pontieri P., Massardo D. R., Prati M. V., De Bellis L., Alifano P. (2007). Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic gamma-proteobacterium. Appl. Environ. Microbiol. 73, 3556–3565 10.1128/AEM.02678-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt J. A. (2002). Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178, 239–249 10.1007/s00203-002-0450-2 [DOI] [PubMed] [Google Scholar]
- Vorholt J. A., Vaupel M., Thauer R. K. (1996). A polyferredoxin with eight [4Fe-4S] clusters as a subunit of molybdenum formylmethanofuran dehydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 236, 309–317 10.1111/j.1432-1033.1996.t01-1-00309.x [DOI] [PubMed] [Google Scholar]
- Walker C., De La Torre J., Klotz M., Urakawa H., Pinel N., Arp D., Brochier-Armanet C., Chain P., Chan P., Gollabgir A., Hemp J., Hügler M., Karr E. A., Köke M., Shin M., Lawton T. J., Lowe T., Martens-Habbena W., Sayavedra-Soto L. A., Lang D., Sievert S. M., Rosenzweig A. C., Manning G., Stahl D. A. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U.S.A. 107, 8818–8823 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. B., Tuit C. B., Jayakumar A., Rich J. J., Moffett J., Naqvi S. W. A. (2008). Organic carbon, and not copper, controls denitrification in oxygen minimum zones of the ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 55, 1672–1683 10.1016/j.dsr.2008.07.005 [DOI] [Google Scholar]
- Ward N., Larsen Ø., Sakwa J., Bruseth L., Khouri H., Durkin A. S., Dimitrov G., Jiang L., Scanlan D., Kang K. H., Lewis M., Nelson K. E., Methé B., Wu M., Heidelberg J. F., Paulsen I. T., Fouts D., Ravel J., Tettelin H., Ren Q., Read T., DeBoy R. T., Seshadri R., Salzberg S. L., Jensen H. B., Birkeland N. K., Nelson W. C., Dodson R. J., Grindhaug S. H., Holt I., Eidhammer I., Jonasen I., Vanaken S., Utterback T., Feldblyum T. V., Fraser C. M., Lillehaug J. R., Eisen J. A. (2004). Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2, e303. 10.1371/journal.pbio.0020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Vajrala N., Hauser L., Sayavedra-Soto L. A., Arp D. J. (2006). Iron nutrition and physiological responses to iron stress in Nitrosomonas europaea. Arch. Microbiol. 186, 107–118 10.1007/s00203-006-0126-4 [DOI] [PubMed] [Google Scholar]
- Wetzel R. G. (2001). Limnology: Lake and River Ecosystems, 3rd Edn San Diego: Academic Press [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. (1982). Nutrition and carbon metabolism of Methanococcus voltae. J. Bacteriol. 149, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Bowen T. L., Boone D. R. (2006). The methanogenic bacteria. Prokaryotes 3, 165–207 10.1007/0-387-30743-5_9 [DOI] [Google Scholar]
- Whittaker M., Bergmann D., Arciero D., Hooper A. B. (2000). Electron transfer during the oxidation of ammonia by the chemolithotrophic bacterium Nitrosomonas europaea. Biochim. Biophys. Acta 1459, 346–355 10.1016/S0005-2728(00)00171-7 [DOI] [PubMed] [Google Scholar]
- Wrage N., Velthof G. L., van Beusichem M. L., Oenema O. (2001). Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33, 1723–1732 10.1016/S0038-0717(01)00096-7 [DOI] [Google Scholar]
- Wu M. L., Ettwig K. F., Jetten M. S., Strous M., Keltjens J. T., van Niftrik L. (2011). A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera.’ Biochem. Soc. Trans. 39, 243–248 10.1042/BST0390243 [DOI] [PubMed] [Google Scholar]
- Yoshinari T. (1985). Nitrite and nitrous oxide production by Methylosinus trichosporium. Can. J. Microbiol. 31, 139–144 10.1139/m85-027 [DOI] [PubMed] [Google Scholar]
- Zahn J. A., Bergmann D. J., Boyd J. M., Kunz R. C., DiSpirito A. A. (2001). Membrane-associated quinoprotein formaldehyde dehydrogenase from Methylococcus capsulatus Bath. J. Bacteriol. 183, 6832–6840 10.1128/JB.183.23.6832-6840.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn J. A., DiSpirito A. A. (1996). Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178, 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandvoort M. H., Osuna M. B., Geerts R., Lettinga G., Lens P. N. (2002). Effect of nickel deprivation on methanol degradation in a methanogenic granular sludge bioreactor. J. Ind. Microbiol. Biotechnol. 29, 268–274 10.1038/sj.jim.7000311 [DOI] [PubMed] [Google Scholar]
- Zandvoort M. H., van Hullebusch E. D., Gieteling J., Lens P. N. L. (2006). Granular sludge in full-scale anaerobic bioreactors: trace element content and deficiencies. Enzyme Microb. Technol. 39, 337–346 10.1016/j.enzmictec.2006.03.034 [DOI] [Google Scholar]
- Zellner G., Winter J. (1987). Growth promoting effect of tungsten on methanogens and incorporation of tungsten-185 into cells. FEMS Microbiol. Ecol. 40, 81–87 10.1111/j.1574-6968.1987.tb01987.x [DOI] [Google Scholar]
- Zerkle A. L., House C. H., Brantley S. L. (2005). Biogeochemical signatures through time as inferred from whole microbial genomes. Am. J. Sci. 305, 467–502 10.2475/ajs.305.6-8.467 [DOI] [Google Scholar]
- Zhang Y., Rodionov D. A., Gelfand M. S., Gladyshev V. N. (2009). Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10, 1–26 10.1186/1471-2164-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirngibl C., van Dongen W., Schwörer B., Bünau R., Richter M., Klein A., Thauer R. K. (1992). H2-forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron-sulfur clusters in methanogenic archaea. Eur. J. Biochem. 208, 511–520 10.1111/j.1432-1033.1992.tb17215.x [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (2005). Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J. Inorg. Biochem. 99, 194–215 10.1016/j.jinorgbio.2004.09.024 [DOI] [PubMed] [Google Scholar]