Abstract
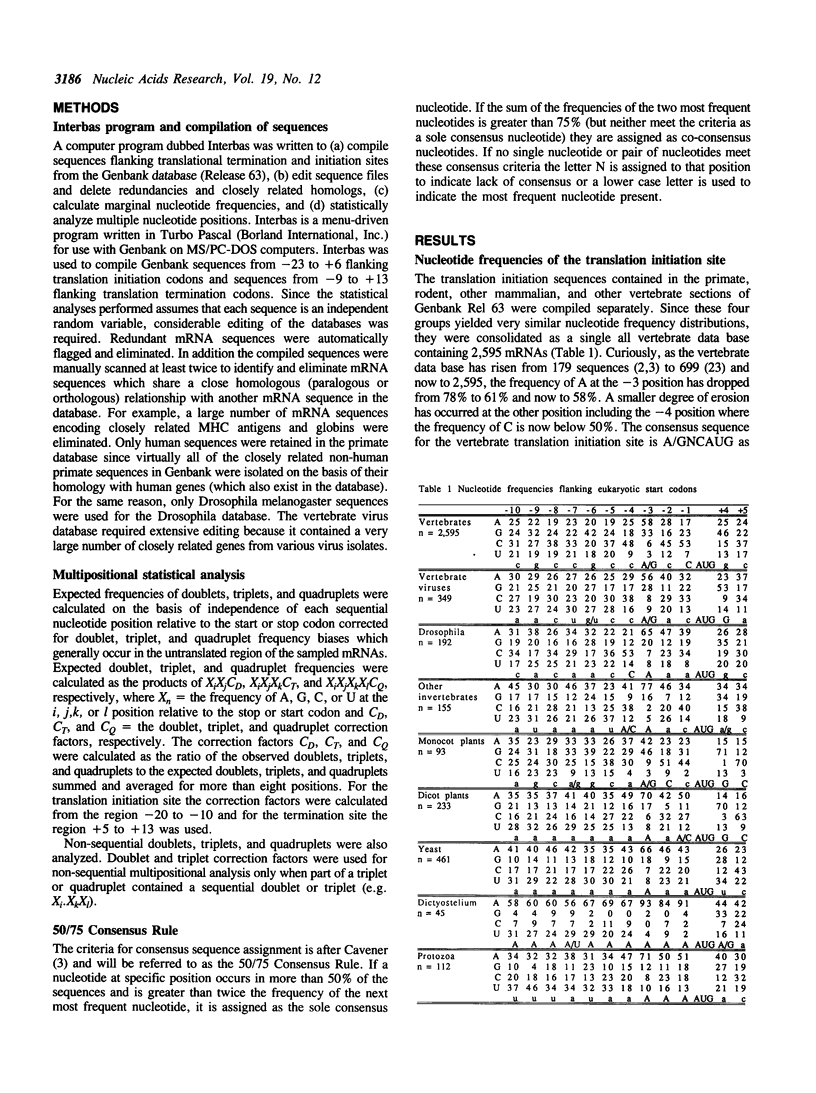
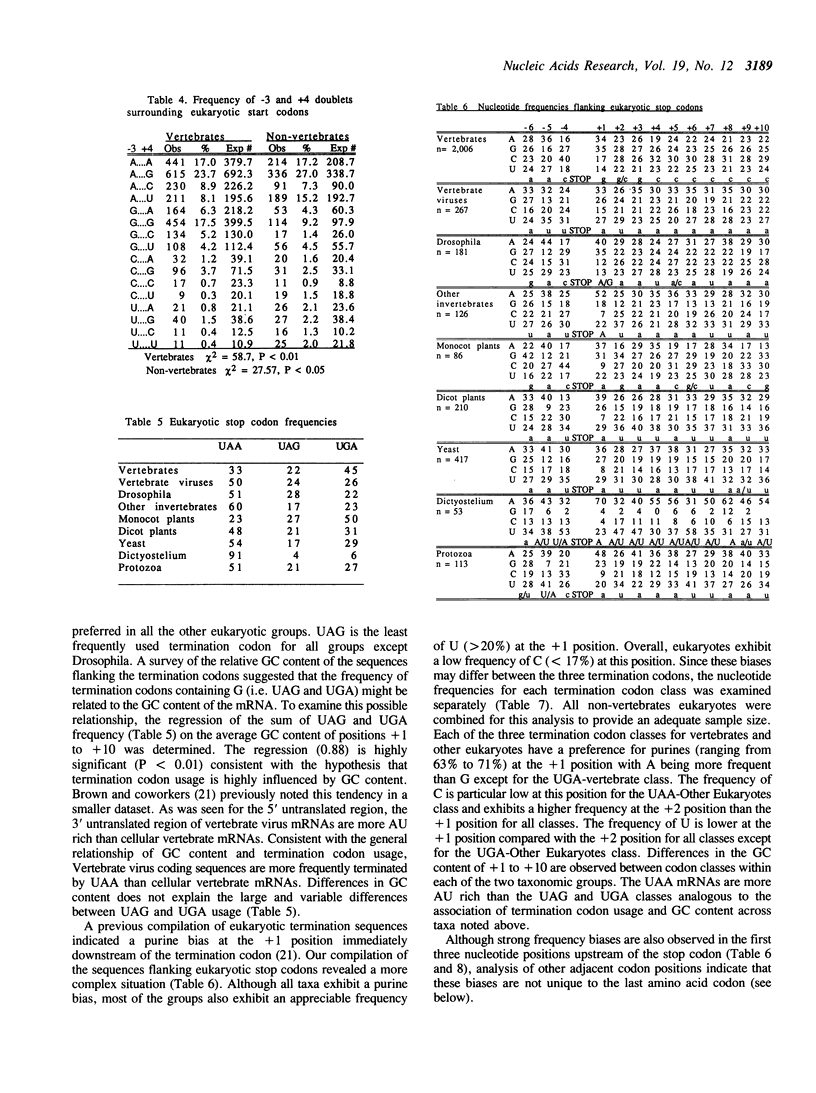
Sequences flanking translational initiation and termination sites have been compiled and statistically analyzed for various eukaryotic taxonomic groups. A few key similarities between taxonomic groups support conserved mechanisms of initiation and termination. However, a high degree of sequence variation at these sites within and between various eukaryotic groups suggest that translation may be modulated for many mRNAs. Multipositional analysis of di-, tri-, and quadrinucleotide sequences flanking start/stop sites indicate significant biases. In particular, strong tri-nucleotide biases are observed at the -3, -2, and -1 positions upstream of the start codon. These biases and the interspecific variation in nucleotide preferences at these three positions have lead us to propose a revised model of the interaction of the 18S ribosomal RNA with the mRNA at the site of translation initiation. Unusually strong biases against the CG dinucleotide immediately downstream of termination codons suggest that they may lead to faulty termination and/or failure of the ribosome to disassociate from the mRNA.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baim S. B., Sherman F. mRNA structures influencing translation in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Apr;8(4):1591–1601. doi: 10.1128/mcb.8.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990 Nov 11;18(21):6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Donahue T. F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Translational frameshifting: where will it stop? Cell. 1987 Jul 3;50(1):1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- Feng Y., Gunter L. E., Organ E. L., Cavener D. R. Translation initiation in Drosophila melanogaster is reduced by mutations upstream of the AUG initiator codon. Mol Cell Biol. 1991 Apr;11(4):2149–2153. doi: 10.1128/mcb.11.4.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hamilton R., Watanabe C. K., de Boer H. A. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987 Apr 24;15(8):3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987 Aug 20;196(4):947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989 Nov;9(11):5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987 Oct;7(10):3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C., Craigen W. J., Muzny D. M., Harlow E., Caskey C. T. Cloning and expression of a mammalian peptide chain release factor with sequence similarity to tryptophanyl-tRNA synthetases. Proc Natl Acad Sci U S A. 1990 May;87(9):3508–3512. doi: 10.1073/pnas.87.9.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. F., Hinnebusch A. G. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989 Aug;3(8):1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Hijazi K. A., Göringer H. U., Dahlberg A. E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Berg P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol. 1986 Jul;6(7):2695–2703. doi: 10.1128/mcb.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986 Jul;6(7):2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargan D. R., Gregory S. P., Butterworth P. H. A possible novel interaction between the 3'-end of 18 S ribosomal RNA and the 5'-leader sequence of many eukaryotic messenger RNAs. FEBS Lett. 1982 Oct 18;147(2):133–136. doi: 10.1016/0014-5793(82)81026-0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle R. P., Morch M. D. Stop making sense: or Regulation at the level of termination in eukaryotic protein synthesis. FEBS Lett. 1988 Aug 1;235(1-2):1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Van Knippenberg P. H. Sequence, modified nucleotides and secondary structure at the 3'-end of small ribosomal subunit RNA. Nucleic Acids Res. 1982 Feb 25;10(4):1149–1158. doi: 10.1093/nar/10.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Feller A., Messenguy F., Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987 Jun 19;49(6):805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- Williams N. P., Mueller P. P., Hinnebusch A. G. The positive regulatory function of the 5'-proximal open reading frames in GCN4 mRNA can be mimicked by heterologous, short coding sequences. Mol Cell Biol. 1988 Sep;8(9):3827–3836. doi: 10.1128/mcb.8.9.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]


