Abstract
In the USA, small animal veterinary hospitals (SAVHs) commonly keep resident cats living permanently as pets within their facilities. Previously, multi-drug resistant (MDR) enterococci were found as a contaminant of multiple surfaces within such veterinary hospitals, and nosocomial infections are a concern. The objectives of this study were to determine whether resident cats carry MDR enterococci and to compare the feline isolates genotypically to those obtained from SAVH surfaces in a previous study. Enterococcal strains (n = 180) were isolated from the feces of six healthy resident cats from different SAVHs. The concentration of enterococci ranged from 1.1 × 105 to 6.0 × 108 CFU g−1 of feces, and the population comprised Enterococcus hirae (38.3 ± 18.6%), E. faecium (35.0 ± 14.3%), E. faecalis (23.9 ± 11.0%), and E. avium (2.8 ± 2.2%). Testing of phenotypic resistance to 14 antimicrobial agents revealed multi-drug resistance (≥3 antimicrobials) in 48.9% of all enterococcal isolates with most frequent resistance to tetracycline (75.0%), erythromycin (50.0%), and rifampicin (36.1%). Vancomycin resistant E. faecalis (3.9%) with vanB not horizontally transferable in in vitro conjugation assays were detected from one cat. Genotyping with pulsed-field gel electrophoresis demonstrated a host-specific clonal population of MDR E. faecalis and E. faecium. Importantly, several feline isolates were genotypically identical or closely related to isolates from surfaces of cage door, thermometer, and stethoscope of the corresponding SAVHs. These data demonstrate that healthy resident cats at SAVHs carry MDR enterococci and likely contribute to contamination of the SAVH environment. Proper disposal and handling of fecal material and restricted movement of resident cats within the ward are recommended.
Keywords: Enterococcus, antimicrobial resistance, resident cats, small animal veterinary hospital, clonal diversity
Introduction
USA households own a large number of dogs (77.5 million) and cats (93.6 million) as pets (APPA, 2009/2010), leading to an average of 2–3 veterinary hospital visits per household per year (AVMA, 2007). Due to high traffic with healthy and sick patients and clients in and out of small animal veterinary hospitals (SAVHs), it is important to maintain good hygiene of these facilities. Previous studies have documented bacterial contamination of surfaces in SAVHs (Aksoy et al., 2010; KuKanich et al., 2012). There are several sources that may contribute to this contamination. Frequent use of antimicrobial agents in veterinary medicine likely results in the selection of resistant bacteria within the gastrointestinal tract of patients that can be spread in the hospital environment by fecal contamination. In addition, in the USA, many SAVHs keep resident pets that freely move around their premises and could act as a carrier/reservoir of resistant bacteria. Other possible sources of contamination include visiting patients, animal owners, hospital personnel as well as lack of efficient infection control measures.
Although enterococci are commensals in the gut microbiota of mammals; in the last decades they become one of the important nosocomial human pathogens (Roberts et al., 2009a). Enterococci can readily acquire genes leading to increased virulence, antimicrobial resistance (Gilmore et al., 2002; Arias and Murray, 2008), and have the ability to survive on inanimate dry hospital surfaces for up to 4 months (Kramer et al., 2006). Therefore, the appropriate use of disinfectants is critical for maintaining proper hygiene standards in hospitals and for reducing nosocomial infections (Hota, 2004). Healthy companion animals have been shown to carry antimicrobial resistant enterococcal lineages associated with human infections and thus may be considered a reservoir shedding these bacteria into the environment (Guardabassi et al., 2004; Damborg et al., 2009; Jackson et al., 2009a). Our recent study has revealed that the dogs leaving the veterinary intensive care unit (ICU) carried a very large population of multi-drug resistant (MDR) enterococci with the capacity for biofilm formation and horizontal transfer of resistance genes. Furthermore, genotyping of canine enterococcal isolates demonstrated that some of them belonged to the human nosocomial clonal complex-17 (CC-17) (Ghosh et al., 2011). In small animals, enterococci have also been implicated in diarrhea, mastitis, urinary tract and catheter-associated infections, and cholangiohepatitis (Helie and Higgins, 1999; Manson et al., 2003; Pressel et al., 2005; Marsh-Ng et al., 2007). With the exception of rare case reports (Boerlin et al., 2001; Benedict et al., 2008), little is known about the nosocomial potential of enterococci in veterinary medicine. Considering the previous study documenting MDR and potentially virulent enterococci in the SAVH environment (KuKanich et al., 2012), further research is needed to better understand bacterial contamination in order to assess and mitigate nosocomial and public health risks.
In the current study, we hypothesized that resident cats in SAVHs acquire and carry antimicrobial resistant enterococci and play a role in dissemination of antimicrobial resistance traits in the hospital environment. The objectives of this study were: (1) to isolate fecal enterococci from six resident cats and to determine their concentration, diversity, clonality, and phenotypic antimicrobial resistance and virulence profiles; (2) to compare the clonality of feline enterococcal isolates to that of isolates from cage doors, thermometers, and stethoscopes in the corresponding hospitals (KuKanich et al., 2012).
Materials and Methods
Sample collection, isolation, and identification of enterococci
Ten privately owned SAVHs (hospitals 1–10) were enrolled in a previous surveillance study that was conducted to evaluate bacterial contamination on surfaces of cage doors, stethoscopes, thermometers, and mouth gags (KuKanich et al., 2012). As a follow-up of the surface study, fecal material was collected from six resident cats (6/10 SAVHs had resident pets) from individual SAVHs. Each cat was considered healthy by its primary veterinarian. Most cats had lived at the hospital premise since they were kittens; four of them had never received any antimicrobial treatment, and the remaining two were not exposed to antimicrobials for at least 1 year prior to our sampling. Detailed information regarding the enrolled cats and their diets is in Table A1 in Appendix. This study was given exempt status from the institutional animal care and use committee because it did not involve any direct contact with animals.
Fresh feces were collected from the litter box and one gram of the feces was re-suspended in 10 ml of phosphate buffered saline (PBS, pH 7.2; MP Biomedicals, Solon, OH, USA). A 100 μl of the suspension was plated in triplicate on mEnterococcus agar (Difco, BD Diagnostic Systems, Sparks, MD, USA), and presumptive enterococcal colonies were selected and confirmed at the genus level as described previously (Macovei and Zurek, 2007). Identification at the species level was carried out for randomly selected 30 isolates per fecal sample using species-specific multiplex PCR (Kariyama et al., 2000) or sequencing of the manganese-dependent superoxide dismutase gene (sodA) (Poyaert et al., 2000). The concentration of enterococci for each sample was calculated in CFU per gram of feces. In order to assess whether the cat diets are the source of MDR enterococci, three out of six commercially available cat diets fed to individual cats were sampled and tested. Two grams of dry food were re-suspended in 20 ml PBS and 100 μl of the suspension was plated on mEnterococcus agar and incubated for 48 h in 37°C.
Antimicrobial susceptibility testing and horizontal gene transfer
The identified isolates were tested for their susceptibility to antimicrobials commonly used in veterinary medicine as well as those targeting human enterococcal infections. Antimicrobial susceptibility was determined by the disk diffusion method on Mueller-Hinton agar (Difco, BD Diagnostic Systems, Sparks, MD, USA) using 10 antimicrobial agents (μg disk−1): ampicillin (10), tetracycline (30), doxycycline (30), gentamicin (120), erythromycin (15), enrofloxacin (5), vancomycin (30), quinupristin/dalfopristin (15), nitrofurantoin (300), and tigecycline (15). Agar dilution technique was carried out to determine resistance to the following antimicrobials (μg ml−1): streptomycin (2,000) on brain heart infusion agar (BBL, BD Diagnostic Systems, Sparks, MD, USA) and chloramphenicol (16), rifampicin (24), and linezolid (8) on Mueller-Hinton agar. Minimum inhibitory concentration (MIC) for vancomycin (8–256 μg ml−1, in double fold increments) and teicoplanin (0.25–8 μg ml−1, in double fold increments) was determined for vancomycin resistant isolates by broth microdilution technique using Mueller-Hinton broth (BBL, BD Diagnostic Systems, Sparks, MD, USA; CLSI, 2008, 2010). Routine quality control of antimicrobial disks was performed using control strains of Enterococcus faecalis ATCC 19433 and E. faecium ATCC 19434. Diameters of zones of inhibition were interpreted in accordance with guidelines of the Clinical and Laboratory Standards Institute for Enterococcus spp. (CLSI, 2008, 2010). The breakpoint for tigecycline is according to manufacturer specifications upon recommendation from the CLSI. Antimicrobial breakpoints for enterococcal species are not all veterinary approved and are therefore not validated in relation to clinical outcome in a veterinary application. Multi-drug resistance was defined as resistance to three or more antimicrobial agents, regardless of class. Linear regression analysis was performed to compare the antimicrobial resistance profiles of each enterococcal species from all animals and animals with no antimicrobial treatment. The antimicrobial resistance profiles of feline isolates were also compared to that of the isolates from hospital surfaces from our previous work (KuKanich et al., 2012) although in that study the isolates were not screened for resistance to tetracycline, streptomycin, linezolid, rifampicin, and chloramphenicol.
PCR amplification of vanA and vanB genes was conducted with primers described previously (Kariyama et al., 2000; Elsayed et al., 2001). Broth and filter conjugation assays were carried out as described by Ike et al. (1998) and Tendolkar et al. (2006), respectively to determine the mobility of the vancomycin resistance trait among E. faecalis strains. The recipients included the following strains with resistance markers: 41–31 (linezolid, MIC = 8 μg ml−1; Qi et al., 2006), OG1SSp (spectinomycin, MIC = 250 μg ml−1; Dunny et al., 1982), and JH2-2 (rifampicin, MIC = 30 μg ml−1; Oancea et al., 2004). Both assays were performed with a donor and recipient ratio of 1:10. The transconjugants were detected on agar supplemented with vancomycin (16 μg ml−1) and the respective antimicrobial markers of the recipient strains with the same antimicrobial concentrations as described above.
Screening for virulence traits by phenotype
Identified isolates were tested for the gelatinase on Todd Hewitt Agar (Difco, BD Diagnostic Systems, Sparks, MD, USA) with 1.5% skim milk, and for the cytolysin (β-hemolysis) on Columbia blood agar base (Difco, BD Diagnostic Systems, Sparks, MD, USA) supplemented with 5.0% human blood (Rockland Immunochemicals, Inc., Gilbertsville, PA, USA) as described previously (Macovei and Zurek, 2006).
Genotyping by pulsed-field gel electrophoresis
The feline enterococcal isolates (E. faecalis, n = 15; E. faecium, n = 18) for pulsed-field gel electrophoresis (PFGE) were selected based on their different antibiotic resistance profile. These were genotyped to assess their clonality within and between cats. Also, in this study, nine E. faecalis isolates from various hospital surfaces were genotyped and compared with the feline isolates while the feline E. faecium isolates were compared to the 42 E. faecium hospital isolates typed in our previous study (KuKanich et al., 2012). PFGE was carried out following the protocols of Murray et al. (1990) and Turabelidze et al. (2000). Briefly, agarose plugs were restriction digested with 20U of SmaI (New England Biolab, Ipswich, MA, USA) for 4 h at 25°C. Digested plugs were run on to a 1.0% SeaKem Gold Agarose (Lonza, Rockland, ME, USA) gel using CHEF Mapper (Bio-Rad Laboratories, Hercules, CA, USA) with initial pulse time for 1 s and final time for 20 s at 200 V for 21 h. Cluster analysis was performed with BioNumerics (Applied Maths, Inc., Austin, TX, USA) by using the band-based Dice correlation coefficient and the unweighted pair group mathematical average algorithm (UPGMA) with position tolerance setting of 1.0% for both optimization and band comparison. The cut off 95% was used to classify the strains as clonal.
Results
Enterococcal concentration and species diversity
The mean enterococcal concentration was 1.1 ± 0.7 × 106 CFU g−1 of feces in five cats, with one outlying cat that had 6.0 × 108 CFU g−1 (Table 1). There was great variation in enterococcal composition at the species level among individual cats (Table 1). Altogether, of the 180 isolates, E. hirae (69/180, 38.3 ± 18.6%) was the most common, followed by E. faecium (63/180, 35.0 ± 14.3%), E. faecalis (43/180, 23.9 ± 11.0%), and E. avium (5/180, 2.8 ± 2.2%). No enterococcal colonies were detected in the cat diets.
Table 1.
Enterococcal concentration and species diversity in the feces of resident cats from small animal veterinary hospitals.
| Sample ID | Hospital ID† | Concentration (CFU g−1) | Diversity (%)* |
|||
|---|---|---|---|---|---|---|
| E. faecalis | E. faecium | E. hirae | E. avium | |||
| Cat-A | 1 | 6.0 × 108 | 50.0 | 36.7 | 0 | 13.3 |
| Cat-B | 2 | 9.1 × 105 | 0 | 100 | 0 | 0 |
| Cat-C | 4 | 2.1 × 105 | 0 | 10.0 | 90.0 | 0 |
| Cat-D | 6 | 1.7 × 105 | 36.7 | 30.0 | 33.3 | 0 |
| Cat-E | 7 | 4.1 × 106 | 56.7 | 33.3 | 6.7 | 3.3 |
| Cat-F | 9 | 1.1 × 105 | 0 | 0 | 100 | 0 |
†Hospital IDs consistent with previous surveillance study (KuKanich et al., 2012).
*Based on 30 isolates from each fecal sample.
Antimicrobial resistance and virulence
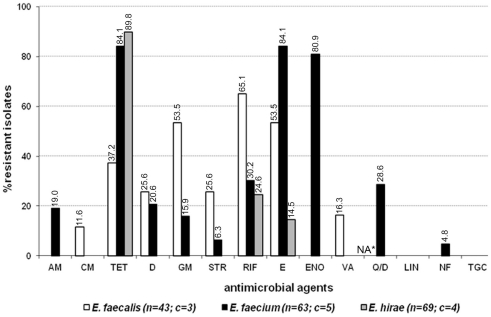
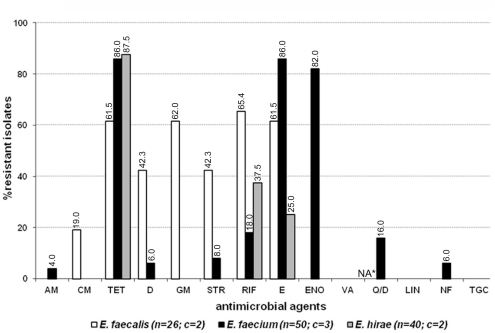
Eighty-eight out of 180 enterococcal isolates (48.9%) from feces of the enrolled healthy cats showed multi-drug resistance (3–8 antimicrobials; Table 2). Overall, E. faecium exhibited high incidence of resistance to tetracycline, erythromycin, and enrofloxacin while resistance to rifampicin, quinupristin/dalfopristin, doxycycline, ampicillin, gentamicin, streptomycin, and nitrofurantoin was less frequent (Figure 1). E. faecalis was frequently resistant to a total of six different antimicrobials, namely rifampicin, erythromycin, gentamicin, tetracycline, doxycycline, and streptomycin whereas resistance to vancomycin, and chloramphenicol was less common (Figure 1). A considerable number of E. hirae were resistant to tetracycline, rifampicin, and erythromycin (Figure 1). Five isolates of E. avium showed resistance to rifampicin, erythromycin, and/or tetracycline (Table A2 in Appendix). No resistance was detected to the newer generation drugs, linezolid and tigecycline, in any enterococcal isolates. The antimicrobial resistance profile for cats that had never been administered antimicrobial agents is depicted in Figure A1 in Appendix. Based on statistical analysis, antimicrobial resistance profiles of enterococcal species from all cats and cats with no antimicrobial treatment were highly similar [R = 0.9458 (E. faecalis), R = 0.9764 (E. faecium), and R = 0.9844 (E. hirae)].
Table 2.
Multiple (≥3) antimicrobial resistance profile among enterococcal isolates from the feces of resident cats from small animal veterinary hospitals.
| Resistance profile | Number of strains (%)* |
Origin | ||
|---|---|---|---|---|
| E. faecalis (n = 43) | E. faecium (n = 63) | E. hirae (n = 69) | ||
| TET, E, ENO | 24 (38.1) | Cat-B | ||
| TET, E, Q/D | 5 (7.9) | Cat-A | ||
| TET, E, RIF | 10 (14.5) | Cat-D | ||
| TET, E, ENO, RIF | 2 (3.2) | Cat-D | ||
| E, ENO, RIF | 7 (11.1) | Cat-D | ||
| E, GM, VA | 7 (16.3) | Cat-E | ||
| TET, E, GM, CM | 3 (6.9) | Cat-A | ||
| TET, E, Q/D, NF | 1 (1.6) | Cat-A | ||
| TET, E, RIF, GM, CM | 2 (4.6) | Cat-A | ||
| TET, E, ENO, RIF, STR | 1 (1.6) | Cat-D | ||
| TET, E, ENO, STR, AM | 1 (1.6) | Cat-A | ||
| TET, E, Q/D, STR, D | 2 (3.2) | Cat-A | ||
| TET, E, Q/D, D, NF | 1 (1.6) | Cat-A | ||
| TET, E, RIF, GM, STR, D | 11 (25.6) | Cat-D | ||
| TET, E, ENO, STR, AM, NF | 1 (1.6) | Cat-A | ||
| TET, E, ENO, Q/D, RIF, GM, AM, D | 10 (15.9) | Cat-E | ||
AM, ampicillin; CM, chloramphenicol; TET, tetracycline; D, doxycycline; GM, gentamicin; STR, streptomycin; RIF, rifampicin; E, erythromycin; ENO, enrofloxacin; VA, vancomycin; Q/D, quinupristin/dalfopristin; NF, nitrofurantoin.
*E. avium are not included as they were not multi-drug resistant.
Figure 1.
Antimicrobial resistance profile of enterococci from the feces of six resident cats from small animal veterinary hospitals. AM, ampicillin; CM, chloramphenicol; TET, tetracycline; D, doxycycline; GM, gentamicin; STR, streptomycin; RIF, rifampicin; E, erythromycin; ENO, enrofloxacin; VA, vancomycin; Q/D, quinupristin/dalfopristin; LIN, linezolid; NF, nitrofurantoin; TGC, tigecycline; c, number of cats that contributed for the total number (n) of isolates. *Not applicable for E. faecalis isolates due to their intrinsic resistance.
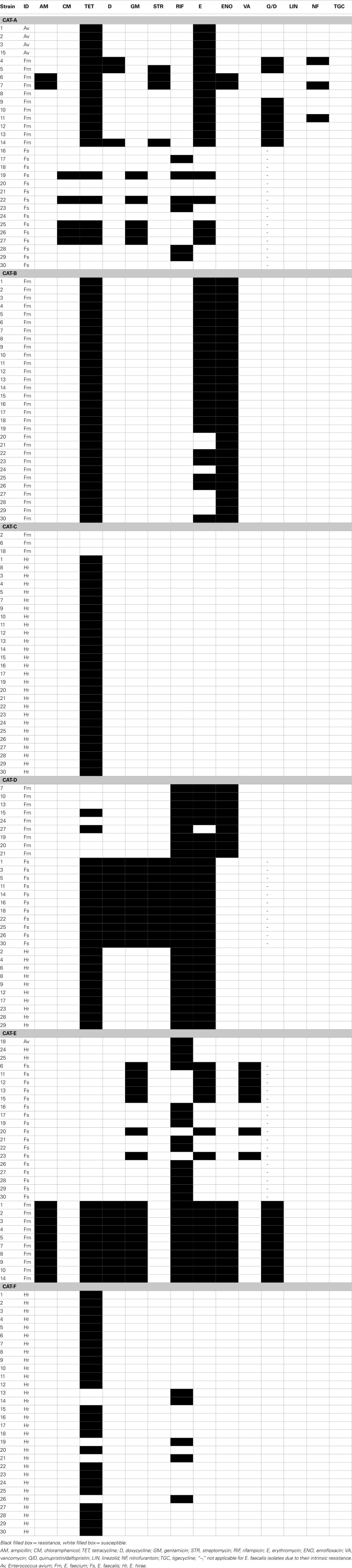
Isolates from feces of individual cats provided a unique profile of antimicrobial resistance. In cat-A, 8 out of 11 E. faecium were resistant to quinupristin/dalfopristin, and several were resistant to high level of streptomycin (4/11), doxycycline (3/11), and nitrofurantoin (3/11; Table A2 in Appendix). E. faecalis strains (5/15) resistant to chloramphenicol and high level of gentamicin were also detected in the feces of this cat. In cat-D, the enterococcal population in the gut was almost equally shared by MDR strains of E. faecalis (11/30), E. hirae (10/30), and E. faecium (9/30). Among these, all E. faecalis were phenotypically resistant to six antimicrobials (tetracycline, doxycycline, erythromycin, rifampicin, gentamicin, and streptomycin) whereas one E. faecium strain was resistant to tetracycline, erythromycin, enrofloxacin, rifampicin, and streptomycin (Table 2; Table A2 in Appendix). Cat-E was colonized with MDR E. faecalis (7/30) and E. faecium (10/30), and all E. faecalis were vancomycin resistant with an MIC of 16–32 μg ml−1, but susceptible to teicoplanin (MIC ≤ 1 μg ml−1). These vancomycin resistant strains harbored the vanB gene but not vanA. Standard in vitro conjugation assays failed to transfer vanB from the feline isolates to several E. faecalis recipient strains. Besides vancomycin, these E. faecalis were also resistant to high level of gentamicin and erythromycin. In addition, all E. faecium from cat-E displayed resistance to 8 antimicrobials (tetracycline, doxycycline, erythromycin, enrofloxacin, quinupristin/dalfopristin, rifampicin, gentamicin, and ampicillin; Table 2, Table A2 in Appendix). Cat-B was colonized with E. faecium (24/30) resistant to enrofloxacin, erythromycin, and tetracycline. E. hirae from cat-C were resistant to tetracycline (27/30) while those from cat-F were resistant to tetracycline (25/30) and rifampicin (5/30) (Table A2 in Appendix).
All feline isolates were screened for two enterococcal virulence factors, gelatinase and cytolysin. The majority of E. faecalis (38/43, 88.4%) showed strong gelatinase phenotype, and several (5/43, 11.6%) were β-hemolytic on human blood agar. E. faecium (58/63, 92.1%) and E. hirae (66/69, 95.6%) showed weak gelatinase phenotype; the remainder were negative for gelatinase activity and none were β-hemolytic.
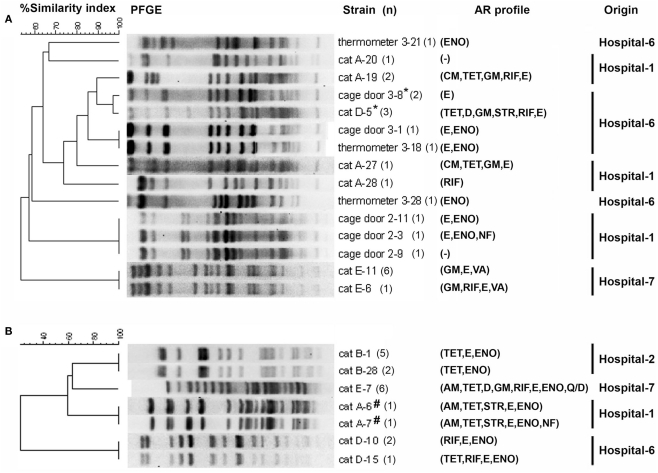
Genotyping of feline enterococcal isolates
Pulsed-field gel electrophoresis analysis revealed that enterococcal isolates from individual cats were host-specific clonal groups (unique pulsotypes in individual cats; Figures 2A,B). The feline enterococcal isolates were also compared to the surface isolates from the corresponding hospitals (Figure 2A; KuKanich et al., 2012). E. faecalis from a cage door of Hospital-6 showed close genotypic similarity (96.9%) to several fecal isolates from the cat (cat-D) residing at the facility (Figure 2A). Furthermore, two E. faecium strain from feces of cat-A were clonally indistinguishable or closely related (96.9%) to isolates from the surfaces (a cage door, a stethoscope, and a thermometer) of Hospital-1 (KuKanich et al., 2012).
Figure 2.
Dendrograms based on SmaI restriction pattern resolved by pulsed-field gel electrophoresis (PFGE) depicting the relationships among representative isolates of (A) E. faecalis (feline, n = 15; surface, n = 9) and (B) E. faecium (feline, n = 18) in small animal veterinary hospitals. Strain numbers, number of strains typed (n), antimicrobial resistance (AR) profiles, and corresponding hospitals are included along each PFGE lane. “*” Indicates 96.9% similarity among E. faecalis isolated from the hospital surface and cat feces. “#” Indicates 100% similarity to E. faecium isolated from surface of a cage door and a stethoscope, and 96.9% similarity to E. faecium isolated from surface of a thermometer of the corresponding hospital (KuKanich et al., 2012); AM, ampicillin; CM, chloramphenicol; TET, tetracycline; D, doxycycline; GM, gentamicin; STR, streptomycin; RIF, rifampicin; E, erythromycin; ENO, enrofloxacin; VA, vancomycin; Q/D, quinupristin/dalfopristin; NF, nitrofurantoin. Hospital surface isolates were not screened for resistance to tetracycline, chloramphenicol, linezolid, streptomycin, and rifampicin.
Discussion
With large numbers of pet animals in the USA, maintaining a healthy environment within SAVHs is highly desirable. Recent studies have reported surface contamination by MDR enterococci and other bacteria in SAVH environment (Aksoy et al., 2010; KuKanich et al., 2012). Our study was designed to assess healthy resident cats in SAVHs as carriers/reservoirs of MDR enterococci.
The mean value of enterococcal population size in resident cats was comparable to that reported from the feces of healthy pets (103–106 CFU g−1 of feces) (Damborg et al., 2009). Though the enterococcal species composition was diverse among individual cats, the overall diversity at the species level indicated a healthy feline gut microbiota and was in congruence with other studies on enterococcal population in healthy cats (Devriese et al., 1992; de Leener et al., 2005). Previously, Poeta et al. (2006) reported an intermediate prevalence of resistance in enterococci with few MDR strains isolated from healthy cats in Portugal. de Leener et al. (2005) and Moyaert et al. (2006) screened cats from diverse populations in Belgium and demonstrated that enterococcal strains in cats from hospitals and catteries had relatively higher prevalence of resistance to several antimicrobial agents (erythromycin, tylosin, lincomycin, tetracycline, chloramphenicol, and kanamycin) compared to that of healthy domestic cats. In the USA, Jackson et al. (2009a) reported antimicrobial resistant E. faecium and E. faecalis from a total of 116 healthy cats visiting veterinary clinics. They detected MDR E. faecalis and E. faecium (3–8 antimicrobials) but no vancomycin or quinupristin/dalfopristin resistant strains. Our results showed that four out of six healthy cats harbored a MDR enterococcal population with E. faecium occurring most frequently. To our knowledge, our study reported for the first time MDR (tetracycline, erythromycin, and rifampicin) E. hirae from a healthy companion animal. All of our enterococcal isolates were susceptible to the relatively new antimicrobial agents linezolid and tigecycline, and this is in agreement with the study of Jackson et al. (2009a). Importantly, the overall antimicrobial resistance profile of the isolates from our healthy resident cats was similar to that of cattery and hospitalized feline isolates rather than the isolates from healthy domestic cats.
Due to limited number of animals and bacterial isolates, we could not assess a correlation between the antimicrobials administered to resident cats and antimicrobial resistance profiles of the feline enterococcal isolates. Clearly, three out of four cats that have never been administered antimicrobials carried a MDR enterococcal population. Vice versa, feces of one of the two cats that received an antimicrobial treatment had the enterococcal community susceptible to all antibiotics tested with the exception of tetracycline. While enterococci are not commonly associated with infections in small animals, due to their ability to horizontally transfer resistance traits to other bacteria including Staphylococcus aureus (Noble et al., 1992; Clewell and Dunny, 2002; Weigel et al., 2003), the enterococcal resistance reported in this study is important especially that to doxycycline, enrofloxacin, and ampicillin. This is because tetracyclines, fluoroquinolones, and β-lactams are very commonly used in small animal veterinary medicine to treat various bacterial infections (Prescott et al., 2002). Due to close physical contact between pets and people, the likelihood of transfer of bacteria is very high and it has been demonstrated for different bacterial taxa including enterococci and staphylococci (Simjee et al., 2002; Guardabassi et al., 2004; Bramble et al., 2011; Chomel and Sun, 2011). Once established in the digestive tract, MDR enterococci can persist for months to years (Byers et al., 2002; Sørum et al., 2006). Detection of chloramphenicol resistant E. faecalis and quinupristin/dalfopristin resistant E. faecium requires attention since these antimicrobials are used to treat human infections caused by vancomycin resistant enterococci (VRE) (Lautenbach et al., 1998; Linden et al., 2001; Arias and Murray, 2008). We also report vancomycin resistant E. faecalis with the vanB gene not transferable in vitro; however, transfer of this trait in natural niches cannot be ruled out. van Belkum et al. (1996) reported that 4/24 (16%) of the rectal swabs from cats that attended a veterinary practice in the Netherlands were positive for vancomycin resistant E. faecium with vanA. Another study reported association of vancomycin resistant E. faecium in a cat with cholangiohepatitis (Pressel et al., 2005). To our knowledge, isolation of vancomycin resistant E. faecalis from a healthy cat in our study is the first of its kind. Interestingly, VRE were not detected in our previous surveillance study (KuKanich et al., 2012) and this antibiotic is also typically not used to treat animals at SAVHs (Weese, 2008). Although the source of VRE in this cat is unknown, it is likely of a human (pet owner, hospital staff) origin since VRE strains are rare in the USA outside of the human clinical environment (Sapkota et al., 2007; Roberts et al., 2009b). The VRE transfer from personnel in human hospitals to patients as well as between pet owners and pets has been suggested in several studies (Hayden, 2000; Guardabassi et al., 2004; Tacconelli and Cataldo, 2008) and it is therefore also possible in SAVHs. Resistance to ampicillin and high level of gentamicin in E. faecium is also a concern, as gentamicin in combination with β-lactams or glycopeptides is typically used to treat human enterococcal infections (Arias and Murray, 2008).
Gelatinase and cytolysin are among the virulence traits implicated in enhancement of enterococcal infections (Gilmore et al., 2002). Gulhan et al. (2006) demonstrated that prevalence of gelatinase and cytolytic activity among E. faecalis and E. faecium were negligible in healthy cats. In contrast, the presence of gelatinolytic as well as several β-hemolytic strains of E. faecalis isolated from the healthy resident cats in our study indicates these isolates have increased virulence potentially resulting in biofilm formation (Ghosh et al., 2011), proliferation in bloodstream, and direct tissue damage (Gilmore et al., 2002). Therefore, our data demonstrate that resident cats in SAVHs are carriers and possible reservoirs of antimicrobial resistant and potentially virulent enterococcal strains. Since fecal samples of these cats were not tested prior to their adoption as resident cats (as kittens), we cannot entirely exclude a rather remote possibility that the cats in this study acquired the enterococci from an outside source before their stay in SAVHs.
Sharing of methicillin-resistant S. aureus (MRSA) clones among hospital staff, surfaces, patients, and resident animals had been documented in a veterinary hospital (Loeffler et al., 2005) as well as in a human geriatric ward (Scott et al., 1988) indicating cross-contamination within the hospital environment. In our study, clonal matches based on PFGE clearly demonstrate cross-contamination between the resident cats and the hospital environment although the directionality of the bacterial transfer requires further investigation. The PFGE dendrograms also revealed that feline enterococcal isolates in our study were host-specific (unique to individual cats) and not diverse although genotyping of more isolates is warranted to assess the diversity to a greater extent. These findings differ from the study by Jackson et al. (2009b) where they found that healthy domestic cats harbored MDR enterococcal strains of diverse clonal origin. We hypothesize that the antimicrobial use in the SAVHs leads to the selection of specific MDR clonal lineages in the patients that contaminate the hospital environment. Consequently, this contamination becomes the source of MDR enterococci for the resident cats that then become carriers/reservoirs of these bacteria and possibly re-contaminate the hospital environment. The cat diet is an unlikely source of the antimicrobial resistant enterococci as all resident cats enrolled in this study were fed commercial cat food (Table A1 in Appendix) and our random screening of these diets failed to detect any enterococcal contamination. Other possible sources of antimicrobial resistant enterococci such as the hospital staff or pet owners cannot be ruled out.
In conclusion, we demonstrate that resident cats in SAVHs in the USA can carry and shed MDR enterococci and this may create risk to people and animals within veterinary hospitals. Further research is warranted to determine direction of enterococcal transmission between resident cats and environmental surfaces of the clinics, and also to evaluate the duration (transient or long-lasting) of such carriage. Nonetheless, proper disposal and handling of fecal material of resident cats, good overall hygiene, effective infection control, and restricted movement of these animals within the ward are recommended.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding and support provided by NIH T35 RR007064, Department of Clinical Sciences Research Grant, College of Veterinary Medicine, Kansas State University, and the Veterinary Research Scholars Program. We thank Michael Haden for technical help. This is contribution no. 12-270-J of the Kansas Agricultural Experiment Station.
Appendix
Table A1.
Information on resident cats from the small animal veterinary hospitals.
| Sample ID | Age (year)/sex/breed | Stay at clinic (year) | Antibiotic(s) administration (year ago) | Diet |
|---|---|---|---|---|
| Cat-A | 11/MC/DSH | 10 | Never | HSD c/d multicat dry |
| Cat-B | 2/MC/DSH | 2 | Never | HSD sensitive stomach maintenance dry |
| Cat-C | 10/FS/DSH | 10 | Penicillin and enrofloxacin (5), amoxicillin–clavulanic acid (4), erythromycin ophthalmic drops (1) | HSD w/d dry |
| Cat-D | 3/FS/DSH | 3 | Never | Felidae maintenance dry |
| Cat-E | 8/FS/DLH | 8 | Amoxicillin (3) | HSD maintenance dry |
| Cat-F | 1.3/MC/DSH | 1.3 | Never | HSD hairball light maintenance dry |
FS, female spayed; MC, male castrated; DSH, domestic short haired; DLH, domestic long haired; HSD, Hill’s science diet.
Table A2.
Antimicrobial resistance profile of enterococcal isolates from the feces of individual resident cats from small animal veterinary hospitals.
Black filled box = resistance, white filled box = susceptible.
AM, ampicillin; CM, chloramphenicol; TET, tetracycline; D, doxycycline; GM, gentamicin; STR, streptomycin; RIF, rifampicin; E, erythromycin; ENO, enrofloxacin; VA, vancomycin; Q/D, quinupristin/dalfopristin; LIN, linezolid; NF, nitrofurantoin; TGC, tigecycline; “–,” not applicable for E. faecalis isolates due to their intrinsic resistance. Av, Enterococcus avium; Fm, E. faecium; Fs, E. faecalis; Hr, E. hirae.
Figure A1.
Antimicrobial resistance profile of enterococcal isolates from the feces of four resident cats from small animal veterinary hospitals that had never been administered any antimicrobial agents. AM, ampicillin; CM, chloramphenicol; TET, tetracycline; D, doxycycline; GM, gentamicin; STR, streptomycin; RIF, rifampicin; E, erythromycin; ENO, enrofloxacin; VA, vancomycin; Q/D, quinupristin/dalfopristin; LIN, linezolid; NF, nitrofurantoin; TGC, tigecycline; c, number of cats that contributed for the total number (n) of isolates. *Not applicable for E. faecalis isolates due to their intrinsic resistance.
References
- Aksoy E., Boag A., Brodbelt D., Grierson J. (2010). Evaluation of surface contamination with staphylococci in a veterinary hospital using a quantitative microbiological method. J. Small Anim. Pract. 51, 574–580 10.1111/j.1748-5827.2010.00994.x [DOI] [PubMed] [Google Scholar]
- APPA (2009/2010). The 2009/2010 APPA National Pet Owners Survey, ed. American Pet Products Association Greenwich: APPA Press [Google Scholar]
- Arias C. A., Murray B. E. (2008). Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 6, 637–655 10.1586/14787210.6.5.637 [DOI] [PubMed] [Google Scholar]
- AVMA. (2007). U.S. Pet Ownership and Demographics Sourcebook, 2nd Edn ed. American Veterinary Medical Association Schaumburg: AVMA Press [Google Scholar]
- Benedict K. M., Morley P. S., van Metre D. C. (2008). Characteristics of biosecurity and infection control programs at veterinary teaching hospitals. J. Am. Vet. Med. Assoc. 233, 767–773 10.2460/javma.233.5.767 [DOI] [PubMed] [Google Scholar]
- Boerlin P., Eugster S., Gaschen F., Straub R., Schawalder P. (2001). Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet. Microbiol. 82, 347–359 10.1016/S0378-1135(01)00396-0 [DOI] [PubMed] [Google Scholar]
- Bramble M., Morris D., Tolomeo P., Lautenbach E. (2011). Potential role of pet animals in household transmission of methicillin-resistant Staphylococcus aureus: a narrative review. Vector Borne Zoonotic Dis. 2010.0025. 10.1089/vbz.2010.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers K. E., Anglim A. M., Anneski C. J., Farr B. M. (2002). Duration of colonization with vancomycin-resistant Enterococcus. Infect. Cont. Hosp. Epidemiol. 23, 207–211 10.1086/502036 [DOI] [PubMed] [Google Scholar]
- Chomel B. B., Sun B. (2011). Zoonoses in the bedroom. Emerg. Infect. Dis. 17, 167–172 10.3201/eid1707.110590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Dunny G. M. (2002). “Conjugation and genetic exchange in enterococci” in The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance, eds Gilmore M. S., Clewell D. B., Courvalin P., Dunny G. M., Murray B. E., Rice L. B. (Washington, DC: ASM Press; ), 265–300 [Google Scholar]
- CLSI (2008). “Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals,” in Approved Standard CLSI Document M31-A3, Vol. 28, 3rd Edn ed. Clinical and Laboratory Standards Institute (Wayne: CLSI Press; ), 1–99 [Google Scholar]
- CLSI. (2010). “Performance standards for antimicrobial susceptibility testing; twentieth informational supplement,” in CLSI Document M100-S20, Vol. 30 ed. Clinical and Laboratory Standards Institute (Wayne: CLSI Press; ), 76–79 [Google Scholar]
- Damborg P., Top J., Hendrickx A. P. A., Dawson S., Willems R. J. L., Guardabassi L. (2009). Dogs are a reservoir of ampicillin-resistant E. faecium lineages associated with human infections. Appl. Environ. Microbiol. 75, 2360–2365 10.1128/AEM.02035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leener E., Decostere A., de Graef E. M., Moyaert H., Haesebrouck F. (2005). Presence and mechanism of antimicrobial resistance among enterococci from cats and dogs. Microb. Drug Resist. 11, 395–403 10.1089/mdr.2005.11.395 [DOI] [PubMed] [Google Scholar]
- Devriese L. A., Colque J. I. C., de Herdt P., Haesebrouck F. (1992). Identification and composition of the tonsillar and anal enterococcal and streptococcal flora of dogs and cats. J. Appl. Microbiol. 73, 421–425 10.1111/j.1365-2672.1992.tb04998.x [DOI] [PubMed] [Google Scholar]
- Dunny G., Yuhasz M., Ehrenfeld E. (1982). Genetic and physiological analysis of conjugation in Streptococcus faecalis. J. Bacteriol. 151, 855–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed S., Hamilton N., Boyd D., Mulvey M. (2001). Improved primer design for multiplex PCR analysis of vancomycin-resistant Enterococcus spp. J. Clin. Microbiol. 39, 2367–2368 10.1128/JCM.39.6.2367-2368.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Dowd S. E., Zurek L. (2011). Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS ONE 6, e22451. 10.1371/journal.pone.0022451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore M. S., Coburn P. S., Nallapareddy S. R., Murray B. E. (2002). “Enterococcal virulence” in The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance, eds Gilmore M. S., Clewell D. B., Courvalin P., Dunny G. M., Murray B. E., Rice L. B. (Washington, DC: ASM Press; ), 301–354 [Google Scholar]
- Guardabassi L., Schwarz S., Lloyd D. H. (2004). Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 54, 321–332 10.1093/jac/dkh332 [DOI] [PubMed] [Google Scholar]
- Gulhan I., Aksakal A., Ekunc U. H. (2006). Virulence factors of Enterococcus faecium and Enterococcus faecalis strains isolated from humans and pets. Turk. J. Vet. Anim. Sci. 30, 477–482 [Google Scholar]
- Hayden M. K. (2000). Insights into the epidemiology and control of infection with vancomycin-resistant enterococci. Clin. Infect. Dis. 31, 1058–1065 10.1086/318126 [DOI] [PubMed] [Google Scholar]
- Helie P., Higgins R. (1999). Diarrhea associated with Enterococcus faecium in an adult cat. J. Vet. Diagn. Invest. 11, 457–458 10.1177/104063879901100511 [DOI] [PubMed] [Google Scholar]
- Hota B. (2004). Contamination, disinfection, and cross-colonization: are hospital surfaces reservoirs for nosocomial infection? Clin. Infect. Dis. 39, 1182–1189 10.1086/424667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Tanimoto K., Tomita H., Takeuchi K., Fujimoto S. (1998). Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J. Bacteriol. 180, 4886–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. R., Fedorka-Cray P. J., Davis J. A., Barrett J. B., Frye J. G. (2009a). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 107, 1269–1278 10.1111/j.1365-2672.2009.04310.x [DOI] [PubMed] [Google Scholar]
- Jackson C. R., Fedorka-Cray P. J., Davis J. A., Barrett J. B., Brousse J. H., Gustafson J., Kucher M. (2009b). Mechanisms of antimicrobial resistance and genetic relatedness among enterococci isolated from dogs and cats in the United States. J. Appl. Microbiol. 108, 2171–2179 [DOI] [PubMed] [Google Scholar]
- Kariyama R., Mitsuhata R., Chow J. W., Clewell D. B., Kumon H. (2000). Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38, 3092–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A., Schwebke I., Kampf G. (2006). How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6, 130. 10.1186/1471-2334-6-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KuKanich K., Ghosh A., Skarbek J., Lothamer K., Zurek L. (2012). Surveillance of bacterial contamination in small animal veterinary hospitals with special focus on antimicrobial resistance and virulence traits of enterococci. J. Am. Vet. Med. Assoc. 240, 437–445 10.2460/javma.240.4.437 [DOI] [PubMed] [Google Scholar]
- Lautenbach E., Schuster M. G., Bilker W. B., Brennan P. J. (1998). The role of chloramphenicol in the treatment of bloodstream infection due to vancomycin-resistant Enterococcus. Clin. Infect. Dis. 27, 1259–1265 10.1086/515002 [DOI] [PubMed] [Google Scholar]
- Linden P. K., Moellering R. C., Jr., Wood C. A., Rehm S. J., Flaherty J., Bompart F., Talbot G. H. (2001). Treatment of vancomycin-resistant Enterococcus faecium infections with quinupristin/dalfopristin. Clin. Infect. Dis. 33, 1816–1823 10.1086/323899 [DOI] [PubMed] [Google Scholar]
- Loeffler A., Boag A. K., Sung J., Lindsay J. A., Guardabassi L., Dalsgaard A., Smith H., Stevens K. B., Lloyd D. H. (2005). Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 56, 692–697 10.1093/jac/dki312 [DOI] [PubMed] [Google Scholar]
- Macovei L., Zurek L. (2006). Ecology of antibiotic resistance genes: characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 72, 4028–4035 10.1128/AEM.00034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei L., Zurek L. (2007). Influx of enterococci and associated antibiotic resistance and virulence genes from ready-to-eat food to the human digestive tract. Appl. Environ. Microbiol. 73, 6740–6747 10.1128/AEM.01444-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson J. M., Keis S., Smith J. M. B., Cook G. M. (2003). Characterization of a vancomycin-resistant Enterococcus faecalis (VREF) isolate from a dog with mastitis: further evidence of a clonal lineage of VREF in New Zealand. J. Clin. Microbiol. 41, 3331–3333 10.1128/JCM.41.7.3331-3333.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh-Ng M. L., Burney D. P., Garcia J. (2007). Surveillance of infections associated with intravenous catheters in dogs and cats in an intensive care unit. J. Am. Anim. Hosp. Assoc. 43, 13–20 [DOI] [PubMed] [Google Scholar]
- Moyaert H., de Graef E. M., Haesebrouck F., Decostere A. (2006). Acquired antimicrobial resistance in the intestinal microbiota of diverse cat populations. Res. Vet. Sci. 81, 1–7 10.1016/j.rvsc.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Murray B. E., Singh K. V., Heath J. D., Sharma B. R., Weinstock G. M. (1990). Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28, 2059–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W. C., Virani Z., Cree R. G. (1992). Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72, 195–198 10.1111/j.1574-6968.1992.tb05089.x [DOI] [PubMed] [Google Scholar]
- Oancea C., Klare I., Witte W., Werner G. (2004). Conjugative transfer of the virulence gene, esp, among isolates of Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 54, 232–235 10.1093/jac/dkh249 [DOI] [PubMed] [Google Scholar]
- Poeta P., Costa D., Rodrigues J., Torres C. (2006). Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. Int. J. Antimicrob. Agents 27, 131–137 10.1016/j.ijantimicag.2005.09.018 [DOI] [PubMed] [Google Scholar]
- Poyaert C., Quesnes G., Trieu-Cuot P. (2000). Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38, 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J. F., Hanna W. J. B., Reld-Smith R., Drost K. (2002). Antimicrobial drug use and resistance in dogs. Can. Vet. J. 43, 107–116 [PMC free article] [PubMed] [Google Scholar]
- Pressel M. A., Fox L. E., Apley M. D., Simutis F. J. (2005). Vancomycin for multi-drug resistant Enterococcus faecium cholangiohepatitis in a cat. J. Feline Med. Surg. 7, 317–321 10.1016/j.jfms.2005.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C., Zheng X., Obias A., Scheetz M. H., Malczynski M., Warren J. R. (2006). Comparison of testing methods for detection of decreased linezolid susceptibility due to G2576T mutation of the 23S rRNA gene in Enterococcus faecium and Enterococcus faecalis. J. Clin. Microbiol. 44, 1098–1100 10.1128/JCM.44.3.1098-1100.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. R., Hota B., Ahmad I., Scott R. D., Foster S. D., Abbasi F., Schabowski S., Kampe L. M., Ciavarella G. G., Supino M., Naples J., Cordell R., Levy S. B., Weinstein R. A. (2009a). Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin. Infect. Dis. 49, 1175–1184 10.1086/605630 [DOI] [PubMed] [Google Scholar]
- Roberts M. C., Soge O. O., Giardino M. A., Mazengia E., Ma G., Meschke J. S. (2009b). Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. J. Appl. Microbiol. 107, 300–307 10.1111/j.1365-2672.2009.04207.x [DOI] [PubMed] [Google Scholar]
- Sapkota A. R., Curriero F. C., Gibson K. E., Schwab K. J. (2007). Antibiotic-resistant enterococci and fecal indicators in surface waters and groundwater impacted by a concentrated swine feeding operation. Environ. Health Perspect. 115, 1040–1045 10.1289/ehp.9770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G. M., Thomson R., Malone-Lee J., Ridgway G. L. (1988). Cross-infection between animals and man: possible feline transmission of Staphylococcus aureus infection in humans? J. Hosp. Infect. 12, 29–34 10.1016/0195-6701(88)90119-3 [DOI] [PubMed] [Google Scholar]
- Simjee S., White D. G., McDermott P. F., Wagner D. D., Zervos M. J., Donabedian S. M., English L. L., Hayes J. R., Walker R. D. (2002). Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J. Clin. Microbiol. 40, 4659–4665 10.1128/JCM.40.12.4659-4665.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørum M., Johnsen P. J., Aasnes B., Rosvoll T., Kruse H., Sundsfjord A., Simonsen G. S. (2006). Prevalence, persistence, and molecular characterization of glycopeptide-resistant enterococci in Norwegian poultry and poultry farmers 3 to 8 years after the ban on avoparcin. Appl. Environ. Microbiol. 72, 516–521 10.1128/AEM.72.1.516-521.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Cataldo M. A. (2008). Vancomycin-resistant enterococci (VRE): transmission and control. Int. J. Antimicrob. Agents 31, 99–106 10.1016/j.ijantimicag.2007.08.026 [DOI] [PubMed] [Google Scholar]
- Tendolkar P. M., Baghdayan A. S., Shankar N. (2006). Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 188, 2063–2072 10.1128/JB.188.6.2063-2072.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turabelidze D., Kotetishvili M., Kreger A., Morris J. G., Jr., Sulakvelidze A. (2000). Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38, 4242–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., van den Braak N., Thomassen R., Verbrugh H., Endtz H. (1996). Vancomycin-resistant enterococci in cats and dogs. Lancet 348, 1038–1039 10.1016/S0140-6736(05)64973-2 [DOI] [PubMed] [Google Scholar]
- Weese J. S. (2008). Issues regarding the use of vancomycin in companion animals. J. Am. Vet. Med. Assoc. 233, 565–567 10.2460/javma.233.4.565 [DOI] [PubMed] [Google Scholar]
- Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., Flannagan S. E., Kolonay J. F., Shetty J., Killgore G. E., Tenover F. C. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302, 1569–1571 10.1126/science.1090956 [DOI] [PubMed] [Google Scholar]