Abstract
The T-box transcription factor Tbx1 is expressed in the otic vesicle and surrounding mesoderm of the periotic mesenchyme (POM) during inner ear development. Mesenchymal Tbx1 is essential for inner ear development, with conditional mutants displaying defects in both the auditory and vestibular systems. We have previously reported that mesodermal Tbx1 loss of function mutants (Mest-KO) have reduced expression of retinoic acid (RA) metabolic genes, Cyp26a1 and Cyp26c1, in the POM, consistent with other studies showing an increase in mesodermal RA reporter expression in Tbx1-/- embryos. However, putative RA effector genes whose expression is altered downstream of increased otic mesenchymal-epithelial RA signaling have remained elusive. Here we report the identification of eighteen retinoic acid responsive genes altered in Mest-KO conditional mutants by microarray gene profiling. Nine were chosen for biological validation including quantitative RT-PCR and in situ hybridization (Otor, Mia, Col2a1, Clu, Adm, Myt1, Dlx3, Itgb3 and Itga2b). This study provides a series of newly identified RA effector genes for inner ear development downstream of mesenchymal Tbx1 that may contribute to the inner ear phenotype observed in Tbx1 loss of function mouse models.
INTRODUCTION
Haploinsufficiency of TBX1, a transcription factor of the T-box family, has been implicated in the etiology of velo-cardio-facial syndrome/DiGeorge syndrome/22q11.2 deletion syndrome (VCFS/DGS/22q11DS). VCFS/DGS patients display a wide range of phenotypes including hearing loss, with 30% of patients suffering from conductive hearing loss subsequent to recurrent middle ear infections and 10% suffering from sensorineural hearing loss (Digilio et al., 1999). Mouse models for Tbx1 have similar phenotypes, with heterozygous animals displaying chronic otitis media and homozygous null mice having sensorineural hearing loss due to arrested development of inner ear structures (Vitelli et al., 2003; Liao et al., 2004).
The mammalian inner ear is composed of the vestibular system, with three semicircular canals for sensing angular acceleration as well as the utricle and saccule for sensing linear acceleration, and the cochlea of the auditory system for processing of sound. Murine Tbx1 is expressed in the otic vesicle (OV) epithelium from which these inner ear structures derive as well as the surrounding mesoderm of the periotic mesenchyme (POM) which gives rise to the cartilaginous otic capsule during development and later the bony labyrinth surrounding the mature inner ear. Homozygous null Tbx1 mutants have a severe inner ear phenotype with an absence of mature inner ear structures. There are no auditory or vestibular system components, and the otic capsule is malformed (Vitelli et al, 2003; Raft et al., 2004).
Due to this severe early phenotype in homozygous null mutants, conditional ablation studies of Tbx1 in the OV and, separately, the POM have been used to study tissue-specific roles of this transcription factor during inner ear development. Conditional inactivation of Tbx1 in the OV epithelium using Pax2-Cre mice phenocopies Tbx1-/- embryos in the inner ear, suggesting a major early role for Tbx1 in the epithelium of the OV (Arnold et al., 2006). Conversely, ablation of Tbx1 in the mesoderm and POM, using mesodermal Cre mouse lines such as Mesp1-Cre and T-Cre, lead to a later, less severe inner ear defect consisting of failed cochlear outgrowth, hypoplastic semicircular canals (SCs), and an enlarged endolymphatic duct (Xu et al., 2007; Braunstein and Monks et al., 2009). This suggests that, as a transcription factor, Tbx1 in the surrounding mesoderm plays a key role in regulating signals from the POM to the OV, referred to as mesenchymal-epithelial signaling, during development of the inner ear.
Retinoic acid (RA) is a major signaling pathway implicated in both epithelial-mesenchymal signaling during otic capsule chondrogensis (Frenz et al., 1997) as well as mesenchymal-epithelial signaling to the OV during cochlear outgrowth (Braunstein and Monks et al., 2009). Earlier studies investigating the role of Tbx1 and RA in heart development demonstrated increased RA reporter activity in Tbx1-/- embryos, including an anterior shift encompassing the developing ear (Guris et al., 2006), concurrent with decreased expression of the RA metabolic genes, Cyp26a1 and Cy26c1, in the POM at E9.5 (Roberts et al., 2006). We have previously demonstrated such dysregulation of RA signaling downstream of Tbx1 is attributable to the POM domain of Tbx1, as loss of Tbx1 in the mesoderm leads to reduction in the expression of Cyp26a1 and Cyp26c1 in the POM, while expression of Cyp26c1 in the ventral OV remains intact (Braunstein and Monks et al., 2009). This suggests that loss of the transcription factor Tbx1 in the mesoderm regulates RA signaling, either directly or indirectly, in the POM during inner ear development (Braunstein and Monks et al., 2009). Additional evidence for mesenchymal-epithelial RA signaling comes from genetic studies using Brn4+/- mice. Brn4 (Pou3f4) is an X-linked gene encoding a POU domain transcription factor required for brain, ear and pancreatic development (Mathis et al., 1992; Phippard et al., 1999; Hussain et al., 2002). Hemizygosity for Brn4 leads to defects in cochlear outgrowth similar to those observed in mesenchymal Tbx1 ablation (Braunstein et al., 2008). Furthermore, Brn4 was shown to genetically interact with Tbx1 for proper cochlear outgrowth, and both are required for Cyp26c1 expression in the POM (Braunstein and Monks et al., 2009).
While RA has been implicated in mesenchymal-epithelial signaling during inner ear development, downstream effector genes for RA signaling in the OV and/or POM have not been identified. Using a gene profiling microarray approach, we have identified genes which may serve as RA targets, based upon published data in other cells or tissues, at the stage of mid-gestational auditory and vestibular system morphogenesis (E12.5).
RESULTS
Gene profiling to identify Tbx1 regulated POM genes
To identify possible downstream genes of mesodermal Tbx1, gene profiling microarray analysis was performed on microdissected periotic regions (POM + OV) of T-Cre mediated conditional Tbx1 mutants (Mest-KO) at E12.5. A total of 161 differentially expressed genes were identified, including 80 up-regulated and 81 down-regulated genes (Supplemental Table 1). Data obtained from this microarray screen were analyzed for changes in common gene networks or signaling pathways so as to explain molecularly how mesenchymal-epithelial signaling might be perturbed in Mest-KO embryos during inner ear morphogenesis. This screen identified eighteen genes that have been shown in other systems to be downstream of RA signaling (Figure 1). All genes were altered in the direction of known response to RA, with Adm (Minamino et al., 1995), Cp (Tice et al., 2002), Myt1 (Franco et al., 1999), Ncam1 (De Laurenzi et al., 2000), Pak1 (Csomos et al., 2010), and Vwf (Hatzopoulos et al., 1998) up-regulated and Aplnr (Zhong et al., 2005), Bhmt (Xu et al., 2010), Col2a1 (Jimenez et al., 2001), Clu (Orlandi et al., 2005), Dlx3 (Acquafreda et al., 2010), Fgf4 (Maerz et al., 1998), Hoxc9 (Mao et al., 2011), Itga2b (Liu et al., 2000), Itgb3 (Csomos et al., 2010), Mia1 (Dietz et al., 1996), Msc (Hishikawa et al., 2005), Otor (Roberts et al., 2000), and S100a8 (Liu et al., 2000) down-regulated, consistent with previously published data in vitro or in other model systems (Table 1).
Figure 1. Altered expression of retinoic acid responsive genes in Mest-KO embryos consistent with up-regulated RA signaling.
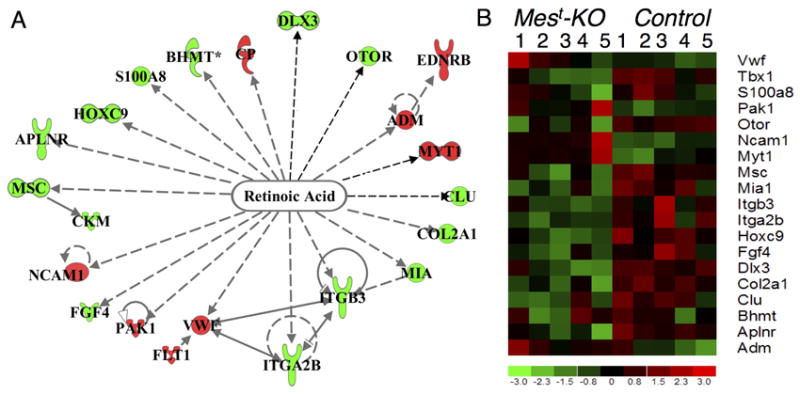
(A) Ingenuity Pathways Analysis identifies expression changes in 19 genes responsive to RA levels, overlayed with expression values from E12.5 microarray analysis. Up-regulated (red) and down-regulated (green) genes are altered in a manner consistent with increased RA levels in Mest-KO embryos. (B) Heat map of E12.5 microarrays representing five Mest-KO (left) and five Control (right) chips with RA genes represented on the right along with the control probe Tbx1, decreased in expression after conditional Tbx1 albation. Genes with negative response to treatment with RA are down-regualted (green) on mutant arrays, while those described to be positively regulated by RA are up-regulated (red) on mutant arrays.
Table 1.
Identification of RA regulated genes by microarray
| Gene Symbol | Gene Name | Fold Change (KO vs Control) | p-value | Predicted response to RA | Reference for RA regulation |
|---|---|---|---|---|---|
| Adm | Adrenomedullin | 1.4 | 0.026 | INCREASE | Minamino et al., 1995 |
| Col2a1 | Collagen II, subunit a1 | -1.47 | 0.006 | DECREASE | Jimenez et al., 2001 |
| Clu | Clusterin | -1.4 | 0.019 | DECREASE | Orlandi et al., 2005 |
| Dlx3 | Distalless-like homeobox 3 | -1.6 | 0.005 | DECREASE | Acquafreda et al., 2010 |
| Itga2b | Integrin alpha 2, subunit b | -1.31 | 0.017 | DECREASE | Liu et al., 2000 |
| Itgb3 | Integrin beta 3 | -1.29 | 0.014 | DECREASE | Csomos et al., 2010 |
| Mia1 | Melanoma inhibitory activity | -1.45 | 0.039 | DECREASE | Dietz et al., 1996 |
| Myt1 | Myelin transcription factor 1 | 1.58 | 0.044 | INCREASE | Franco et al., 1999 |
| Otor | Otoraplin | -1.28 | 0.009 | DECREASE | Roberts et al., 2000 |
| Cp | Ceruloplasmin | 1.37 | 0.017 | INCREASE | Tice et al., 2002 |
| Bhmt | Betaine-homocysteine methyltransferase | -1.27 | 0.002 | DECREASE | Xu et al., 2010 |
| S100a8 | S100 calcium binding protein A8 | -1.29 | 0.025 | DECREASE | Liu et al., 2000 |
| Hoxc9 | Homeobox C9 | -1.3 | 0.001 | DECREASE | Mao et al., 2011 |
| Aplnr | Apelin receptor | -1.27 | 0.2 | DECREASE | Zhong et al., 2005 |
| Msc | Musculin | -1.34 | 0.037 | DECREASE | Hishikawa et al., 2005 |
| Ncam1 | Neural cell adhesion molecule 1 | 1.26 | 0.016 | INCREASE | De Laurenzi et al., 2000 |
| Fgf4 | Fibroblast growth factor 4 | -1.26 | 0.048 | DECREASE | Maerz et al., 1998 |
| Pak1 | p21 protein activated kinase 1 | 1.3 | 0.049 | INCREASE | Csomos et al., 2010 |
| Vwf | Von Willebrand factor | 1.32 | 0.0235 | INCREASE | Hatzopoulos et al., 1998 |
Microarray analysis of Mest-KO E12.5 periotic tissue (OV and surrounding POM) reveals changes in genes regulated by retinoic acid (RA). Gene expression changes are consistent with previously published data implicating either activation or repression by treatment with retinoic acid in cell culture or other model organisms, as indicated. Fold change versus control and p-values from microarray analysis are presented for these selected genes. For full microarray expression data, see Supplemental Table 1
Nine genes from the RA pathway were selected for validation by quantitative RT-PCR (qRT-PCR), based on known ear expression or strongest fold-change values by microarray. The nine selected genes were Adm, Clu, Col2a1, Dlx3, Itga2b, Itb3, Mia1, Myt1, and Otor. Validation by qRT-PCR confirmed significantly altered expression of these RA responsive genes with the exception of Col2a1 and Mia1, which showed a trend of down-regulation but were not significant (Figure 2).
Figure 2. qPCR validation of RA target genes.
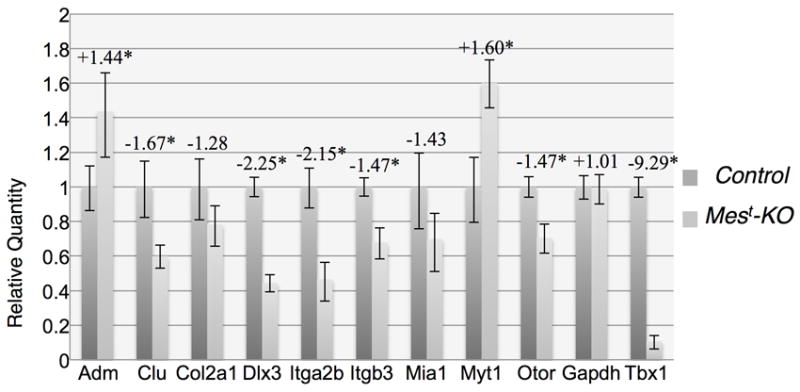
Genes shown to be regulated by RA in other systems (Table 1) were validated by qPCR on microdissected Mest-KO periotic regions (OV+POM) using TaqMan probes. As expected, Adm, Clu, Dlx3, Itga2b, Itgb3, Myt1, Otor, and control probe Tbx1 were significantly changed in T-Cre KO versus control ears. Col2a1, Mia1, and control probe Gapdh were not significantly changed, though both Col2a1 and Mia1 show a trend towards down-regulation that is not statistically significant. Fold change values are shown above (Mest-KO vs. Control) and asterisk indicates significance with p<0.05.
Localization of RA regulated genes to developing inner ear
While RA regulation of these downstream genes has been implicated or established, most of these studies were conducted in either cell culture systems or in the development of organs other than the ear (Table 1). For this reason, specific temporo-spatial expression data for ear development is largely unavailable. In order to localize expression patterns of these genes to the otic epithelium and/or POM during inner ear development, in situ hybridization on microdissected E12.5 periotic regions, containing the OV, POM, cochleovestibular ganglion (CVG), and surrounding head mesenchyme, was performed.
We have previously reported that the E11.5 Mest-KO inner ear begins to show signs of hypoplasia, specifically in the ventral region that gives rise to the cochlea, and that E15.5 Mest-KO ears display a shortened cochlea, semicircular canal hypoplasia, and an enlarged endolymphatic duct. (Braunstein and Monks et al., 2009). In order to determine the presence and morphology of these structures at the stage of microarray analysis, E12.5 Mest-KO and control embryos were analyzed by paint filling. At E12.5, the Mest-KO cochlea is present, but shortened, as compared to the cochlea in control embryos, conditional Tbx1 heterozygotes, at this stage (green arrows, Figure 3A,D). Dorsal structures, including the endolymphatic duct (red arrow) and canal outpouches, which will give rise to the semicircular canals of the vestibular system (yellow arrows), are present and appear morphologically normal at E12.5.
Figure 3. Tissue localization of RA target genes in the developing ear.
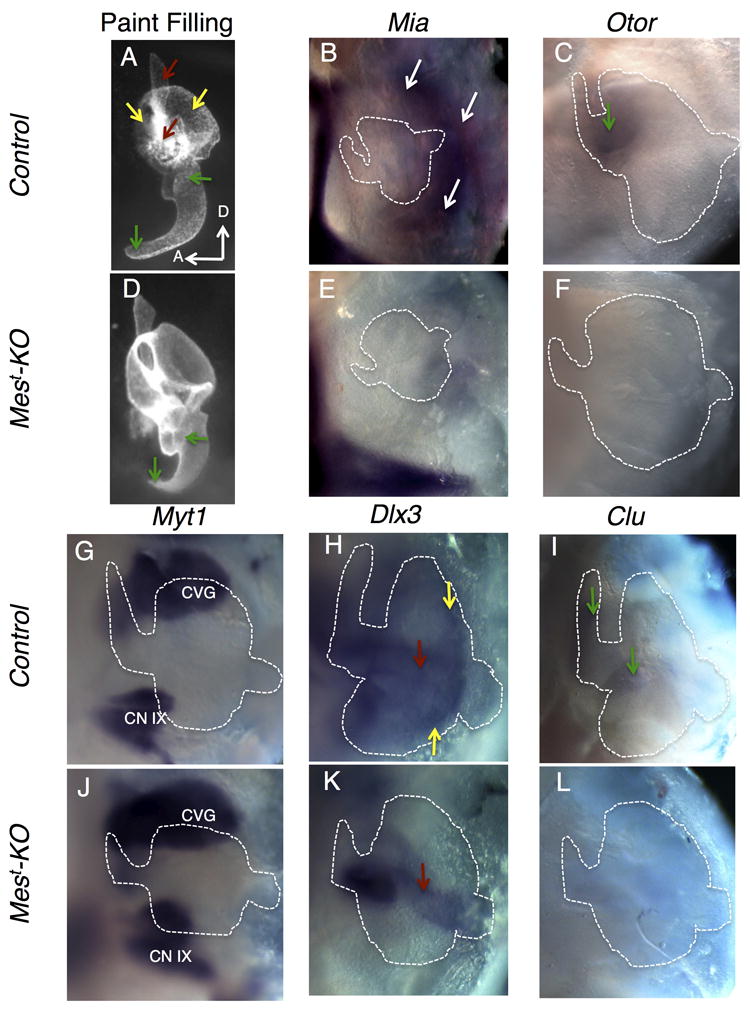
In situ hybridization on microdissected periotic tissue was performed to localize expression changes to the OV, POM, or CVG during ear development. Structures are labeled with color-coded arrows; red representing the endolymphatic duct (ED), yellow representing the canal outpouches of the vestibular system, and green representing the cochlea. Mest-KO embryos display a shortened cochlea as early as E12.5, while the developing endolymphatic duct and canal outpouches (OP) appear unaffected at this stage in Mest-KO embryos (A,D). Mia1 is expressed in the developing POM surrounding the dorsal OV (arrows, B), with nearly complete loss of expression in mutants (E). Otoraplin, closely related to Mia1, is expressed in the base of the developing cochlea (arrow, C), and expression is completely absent in mutants (F). Myt1 marks the developing CVG (CN VIII), as well as the inferior CN XI ganglion, in both mutants and controls (G,J) inner ear is outlined for reference, but out of the focal plane. Dlx3 is expressed in the outpouches and ED of the vestibular system of control embryos (H), with loss of OP expression in mutants, though ED expression is in tact (arrow, K). Clu displays specific expression from the base to the apex of the cochlea (arrows, I). Mest-KO embryos display loss of expression throughout the cochlea (L). OV otic vesicle, POM periotic mesenchyme, CVG cochleovestibular ganglion, CN IX inferior cranial nerve IX ganglion, OP canal outpouches.
The mesenchymal gene Melanoma inhibitory activity 1 (Mia1), also known as cartilage-derived retinoic acid sensitive protein (CD-RAP), was identified by microarray analysis as down-regulated in Mest-KO embryos (Table 1). Validation by qRT-PCR supported a trend of down-regulation, but was not statistically significant (Figure 2). Supporting the microarray findings, in situ hybridization revealed expression of Mia1 in the dorsal POM of control embryos, with nearly complete loss in mutant embryos (arrows, Figure 3B,E). Such differences in validation by qRT-PCR and in situ hybridization might be due to statistical noise caused by slight differences in the region of microdissected tissue included in qRT-PCR analysis. This seems likely for Mia1, as it is expressed in POM lying at the dorsal endpoint of microdissection for qRT-PCR. Microdissected tissue for in situ analysis contains more tissue including head mesenchyme and surface ectoderm surrounding the developing ear, making expression changes easier to visualize. Mia1 was identified in cell culture of bovine chondrocytes, where its expression was significantly reduced upon treatment with RA, consistent with our microarray and in situ hybridization data (Dietz et al., 1996).
Otoraplin (Otor) was initially discovered as a homologue to CD-RAP/MIA expressed in a cochlear cDNA library, and was accordingly classified as negatively regulated by treatment with all-trans RA (atRA). Its expression was localized to the chicken cochlea by northern blot analysis, but previous attempts at in situ hybridization were unsuccessful (Robertson et al, 2000). Through tissue specific in situ hybridization, Otor expression was detectable in the developing mouse cochlea at E12.5. Otor was expressed in the base of the cochlea of control embryos, but strikingly absent from the cochlea of Mest-KO embryos, consistent with qRT-PCR results (green arrow, Figure 3C,F). No Otor expression was detected in other regions of the ear, consistent with previous data (Robertson et al., 2000).
Myelin transcription factor 1, Myt1, encodes a zinc finger transcription factor implicated in neurogenesis and pancreatic islet cell development (Romm et al., 2005). In Xenopus, XMyT1 expression is expanded in the neural ectoderm upon treatment with RA, while inhibition of RA signaling leads to decreased expression (Franco et al., 1999). Consistent with a role in neurogenesis, Myt1 was localized to the developing cochleovestibular ganglion (CVG), as well as the inferior vagal ganglion (CN IX) by in situ hybridization. Microarray and qRT-PCR analysis confirms up-regulation in Mest-KO embryos. However, due to the complex morphology of the CVG at E12.5, such up-regulation was not detected by in situ hybridization (Figure 3G,J).
Transcription factors of the distaless homeobox family are known to play important roles in the developing vestibular system of the inner ear (Robledo et al., 2006). One member of this family, Dlx3, was down-regulated by microarray and qRT-PCR analysis. Dlx3 is expressed in the dorsal region of the OV at E12.5 of control embryos, including the developing endolymphatic duct and canal outpouches (yellow arrows, Figure 3H), consistent with previously reported dorsal expression in chick (Brown et al., 2005). Expression was lost in the developing semicircular canals (SC) of Mest-KO embryos, while expression in the endolymphatic duct remained intact (red arrow, Figure 3K). A recent report using an oral squamous cell carcinoma line treated with atRA suggests that Dlx3 is negatively regulated by RA (Acquafreda et al., 2010).
Clusterin (Clu) is a widely expressed protein serving both pro- and anti-apoptotic functions during development and in mature tissues, with major functions in prostate development and Alzheimer’s disease (Ahuja et al., 1994; Calero et al., 2005). Expression in the developing inner ear has been described, with early widespread OV expression (Ahuja et al., 1994) and later localization to the ventral cochlear epithelium (Ficker et al., 2004). Evidence for RA regulation of Clu comes from studies in vascular smooth muscle cells, where treatment with atRA led to decreased Clu expression (Orlandi et al., 2005). Consistent with this data, E12.5 expression is localized to the ventral cochlear portion of the OV and is completely absent in Mest-KO embryos at this stage (green arrows, Figure 3I,L).
Localization by in situ hybridization for three RA-responsive genes encoding Adrenomedullin (Adm), Integrin alpha 2b (Itga2b), and Integrin beta 3 (Itgb3), though attempted, failed due to lack of ability of probes to detect specific expression. Adm is a vasodilator peptide expressed in smooth muscle cells, where its expression is positively regulated by RA exposure (Minamino et al., 1995). Consistent with these reports, Adm expression was increased 1.44-fold in Mest-KO embryos by qRT-PCR (Figure 2). Adrenomedullin expression was previously identified in the mouse cochlea by RT-PCR and further localized to the stria vascularis by immunohistochemistry (Tono et al., 2002). Itga2b and Itgb3 are known to be down-regulated by RA in a leukemia cell line (Liu et al., 2000; Csomos et al., 2010), though expression during inner ear development has not been characterized. However, integrins are known to play important roles in mesenchymal condensation and chondrogenesis, suggesting possible expression of both Itga2b and Itgb3 in the POM (Schubert et al., 2010). In summary, all gene expression changes detected by microarray analysis were in the direction predicted downstream of increased RA levels, some of which may be effectors of cochlear or vestibular morphogenesis based on tissue expression patterns.
Clu-/- embryos display normal cochlear morphology
Due to the strong expression of Clu in control cochleae and a striking loss of expression in Mest-KO embryos at E12.5 (Figure 3I,L) and E11.5 (Figure 4A-B), loss of ventral Clu is an attractive candidate for the cochlear phenotype of mesodermal Tbx1 conditional knockout embryos (Xu et al., 2007; Braunstein and Monks et al., 2009). To determine if loss of Clu leads to defects in gross cochlear morphology, E15.5 Clu homozygous null embryos were analyzed via paint filling. No defects in inner ear morphology were detectible in these mutants as compared to Tbx1 heterozygous control embryos at the same stage (Figure 4C-D), suggesting that loss of Clu is either compensated for by another, to date unidentified, gene, or that Clu plays a minor role, if any, in the cochlear outgrowth phenotype observed in Mest-KO embryos.
Figure 4. Loss of Clu is not responsible for defects in cochlear morphology.
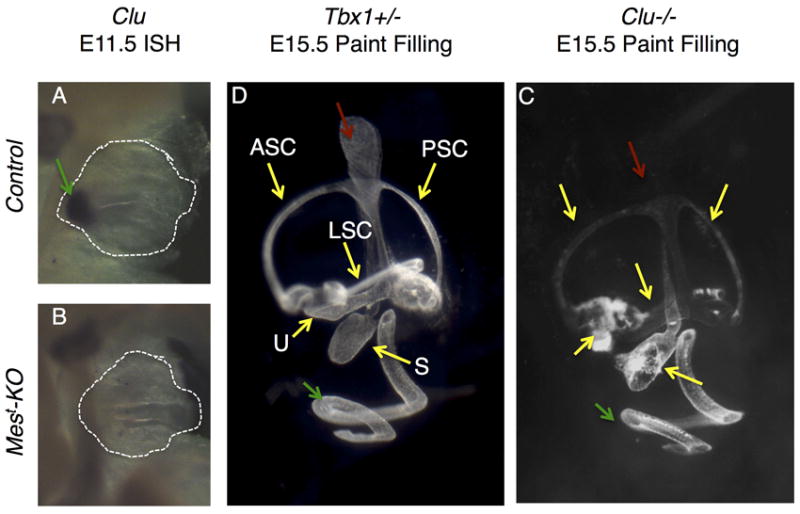
(A-B) In situ hybridization for Clu shows reduced expression in Mest-KO embryos as early as E11.5 (B) as compared to conditional Tbx1 heterozygous controls, where expression is already apparent in the ventral OV which will form the cochlea (arrow, A). (C-D) Homozygous knockout of Clu leads to no morphological defects in the cochlea or other inner ear structures at E15.5, as assayed by paint filling analysis and compared to Tbx1 heterozygous controls. ED endolymphatic ducts (red arrow), ASC/PSC/LSC anterior/posterior/lateral semicircular canals (yellow arrows), U utricle (yellow arrows), S saccule (yellow arrows), CD cochlear duct (green arrow).
DISCUSSION
RA has previously been implicated in earlier stages of inner ear development, such as otic induction, where increased signaling expands competence to form otic placode and is necessary for proper Fgf signaling from the hindbrain in zebrafish (Hans and Westerfield, 2007). A role for RA during early murine inner ear development has also been described, with RA treatment prior to E9.0 leading to widespread morphological defects and embryonic resorption and later exposure at E10 in vivo (Frenz et al., 1996) and at E12.5-E14 leading to specific cochlear defects in an explant culture system (Frenz et al., 1997). More recently, a specific temporo-spatial role for RA in establishing anterior-posterior axis identity in the avian OV has been established, with early exposure (E7.75-8.25) leading to posteriorization, marked by expanded Tbx1 expression, and exposure at E8.5 resulting in fewer gene expression and morphology changes (Bok et al., 2011). RA is also known to play a role in condensation of the POM to form the cartilaginous otic capsule, with treatment of cultured POM+OV unable to form cartilage in the presence of exogenous atRA (Frenz et al., 2000).
However, elucidating the in vivo roles of RA in later stage murine ear development, during mid-gestation, has been difficult due to widespread teratogenicity of RA treatment. This is particularly important in studying events such as cochlear coiling, as avian cochleae do not form a coiled structure as mice and higher vertebrates do. Mesodermal knockout of Tbx1 provides an excellent system to bypass early RA teratogenicity and study the slightly later, tissue-specific roles of RA during inner ear morphogenesis in the mouse system.
Previous studies have revealed increased RA signaling in the absence of Tbx1 expression, including the region of the developing E9.5 inner ear (Roberts et al., 2006; Guris et al., 2006). This early increase in RA signaling appears to be mediated in part by reduction in the genes encoding cytochrome P450 enzymes responsible for RA degradation, Cyp26a1 and Cyp26b1. We have previously reported a similar reduction in Cyp26a1 and Cyp26c1 gene expression in the periotic mesenchyme when Tbx1 is ablated in the same cell population using the mesodermal T-Cre line (Braunstein and Monks et al., 2009).
Consistent with decreased expression of Cyp26a1 and Cyp26c1 enzymes in Mest-KO embryos, two putative T-sites were identified in the region of mouse chromosome 19 containing these contiguous genes (see Materials and Methods). The first lies approximately 50 bp upstream of the published Cyp26a1 promoter (Loudig et al., 2000) while the second is approximately 500 bp upstream of the Cyp26c1 coding sequence, with the distance between these two putative T-sites spanning a total interval of less than 20kb. Given this small genomic interval, TBX1 protein binding to either of these T-sites could regulate transcription of one or both Cyp26 genes. In order to determine if TBX1 can bind these putative T-sites, electrophoretic mobility shift assays were performed using a GST-tagged TBX1 protein and oligonucleotides encompassing the putative T-sites. Neither putative T-site was found to bind TBX1 in this assay, suggesting that regulation of Cyp26a1 and Cyp26c1 occurs either indirectly or via unidentified T-sites in the region (Data not shown).
In the E12.5 Mest-KO conditional Tbx1 mutant, we have identified changes in over 160 genes. After pathways analysis, we identified eighteen genes previously described as regulated by retinoic acid using either in vitro or in vivo approaches in other model organisms. All eighteen genes show changes in transcriptional up- or down-regulation in the direction predicted by the presence of increased RA signaling in Mest-KO embryos by microarray, while seven out of nine genes assayed remained significant after qRT-PCR validation. The expression changes, while significant and obvious upon in situ hybridization, do not display large fold change values by these quantitative methods. This is likely due to the large amounts of tissue present in the E12.5 periotic region used for these studies as well as the specific expression patterns in very small portions of that tissue, thus diluting gene expression levels. Taking this into account, it is likely that changes in genes such as Mia1 might actually have physiologic significance, as suggested by clearly reduced expression in the small region of dorsal POM by in situ hybridization, despite the small fold-change on the microarray and/or statistical insignificance upon qRT-PCR validation. It is also important to note that, due to these dilution effects, there are likely many more genes changed in the periotic region of Mest-KO mutants than have been identified by this approach. The list presented, however, represents those genes identified to have significant expression changes in the periotic region in addition to previous data suggesting regulation by RA signaling.
The significant gene changes can be divided into two broad expression domains: the otic epithelium, giving rise to the inner ear proper, and the periotic mesenchyme, giving rise to the bony labyrinth. Genes expressed specifically in the otic epithelium, such as Adm, Clu, Dlx3, and Otor, may play a role in the inner ear phenotype observed in Mest-KO mutants, with the exception of Clu, which showed no cochlear phenotype in our analysis. Similarly, genes expressed in the POM, such as Mia, may contribute more directly downstream of mesenchymal Tbx1 to defects in the otic capsule or to inner ear morphogenesis. Itga2b and Itgb3 were unable to be localized by in situ hybridization due to unavailability of specific probes and absence of previous expression data during inner ear development. However, integrins are known to play a key role in mesenchymal condensation and chondrogenesis via an interaction with Mia1 (Schubert et al., 2010), suggesting the possibility that these two genes interact and contribute to inner ear and/or otic capsule defect observed in Mest-KO embryos (Braunstein and Monks et al., 2009).
Interestingly, exogenous RA signaling has been shown to act upstream of the otic epithelial expression domain of tbx1 in zebrafish (Radosevic et al., 2011). Thus, RA might act both upstream and downstream of tbx1. In our previous studies, alteration of Tbx1 expression was not detected in the OV domain of Mest-KO embryos, concurrent with increased RA (Braunstein and Monks et al., 2009), suggesting that there may be species-specific or temporal differences in regulation of Tbx1 downstream of RA signaling.
Failed cochlear outgrowth is perhaps the most striking phenotype observed in Mest-KO embryos. In order to investigate the mechanism of this defect, we sought knockout mouse models for Otor and Clu to provide further genetic data for this phenotype. The only available mouse model, Clusterin, though expressed specifically in the developing cochlea and strikingly lost in Mest-KO embryos, failed to produce a cochlear phenotype when inactivated. Development of future mouse models for genes such as Otor might provide the missing link between altered RA signaling and cochlear morphology defects observed in Mest-KO embryos. Alternatively, the cochlear phenotype might arise due to expression changes undetected in our microarray analysis or a combination of changes in multiple genes and signaling pathways.
Together, this data aids in the construction of a putative model for RA mesenchymal-epithelial signaling downstream of Tbx1 (Figure 5). In this model, Tbx1 in the POM regulates the Cyp26a1 and Cyp26c1 RA metabolic genes in the same tissue via an indirect mechanism or via unidentified T-sites, as evidenced by previous data demonstrating altered Cyp gene expression in the POM, but not OV, of Mest-KO mutants (Braunstein and Monks et al., 2009) and the inability of Tbx1 to bind to the two putative T-sites identified to date). Reduced RA breakdown, through diminished Cyp26 expression in the POM, leads to increased RA in the periotic region at E12.5, as supported by increased RARE-LacZ reporter activity in Tbx1-/- embryos where Cyp26a1 and Cyp26c1 expression is similarly reduced in the POM (Guris et al., 2006). Expression of RA-responsive genes in the OV (Dlx3, Adm, Clu, Otor), and POM itself (Mia) is affected in response to increase RA signaling in Mest-KO embryos at E12.5, further supporting this model.
Figure 5. Model of RA target gene regulation by mesenchymal Tbx1.
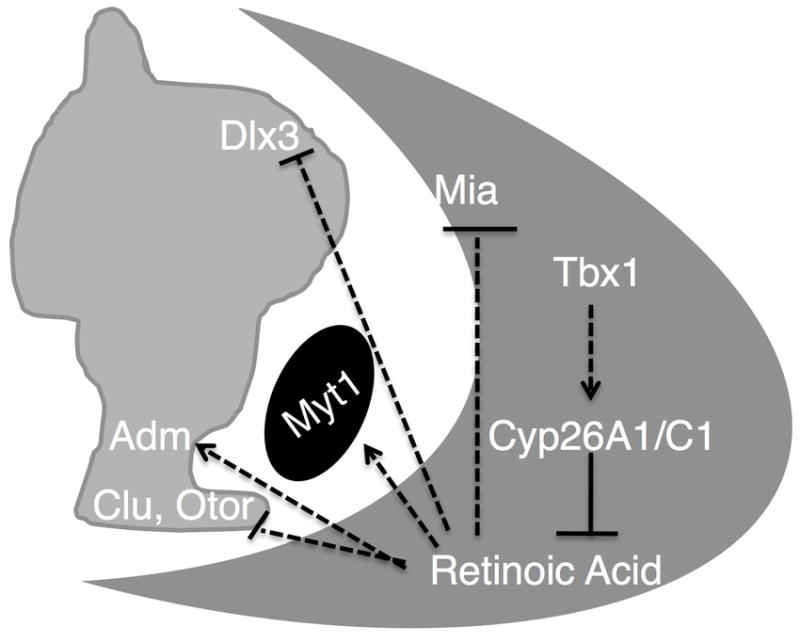
Tbx1 in the POM (dark gray) indirectly promotes expression of RA metabolic genes Cyp26a1 and Cyp26c1 which, in turn, breakdown/inhibit RA signaling to the OV (light gray). During inner ear development, RA activates the expression of Adm in the CD and Myt1 in the CVG (black) while inhibiting expression of Dlx3 in the developing vestibular system and Otor and Clu expression in the CD. CD cochlear duct, CVG cochleovestibular ganglion, POM periotic mesenchyme.
As an important environmental and dietary teratogen, RA signaling plays a role in countless organs during vertebrate development. Early perturbation of RA signaling surrounding the developing ear leads to defects in otic placode specification and subsequently major defects in otic development. Timed exposure to RA has not been well studied in mice due to the complexity of such experiments, and can also lead to widespread teratogenic effects. However, loss of mesenchymal Tbx1 provides a useful model by which the later actions of physiologic RA can be studied in ear development. More importantly, the identification of RA target gene changes in the developing ear and periotic mesenchyme, taken with previous data reporting altered Cyp26 gene expression in mesenchymal Tbx1 mutants, adds specific RA target genes to the only mesenchymal-epithelial signaling pathway thus far identified during inner ear development.
EXPERIMENTAL PROCEDURES
Mouse Matings
Conditional Tbx1 mutants were obtained by crossing T-Cre;Tbx1+/- males with Tbx1flox/flox females, resulting in T-Cre;Tbx1flox/+ (Control) and T-Cre:Tbx1flox/- (Mest-KO) embryos as previously described (Braunstein and Monks et al., 2009). Clu (Clusterin) heterozygotes (Jackson Laboratories, Maine, USA: Stock#00562) were intercrossed to obtain Clu-/- embryos, and crossed into the Tbx1+/- (Arnold et al., 2006) background to obtain Clu-/-;Tbx1+/- embryos for paint filling analysis.
Paint Filling Analysis
Embryos of appropriate genotype were dissected at E12.5 or E15.5 in cold PBS, cut below the forelimb, fixed overnight in Bodian Fixative, washed in 100% Ethanol, cleared in methyl salicylate and injected with 0.1% white correction fluid in methyl salicylate as previously described (Braunstein and Monks et al., 2009).
Microdissection and RNA isolation
Freshly dissected embryos were immediately transferred to ice cold PBS and cut below the forelimb and above the mandible to retain the periotic tissue. This tissue was then hemisected along the neural tube and neural tissue was removed. Forceps were used to remove the remaining jaw and tissue surrounding the periotic region, leaveing only otic epithelium and surrounding, condensed, periotic mesenchyme. Left and right periotic tissue was combined and homogenized by vortexing in Qiagen Buffer RLT with ß-mercaptoethanol and stored at -80°C until all samples were dissected. RNA was then isolated according to the manufacturer’s protocol (Qiagen RNeasy Micro Kit).
Microarray Analysis
cDNA was synthesized using the NuGen Ovation kit from pooled left and right ears of the same embryo. Affymetrix Mouse Gene ST 1.0 microarrays were performed on 5 Mest-KO embryos and 5 Control (conditional heterozygous) embryos. Data was normalized via the GC-RMA method and a t-test was performed. Samples with p values <0.05 were considered significant and filtered for fold change greater than +/-1.25, resulting in 68 upregulated and 64 downregulated genes in Mest-KO vs. conditional Tbx1 heterozygous controls. Raw data and analysis details have been submitted to the NCBI Gene Expression Omnibus database to comply with MIAME guidelines.
qRT-PCR Validation
RNA from 3 separate Mest-KO and 3 Control embryos was isolated with samples for microarray analysis but used for qRT-PCR validation. Validation was carried out in triplicate for each sample using TaqMan based assays (Applied Biosystems) for Col2a1 (Mm01309652_g1), Clu (Mm00442773_m1), Adm (Mm00437438_g1), Mia1 (Mm00444563_m1), Otor (Mm00498571_m1), Itga2b (Mm00439741_m1), Itgb3 (Mm00443980_m1), and Myt1 (Mm00456190_m1) as well as Gapdh and Tbx1 as controls (Mm00448948_m1) on an Applied Biosystems 7900HT. Data analysis was performed by SDS 2.2 and RQ Manager (Version 1.2) software (Applied Biosystems) with a confidence interval of 95%.
Microdissected in situ hybridization
Microdissected tissue-specific in situ hybridization was performed as previously described, with minor modifications (Nichols et al., 2008). Briefly, embryos were fixed in 4% PFA for 24hrs, dehydrated in a graded methanol series, and stored at -20°C until use. Embryos were then rehydrated in a methanol series to PBS, placed in 0.4% PFA, and microdissected in a manner similar to that used for microarray analyses, except that head mesoderm surrounding the periotic region could not be removed due to changes in tissue architecture caused by fixation. Primer sequences and conditions for PCR-generated riboprobes are provided in Supplemental Table 2. All in situ hybridizations were carried out on periotic regions from at least 5 separate embryos of each genotype.
Electrophoretic Mobility Shift Assay
Two conserved putative T-sites were identified in the genomic interval of mouse chromosome 19 containing Cyp26a1 and Cyp26c1 loci by manual inspection; one 50bp upstream of the Cyp26a1 promoter (UCSC Genome Browser; NCBI37/mm9; chr19:37,722,095) and the other 500bp upstream of the Cyp26c1 transcriptional start site (chr19;37,760,086). Electrophoretic mobility shift assay for putative T-sites, mutated versions of those T-sites, and Pitx2, a positive control previously identified as a direct target of TBX1 in heart development, was performed as previously described (Nowotschin et al., 2006), except that purified GST-tagged TBX1 was used.
Supplementary Material
Gene expression changes from the E12.5 OV+POM microarray carried out on the Affymetrix Mouse Gene ST 1.0 platform. Genes with fold-changed >+/-1.25 and p-value <0.05 are represented
Primers for PCR amplification of E12.5 cDNA are listed. T3 polymerase and T7 polymerase site sequences were add to the forward and reverse primers, respectively, for the synthesis of sense and anti-sense riboprobes. All reactions were carried out with 2.5mM MgCl2 and without the addition of DMSO or Betaine for 35 cycles of 1 minute each. Probe sizes and individual annealing temperatures are provided.
Acknowledgments
We would like to thank Dr. Tingwei Guo (Albert Einstein College of Medicine) for his assistance generating the microarray heatmap, as well as Dr. Bernd Fritzsch (University of Iowa) for his training and assistance in microdissected in situ hybridization techniques. We acknowledge support of this research by NIH grant DC05186 to B.E.M.
References
- Acquafreda T, Nunes FD, Soprano DR, Soprano KJ. Expression of homeobox genes in oral squamous cell carcinoma cell lines treated with all-trans retinoic acid. Journal of Cellular Biochemistry. 2010;111:1437–1444. doi: 10.1002/jcb.22871. [DOI] [PubMed] [Google Scholar]
- Ahuja HS, Tenniswood M, Lockshin R, Zakeri ZF. Expression of clusterin in cell differentiation and cell death. Biochemistry and Cell Biology = Biochimie et Biologie Cellulaire. 1994;72:523–530. doi: 10.1139/o94-070. [DOI] [PubMed] [Google Scholar]
- Arnold JS, Braunstein EM, Ohyama T, Groves AK, Adams JC, Brown MC, Morrow BE. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Human Molecular Genetics. 2006;15:1629–1639. doi: 10.1093/hmg/ddl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J, Raft S, Kong KA, Koo SK, Drager UC, Wu DK. Transient retinoic acid signaling confers anterior-posterior polarity to the inner ear. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:161–166. doi: 10.1073/pnas.1010547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein EM, Crenshaw EB, 3rd, Morrow BE, Adams JC. Cooperative function of Tbx1 and Brn4 in the periotic mesenchyme is necessary for cochlea formation. Journal of the Association for Research in Otolaryngology : JARO. 2008;9:33–43. doi: 10.1007/s10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein EM, Monks DC, Aggarwal VS, Arnold JS, Morrow BE. Tbx1 and Brn4 regulate retinoic acid metabolic genes during cochlear morphogenesis. BMC Developmental Biology. 2009;9:31. doi: 10.1186/1471-213X-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ST, Wang J, Groves AK. Dlx gene expression during chick inner ear development. The Journal of Comparative Neurology. 2005;483:48–65. doi: 10.1002/cne.20418. [DOI] [PubMed] [Google Scholar]
- Calero M, Rostagno A, Frangione B, Ghiso J. Clusterin and Alzheimer’s disease. Sub-cellular Biochemistry. 2005;38:273–298. [PubMed] [Google Scholar]
- Csomos K, Nemet I, Fesus L, Balajthy Z. Tissue transglutaminase contributes to the all-trans-retinoic acid-induced differentiation syndrome phenotype in the NB4 model of acute promyelocytic leukemia. Blood. 2010;116:3933–3943. doi: 10.1182/blood-2010-01-266064. [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Raschella G, Barcaroli D, Annicchiarico-Petruzzelli M, Ranalli M, Catani MV, Tanno B, Costanzo A, Levrero M, Melino G. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. The Journal of Biological Chemistry. 2000;275:15226–15231. doi: 10.1074/jbc.275.20.15226. [DOI] [PubMed] [Google Scholar]
- Dietz UH, Sandell LJ. Cloning of a retinoic acid-sensitive mRNA expressed in cartilage and during chondrogenesis. The Journal of Biological Chemistry. 1996;271:3311–3316. doi: 10.1074/jbc.271.6.3311. [DOI] [PubMed] [Google Scholar]
- Digilio MC, Pacifico C, Tieri L, Marino B, Giannotti A, Dallapiccola B. Audiological findings in patients with microdeletion 22q11 (di George/velocardiofacial syndrome) British Journal of Audiology. 1999;33:329–333. doi: 10.3109/03005369909090116. [DOI] [PubMed] [Google Scholar]
- Ficker M, Powles N, Warr N, Pirvola U, Maconochie M. Analysis of genes from inner ear developmental-stage cDNA subtraction reveals molecular regionalization of the otic capsule. Developmental Biology. 2004;268:7–23. doi: 10.1016/j.ydbio.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Franco PG, Paganelli AR, Lopez SL, Carrasco AE. Functional association of retinoic acid and hedgehog signaling in Xenopus primary neurogenesis. Development. 1999;126:4257–4265. doi: 10.1242/dev.126.19.4257. [DOI] [PubMed] [Google Scholar]
- Frenz DA, Liu W, Galinovic-Schwartz V, Van De Water TR. Retinoic acid-induced embryopathy of the mouse inner ear. Teratology. 1996;53:292–303. doi: 10.1002/(SICI)1096-9926(199605)53:5<292::AID-TERA3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Frenz DA, Liu W. Effect of retinoic acid on otic capsule chondrogenesis in high-density culture suggests disruption of epithelial-mesenchymal interactions. Teratology. 1997;56:233–240. doi: 10.1002/(SICI)1096-9926(199710)56:4<233::AID-TERA1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Frenz DA, Liu W. Treatment with all-trans-retinoic acid decreases levels of endogenous TGF-beta(1) in the mesenchyme of the developing mouse inner ear. Teratology. 2000;61:297–304. doi: 10.1002/(SICI)1096-9926(200004)61:4<297::AID-TERA9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Developmental Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Hans S, Westerfield M. Changes in retinoic acid signaling alter otic patterning. Development. 2007;134:2449–2458. doi: 10.1242/dev.000448. [DOI] [PubMed] [Google Scholar]
- Hatzopoulos AK, Folkman J, Vasile E, Eiselen GK, Rosenberg RD. Isolation and characterization of endothelial progenitor cells from mouse embryos. Development. 1998;125:1457–1468. doi: 10.1242/dev.125.8.1457. [DOI] [PubMed] [Google Scholar]
- Hishikawa K, Marumo T, Miura S, Nakanishi A, Matsuzaki Y, Shibata K, Ichiyanagi T, Kohike H, Komori T, Takahashi I, Takase O, Imai N, Yoshikawa M, Inowa T, Hayashi M, Nakaki T, Nakauchi H, Okano H, Fujita T. Musculin/MyoR is expressed in kidney side population cells and can regulate their function. The Journal of Cell Biology. 2005;169:921–928. doi: 10.1083/jcb.200412167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MA, Miller CP, Habener JF. Brn-4 transcription factor expression targeted to the early developing mouse pancreas induces ectopic glucagon gene expression in insulin-producing beta cells. The Journal of Biological Chemistry. 2002;277:16028–16032. doi: 10.1074/jbc.M107124200. [DOI] [PubMed] [Google Scholar]
- Jimenez MJ, Balbin M, Alvarez J, Komori T, Bianco P, Holmbeck K, Birkedal-Hansen H, Lopez JM, Lopez-Otin C. A regulatory cascade involving retinoic acid, Cbfa1, and matrix metalloproteinases is coupled to the development of a process of perichondrial invasion and osteogenic differentiation during bone formation. The Journal of Cell Biology. 2001;155:1333–1344. doi: 10.1083/jcb.200106147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Human Molecular Genetics. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- Liu TX, Zhang JW, Tao J, Zhang RB, Zhang QH, Zhao CJ, Tong JH, Lanotte M, Waxman S, Chen SJ, Mao M, Hu GX, Zhu L, Chen Z. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood. 2000;96:1496–1504. [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Molecular Endocrinology. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Maerz WJ, Baselga J, Reuter VE, Mellado B, Myers ML, Bosl GJ, Spinella MJ, Dmitrovsky E. FGF4 dissociates anti-tumorigenic from differentiation signals of retinoic acid in human embryonal carcinomas. Oncogene. 1998;17:761–767. doi: 10.1038/sj.onc.1201992. [DOI] [PubMed] [Google Scholar]
- Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W, Cui H, Ding HF. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Research. 2011;71:4314–4324. doi: 10.1158/0008-5472.CAN-11-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis JM, Simmons DM, He X, Swanson LW, Rosenfeld MG. Brain 4: a novel mammalian POU domain transcription factor exhibiting restricted brain-specific expression. The EMBO Journal. 1992;11:2551–2561. doi: 10.1002/j.1460-2075.1992.tb05320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino N, Shoji H, Sugo S, Kangawa K, Matsuo H. Adrenocortical steroids, thyroid hormones and retinoic acid augment the production of adrenomedullin in vascular smooth muscle cells. Biochemical and Biophysical Research Communications. 1995;211:686–693. doi: 10.1006/bbrc.1995.1866. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, Fritzsch B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell and Tissue Research. 2008;334:339–358. doi: 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- Orlandi A, Pucci S, Ciucci A, Pichiorri F, Ferlosio A, Spagnoli LG. Modulation of clusterin isoforms is associated with all-trans retinoic acid-induced proliferative arrest and apoptosis of intimal smooth muscle cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2005;25:348–353. doi: 10.1161/01.ATV.0000152609.28569.e1. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Phippard D, Lu L, Lee D, Saunders JC, Crenshaw EB., 3rd Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 1999;19:5980–5989. doi: 10.1523/JNEUROSCI.19-14-05980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radosevic M, Robert-Moreno A, Coolen M, Bally-Cuif L, Alsina B. Her9 represses neurogenic fate downstream of Tbx1 and retinoic acid signaling in the inner ear. Development. 2011;138:397–408. doi: 10.1242/dev.056093. [DOI] [PubMed] [Google Scholar]
- Raft S, Nowotschin S, Liao J, Morrow BE. Suppression of neural fate and control of inner ear morphogenesis by Tbx1. Development. 2004;131:1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Human Molecular Genetics. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- Robertson NG, Heller S, Lin JS, Resendes BL, Weremowicz S, Denis CS, Bell AM, Hudspeth AJ, Morton CC. A novel conserved cochlear gene, OTOR: identification, expression analysis, and chromosomal mapping. Genomics. 2000;66:242–248. doi: 10.1006/geno.2000.6224. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Lufkin T. Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis. 2006;44:425–437. doi: 10.1002/dvg.20233. [DOI] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. Journal of Neurochemistry. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert T, Schlegel J, Schmid R, Opolka A, Grassel S, Humphries M, Bosserhoff AK. Modulation of cartilage differentiation by melanoma inhibiting activity/cartilage-derived retinoic acid-sensitive protein (MIA/CD-RAP) Experimental & Molecular Medicine. 2010;42:166–174. doi: 10.3858/emm.2010.42.3.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice DA, Szeto W, Soloviev I, Rubinfeld B, Fong SE, Dugger DL, Winer J, Williams PM, Wieand D, Smith V, Schwall RH, Pennica D, Polakis P. Synergistic induction of tumor antigens by Wnt-1 signaling and retinoic acid revealed by gene expression profiling. The Journal of Biological Chemistry. 2002;277:14329–14335. doi: 10.1074/jbc.M200334200. [DOI] [PubMed] [Google Scholar]
- Tono T, Shimozono M, Kawano H, Asada Y, Kitamura K, Komune S. Expression and immunohistochemical localization of adrenomedullin in the mouse cochlea. ORL; Journal for Oto-rhino-laryngology and its Related Specialties. 2002;64:169–172. doi: 10.1159/000058020. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Viola A, Morishima M, Pramparo T, Baldini A, Lindsay E. TBX1 is required for inner ear morphogenesis. Human Molecular Genetics. 2003;12:2041–2048. doi: 10.1093/hmg/ddg216. [DOI] [PubMed] [Google Scholar]
- Xu H, Viola A, Zhang Z, Gerken CP, Lindsay-Illingworth EA, Baldini A. Tbx1 regulates population, proliferation and cell fate determination of otic epithelial cells. Developmental Biology. 2007;302:670–682. doi: 10.1016/j.ydbio.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YY, Guan DY, Yang M, Wang H, Shen ZH. All-trans-retinoic acid intensifies endoplasmic reticulum stress in N-acetylglucosaminyltransferase V repressed human hepatocarcinoma cells by perturbing homocysteine metabolism. Journal of Cellular Biochemistry. 2010;109:468–477. doi: 10.1002/jcb.22423. [DOI] [PubMed] [Google Scholar]
- Zhong JC, Huang DY, Liu GF, Jin HY, Yang YM, Li YF, Song XH, Du K. Effects of all-trans retinoic acid on orphan receptor APJ signaling in spontaneously hypertensive rats. Cardiovascular Research. 2005;65:743–750. doi: 10.1016/j.cardiores.2004.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression changes from the E12.5 OV+POM microarray carried out on the Affymetrix Mouse Gene ST 1.0 platform. Genes with fold-changed >+/-1.25 and p-value <0.05 are represented
Primers for PCR amplification of E12.5 cDNA are listed. T3 polymerase and T7 polymerase site sequences were add to the forward and reverse primers, respectively, for the synthesis of sense and anti-sense riboprobes. All reactions were carried out with 2.5mM MgCl2 and without the addition of DMSO or Betaine for 35 cycles of 1 minute each. Probe sizes and individual annealing temperatures are provided.


