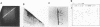Abstract
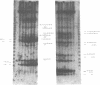
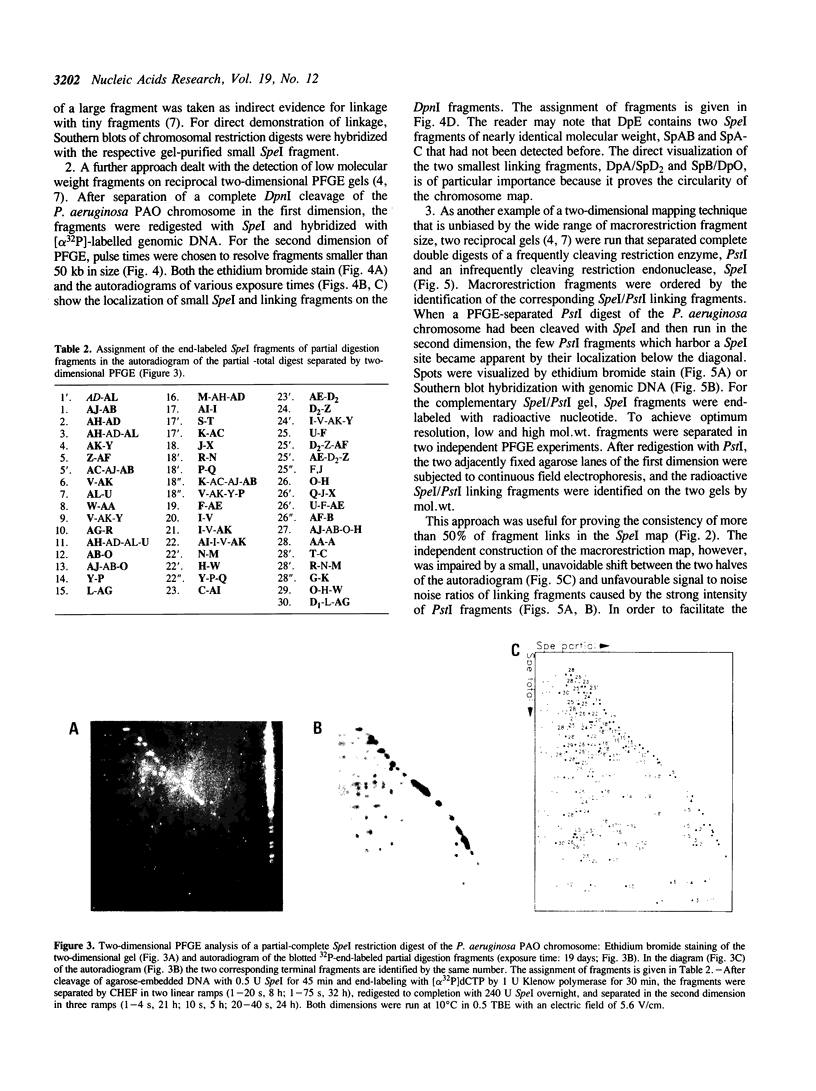
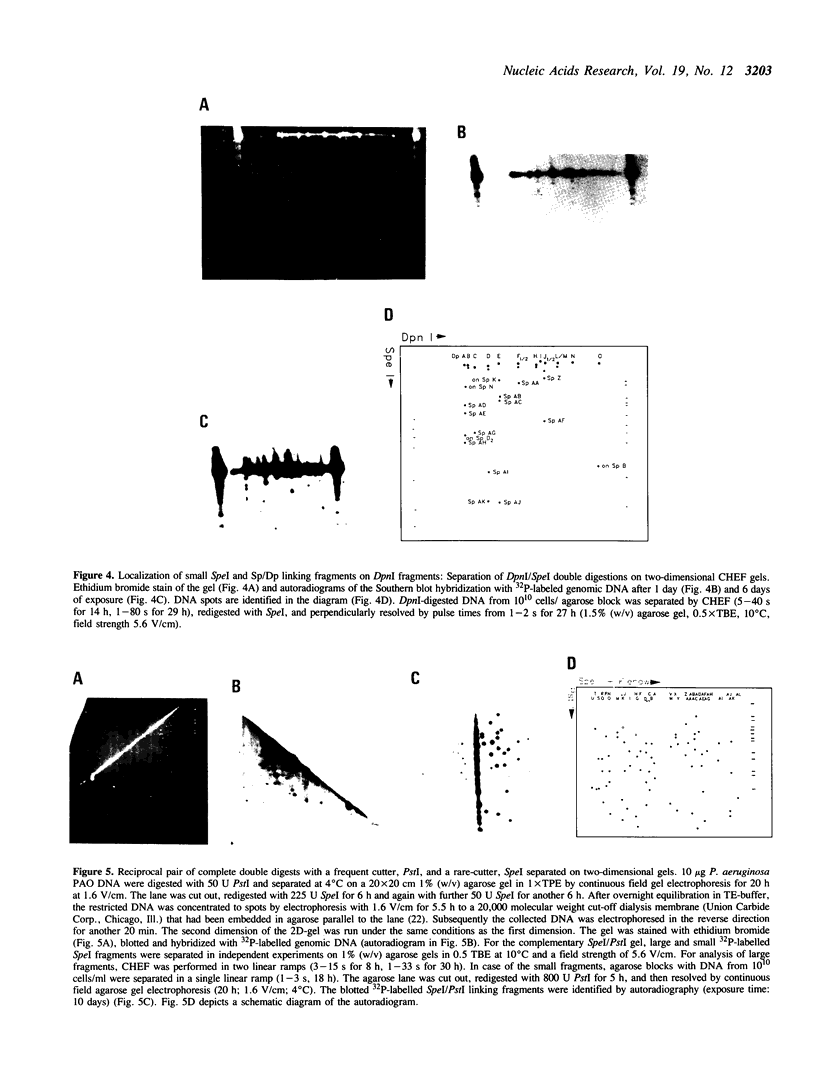
The SpeI/DpnI map of the 5.9 Mb Pseudomonas aeruginosa PAO (DSM 1707) genome was refined by two-dimensional (2D) pulsed-field gel electrophoresis techniques (PFGE) which allow the complete and consistent physical mapping of any bacterial genome of interest. Single restriction digests were repetitively separated by PFGE employing different pulse times and ramps in order to detect all bands with optimum resolution. Fragment order was evaluated from the pattern of 2D PFGE gels: 1. Partial-complete digestion. A partial restriction digest was separated in the first dimension, redigested to completion, and subsequently perpendicularly resolved in the second dimension. 2D-gel comparisons of the ethidium bromide stain of all fragments and of the autoradiogram of end-labeled partial digestion fragments was nearly sufficient for the construction of the macrorestriction map. 2. Reciprocal gels. A complete restriction digest with enzyme A was run in the first dimension, redigested with enzyme B, and separated in the second orthogonal direction. The order of restriction digests was reverse on the second gel. In case of two rare-cutters, fragments were visualized by ethidium bromide staining or hybridization with genomic DNA. If a frequent and a rare cutter were employed, linking fragments were identified by end-labeling of the first digest. 3. A few small fragments were isolated by preparative PFGE and used as a probe for Southern analysis.--38 SpeI and 15 DpnI fragments were positioned on the map. The zero point was relocated to the 'origin of replication'. The anonymous mapping techniques described herein are unbiased by repetitive DNA, unclonable genomic regions, unfavourable location of restriction sites, or cloning artifacts as frequently encountered in other top-down or bottom-up approaches.
Full text
PDF
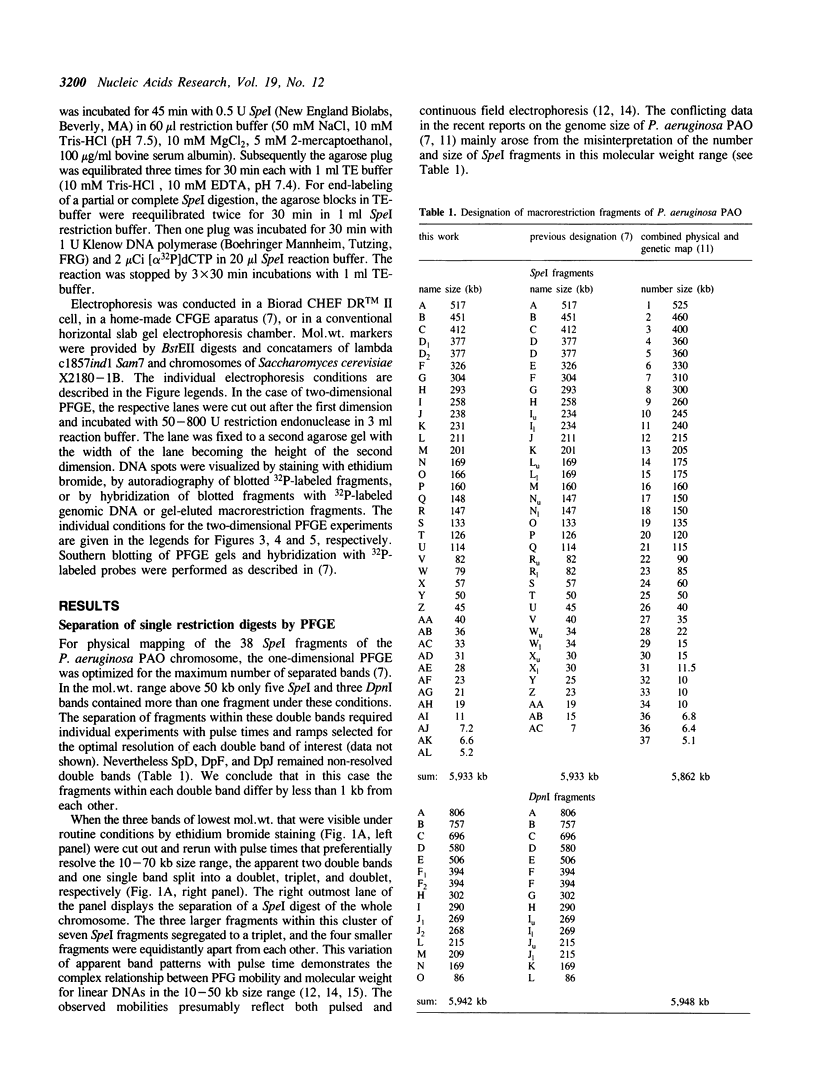



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bautsch W. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 1988 Dec 23;16(24):11461–11467. doi: 10.1093/nar/16.24.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock C. J. Parameters of field inversion gel electrophoresis for the analysis of pox virus genomes. Nucleic Acids Res. 1988 May 25;16(10):4239–4252. doi: 10.1093/nar/16.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. D., Moxon E. R. A physical map of the genome of Haemophilus influenzae type b. J Gen Microbiol. 1990 Dec;136(12):2333–2342. doi: 10.1099/00221287-136-12-2333. [DOI] [PubMed] [Google Scholar]
- Cocks B. G., Pyle L. E., Finch L. R. A physical map of the genome of Ureaplasma urealyticum 960T with ribosomal RNA loci. Nucleic Acids Res. 1989 Aug 25;17(16):6713–6719. doi: 10.1093/nar/17.16.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P., Wilde A., Yelverton E. M., Figurski D., Hedges R. W. Structure and regulation of the anthranilate synthase genes in Pseudomonas aeruginosa: II. Cloning and expression in Escherichia coli. Mol Biol Evol. 1986 Sep;3(5):449–458. doi: 10.1093/oxfordjournals.molbev.a040409. [DOI] [PubMed] [Google Scholar]
- Douglas C. M., Guidi-Rontani C., Collier R. J. Exotoxin A of Pseudomonas aeruginosa: active, cloned toxin is secreted into the periplasmic space of Escherichia coli. J Bacteriol. 1987 Nov;169(11):4962–4966. doi: 10.1128/jb.169.11.4962-4966.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. B., Chikashige Y., Smith C. L., Niwa O., Yanagida M., Cantor C. R. Construction of a Not I restriction map of the fission yeast Schizosaccharomyces pombe genome. Nucleic Acids Res. 1989 Apr 11;17(7):2801–2818. doi: 10.1093/nar/17.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauc L., Mitchell M., Goodgal S. H. Size and physical map of the chromosome of Haemophilus influenzae. J Bacteriol. 1989 May;171(5):2474–2479. doi: 10.1128/jb.171.5.2474-2479.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Blake D. J., Brownlee G. G. Completion of the detailed restriction map of the E. coli genome by the isolation of overlapping cosmid clones. Nucleic Acids Res. 1989 Aug 11;17(15):5901–5912. doi: 10.1093/nar/17.15.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Krawiec S., Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990 Dec;54(4):502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O., Redfield R. J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989 Jun;171(6):3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M. K., Hui C. F., Smith C. L., Cantor C. R. High-resolution separation and accurate size determination in pulsed-field gel electrophoresis of DNA. 4. Influence of DNA topology. Biochemistry. 1988 Dec 27;27(26):9222–9226. doi: 10.1021/bi00426a022. [DOI] [PubMed] [Google Scholar]
- Pyle L. E., Finch L. R. A physical map of the genome of Mycoplasma mycoides subspecies mycoides Y with some functional loci. Nucleic Acids Res. 1988 Jul 11;16(13):6027–6039. doi: 10.1093/nar/16.13.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaningsih E., Dharmsthiti S., Krishnapillai V., Morgan A., Sinclair M., Holloway B. W. A combined physical and genetic map of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990 Dec;136(12):2351–2357. doi: 10.1099/00221287-136-12-2351. [DOI] [PubMed] [Google Scholar]
- Römling U., Grothues D., Bautsch W., Tümmler B. A physical genome map of Pseudomonas aeruginosa PAO. EMBO J. 1989 Dec 20;8(13):4081–4089. doi: 10.1002/j.1460-2075.1989.tb08592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Suwanto A., Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989 Nov;171(11):5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel R., Herrmann R. Physical mapping of the Mycoplasma pneumoniae genome. Nucleic Acids Res. 1988 Sep 12;16(17):8323–8336. doi: 10.1093/nar/16.17.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley J. C., Muto A., Finch L. R. A physical map for Mycoplasma capricolum Cal. kid with loci for all known tRNA species. Nucleic Acids Res. 1991 Jan 25;19(2):399–400. doi: 10.1093/nar/19.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T., Lai E., Kronenberg M., Hood L. Mapping genomic organization by field inversion and two-dimensional gel electrophoresis: application to the murine T-cell receptor gamma gene family. Nucleic Acids Res. 1988 May 11;16(9):3863–3875. doi: 10.1093/nar/16.9.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T. W., Smith D. W. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1278–1282. doi: 10.1073/pnas.87.4.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]