Abstract
Heterogeneity of cellular systems has been widely recognized but only recently have tools become available that allow probing of genes and proteins in single cells to understand it. While the advancement in single cell genomic analysis has been greatly aided by the power of amplification techniques (e.g., PCR), analysis of proteins in single cells has proven to be more challenging. However, recent advances in multi-parameter flow cytometry, microfluidics and other techniques have made it possible to measure wide variety of proteins in single cells. In this review, we highlight key recent developments in analysis of proteins in a single cell, and discuss their significance in biological research.
Introduction
Proteins are central to all cellular processes - including providing structure to cells, transporting molecules across cell membranes, controlling cell growth and adhesion, catalyzing biochemical processes by functioning as enzymes and regulating signal transduction. Characterizing the quantity and activity of proteins is therefore critical for understanding molecular mechanisms of cellular processes including those involved in disease progression, cell differentiation and fate, and for targeted discovery and development of novel therapeutics, vaccines and diagnostics. Measuring DNA and RNA can provide qualitative information on gene-products (proteins) but cannot provide information on protein location, post-translational modifications (PTMs) or interactions with other proteins and hence, we need tools and assays to directly measure proteins, their modifications and interactions. Numerous analytical methods have been developed to analyze proteins such as gel electrophoresis, immunoassays, chromatography and mass spectrometry. However, these methods require a large number of cells for analysis, resulting in a population-averaged measurement. Cells are heterogeneous in nature and hence, population-averaged data can mask the underlying molecular mechanisms; more desirable data in many instances could be data at the level of single cells[1–5]. A well-known example is response of bacteria to antibiotics, at certain doses many die but some survive. Similarly, one of the unanswered questions in cancer therapy has been why essentially identical cells respond differently to a drug. Single-cell level measurement of proteins (and other molecules) has provided valuable insight into mechanisms that dictate heterogeneity in cellular response to drugs and other internal and external stimuli. For example, it was reported that dynamic response of tumor suppressor protein p53 network derived from population studies was misleading [4]. Instead of damped oscillations seen in population-averaged data, individual cells show series of undamped p53 pulses with fixed amplitude and duration, independent of the amount of γ-irradiation. Similarly, real-time imaging of transcription factor RelA translocation in single cells revealed variability in the oscillatory dynamics of RelA translocation among single cells, and that RelA translocation dynamics determined the degree and timing of downstream gene expression[3]. Usefulness of single cell measurements is obvious for stem cell research as decisions in individual cells determines their fate. For haematopoietic stem cells, studying the varying levels of Sca-1 protein in individual cells revealed that Sca-1 protein abundance determines the timing and type of differentiation[6]. In a clinical context, single-cell level examination of T cell populations previously thought to be homogeneous were found to contain subpopulations with different cytokine profiles [25], and these differences may serve to predict patient immune response to drug therapy.
Challenges in single-cell protein analysis
The biggest challenges to measuring proteins in single cells (assuming they have been successfully isolated from a tissue, microbial community or culture) are vanishingly small amount of protein in a single cell and the enormous complexity of the proteome. Adapting traditional bulk protein analysis methods for single-cell level applications has met with varied degrees of success, with quantitative analysis being especially elusive. Proteomic measurements can be quite complex as there are various types of measurements to be made–protein abundance or expression levels, post-translational modifications, protein translocation, interactions with other proteins, DNA etc., and protein activity (Figure 1). No single analytical method can measure all of these protein parameters in a single cell hence, a suite of biochemical and biophysical methods have been developed as discussed in the following sections.
Figure 1. Protein analysis in single cells.
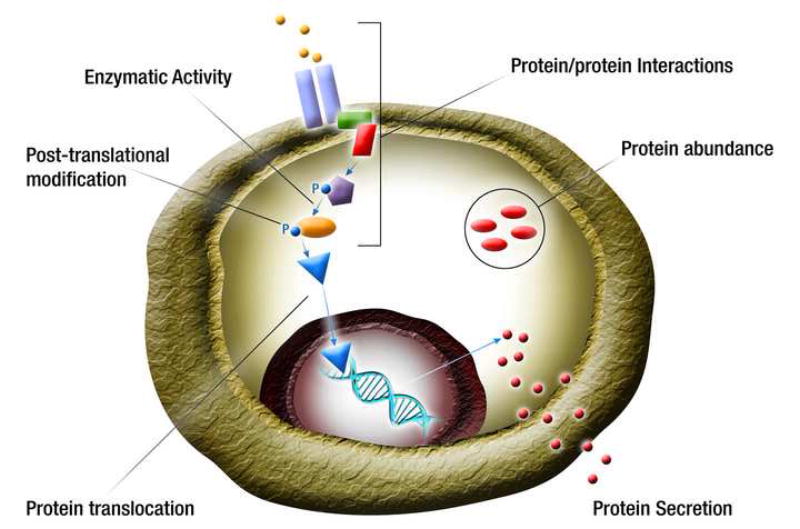
Proteome of a cell is complex and requires measurements of many attributes including abundance or concentration of cytosolic, membrane-bound and secreted proteins, protein interactions with other proteins and molecules, inter- and intra-organelle translocation of proteins, post-translational modifications, and enzymatic and other activities of a protein. Colored shapes represent different proteins. P – phosphorylation.
Single-cell protein analysis by flow cytometry
The most established and user-friendly method for single cell protein analysis is flow cytometry. Its effectiveness derives from the fact that the while absolute protein amounts in a cell can be vanishingly small, the localized protein concentrations can be larger and measurable if the cells are kept intact. Since its invention in late sixties, flow cytometry has been transformed from a technique limited to measuring 1–2 fluorescent species in a cell to 10–15 species today, allowing profiling of entire pathways in single cells. This improvement has been enabled by advancements in both instrumentation and availability of highly specific antibodies. Roederer [7,8] and Nolan[9] groups pioneered the use of multi-parameter analysis using multi-color flow cytometers to measure 10–15 key proteins in signaling pathways simultaneously in single cells. The ability to perform correlated measurements of multiple proteins in single cells has turned cytometry into a powerful tool to semi-quantitatively analyze pathways underlying many diseases [10,11]. Tyramide signal amplification, which has long been used as a means to amplify nucleic acid detection in in-situ hybridization protocols, has been added to traditional antibody staining techniques for analysis of low abundance proteins [12]. While multi-parameter cytometry allowed high content screening (for example, of multiple kinases) in cells, its throughput was still too limited to be useful for drug screening. This limitation was overcome by developing a barcoding method where differently treated cells are tagged with a combination of three dyes [13]. Each dye, depending upon dilution, permits up to 7 intensity levels and hence, combination of the 3 dyes allows processing of up to 343 samples from one pool of cells.
While flow cytometry has been most commonly used for analysis of kinases and phosphatases, it is also useful for other types of protein measurements such as glycosylation levels and cytokine production. Venable et al. used a panel of 14 lectins to characterize glycans present on cell surface as potential markers of pluripotency in human embryonic stem cells[14].
A factor limiting the wider application of flow cytometry to glycolsylated protein is that there are very few lectins available. Flow cytometry can also be used for analysis of secreted proteins such as cytokines. This requires treating cells with a vesicle formation inhibitor to trap synthesized cytokines in the Golgi, followed by fixation and permeabilization to stain the trapped cytokines with fluorescent antibodies for flow cytometric analysis[15].
Microfluidic flow cytomtery
While commercial flow cytometers allow interrogation of cells one at a time, the sample preparation is still done manually and therefore, requires a large numbers of cells. This makes it hard to analyze samples that are limited in amount such as cells recovered from a biopsy sample, tissue specimens or small volumes of blood. To permit analysis of small number of cells (100–1000), microfluidic platforms have been developed that integrate sample handling with flow cytometry and sorting [16,17]. Srivastava et. al. [18] developed an integrated microfluidic device, retro-fitted to commercial microscopes, that can perform cell culture, stimulation and sample preparation in combination with conventional fluorescence imaging and microfluidic flow cytometry (Figure 2) to monitor immune response in macrophages. This microfluidic device not only drastically reduced the amount of sample and reagent required, it also provided a means to perform two orthogonal modes of measurements, imaging and cytometry, in one experiment. Similarly, McKenna et al. took advantage of microfluidics to implement a 384-channel flow cytometer for imaging cells. This approach overcomes the low throughput of CCD-based imaging flow cytometers by collecting one-dimensional, low-pixel images (~1/1000 information compared to a CCD image) in up to 384 fluidic channels simultaneously[19].
Figure 2. Microfluidic Platform for single cell protein analysis.
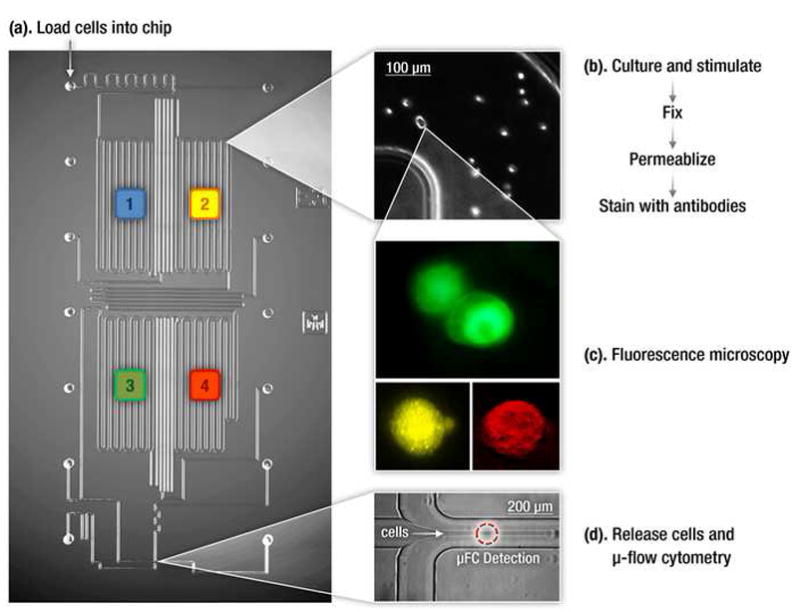
An integrated microfluidic platform that integrates cell culture and sample preparation with two orthogonal single-cell resolution techniques- flow cytomtery and fluorescence microscopy. [18].
Single-cell protein analysis by mass cytometry
The flow cytometry based analysis of single cells has permitted analysis of as many as 15 proteins simultaneously as described earlier. However, system-level interrogation of biological pathways requires the ability to do many more correlated measurements. A promising development has been an approach called mass cytometry where the throughput of flow cytometry is combined with the ultra-high dimensionality and sensitivity of mass spectrometry [20]. In mass cytomtery, cells are stained with 20–30 antibodies conjugated to different metal isotope containing polymers[21]. The labeled cells are injected and nebulized and the metal tags are quantified using inductively coupled plasma time-of-flight mass spectrometry. This powerful technique was used to simultaneously profile 34 parameters in single bone marrow cells, including binding of 31 antibodies, viability, DNA content, and relative cell size[22]. This technique should be capable of measuring even higher numbers of markers simultaneously considering that the precision of mass spectrometry detection overcomes the issue of spectral overlap confounding fluorescence measurements.
Affinity arrays for single-cell protein analysis
Surface immobilized antibody platforms can also be adapted to detect secreted proteins from single cells. For example, commercially-available ELISpot method [23], which relies on antibody-coated PVDF membrane to bind target protein followed by detection with a second antibody, was modified to use fluorescence based detection of cytokines secreted from single cells[24]. However, it can take many hours, even days before cytokines are detectable and multiplexing is limited to 1–3 cytokines. A major stride towards multiplexing was made recently in the form of high-throughput single-cell barcoded chip[25]. The chip contains 1040 3-nl volume microwells containing DNA-barcoded antibodies to more than 10 cytokines, with the quantitation standards at protein copy number resolution built into the antibody barcodes. 1 to 40 cells are isolated in each microwell, and their cytokine secretions are assessed by fluorescence immunosandwich assay. The chip was successfully used to profile cytokine secretion from tumor antigen-specific cytotoxic T cells and could be useful for profiling of other immune pathways. Another improvement to the affinity array was made by Choi et. al., where cytokines released from single cells were detected by antibodies with covalently attached fluorescent oligomers that can be amplified to increase detection sensitivity up to 200-fold [26].
Mass spectrometry based single-cell protein analysis
Single cell mass spectrometry (MS) has the potential to provide label-free quantitative analysis of the entire proteome of a single cell, including proteins, peptides and PTMs. The advantage of MS is that there is no need for molecular labels, and femtomolar sensitivity is routinely achieved for pure proteins. Mass spectrometry techniques used for single cell studies include electrospray MS, laser/desorption/ionization (LDI-MS), and secondary ion MS (SIMS). Matrix-assisted laser desorption (MALDI) -MS has been used to analyze neuropeptides in single neurons[27]. Using MS to study proteins in the single cells has its limitations. The biggest drawback is the lack of sensitivity (low signal/noise) to detect low amount of proteins typically found in single cells. Sample preparation methods that can fractionate proteins prior to mass spectrometry can help. In a recent article, integrating microfluidic cell lysis and capillary electrophoretic separation with electrospray mass spectrometry was used for high throughput detection of hemoglobin in individual erythrocytes[28]. A recent report combining microarray with MS analysis holds promise for increasing the throughput of single cell protein analysis[29].
Separation based single-cell protein analysis
A cell typically has thousands of expressed proteins and high-resolution separation of the cell content into individual proteins by electrophoresis or chromatography can greatly aid measurements of those proteins. The advantage of separation methods, especially multidimensional separations, is that they allow unbiased measurement of the entire proteome in one experiment. This enables researchers to monitor changes in a cell’s proteome at a global level as a function of an external stimulus. Conventional scale separations such as HPLC or slab gel electrophoresis are impractical for single cells because of their inability to process minute amounts of proteins. Capillary electrophoresis, implemented in capillaries that mimic the dimensions of cells (10–100s of μm i.d.) or more recently in microfluidic chips with micron-sized channels, is more promising as it has the potential to separate and analyze proteome of a single cell, especially the large mammalian cells[30]. To improve peak capacity, two-dimensional separation has been attempted to profile changes in cancer cells [31]. A microfluidic device that integrates capture, lysis, capillary electrophoresis of single cells with single-molecule fluorescence counting was used to quantify rare protein species (<1000 copies per cell)[32]. Despite these advances, separation based methods are not routinely used in analysis of single cells because of two major factors—the capillary- or microfluidic chip-based separations do not have the peak capacity to resolve thousands of proteins in a cell and most of the detection methods used do not have the ability to detect low-abundant proteins.
Genetic and chemical probes for single-cell protein analysis
The accuracy and sensitivity of protein measurements rely largely upon the availability and functionality of the molecular probes that can selectively bind to proteins of interest. Antibodies enjoy widespread popularity due to their high specificity, but they are not the universal remedy for all protein analysis applications. Antibodies do not enter live cells, so they cannot be used in live-cell studies. They are also cumbersome to generate against new antigens, and too expensive when many proteins are to be profiled simultaneously. Alternatives to antibodies include genetic probes – the GFP family of fluorescent proteins that can be genetically fused to the protein of interest, and translated into fluorescent fusion proteins that are detectable and quantifiable, even for large scale proteomic profiling[33]. The drawbacks of GFP and other genetic probes are a) that they require genetic engineering of the cell, limiting their application to cultured cells, and b) their large size can interfere with protein function. To tag proteins with smaller probes in a living cell, a new class of chemical molecules, referred to as bioorthogonal probes, has been developed. Bioorthogonal chemistry utilizes the cell’s own machinery to covalently incorporate abiotic probes to specific cellular components including proteins and their post-translational modifications. Some of the highlights in bioorthogonal probe development include the biarsenical fluorescent dye FLAsH, which specifically reacts with target proteins genetically fused with the tetracysteine motif[34]; the carbonyl condensation of ketone-functionalized amono acids into proteins and detection by a hydrazide-funcitonalized fluorescent probe[35]; and the selective bioorthogonal chemical probes for metabolic labeling of glycans [36].
Future perspective
The future of single cell protein analysis will depend on several factors. First, a larger repertoire of high affinity probes (other than monoclonal antibodies) are needed to detect low abundance proteins , post-tranlsational modifications and protein interactions. Some promising candidates include aptamers, single-chain variable fragments, and biorthogonal reporters. Second, techniques that can effectively and reliably isolate single cells from culture, tissues, clinical and environmental samples will need to be further improved in their yield, ease-of-use and reproducibility. Third, most of the techniques described above use experiments at the population level followed by analysis at the single cell level. It will also be useful to have technologies that enable experimentation as well as subsequent analysis at the single cell level. For example, introducing one virus to one immune cell and recording the interaction in real time. While such experiments do not necessarily represent biological phenomena in vivo, they can simplify the interactions sufficiently to make basic observations that are otherwise hidden in a population experiment. Finally, in order for new single- cell technologies to become widely used, we also need to develop informatics and modeling tools that are customized for single cells and can integrate the protein measurements with other types of single cell measurements such as genomics, transcriptomics, and metabolomics.
Figure 3. Mass cytometry measurement of immune cell response.
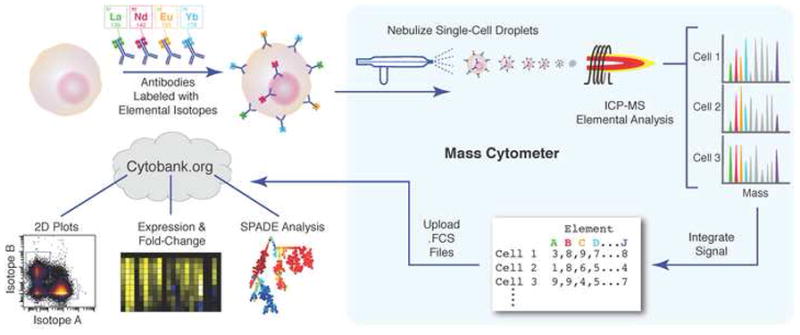
Cells are stained with epitope-specific antibodies conjugated to transition element isotope reporters of different masses. Cells are nebulized into single-cell droplets, and an elemental mass spectrum is acquired for each. The integrated integrated elemental reporter signals can then be interpreted using conventional flow cytometry data analysis methods and plotted as heat maps or tree plots. (Reproduced with permission from Bendall et. al. [21])
Acknowledgments
Financial support for preparation and some of the work included was provided by the grants: R01 DE020891, funded by the NIDCR; P50GM085273 (the New Mexico Spatiotemporal Modeling Center) funded by the NIGMS; and ENIGMA, a Lawrence Berkeley National Laboratory Scientific Focus Area Program supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research. Sandia is a multi-program laboratory operated by Sandia Corp., a Lockheed Martin Co., for the United States Department of Energy under Contract DE-AC04-94AL85000.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• Special Interest
• • Outstanding Interest
- 1.Spiller DG, Wood CD, Rand DA, White MR. Measurement of single-cell dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 2.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor RJ, Falconnet D, Niemisto A, Ramsey SA, Prinz S, Shmulevich I, Galitski T, Hansen CL. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc Natl Acad Sci U S A. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •7.De Rosa SC, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. doi: 10.1038/84701. First report of multiparameter flow cytometry, the beginning of a very prolific field of single cell analysis. [DOI] [PubMed] [Google Scholar]
- 8.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- ••9.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20:155–162. doi: 10.1038/nbt0202-155. Multi-parameter flow cytometry applied to analyzing phosphorylation states in a signaling cascade, landmark paper in the field of phosphoprofiling. [DOI] [PubMed] [Google Scholar]
- 10.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 11.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Clutter MR, Heffner GC, Krutzik PO, Sachen KL, Nolan GP. Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry A. 2010;77:1020–1031. doi: 10.1002/cyto.a.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •13.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. Simple, yet effective method to increase the number of parameters that can be multiplexed using one cell population by barcoding cells with varying intensities of fluorescent dyes prior to phosphostaining. [DOI] [PubMed] [Google Scholar]
- 14.Venable A, Mitalipova M, Lyons I, Jones K, Shin S, Pierce M, Stice S. Lectin binding profiles of SSEA-4 enriched, pluripotent human embryonic stem cell surfaces. BMC Dev Biol. 2005;5:15. doi: 10.1186/1471-213X-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson AC, Martin JN, Younger SR, Bredt BM, Epling L, Ronquillo R, Varma A, Deeks SG, McCune JM, Nixon DF, et al. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003;283:141–153. doi: 10.1016/j.jim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Lindstrom S, Andersson-Svahn H. Overview of single-cell analyses: microdevices and applications. Lab Chip. 2010;10:3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- 17.Perroud TD, Kaiser JN, Sy JC, Lane TW, Branda CS, Singh AK, Patel KD. Microfluidic-based cell sorting of Francisella tularensis infected macrophages using optical forces. Anal Chem. 2008;80:6365–6372. doi: 10.1021/ac8007779. [DOI] [PubMed] [Google Scholar]
- ••18.Srivastava N, Brennan JS, Renzi RF, Wu M, Branda SS, Singh AK, Herr AE. Fully integrated microfluidic platform enabling automated phosphoprofiling of macrophage response. Anal Chem. 2009;81:3261–3269. doi: 10.1021/ac8024224. Fully integrated microfluidic platform that combines cell culture, stimulation, preparation, and two methods for analysis. [DOI] [PubMed] [Google Scholar]
- •19.McKenna BK, Evans JG, Cheung MC, Ehrlich DJ. A parallel microfluidic flow cytometer for high-content screening. Nat Methods. 2011;8:401–403. doi: 10.1038/nmeth.1595. Simple 1D imaging cytometer demonstrated on yeast cells expressing GFP fusion proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. The development of the mass cytometry technique, including schematics of the instrument. [DOI] [PubMed] [Google Scholar]
- 21.Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. Application of the newly available mass cytometry technique in primary stem cell profiling. Over 30 parameters profiled, and excellent use of bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods. 1983;65:109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 24.Casey R, Blumenkrantz D, Millington K, Montamat-Sicotte D, Kon OM, Wickremasinghe M, Bremang S, Magtoto M, Sridhar S, Connell D, et al. Enumeration of functional T-cell subsets by fluorescence-immunospot defines signatures of pathogen burden in tuberculosis. PLoS One. 2010;5:e15619. doi: 10.1371/journal.pone.0015619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••25.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;17:738–743. doi: 10.1038/nm.2375. Single-cell barcoded chip that can multiplex the measurement of up to 10 released cytokines. Patient T cells were profiled using the chip, and cytokine profiles proved heterogenous in otherwise phenotypically similar T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi J, Love KR, Gong Y, Gierahn TM, Love JC. Immuno-hybridization chain reaction for enhancing detection of individual cytokine-secreting human peripheral mononuclear cells. Ana Chem. 2011;17:6890–5. doi: 10.1021/ac2013916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal Chem. 2010;82:967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Urban PL, Jefimovs K, Amantonico A, Fagerer SR, Schmid T, Madler S, Puigmarti-Luis J, Goedecke N, Zenobi R. High-density micro-arrays for mass spectrometry. Lab Chip. 2010;10:3206–3209. doi: 10.1039/c0lc00211a. Combination of high-throughput microarray platform with MALDI-MS to profile single cells or proteins. [DOI] [PubMed] [Google Scholar]
- •30.Hu S, Le Z, Newitt R, Aebersold R, Kraly JR, Jones M, Dovichi NJ. Identification of proteins in single-cell capillary electrophoresis fingerprints based on comigration with standard proteins. Anal Chem. 2003;75:3502–3505. doi: 10.1021/ac034154j. 2D capillary electrophoresis performed on single breast cancer cells. [DOI] [PubMed] [Google Scholar]
- 31.Sobhani K, Fink SL, Cookson BT, Dovichi NJ. Repeatability of chemical cytometry: 2-DE analysis of single RAW 264. 7 macrophage cells. Electrophoresis. 2007;28:2308–2313. doi: 10.1002/elps.200700017. [DOI] [PubMed] [Google Scholar]
- •32.Huang B, Wu H, Bhaya D, Grossman A, Granier S, Kobilka BK, Zare RN. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. A microfluidic device that can lyse and separate protein contents from a single cell, and quantify the fluorescent antibody labeled proteins by using single-molecule fluoresence counting. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 35.Cornish VWK, Hahn M, Schultz PG. Probing Protein Structure and Function with an Expanded Genetic Code. J Am Chem Soc. 1996;118:8150–8151. [Google Scholar]
- ••36.Boyce M, Bertozzi CR. Bringing chemistry to life. Nat Methods. 2011;8:638–642. doi: 10.1038/nmeth.1657. Comprehensive review of bioorthogonal chemistry and probes. [DOI] [PMC free article] [PubMed] [Google Scholar]


