Abstract
To study mGluR6 expression, the authors investigated two transgenic mouse lines that express enhanced green fluorescent protein (GFP) under control of mGluR6 promoter. In retina, GFP was expressed exclusively in all ON bipolar cell types, either uniformly across all cells of this class (line 5) or in a mosaic (patchy) fashion (line 1). In brain, GFP was found in certain cortical areas, superior colliculus, axons of the corpus callosum, accessory olfactory bulb, and cells of the subcommissural organ. Outside the nervous system, GFP was seen in the corneal endothelium, testis, the kidney’s medulla, collecting ducts and parietal layer that surround the glomeruli, and B lymphocytes. Furthermore, RT-PCR showed that most tissues that expressed GFP in the transgenic mouse also transcribed two splice variants of mGluR6 in the wild-type mouse. The alternate variant was lacking exon 8, predicting a protein product of 545 amino acids that lacks the 7-transmembrane domains of the receptor. In cornea, immunostaining for mGluR6 gave strong staining in the endothelium, and this was stronger in wild-type than in mGluR6-null mice. Furthermore, calcium imaging with Fura-2 showed that application of L-AP4, an agonist for group III metabotropic glutamate receptors including mGluR6, elevated calcium in endothelial cells.
Keywords: retina, ON bipolar cells, kidney, cornea, subcommissural organ, accessory olfactory bulb, lymphocytes
Glutamate serves as the most important fast excitatory neurotransmitter in the brain, and it is essential for neuronal communication. Glutamatergic excitation is mediated primarily by ionotropic glutamate receptors iGluR1-7 and NMDA receptors (NR1 and NR2). On a slightly slower time scale, glutamate also modulates synaptic transmission via G-protein coupled receptors or metabotropic glutamate receptors (mGluRs) types 1–8 (recently reviewed by Nakanishi 1992; Bockaert et al. 1993; Pin and Duvoisin 1995; Niswender and Conn 2010). Among mGluRs, type 6 is unique in its particularly high sensitivity to glutamate and relatively slow desensitization (Nakajima et al. 1993). It belongs to group III metabotropic glutamate receptors characterized by high sensitivity to L-AP4, which is a specific agonist. In retina, mGluR6 is required for the mediation of the light response of a specific class of bipolar cells (ON bipolar cells) that reverse the sign of the signal from photoreceptors. In the dark, photoreceptors continuously release glutamate, and mGluR6 located at the tips of ON bipolar cell dendrites continuously senses this glutamate. This in turn leads to closure of TRPM1, a non-specific cation channel, and to hyperpolarization of the cell (Nawy and Jahr 1990; Shiells and Falk 1990; Morgans et al. 2009; Shen et al. 2009; Koike et al. 2010). When light intensity increases, glutamate release decreases and mGluR6 deactivates to open the cation channels and depolarize the ON bipolar cells.
Although the primary function of glutamate as a neurotransmitter is exerted by neurons, numerous recent reports document the expression of glutamate receptors also in non-neuronal tissues. For example, rat oesteoclasts express NMDA receptors (Chenu et al. 1998; Gu et al. 2002); rat and human testis express mGluR1 and 5 (Storto et al. 2001); rat pancreatic islets express mGluR8 (Tong et al. 2002); and lymphocytes have both NMDA receptors and certain types of mGluRs (reviewed by Boldyrev et al. 2005). It is reasonable to assume that the glutamate receptors expressed in non-neuronal systems function to receive glutamate signaling. This idea gains support from the existence of other glutamate-associated proteins, such as membrane glutamate transporters and vesicular glutamate transporters, in some of these tissues (Mason et al. 1997; reviewed by Skerry and Genever 2001; Hayashi et al. 2003; reviewed by Hinoi, Takarada, Ueshima et al. 2004). While it is becoming clear that glutamate serves as an important communicator in these tissues, the extent of its receptor distribution and the functional significance of these communications are largely unknown.
In an effort to study retinal ON bipolar cells, we produced a transgenic mouse that expresses green fluorescent protein (GFP) under the control of mGluR6 promoter (Morgan et al. 2006; Dhingra et al. 2008). In four out of six lines that were produced, GFP in retina was restricted to ON bipolar cells, the cell class that is known to express mGluR6. In line 5, GFP fluorescence was strong enough to follow its expression, study the development of the cells throughout retinal development (Morgan et al. 2006), and sort the cells to create an ON bipolar cell cDNA library (Dhingra et al. 2008). The fact that GFP expression was specific in several lines and that glutamate signaling occurs also outside the nervous system prompted us to investigate GFP localization in this transgenic mouse. It should be noted that although a variety of glutamate receptors and transporters were previously found in many organs, mGluR6 expression has not been reported in non-neural cells, except for prostate cancer cell line or microglial culture (Taylor et al. 2003; Pissimissis et al. 2009). Here we investigated the extent of GFP expression in two transgenic lines (lines 1 and 5) and found it to be abundant in many organs outside the brain. We further found that most organs that express GFP in the transgenic mouse transcribe two splice variants of mGluR6 in the wild-type mouse. For one tissue, cornea, we also investigated if there is any detectable expression of mGluR6 protein. Protein assays with antibodies did not clearly detect it, but corneal endothelium did respond to the mGluR group III agonist, L-AP4.
Methods
Mouse Models
Work was performed on C57BL/6J wild-type mice (Charles River laboratories), Grm6-GFP mice, and Grm6-null mice. Grm6:GFP mouse lines were previously described in detail (Dhingra et al. 2008). Briefly, mGluR6 promoter sequence that was extracted from pN2N (a generous gift of Y. Nakajima and S. Nakanishi) was ligated upstream to a GFP coding sequence in pEGFP1 (Clontech, Cambridge, UK). A 10.5-kb sequence consisting of mGluR6 promoter fused to GFP coding sequence was extracted and used for microinjection into fertilized eggs (performed by the University of Pennsylvania Transgenic and Chimeric Mouse Facility). The Grm6-null mouse was obtained from Dr David Copenhagen (University of California, San Francisco) with permission from Dr Nakanishi and was previously described (Masu et al. 1995). Animals were treated in compliance with federal regulations and University of Pennsylvania policies.
GFP Imaging and Immunocytochemistry
Mice were deeply anesthetized by intraperitoneal injection (85 µg/g of ketamine plus 13 µg/g of xylazine), and a variety of tissues and organs were dissected out. Mice were sacrificed by anesthetic overdose (3× initial dose). For Western blotting and RT-PCR, tissue was immediately frozen in liquid nitrogen. For imaging and immunocytochemistry, tissue was fixed by immersion in buffered 4% paraformaldehyde for 10–60 min. It was then rinsed in buffer, soaked overnight in 30% buffered sucrose, and embedded in a mixture of two parts 20% sucrose in phosphate buffer and one part Tissue Freezing Medium (OCT compound, EMS). Tissue blocks were then frozen in liquid nitrogen, and stored in −80C. Blocks were cryosectioned at 10–15 µm thickness on glass slides, dried for 20 min, and again stored in −80C. Sections were defrosted in air and either embedded directly for imaging (by hydrating them with phosphate buffer and then applying Vectashield) or immunostained according to a standard protocol. Sections were soaked in 50 µl of diluent containing 10% normal goat serum, 5% sucrose, and 0.3% Triton X-100 in phosphate buffer. Whole mount cornea was incubated in the same solution free floating in a well on a slow shaker. Sections or corneal pieces were then incubated in a primary antibody overnight at 4C, washed, incubated (3 hr) in a secondary antibody conjugated to a fluorescent marker, rinsed, and then mounted in Vectashield. Sections were imaged with a Leica confocal microscope (in early work) or an Olympus confocal microscope (FV-1000) under 40× (NA 1.3) and 60× (NA 1.42) with a zoom of two or three. For display, images were contrast adjusted with Adobe Photoshop and prepared with Adobe Illustrator. Two antibodies against mGluR6 were used. Both were raised against the C-terminus: one was raised in rabbit (gift of Dr Shigetada Nakanishi) (Nomura et al. 1994) and was used at a dilution of 1:500; the other was raised in sheep (gift of Catherine Morgans; Oregon Health and Science University) (Morgans et al. 2007) and was used at 1:200. Figures shown here used the anti-rabbit antibody. In retina, the antibodies against mGluR6 are specific since staining is eliminated in the mGluR6-null mouse. In cornea, these antibodies seem to recognize other proteins as well (this study). The antibody against Na+/K+/Cl- cotransporter (NKCC1) was raised in rabbit against the C-terminus of mouse NKCC1 (gift of Eric Delpire, Nashville, Tennessee) and was used at a dilution of 1:50. Specificity of this antibody was proven by the abolished staining in an NKCC1-null retina (Zhang et al. 2007).
For Cell Sorting
Single-cell suspensions were prepared from GFP-expressing lymph nodes, and cells were surface stained with the PE (phycoerythrin)–labeled Abs (CD3 or B220; eBioscience; San Diego, CA) and then used for flow cytometry. Cells were sorted by the University of Pennsylvania Flow Cytometry Core using a three-laser, six-color FACSAria (BD Biosciences). FlowJo 8.7.1 (Tree Star) was used to analyze data. B-lymphocytes were identified with B220 antibody and T-lymphocytes were identified with CD3 antibody.
RT-PCR
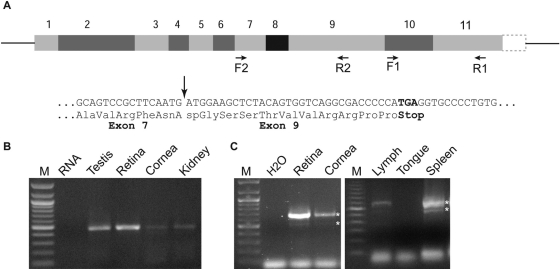
Segments of mGluR6 transcripts were amplified by RT-PCR from the RNA of either whole retina or other tissues, prepared using Nucleospin RNA II kit (Clontech, Mountain View, CA). Reverse transcription was performed on ~1 µg of total RNA with oligo dT primers using Moloney murine leukemia virus reverse transcriptase (Clontech, Mountain View, CA). PCR reaction (95C for 2 min; 35 cycles of 95C for 30 sec, 58.5C for 30 sec, 72C for 1 min; 72C for 7 min) was performed on a programmable thermocycler (PerkinElmer LifeSciences, Boston, MA). Occasionally, when additional product was needed for sequencing, a second amplification was performed. In a few experiments, positive control using the ON bipolar cDNA library (see Dhingra et al. 2008) was used. Typically though, to avoid tissue contamination, PCR was performed only on test tissues. Two sets of primers were used to amplify the Grm6 transcript. For the first set, F1 forward primer: 5′ 2278 GGGAGTGATAGCGTGGTTGG is located in exon 10, and R1 reverse primer 5′ 2737 CGTGGAGGTCTTCTT-GAGGC is located in exon 11 (accession NM_173372.2). The expected product size was 460 bp. For the second set, F2 forward primer 5′ 1317 GCCAGGACTCCACCTATGAA is located in exon 7 and R2 reverse primer 5′ 1998 CGGAC-TATGGGAGTGTCGTT is located in exon 9. The expected product size was 682 bp. Note that Ensembl (ENSMUS-G00000000617) shows only 10 exons while NCBI shows 11 exons.
Western Blotting
Retinal or corneal tissue was homogenized in 0.2–1 ml of ice cold homogenization buffer (in mM: 320 sucrose, 5 Tris-HCl, 2 EDTA, 2.5 β-mercaptoethanol, pH adjusted to 7.4 at room temperature) containing protease inhibitor cocktail (P8340; Sigma Aldrich, St. Louis, MO). It was incubated on ice for 1 hr and centrifuged at 6,000 × g for 20–30 min in the cold room. The supernatant was then collected, and the protein assay was carried out using BCA protein reagent (Biorad). Twenty µg of protein was mixed with NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA), incubated for 5 min at 70C, then loaded onto 4–15% Tris-HCl gradient gel (Bio-Rad). Electrophoresis was performed with a mini-protean II electrophoresis cell (Bio-Rad) under reduced denaturing conditions. The resolved proteins were transferred to a nitrocellulose membrane (Bio-Rad) in transfer buffer (192 mM glycine, 25 mM Tris-Cl, pH 8.3, and 20% methanol) for 60 min at 80V (26–40 Amps) using a Bio-Rad Trans-Blot semidry transfer cell. The membrane was washed 3 times with TBST (10 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20, pH 7.4), blocked with 7% skim milk in TBST for 1 hr, and incubated with sheep anti-mGluR6 antibody (1:2000) overnight at 4C in TBST-milk. After extensive washes in TBST, the membrane was incubated with the secondary antibody conjugated to peroxidase (anti-sheep HRP, 1:25,000) in 3% TBST-milk for 1.5 hr at room temperature. After more extensive washes in TBST, bound antibody was detected using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL).
Calcium Imaging
A cornea was cleaned and cut into 1–2 pieces. Corneal pieces were mounted on a filter paper with endothelium side facing up and maintained in bicarbonate-based Ames medium saturated with carbogen (95% O2, 5% CO2). Corneal pieces were loaded in 5–10 µM of fura-2 AM (Molecular Probes; Eugene, OR) in oxygenated Ames medium for 1 hr at 27–30C. A cornea was placed in an optical recording chamber on a fixed-stage, upright microscope (Olympus BX51WI; New York, NY) and continuously superfused (3–4 ml/min) with Ames medium preheated to 30–33C. Fura-2 was excited alternately at 340 and 380 nm with a xenon arc lamp in lambda DG4 (Sutter Instrument; Novato, CA) and attenuated with a neutral density filter (0.25). Fluorescent images were viewed with a 40×-water immersion lens (NA, 0.8, Olympus; Tokyo, Japan), filtered with a 510/40 nm emission filter (set 71000, Chroma; Rockingham, VT), and captured with a Hamamatsu Orca II ER digital CCD camera (C4742-98-24ER, Hamamatsu; Hamamatsu city, Japan) at an interval of 4 sec. Wavelength switch and imaging capture were controlled by Openlab software (Improvision; Lexington, MA) running under Mac G4. To minimize potential UV phototoxicity, the illuminated corneal area was restricted by closing the field aperture to match the CCD imaging field (216 × 165 m), and the pixels were binned (2×2). Imaging and statistical analysis were done with Openlab and Excel (Microsoft; Seattle, WA). A region of interest was drawn around the cytoplasm of a dye-loaded endothelial cell, and the cell’s average fluorescence was measured at each time point. A constant background was subtracted from the fluorescence intensity of each wavelengh. We computed the calcium-sensitive ratio of the cell’s fluorescence at 340 nm excitation to that at 380 nm (F340/F380).
Results
GFP Is Expressed in Mouse Retina under Promoter of mGluR6
We developed transgenic mice to express enhanced GFP under control of mGluR6 promoter (Morgan et al. 2006; Dhingra et al. 2008). In one transgenic line (line 5), GFP was expressed exclusively in all ON bipolar cells of the retina, the only retinal cell class known to express mGluR6 (Fig. 1A). Several antibodies against mGluR6 stained the dendritic terminals of GFP-expressing ON bipolar cells (Fig. 1B). Thus, GFP expression in retina appears to be a good indicator of mGluR6 localization. So we wondered if presence of GFP in non-neuronal tissue indicates expression of mGluR6 elsewhere in the brain and body.
Figure 1.
In transgenic mouse retina, green fluorescent protein (GFP) under control of mGluR6 promoter is expressed in all ON bipolar cells and in only these cells (line 5). (A) A radial view, visualized by confocal microscopy, shows that GFP is expressed only in ON bipolar cells terminating in the ON sublamina of the IPL. (B) Immunoreactivity for mGluR6 receptor (red) is seen at the dendritic tips of ON bipolar cells. OPL, outer plexiform layer; INL, inner nuclear layer; IPL inner plexiform layer.
GFP Is Expressed in Tissues Other Than Retina
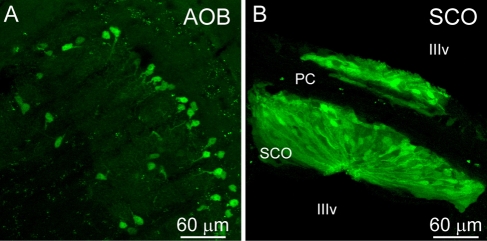
To our surprise, GFP expression under control of mGluR6 promoter was seen in a multitude of tissues throughout the transgenic mouse. There was faint but broad GFP expression throughout many regions of mouse brain—for example, the cortex, superior colliculus, and axons of the corpus callosum (not shown). In addition, there was dense, bright expression in the cells of the accessory olfactory bulb that was possibly specific to the mitral cell layer (Fig. 2A). Very bright expression was also seen in the subcommissural organ cells (Fig. 2B), a non-neuronal structure that bridges across the third ventricle.
Figure 2.
Certain brain regions express green fluorescent protein (GFP). (A) The accessory olfactory bulb (AOB) shows expression that localizes most likely to the mitral cell layer (line 5). (B) Expression is seen in the subcommissural organ (SCO) cells that line the posterior commissure (pc) and are located within the third ventricle (IIIv). No expression is seen in the posterior commissure.
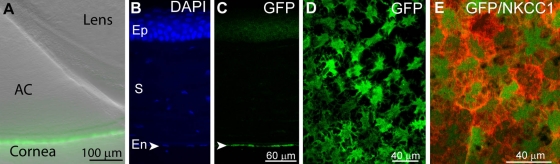
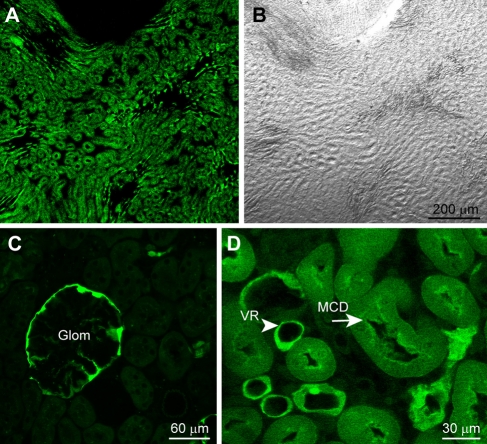
Outside the nervous system, numerous regions expressed GFP. In the cornea, it was seen in the corneal endothelium (Fig. 3B, C), a monolayer of specialized flattened cells that govern fluid and solute transport. Immunostaining a whole-mount cornea for NKCC1, a sodium potassium chloride cotransporter that outlines the plasma membrane of the endothelial cells (Kuang et al. 2001) shows that GFP fluorescence is stronger in the middle of the cells and radiates out (Fig. 3D, E). In the kidney, fluorescence is most pervasive in the inner medulla near the renal pelvis (Fig. 4A, B) and is barely expressed in the renal cortex. In the cortex, the brightest structure is the parietal layer surrounding the glomerulus (Fig. 4C). As one proceeds from the cortex toward the outer medulla, near the cortex, some tubes start to be stained, and they are small. Moving further through the medulla (going toward center), the fluorescing tubes get larger in size. In the inner medulla, toward the renal pelvis, fluorescence gets more pervasive, the stained tubes are bigger, and there are more of them. We thus presume that these are collecting tubes (Fig. 4D). Intermingled with these tubes are brightly stained, much smaller tubes with thinner walls; these seem to be vasa recta.
Figure 3.
Cornea expresses green fluorescent protein (GFP) exclusively in the corneal endothelium. (A) A low-magnification image (differential interference contrast image merged with fluorescent image) of the front of the eye shows a section of the lens, the anterior chamber (AC), and the cornea where GFP expression is localized. (B) A radial section of the cornea stained with the nuclear stain DAPI shows tissue morphology. Ep, epithelial cells; S, stroma; En, endothelial cell layer. (C) The same radial section shows that GFP expression is restricted to the endothelium (en, arrowhead). (D) A tangential view of the corneal endothelial layer shows GFP expression in these cells. (E) A tangential view of a different corneal piece focusing on the endothelium shows NKCC1 (red) labeling of the cell’s plasma membrane (tangential view). GFP expression concentrates near the somas and radiates outwards.
Figure 4.
Kidney expresses green fluorescent protein (GFP) in medullary collecting ducts, vasa recta, and glomeruli. Low magnification images of a kidney section in the deep medulla: (A) the expressed GFP; (B) the same section photographed under differential interference contrast. (C) Kidney cortex shows that in this region expression is mainly in the parietal layer surrounding glomeruli (Glom). (D) Kidney medulla shows GFP expression localizes to the medullary collecting ducts (MCD, arrow) and vasa recta (VR, arrowhead).
Throughout the body, GFP was expressed in B lymphocytes. We determined this by subjecting lymph nodes to flow cytometry and staining for specific markers. We found that GFP-expressing cells were positive for B220 (a marker for B lymphocytes) but not CD3 (a marker for T lymphocytes) (Fig. 5A–D). In addition, GFP could be clearly seen in areas known to house dense B lymphocyte populations, such as the white pulp of the spleen (Fig. 5E, F, H), the germinal center of Peyer’s patches (not shown), and the ribcage bone marrow (Fig. 5G, I). GFP expression was also seen in testis, conjunctiva, spinal cord, and eyelid but not in most of the intestine, tongue, bone, or salivary gland (not illustrated).
Figure 5.
Immune system expresses green fluorescent protein (GFP). (A–D) B-lymphocytes express GFP. GFP fluorescence level (GFP-FL) in FACS-sorted dissociated lymph nodes vs marker fluorescence. (A, C) For control, cells were isolated from a wild-type mouse. (B, D) Cells were isolated from line 5 transgenic mouse. (B) Cells that express a high level of GFP also express a high level of the marker for B-lymphocytes (B220-PE). (D) Cells that express a high level of GFP do not express a high level of the marker for T-lymphocytes (CD3-PE). (E) A low magnification differential contrast image of the spleen shows that red pulp (RP) and white pulp (WP) can easily be differentiated. (F) Same section shows that GFP colocalizes with the white pulp. (G) Bone marrow shows GFP expression in certain cells. (H, I) High magnification images of spleen (S) and bone marrow (BM).
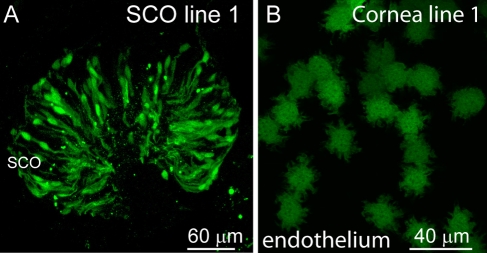
To test if the observed GFP expression was due solely to the random integration of mGluR6 promoter in the genome, we also looked at the expression pattern in a second line of transgenic mice that was created in tandem with the line 5 mouse illustrated above. In the retina of this line 1 mouse, GFP is also exclusively expressed in ON bipolar cells, but the expression is patchy (Dhingra et al. 2008). We found that expression for the subcommissural organ (SCO) (Fig. 6A), cornea (Fig. 6B), kidney, and intestine matches the expression in line 5 mice in both location and pattern. However, as was seen in line 1 ON bipolar cells, the expression was patchy.
Figure 6.
Expression in a different line (line 1) is observed in the same organs, tissues, or cell types as in line 5, but it is patchy. Examples are shown for subcommissural organ (SCO; A) and corneal endothelium (B).
Tissues That Express GFP Also Transcribe mGluR6
To further investigate if GFP expression indicates mGluR6 expression, we used reverse transcriptase followed by RT-PCR on tissues with strong, localized GFP expression. For negative control, we used either water or retinal tissue that was not reverse transcribed. The first set of primers was designed close to the 3′ end on exons 10 and 11 (Fig. 7A). Using these primers, we amplified mGluR6 transcript from retina (positive control), cornea (9 out of 13 attempts), kidney (8 out of 12), spleen (7 out of 8), testis (6 out of 10), SCO (9 out of 11), and lymph node (4 out of 4) (Fig. 7B, C). PCR products from each tissue were sequenced and confirmed to be mGluR6. Thus, most tissues that express GFP in the transgenic mouse transcribe mGluR6 in the wild-type mouse.
Figure 7.
Many green fluorescent protein–expressing tissues transcribe two splice variants of mGluR6 transcript. (A) Top: Grm6 gene structure with 11 exons (according to NCBI) and the location of the two sets of primers (F1 and R1 with expected PCR product of 460 bp and F2 and R2 with expected PCR product of 682 bp). Bottom: sequence of the short variant in which exon 8 is spliced out, along with the predicted change in translation. Arrow points to the transition from exon 7 to exon 9. (B) Example of RT-PCR products using primers F1 and R1 on the tissues indicated. (C) Example of RT-PCR products using primers F2 and R2; these produced the expected band and an additional faint band (stars). In spleen, two clear bands were revealed after reamplification (stars). RNA, H2O, or tongue was used as negative control.
In rat, a splice variant has been described that retains an intron (between exons 7 and 8) (Valerio, Ferraboli, et al. 2001; Valerio, Zoppi, et al. 2001). Since there is 92% nucleotide conservation between rat and mouse, we developed primers F2 and R2 to flank the corresponding region in mouse. Using this set, we expected to see a band size of 682 bp, and indeed we did. However, we frequently observed an additional smaller band at about 530 bp. Both of these bands were observed in retina, cornea, kidney, testis, SCO, lymph node, and spleen (Fig. 7C). DNA sequence analysis confirmed that the 680-bp band was mGluR6, and it showed that the 530-bp band was a result of deleting exon 8 (which has 146 nucleotides) from an otherwise-undisturbed nucleotide sequence for mGluR6. If translated, the deletion of exon 8 would cause a midway frame shift that resulted in a stop codon (TGA) in exon 9 (Fig. 7A). The predicted truncated version of mGluR6 would contain 545 amino acids and would lack the 7-transmembrane domains of the receptor.
Cornea Expresses a Group III Metabotropic Receptor
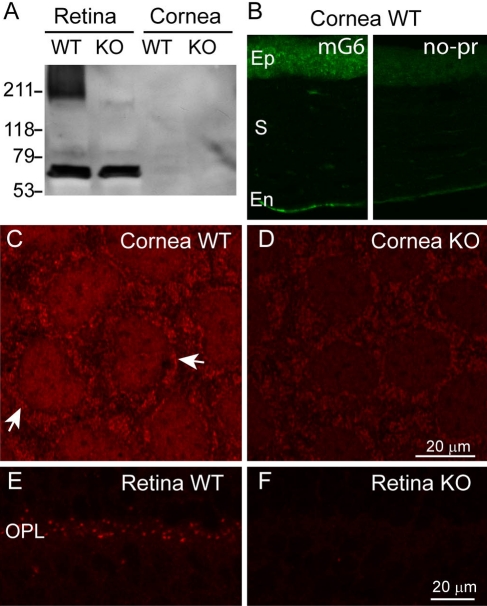
To test if mGluR6 mRNA is translated outside of retina, we concentrated on the cornea. We first performed Western blotting with an antibody against the C-terminus of mGluR6. This antibody should recognize the full-length mGluR6 but not the truncated version. Western blots of retina and cornea often revealed nonspecific bands, which also appeared in the mGluR6-deficient retina (not shown). In one experiment we did see a “smeared” band at about 210 kDa, and this did not appear in the mGluR6-null retina. In this experiment, the cornea did not show this band (Fig. 8A). Given the difficulties in revealing mGluR6 expression by Western blots, we tried immunostaining. In cornea, faint staining for mGluR6 was present in most cells with the endothelium showing a stronger stain (Fig. 8B, left). This stain did not appear when incubating cornea only with the secondary antibody (Fig. 8B, right). In whole mount cornea, when the endothelial cells appear in two dimensions, it was possible to see a diffused stain over the cell’s center and a punctate stain surrounding it (arrows), possibly associated with the plasma membrane (Fig. 8C). In the mGluR6-null cornea, staining of the endothelium was consistently lighter (Fig. 8D), but the pattern appeared similar to that of the wild-type with lighter diffuse and punctate staining. Thus, we regard this result as ambiguous. In retina, the staining in the mGluR6-deficient mouse was diminished (Fig. 8E, F). Thus it appears that the mGluR6 antibody, when applied outside retina, shows cross reactivity that does not permit reliable detection of the protein expressed at low levels. To further explore the possibility that cornea expresses a small amount of mGluR6 that is difficult to detect using antibodies, we reasoned that possibly, as in retina, activation of mGluR6 leads to a change in intracellular free calcium concentration. We thus imaged the responses of corneal endothelium to the group III metabotropic agonist L-AP4 (130 µM) after loading with the ratiometric calcium sensor Fura-2. Thirteen out of 18 analyzed regions of interest showed calcium elevation with highly correlated activity (two experiments). A region of interest probably represents membranes of two apposing cells because the high density of the cells does not permit resolving them. The average activity of several adjacent cells in one experiment is shown in Figure 9.
Figure 8.
mGluR6 may be present in cornea. (A) A Western Blot of retinal (20 µg/lane) and corneal tissues (10 µg) from wild-type (WT) and mGluR6-deficient (KO) retinas. A thick band that probably corresponds to mGluR6 dimer is seen at around 211 kDa in the wild-type retina but not in the null retina or in the wild-type cornea. (B) Immunostaining for mGluR6 in wild-type cornea (left, mG6) and when omitting the primary antibody (right, no-pr) (radial sections). Ep, epithelium; S, stroma; En, endothelium. (C, D) Immunostaining pattern for mGluR6 in wild-type and mGluR6-deficient corneal endothelium is similar to that of the wild-type in a whole mount preparation, but the stain (arrows) appears stronger in the wild-type. (E, F) Immunostaining for mGluR6 in wild-type and mGluR6-deficient retinas shows that the punctate staining in the outer plexiform layer (OPL) is diminished in the knockout.
Figure 9.
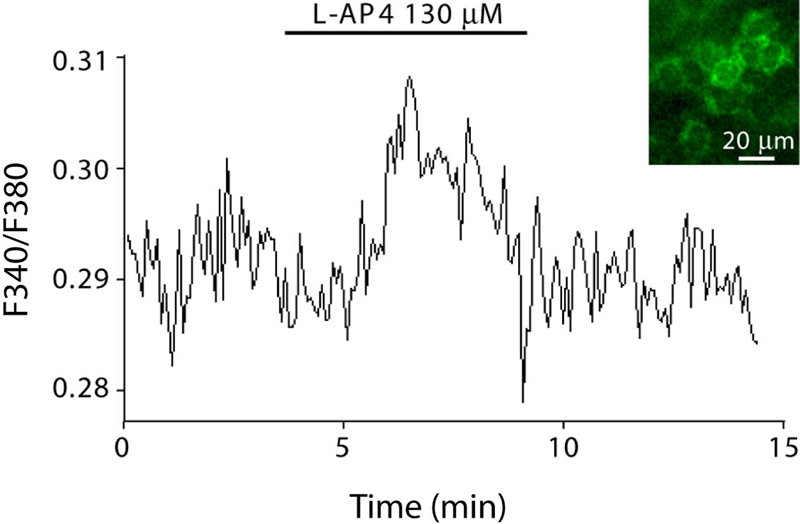
Endothelial cells respond to mGluR6 agonist. Fura-2 imaging of whole mount cornea with endothelium side up. Cornea was maintained in Ames medium at 37C. Bath application of metabotropic group III agonist (L-AP4; 130 µM) elicited calcium rise. Inset: fluorescence image of endothelial cells viewed with 340 nm of excitation light. Data are average of 5 cells.
Discussion
We have generated a transgenic mouse in which GFP is expressed under the control of mGluR6 receptor. As expected from such design, GFP in retina is expressed in all types of ON bipolar cells known to express mGluR6 (Morgan et al. 2006; Dhingra et al. 2008). Furthermore, it is expressed solely in these cells. Here, we showed that GFP is also expressed outside the retina in several brain regions and outside the nervous system in several organs, such as cornea, testis, and lymph nodes. Since the expression level of GFP is high and can easily be seen without amplification, this mouse can be useful for a variety of studies in these tissues.
Glutamate Signaling Outside the Brain
We believe that the GFP expression found in the tissues and organs of our transgenic mouse reflects transcription of the mGluR6 gene for two reasons. First, the same set of tissues and organs that express GFP in one mouse line (line 5 in which GFP is expressed in all cells of a certain type) also express it in a second mouse line (line 1 in which the expression takes a mosaic pattern). Second, RT-PCR using two sets of primers on these tissues amplified one or two bands, thus indicating transcription of two splice variants. Whether transcription of mGluR6 in these tissues actually indicates translation of the protein is unclear. It has been assumed in the past that if mRNA is transcribed, its protein is translated. This dogma has recently been tested vigorously and was proven correct only in general terms, especially for housekeeping proteins (Fu et al. 2007; Shankavaram et al. 2007; Gry et al. 2009). In retinal ON bipolar cells, we have shown a multitude of transcripts (e.g., TRPC2 and CatSper) that are probably not translated (Dhingra et al. 2008). In this study, though we clearly amplified mGluR6 transcript from many tissues, we were unable to unambiguously demonstrate expression of the mGluR6 protein outside the retina. However, it is possible that these non-retinal tissues do indeed express a small amount of protein that went undetected, perhaps due to the high background created by the antibodies used. In fact, in corneal endothelium, we obtained an independent evidence to support mGluR6 expression since L-AP4 affected calcium concentrations in these cells.
Glutamate signaling in non-neuronal tissues has been demonstrated frequently. A well-studied example is bone tissue where iGluRs, mGluRs, and glutamate transporters are expressed and communicate to affect bone homeostasis (e.g. Mason et al. 1997; Chenu et al. 1998; Gu et al. 2002; reviewed by Hinoi, Takarada, Yoneda 2004). Another example is the lymphatic system where lymphocytes have been shown to have both NMDA receptors and group III mGluRs. In this case, both receptor types act synergistically to affect cell death and proliferation (Boldyrev et al. 2004; reviewed by Boldyrev et al. 2005). Given this widespread occurrence of glutamate signaling in non-neuronal tissues, it would not be too surprising if tissues that transcribe mGluR6 do use this receptor and other glutamate receptors to communicate with their environment. If so, what might be the function of this signaling? It is safe to hypothesize that extracellular glutamate concentration in these tissues is lower than at a synapse. mGluR6 would be optimal for sensing this low glutamate level because it is highly sensitive to glutamate (EC50 ranges around 10 µM vs 500 µM for the AMPA receptor), and it does not easily desensitize. Both these features would enable it to provide a sustained signal that might benefit slow modulatory function. Interestingly, we noticed that many of the cells in our study that express GFP are secretory (e.g., corneal endothelium, kidney epithelial cells, SCO). Thus, perhaps the activation of mGluR6 regulates transporters.
It is harder to speculate what mGluR6 transcript can do if it is not translated. It is possible that these transcripts are not functional and that they result from incidental interaction (or cross talk) of the mGluR6 promoter with the transcription factors required in these tissues for other transcripts. It is also possible that mGluR6 is transcribed at low levels in certain cells and that its expression is elevated after injury, presumably as a protective mechanism (Faden et al. 1997). Interestingly, retinal ganglion cells, which transcribe mGluR6 during development (but not in adulthood), transcribe it again after axotomy (Tehrani et al. 2000). However, it is not known if the protein is expressed as well.
Possible Function of Short Splice Variants
Another question raised by our study is the possible function of the short splice variant in retina. It has been shown that both rat and human retinas transcribe an mGluR6 splice variant that would lead to a truncated mGluR6 (Valerio, Ferraboli, et al. 2001; Valerio, Zoppi, et al. 2001). Interestingly, the precise splicing pattern in rat and human is different. In rat, an intron is retained between exons 7 and 8. In human, two additional splice variants exist where one is missing 97 bp from exon 6 and the other has an extra 5 bp from the intron between exons 5 and 6. These predicted truncated proteins range between 405 and 425 (human) to 508 amino acids (rat). Here we showed that mouse retina (as well as other tissues that transcribe mGluR6) splice out exon 8. This results in a stop codon in exon 9 and a predicted truncated protein of 545 amino acids. Thus, even though different species employ different strategies to splice the transcript, in all cases the splice variants would encode a similarly truncated, small soluble protein that has an N-terminus with the ligand recognition site but without the transmembrane receptor domains.
Splice variants that lead to truncated channels and receptors have been demonstrated for several proteins. For example, the human TRPM1 gene has at least 5 splice variants (Oancea et al. 2009). Long transcripts encode a cation channel known to mediate the light response in ON bipolar cells (Morgans et al. 2009; Shen et al. 2009; Koike et al. 2010). Truncated variants lack the transmembrane domains and are expressed in melanocytes (Duncan et al. 1998; Duncan et al. 2001). When the function of the short variant was tested in HEK cells, it was shown to inhibit trafficking of the TRPM1 channel to the plasma membrane (Xu et al. 2001).
It is not trivial to determine if the truncated splice variant of mGluR6 is expressed in retina. First, Western blots for mGluR6 typically reveal several extra bands and often do not reveal the correct size band. Second, available antibodies are directed toward the C-terminus, and this would be truncated in the short variant. If the short variant is translated in retina, it is likely to act as a glutamate sensor or glutamate scavenger since it likely retains its glutamate binding properties as has been shown for mGluR1 truncated protein (Okamoto et al. 1998). If this is the case, we predict that truncated mGluR6 reduces responses to glutamate, possibly in a prolonged dark period. This action could be supported by at least two simple mechanisms. First, the truncated glutamate sensor/scavenger could compete with the actual mGluR6 receptor in binding glutamate in the extracellular cleft. Second, the truncated mGluR6 could dimerize with full length mGluR6 and reduce its surface expression or its trafficking to the dendritic tips.
Acknowledgments
We thank Miriam Meister for help in the initial phase of this work; Drs Bruce Freedman and Peimin Zhu for help with identifying the B-lymphocytes; Drs John Tomaszewski and Lawrence Holzman for help in identifying stained structures in kidney; Drs Shigetada Nakanishi, Yoshihiko Tsukamoto, and Catherine Morgans for donating antibodies against mGluR6; and Drs Shigetada Nakanishi and David Copenhagen for sharing the mGluR6-null mouse. Supported by NIH EY11105 (NV), NIH NEI P30 EY01583 (University of Pennsylvania), and the University of Pennsylvania Research Foundation.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the authorship and publication of this article.
The author(s) received no financial support for the research and authorship of this article.
References
- Bockaert J, Pin J, Fagni L. 1993. Metabotropic glutamate receptors: an original family of G protein-coupled receptors. Fundam Clin Pharmacol. 7:473–485 [DOI] [PubMed] [Google Scholar]
- Boldyrev AA, Carpenter DO, Johnson P. 2005. Emerging evidence for a similar role of glutamate receptors in the nervous and immune systems. J Neurochem. 95:913–918 [DOI] [PubMed] [Google Scholar]
- Boldyrev AA, Kazey VI, Leinsoo TA, Mashkina AP, Tyulina OV, Johnson P, Tuneva JO, Chittur S, Carpenter DO. 2004. Rodent lymphocytes express functionally active glutamate receptors. Biochem Biophys Res Commun. 324:133–139 [DOI] [PubMed] [Google Scholar]
- Chenu C, Serre CM, Raynal C, Burt-Pichat B, Delmas PD. 1998. Glutamate receptors are expressed by bone cells and are involved in bone resorption. Bone. 22:295–299 [DOI] [PubMed] [Google Scholar]
- Dhingra A, Sulaiman P, Xu Y, Fina ME, Veh RW, Vardi N. 2008. Probing neurochemical structure and function of retinal ON bipolar cells with a transgenic mouse. J Comp Neurol. 510:484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. 2001. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 19:568–576 [DOI] [PubMed] [Google Scholar]
- Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. 1998. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 58:1515–1520 [PubMed] [Google Scholar]
- Faden AI, Ivanova SA, Yakovlev AG, Mukhin AG. 1997. Neuroprotective effects of group III mGluR in traumatic neuronal injury. J Neurotrauma. 14:885–895 [DOI] [PubMed] [Google Scholar]
- Fu N, Drinnenberg I, Kelso J, Wu JR, Paabo S, Zeng R, Khaitovich P. 2007. Comparison of protein and mRNA expression evolution in humans and chimpanzees. PLoS One. 2:e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, Nilsson P. 2009. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 10:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Genever PG, Skerry TM, Publicover SJ. 2002. The NMDA type glutamate receptors expressed by primary rat osteoblasts have the same electrophysiological characteristics as neuronal receptors. Calcif Tissue Int. 70:194–203 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Morimoto R, Yamamoto A, Moriyama Y. 2003. Expression and localization of vesicular glutamate transporters in pancreatic islets, upper gastrointestinal tract, and testis. J Histochem Cytochem. 51:1375–1390 [DOI] [PubMed] [Google Scholar]
- Hinoi E, Takarada T, Yoneda Y. 2004. Glutamate signaling system in bone. J Pharmacol Sci. 94:215–220 [DOI] [PubMed] [Google Scholar]
- Hinoi E, Takarada T, Ueshima T, Tsuchihashi Y, Yoneda Y. 2004. Glutamate signaling in peripheral tissues. Eur J Biochem. 271:1–13 [DOI] [PubMed] [Google Scholar]
- Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, et al. 2010. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci U S A. 107:332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang K, Li Y, Wen Q, Wang Z, Li J, Yang Y, Iserovich P, Reinach PS, Sparrow J, Diecke FP, et al. 2001. Corneal endothelial NKCC: molecular identification, location, and contribution to fluid transport. Am J Physiol Cell Physiol. 280:C491–499 [DOI] [PubMed] [Google Scholar]
- Mason DJ, Suva LJ, Genever PG, Patton AJ, Steuckle S, Hillam RA, Skerry TM. 1997. Mechanically regulated expression of a neural glutamate transporter in bone: a role for excitatory amino acids as osteotropic agents? Bone. 20:199–205 [DOI] [PubMed] [Google Scholar]
- Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y, Shigemoto R. 1995. Specific deficit on the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 80:757–765 [DOI] [PubMed] [Google Scholar]
- Morgan JL, Dhingra A, Vardi N, Wong RO. 2006. Axons and dendrites originate from neuroepithelial-like processes of retinal bipolar cells. Nature Neurosci. 9:85–92 [DOI] [PubMed] [Google Scholar]
- Morgans CW, Wensel TG, Brown RL, Perez-Leon JA, Bearnot B, Duvoisin RM. 2007. Gbeta5-RGS complexes co-localize with mGluR6 in retinal ON-bipolar cells. Eur J Neurosci. 26:2899–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Zhang J, Jeffrey BG, Nelson SM, Burke NS, Duvoisin RM, Brown RL. 2009. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc Natl Acad Sci U S A. 106:19174–19178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Iwakabe H, Akazawa C, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. 1993. Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for L-2-amino-4-phosphonobutyrate. J Biol Chem. 268:11868–11873 [PubMed] [Google Scholar]
- Nakanishi S. 1992. Molecular diversity of glutamate receptors and implications for brain function. Science. 258:597–603 [DOI] [PubMed] [Google Scholar]
- Nawy S, Jahr CE. 1990. Suppression by glutamate of cGMP- activated conductance in retinal bipolar cells. Nature. 346: 269–271 [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. 2010. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 50:295–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura A, Shigemoto R, Nakamura Y, Okamoto N, Mizuno N, Nakanishi S. 1994. Developmentally-regulated postsynaptic localization of a metabotropic glutamate-receptor in rat rod bipolar cells. Cell. 77:361–369 [DOI] [PubMed] [Google Scholar]
- Oancea E, Vriens J, Brauchi S, Jun J, Splawski I, Clapham DE. 2009. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci Signal. 2:ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Sekiyama N, Otsu M, Shimada Y, Sato A, Nakanishi S, Jingami H. 1998. Expression and purification of the extracellular ligand binding region of metabotropic glutamate receptor subtype 1. J Biol Chem. 273:13089–13096 [DOI] [PubMed] [Google Scholar]
- Pin JP, Duvoisin RM. 1995. Neurotransmitter receptors I: the metabotropic glutamate receptors. Structure and functions. Neuropharmacology. 34:1–26 [DOI] [PubMed] [Google Scholar]
- Pissimissis N, Papageorgiou E, Lembessis P, Armakolas A, Koutsilieris M. 2009. The glutamatergic system expression in human PC-3 and LNCaP prostate cancer cells. Anticancer Res. 29:371–377 [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, Reimers MA, Scherf U, Kahn A, Dolginow D, et al. 2007. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 6:820–832 [DOI] [PubMed] [Google Scholar]
- Shen Y, Heimel JA, Kamermans M, Peachey NS, Gregg RG, Nawy S. 2009. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J Neurosci. 29:6088–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiells RA, Falk G. 1990. Glutamate receptors of rod bipolar cells are linked to a cyclic GMP cascade via a G-protein. Proc R Soc Lond B Biol Sci. 242:91–94 [DOI] [PubMed] [Google Scholar]
- Skerry TM, Genever PG. 2001. Glutamate signalling in non-neuronal tissues. Trends Pharmacol Sci. 22:174–181 [DOI] [PubMed] [Google Scholar]
- Storto M, Sallese M, Salvatore L, Poulet R, Condorelli DF, Dell’Albani P, Marcello MF, Romeo R, Piomboni P, Barone N, et al. 2001. Expression of metabotropic glutamate receptors in the rat and human testis. J Endocrinol. 170:71–78 [DOI] [PubMed] [Google Scholar]
- Taylor DL, Diemel LT, Pocock JM. 2003. Activation of microglial group III metabotropic glutamate receptors protects neurons against microglial neurotoxicity. J Neurosci. 23:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani A, Wheeler-Schilling TH, Guenther E. 2000. Coexpression patterns of mGLuR mRNAs in rat retinal ganglion cells: a single-cell RT-PCR study. Invest Ophthalmol Vis Sci. 41: 314–319 [PubMed] [Google Scholar]
- Tong Q, Ouedraogo R, Kirchgessner AL. 2002. Localization and function of group III metabotropic glutamate receptors in rat pancreatic islets. Am J Physiol Endocrinol Metab. 282:E1324–1333 [DOI] [PubMed] [Google Scholar]
- Valerio A, Ferraboli S, Paterlini M, Spano P, Barlati S. 2001. Identification of novel alternatively-spliced mRNA isoforms of metabotropic glutamate receptor 6 gene in rat and human retina. Gene. 262:99–106 [DOI] [PubMed] [Google Scholar]
- Valerio A, Zoppi N, Ferraboli S, Paterlini M, Ferrario M, Barlati S, Spano P. 2001. Alternative splicing of mGlu6 gene generates a truncated glutamate receptor in rat retina. Neuroreport. 12:2711–2715 [DOI] [PubMed] [Google Scholar]
- Xu XZ, Moebius F, Gill DL, Montell C. 2001. Regulation of melastatin, a TRP-related protein, through interaction with a cytoplasmic isoform. Proc Natl Acad Sci U S A. 98:10692–10697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LL, Delpire E, Vardi N. 2007. NKCC1 does not accumulate chloride in developing retinal neurons. J Neurophysiol. 98:266–277 [DOI] [PubMed] [Google Scholar]