Abstract
Cornelia de Lange syndrome (CdLS; or Brachmann-de Lange syndrome) is a dominantly inherited congenital malformation disorder with features that include characteristic facies, cognitive delays, growth retardation and limb anomalies. Mutations in nearly 60% of CdLS patients have been identified in NIPBL, which encodes a regulator of the sister chromatid cohesion complex. NIPBL, also known as delangin, is a homolog of yeast and amphibian Scc2 and C. elegans PQN-85. Although the exact mechanism of NIPBL function in sister chromatid cohesion is unclear, in vivo yeast and C. elegans experiments and in vitro vertebrate cell experiments have demonstrated that NIPBL/Scc2 functionally interacts with the MAU2/Scc4 protein to initiate loading of cohesin onto chromatin. To test the significance of this model in the clinical setting of CdLS, we fine-mapped the NIBPL–MAU2 interaction domain and tested the functional significance of missense mutations and variants in NIPBL and MAU2 identified in these minimal domains in a cohort of patients with CdLS. We demonstrate that specific novel mutations at the N-terminus of the MAU2-interacting domain of NIBPL result in markedly reduced MAU2 binding, although we appreciate no consistent clinical difference in the small group of patients with these mutations. These data suggest that factors in addition to MAU2 are essential in determining the clinical features and severity of CdLS.
Keywords: Cornelia de Lange syndrome, cohesin, NIPBL, MAU2, SCC4, sister chromatid cohesion
INTRODUCTION
CdLS
Cornelia de Lange syndrome (CdLS; OMIM 122740, 300590 and 610759) is a dominantly inherited clinically heterogeneous developmental disorder. Patients have specific facial features that include synophrys, long eyelashes, a depressed nasal bridge and a thin upper lip. In addition, patients demonstrate reduction defects of the upper extremities that range from small hands to severe malformations, growth and cognitive retardation, gastrointestinal abnormalities and cardiac defects.1, 2 The mental retardation in CdLS is often severe, with a mean IQ of 53.3 Despite some of the characteristic features, CdLS has a variable phenotype with a broad range from severely to very mildly affected patients.
Mutations in NIPBL, SMC1A and SMC3 cause CdLS
Three genes have been reported to have mutations in patients with CdLS and similar phenotypes. Approximately 60% of the typical CdLS patients have mutations in the NIPBL gene.4, 5, 6, 7, 8, 9 The phenotypic spectrum of CdLS patients with NIPBL mutations can range from mildly to severely affected, although patients with deletions or truncating mutations of NIPBL usually show a more severe phenotype with limb, cardiac and visceral anomalies and those with missense mutations present milder cognitive features, fewer organ defects and rare limb anomalies.4, 5, 10
In addition to NIPBL, about 5% of the patients with CdLS have mutations in genes coding for the cohesin subunits SMC1A and SMC3. Mutations in SMC1A and SMC3 result in a phenotype with few structural anomalies, but moderate-to-severe mental retardation.11, 12, 13, 14
NIPBL function
The NIPBL gene encodes a highly conserved protein, NIPBL or delangin, which contains HEAT-repeat protein interaction motifs, a glutamine-rich domain and a predicted bipartite nuclear localization signal.6, 8, 15 Upon the identification of its role in CdLS, it was noted to have homology to Drosophila Nipped-B,16 identified as a transcriptional regulator, and yeast Scc2, a key component of sister chromatid cohesion.17 It was subsequently demonstrated that NIPBL/Scc2 regulates cohesin,18, 19 a complex composed of four highly conserved subunits: SMC1, SMC3, SCC1/RAD21 and SCC3/STAG.20
Although initially identified for its role in sister chromatid cohesion, recent evidence has demonstrated additional functions. Key roles for cohesin have been shown in a number of cellular processes including DNA repair,21, 22 chromatin modification,23 DNA condensation24 and transcriptional regulation in Drosophila,25, 26, 27, 28 fish,29, 30 mouse31, 32 and human cells.33, 34
MAU2
Yeast17, 35 and vertebrate data36, 37, 38 have demonstrated that the function of Scc2/NIPBL in sister chromatid cohesion is spatially and temporally associated with that of Scc4/MAU2. The C. elegans homolog mau-2 was originally identified for its role in axon guidance39 and has subsequently been show to have a role in mitotic chromosome segregation.37 Human MAU2/SCC4 was identified by homology searches of yeast Scc436 and by mass spectrometric identification of proteins that co-immunoprecipitated with Drosophila Nipped-B, followed by homology searching to identify the human homolog.37 Although in humans it has been referred to previously by several names (KIAA0892, MGC75361, mau-2, MAU2L, SCC4), to maintain clarity, in this work, we will refer to gene and protein as MAU2 and MAU2, respectively.
MAU2 and NIPBL have been demonstrated to interact via their N-terminal regions, both of which are highly conserved in evolution.37, 38 Although the exact mechanism of cohesin binding through the action of the NIPBL/MAU2 heterodimer is poorly understood, there are clear data demonstrating the combined necessity of Scc2 and Scc4 in yeast,17, 35, 40 PQN-85 and MAU-2 in C. elegans,37 and NIPBL and MAU2/SCC4 in human37 and other vertebrate cells,36 where their interaction has been shown to be necessary for chromatin binding.38 In this work, we sought to facilitate our understanding of the role of NIBPL and MAU2 interactions in human developmental disorders. We hypothesized that disruption of the NIPBL–MAU2 interaction may represent a molecular mechanism of CdLS because of the loss of this important interaction. To accomplish this, we finely mapped the interaction domains of NIPBL and MAU2, and tested the role of mutations identified in patients with CdLS and related phenotypes from these defined domains to assess the dependence of NIPBL–MAU2 binding in the determination of human phenotype.
MATERIALS AND METHODS
Patients
The collective cohorts of patients with CdLS included in this study amounted to over 750 individuals, all having been assessed by a clinician skilled in the diagnosis of CdLS. Each individual was enrolled in an institutionally approved study of informed consent by one of the authors (MAD, IDK, GG-K, AK, AT, RH, JW). All patients were screened for mutations in NIPBL. Subsequently, 184 patients who were negative for mutations in NIPBL, SMC1A and SMC3 were screened for mutations in MAU2. For this work, we focused on those patients with missense mutations in the NIPBL–MAU2 interaction domains defined below.
Yeast two-hybrid assays
NIPBL and MAU2 fragments were PCR-amplified (primers available upon request) and subcloned into the Matchmaker GAL4 Two-Hybrid System 3 (Clontech-Takara, Saint-Germain-en-Laye, France) pGBKT7 and pGADT7 plasmids, respectively, to obtain NIPBL–GAL4 BD or MAU2–GAL4 AD fusion proteins. Yeast cells (AH109) were cotransformed with the NIPBL and MAU2 fusion fragments, according to the Matchmaker 3 manual. Proper expression was verified by western blotting analysis using anti-GAL4-BD, anti-GAL4-AD antibodies or anti-c-myc and anti-HA antibodies (Santa Cruz Bio, Santa Cruz, CA, USA; Roche, Mannheim, Germany). Growth selection assays were performed using SD agar plates lacking Trp, Ade, His and Leu to detect interacting transformants. Liquid β-galactosidase assays were performed as per the manufacturer's protocol (Matchmaker 3, Clontech). Briefly, overnight cultures were measured at 600 nm, harvested and resuspended in buffer Z (60 m, Na2HPO4, 40 m NaH2PO4, 10 m KCl, 1 m MgSO4, 50 m 2-mercaptoethanol, pH 7.0). An aliquot (V) was taken and o-nitrophenyl-β--galactopyranoside was added. After a defined incubation period (t), the reaction was stopped by the addition of 1 Na2CO3. The solutions were cleared of insoluble material by centrifugation and the OD was measured at 420 nm. The β-galactosidase activity (U) was calculated by the following equation: U=1000 × OD400/(t × V × OD600). All β-galactosidase activities represent the result of at least six independent yeast transformants analyzed in triplicate.
Mammalian two-hybrid quantitative reporter assays
A fragment of NIPBL containing amino acids 1–300 was inserted into the pCMV-BD expression plasmid (Invitrogen, Karlsruhe, Germany). Mutant constructs were generated by site-directed in vitro mutagenesis using appropriate primer sets (Stratagene, Amsterdam, The Netherlands, primers available upon request). The full-length open reading frame of MAU2 was cloned in-frame into the pCMV-AD plasmid. HeLa cells were transiently transfected in 24-well plates (Sarstedt, Nümbrecht, Germany) with FuGene-HD (Roche), according to the manufacturer's instructions, and expression of fusion proteins was verified by western blotting analysis using an anti-GAL4 (DBD) antibody (Santa Cruz). The phRG-TK Renilla expression vector (Promega, Mannheim, Germany) was used as a transfection control. Following incubations of 24 h, activity of firefly and Renilla luciferase was measured with the Dual Luciferase Reporter Assay System (Promega) in a Berthold Luminometer (Berthold, Pforzheim, Germany). All measurements were recorded for at least three independent experiments analyzed in triplicate. Relative luciferase activity was determined as the average firefly:Renilla luciferase activity.
Mutation screening
All the patients (see above) were screened for mutations in all exons of NIPBL as described previously using CSGE,5, 6 HPLC,4 high-resolution melt curve analysis,41 or sequencing of genomic DNA or cDNA.8, 12 Patients were screened for mutations in MAU2 by sequencing of genomic DNA or by high-resolution melt curve analysis. Primer sequences and PCR conditions are available upon request.
Precocious sister chromatid separation assays
We assayed sister chromatid cohesion in lymphoblastoid cell lines (LCLs) derived from patients, as described previously.42 Briefly, cells were arrested at metaphase with 0.8 μg/ml Colchicine (Sigma-Aldrich, St Louis, MO, USA) for 20 min at 37 °C, treated with hypotonic 0.075 KCl at room temperature and fixed with three parts of methanol and one part of acetic acid. The slides were stained with Wright's Stain (Fisher Scientific, Pittsburgh, PA, USA). A minimum of 50 metaphases was microscopically examined. Precocious sister chromatid separation (PSCS) was designated when no connection at the centromere was noted.
RESULTS
Refinement of NIBPL–MAU2/Scc4 interaction domain
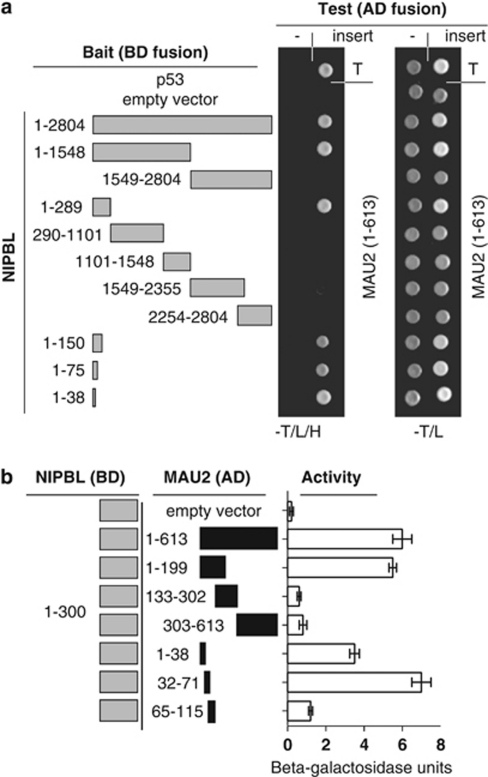
To begin to understand the role of NIPBL–MAU2 function in CdLS, we first confirmed the interaction of NIPBL with MAU2 using yeast two-hybrid selective growth and semiquantitative β-galactosidase assays. We confirmed, as previously reported,36, 37, 38 that the N-terminal 300 amino acids of NIPBL are sufficient for its interaction with MAU2. In addition, full-length NIPBL also interacts with MAU2, clarifying that there is no inhibition of MAU2 binding by the C-terminus of NIPBL, a finding not previously clarified. Additional deletion fragments of NIPBL refined the domain sufficient for MAU2 binding to within the N-terminal 38 amino acids (Figure 1a).
Figure 1.
Localization of NIPBL–MAU2 interaction. (a) NIPBL fragments used in confirmation and localization of NIPBL binding to MAU2 are demonstrated in gray to the left. Amino-acid residues included are indicated. The right panel demonstrates yeast two-hybrid colony assays, indicating interaction-dependent growth on tryptophan-, leucine- and histidine-deficient media (−T/L/H) of the positive control p53 with SV40TAg (T) in the uppermost row. Interaction-independent growth on tryptophan- and leucine-deficient plates is indicated to the right. NIBPL clones were tested with empty (-) and MAU2 containing AD fusion vectors. (b) The left panel depicts the NIPBL (aa 1–300) and MAU2 deletion constructs used for liquid β-galactosidase assay. The right panel indicates the interaction of the NIPBL/delangin fragment (1–300) and the different MAU2 protein fragments by liquid β-galactosidase assay.
MAU2 interacts with NIPBL through its N terminus
To reciprocally map the MAU2 domain essential for binding, overlapping fragments spanning the open reading frame of MAU2 were tested for interaction with a fragment of NIPBL encoding the essential N-terminal 300 amino acids using a semiquantitative β-galactosidase liquid assay. This confirmed that the NIPBL interaction domain is located within the first 199 amino acids of MAU2.37, 38 To refine this critical interaction region, we evaluated additional fragments (amino acids 1–38, 32–71 and 65–115). These results demonstrate strong interaction with the aa 32–71 fragment, reduced interaction with the aa 1–38 fragment and no interaction with the aa 65–115 fragment. These data suggest that the most important segment of MAU2 for interaction with NIPBL is the 40-amino-acid segment from aa 32 to 71 (Figure 1b).
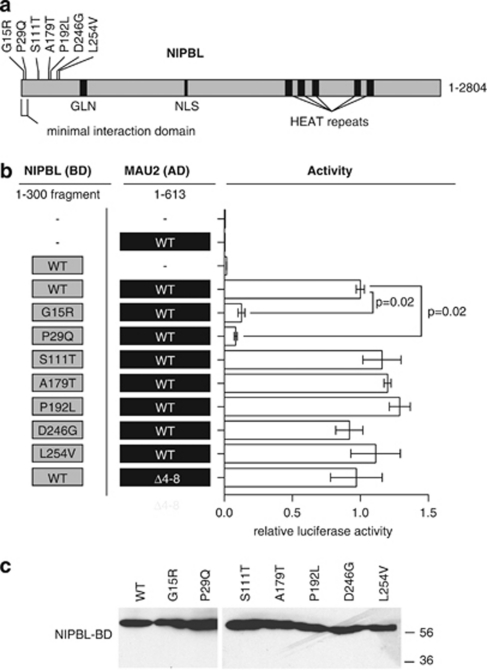
Missense mutations in NIPBL disrupt the interaction of NIPBL and MAU2
To clarify the significance of the NIPBL–MAU2 interaction in CdLS, we evaluated the effect of novel and previously identified missense mutations that reside in the first 300 amino acids of NIPBL. We identified seven novel mutations and further analyzed functional and clinical data for two previously reported mutations as noted in Table 1.4, 8, 43
Table 1. NIBPL mutations and MAU2 rare variants tested.
| Nucleotide | Protein notation | Short notation | Reference |
|---|---|---|---|
| NIPBL | |||
| c.43G>A | p.Gly15Arg | p.G15R | This report |
| c.86C>A | p.Pro29Gln | p.P29Q | Bhuiyan et al, 2005 |
| c.332G>C | p.Ser111Thr | p.S111T | This report |
| c.535G>A | p.Ala179Thr | p.A179T | This report |
| c.535G>T | p.Ala179Ser | p.A179S | This report |
| c.575C>T | p.Pro192Leu | p.P192L | This report |
| c.737A>G | p.Asp246Gly | p.D246G | Yan et al, 2006 |
| c.760C>G | p.Leu254Val | p.L254V | This report |
| MAU2 | |||
| c.9_23del | p.Gln4_Ala8del | pQ4_A8del | This report |
As no mutations had been previously identified for MAU2 in 18 patients with classical CdLS,37 we hypothesized that MAU2 mutations might be rare in CdLS or might result in variant phenotypes. Subsequently, we first screened for mutations in MAU2 in 184 NIPBL, SMC1A and SMC3 mutation-negative patients. We identified several common polymorphisms and several rare variants (Supplementary Table S1), one that was shared by two patients (p.Q4_A8del). These variants were not seen in 192 normal controls. However, the p.Q4_A8del is unlikely to be the sole causative mutation for either patient as it is also seen in one proband's unaffected mother (CDL046), and the other proband was subsequently found to have a 5-Mb 2q23 deletion that is more likely the cause of her overlapping features (CDL383; no parental samples available). Nevertheless, as it is located near the NIPBL interaction domain, we assessed the functional significance of this rare variant on NIBPL binding.
To investigate whether the identified missense mutations in NIPBL directly affect the heterodimerization of NIPBL with MAU2, we introduced these mutations into constructs to perform quantitative mammalian two-hybrid interaction assays in HeLa cells. Constructs encoding the N-terminal 300 amino acids of NIPBL as wild-type or with patient-specific missense mutations were tested for interaction with the wild-type full-length MAU2. In addition, constructs with identified patient MAU2 variant p.Q4_A8del were tested with wild-type NIPBL 1–300 (Figures 2a and b). Missense mutations and rare variants in NIPBL and MAU2 that were assessed are indicated in Table 1.
Figure 2.
The effect of NIPBL mutations on NIPBL/delangin–MAU2 interaction. (a) NIPBL/delangin motifs include a glutamine-rich domain (GLN), nuclear localization signal (NLS) and HEAT-repeat motifs. The minimal MAU-2 interaction fragment used (aa 1–38) and relative positions of mutations tested are demonstrated. (b) Constructs tested are indicated in the right panel and interaction data from mammalian two-hybrid assays are indicated in relative luciferase units for each CdLS mutation tested at the left. Significance of difference in activity of the G15R and P29Q versus wild-type (WT) are indicated. (c) Western blotting of NIPBL-BD 1–300 fragments with missense mutations from two-hybrid assays, using anti-GAL4 (DBD) antibody (Santa Cruz). All proteins are expressed at similar levels.
These data demonstrate that NIPBL mutations S111T, A179T, P192L, D246G and L254V did not significantly affect the interaction of NIPBL with MAU2. However, the NIPBL amino-acid substitutions G15R and P29Q nearly abolish the heterodimerization to 8 and 12% of interaction activity, respectively (Figure 2b). We verified that the effect was not because ofprotein instability caused by these mutations (Figure 2c). The MAU2 variant p.Q4_A8del had no significant effect on the interaction of the proteins, consistent with its location outside of MAU2 amino acids 32–71 (Figure 2b).
Clinical features of patients with N-terminal NIPBL mutations and MAU2 variations
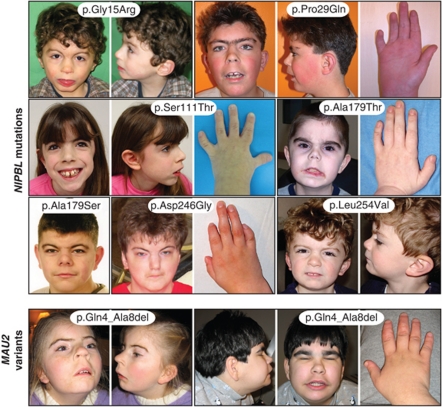
On the basis of the findings that missense mutations G15R and P29Q resulted in significant reductions of NIPBL binding to MAU2, we performed a detailed analysis of patient clinical information to assess whether mutations in this region correlated with clinical features or severity. Photos of the faces and limbs of all patients with mutations tested in the interaction assays are demonstrated in Figure 3 and clinical findings are tabulated in Supplementary Table S2. This analysis revealed that the patients with the G15R and P29Q mutations demonstrate a typical CdLS facial phenotype, consistent with other NIPBL missense mutations.
Figure 3.
Clinical features of patients. Frontal and profile face photos and photos of hands are shown for the patients described. Images for an individual patient are outlined by a black box, with the patient's mutation indicated.
The patient with the G15R mutation is more cognitively affected than the patient with the P29Q mutation. This, however, does not correlate with any loss of NIPBL binding and his features do not vary greatly from that of the other patients with missense mutation in this region. For example, the girl with the S111T mutation has extremely mild cognitive involvement and the boy with the D246G mutation is more severely intellectually affected.
Although it is not statistically significant, it is of interest that neither of the patients with NIPBL mutations that abolish MAU2 binding have cardiac defects, whereas four of six patients with mutations in amino acids 30–300 of NIPBL with normal MAU2 binding have heart defects ranging from Tetralogy of Fallot to a patent ductus arteriosus. Despite this finding, in general, these patients with missense mutations in the N-terminal 300 amino acids of NIPBL represent a similar clinical spectrum to the larger group of patients with missense mutations in NIPBL. For example, none of the patients in this report have severe limb anomalies and tend to trend toward milder cognitive involvement for CdLS. Thus, we were unable to observe a consistent trend between clinical severity and subsets of clinical features for the patients with mutations that disrupt MAU2 interaction with NIPBL.
DISCUSSION
Previous work has demonstrated that NIPBL is mutated in ∼60% of patients with classical CdLS.4, 5, 6, 7, 8, 9 In addition, yeast and C. elegans homologs of NIPBL have been shown to genetically interact with homologs of MAU2 in the regulation of sister chromatid cohesion.17, 36 Furthermore, NIPBL and MAU2 physically interact in yeast and vertebrate cellular experiments.36, 37 In this work, we sought to assess the role of NIPBL and MAU2 interaction in CdLS.
Effect of NIPBL and MAU2 mutations on interaction
We extended previous analyses36, 37 and refined the NIPBL–MAU2 interaction domain to amino acids 1–38 and 32–71, respectively. Furthermore, we did not detect any evidence of domains within NIPBL or MAU2 that negatively regulate this interaction, as in both cases the full-length proteins interact with similar activity as the minimal fragment. This allowed us to limit our analysis to the regions of interaction and reduced the need for testing the effects of mutations in other regions of NIPBL or MAU2. We subsequently analyzed the effect of seven NIPBL missense mutations on the interaction between MAU2 and NIPBL. Five were newly identified and two were previously reported. Of these, two amino-acid substitutions located toward the N terminus of the NIPBL–MAU2 interaction domain (NIBPL residues 15 and 29) nearly abolished the NIPBL–MAU2 interaction, whereas the other six showed no significant alteration. This establishes that the most important region of NIPBL interaction with MAU2 includes the first 38 amino acids of NIPBL.
Rare MAU2 variants are not consistent with causing CdLS
In this work, we analyzed a cohort of 184 mutation-negative patients with CdLS for mutations in MAU2. In addition to several common polymorphisms (Supplementary Table S1), we identified two rare variants with allele frequencies <1% in three patients. Although it is possible that these might represent recessive alleles, further data suggest that they are unlikely to have a role in these patients. None of the three patients were noted to have a second mutation or a suspicious variant. Furthermore, one of these (p.Gln4_Ala8del) results in a five-amino-acid.deletion adjacent to the NIPBL–MAU2 interaction domain and was further analyzed for a role in this binding, where it did not appear to have a significant role (Figure 2b). Interestingly, although these amino acids are conserved, repeats of these five amino acids (QAAAA) have arisen in the vertebrate lineage resulting in two copies in human and three in mouse (Supplementary Figure S1). This also suggests that one copy of this repeat may be dispensable. Furthermore, of the two patients identified with this deletion, one was inherited from a normal mother (CDL046) and, although the parents are unavailable for the other (CDL383), her features are somewhat atypical for CdLS and she was subsequently found to have a 5-Mb deletion on chromosome 2q23 that contains the MBD5 gene, for which haploinsufficiency has been associated with mental retardation and microcephaly.44
Clinical features of patients
We were able to define a functional consequence of two NIPBL mutations on MAU2 binding. However, with the exception of a weak trend toward a reduced incidence of cardiac defects, we did not detect a significant phenotypic correlation with these mutations. These two patients, similar to the other six with mutations that do not change MAU2 binding, fit within the general spectrum of patients with missense mutations in NIBPL.5
As with many studies of rare disorders, one of the main shortcomings of this work is the paucity of identified human mutations in this region. However, these mutations were identified from the collective efforts of our groups and others who have published on CdLS, representing 8 CdLS research groups and nearly 900 patients (Bhuiyan et al4; Gillis et al5; Yan et al8; Selicorni et al9; Pie et al10; Borck et al45; Schoumans et al46 and unpublished data). With a recent estimated European incidence of CdLS of 1 in 81 000 births,47 this represents an evaluated population of nearly 70 million individuals. Without coordinated global efforts, it is unlikely that higher saturation of NIPBL will be obtained.
In previous work, LCLs from patients with CdLS have been shown to have a small increase in PSCS42 that did not correlate with mutation status or severity, although this has not been appreciated in additional studies.48 We have performed PSCS assays on available LCLs from patients with the NIPBL p.Gly15Arg, NIPBL p.Leu254Val and MAU2 p.Gln4_Ala8del and did not appreciate any abnormal sister chromatid separation.
Implications for the understanding of NIPBL and MAU2
Although MAU2 homologs clearly interact functionally and genetically in yeast, C. elegans, Drosophila and vertebrate cell models of sister chromatid cohesion, we were unable to appreciate a definitive functional interrelationship using the clinical phenotype of CdLS.
One possible explanation is that NIPBL may interact with multiple proteins to establish its function, one of which could be MAU2. Any mutation that results in loss of function, including one that disrupts MAU2 binding, would disrupt function, as well as any other mutation in NIBPL that disrupts its interaction with other key proteins. In this context, it may be difficult to distinguish a differential effect of a MAU2 interaction-disrupting mutation on the CdLS phenotype.
A second plausible explanation is that the role of MAU2 homologs may be limited to a role in sister chromatid cohesion. It may be that it has little part in the role that NIPBL has in chromatin reorganization or transcriptional regulation, a role largely speculated to be the predominant defect in CdLS.28, 33, 49 It has yet to be defined whether these additional functions of NIPBL utilize MAU2.
Summary
To further understand the cellular and developmental basis of CdLS, and the role of the NIPBL–MAU2 interaction in this process, we refined the interaction domains of NIPBL and MAU2, and evaluated the effect of mutations and rare variants in these domains. Subsequently, we compared clinical features of the patients with NIPBL mutations that result in a significant reduction of NIPBL–MAU2 interaction, and noted that this disruption does not appear to correlate with clinical phenotype. These data and previous work37 suggest that if MAU2 has a role in the etiology of CdLS, more work will need to be done to elucidate this mechanism. Perhaps as more knowledge is gained regarding the role of the cohesin complex and its regulatory proteins in human disease, this discrepancy will be clarified.
Acknowledgments
We are exceptionally grateful to the patients and families with Cornelia de Lange Syndrome who participated in this study, as well as to the referring physicians and colleagues who have contributed samples and clinical information. We are indebted to the continued support of the many national and the International Cornelia de Lange Syndrome Foundations. This work was supported by the National Institutes of Health Grants K08HD055488 (NICHD, MAD), P01 HD052860 (NICHD, IDK), research grants from the USA CdLS Foundation and institutional funds from the Children's Hospital of Philadelphia.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Kline AD, Krantz ID, Sommer A, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143:1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- Ireland M. Cornelia de Lange syndrome: clinical features, common complications and long-term prognosis. Curr Paediatr. 1996;6:69–73. [Google Scholar]
- Kline AD, Stanley C, Belevich J, Brodsky K, Barr M, Jackson LG. Developmental data on individuals with the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1053–1058. doi: 10.1002/ajmg.1320470724. [DOI] [PubMed] [Google Scholar]
- Bhuiyan Z, Klein M, Hammond P, et al. Genotype-Phenotype correlations of 39 patients with Cornelia de Lange syndrome: the Dutch experience. J Med Genet. 2005;43:568–575. doi: 10.1136/jmg.2005.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis LA, McCallum J, Kaur M, et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Yan J, Saifi GM, Wierzba TH, et al. Mutational and genotype-phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am J Med Genet A. 2006;140:1531–1541. doi: 10.1002/ajmg.a.31305. [DOI] [PubMed] [Google Scholar]
- Selicorni A, Russo S, Gervasini C, et al. Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet. 2007;72:98–108. doi: 10.1111/j.1399-0004.2007.00832.x. [DOI] [PubMed] [Google Scholar]
- Pie J, Gil-Rodriguez MC, Ciero M, et al. Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am J Med Genet A. 2010;152A:924–929. doi: 10.1002/ajmg.a.33348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Zarhrate M, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Incidence and clinical features of X-linked Cornelia de Lange syndrome due to SMC1L1 mutations. Hum Mutat. 2007;28:205–206. doi: 10.1002/humu.9478. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Feldman R, Zhang Z, et al. SMC1A expression and mechanism of pathogenicity in probands with X-Linked Cornelia de Lange syndrome. Hum Mutat. 2009. [DOI] [PMC free article] [PubMed]
- Musio A, Selicorni A, Focarelli ML, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Tonkin E, Smith M, Eichhorn P, et al. A novel gene is disrupted by a Cornelia de Lange-associated translocation breakpoint at 3q26.3. Am J Hum Genet. 2001;69:A618. doi: 10.1007/s00439-004-1134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, et al. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- Musio A, Montagna C, Mariani T, et al. SMC1 involvement in fragile site expression. Hum Mol Genet. 2005;14:525–533. doi: 10.1093/hmg/ddi049. [DOI] [PubMed] [Google Scholar]
- Jahnke P, Xu W, Wulling M, et al. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008;36:6450–6458. doi: 10.1093/nar/gkn688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, et al. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187:455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol. 2010;30:4940–4951. doi: 10.1128/MCB.00642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, et al. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, et al. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344:637–649. doi: 10.1016/j.ydbio.2010.05.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, et al. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, et al. Metazoan Scc4 Homologs link sister chromatid cohesion to cell and axon migration Guidance. PLoS Biol. 2006;4:1411–1425. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard CY, Kebir H, Takagi S, Hekimi S. mau-2 acts cell-autonomously to guide axonal migrations in Caenorhabditis elegans. Development. 2004;131:5947–5958. doi: 10.1242/dev.01433. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerson ML, Warburton T, Nelis E, et al. Mutation scanning the GJB1 gene with high-resolution melting analysis: implications for mutation scanning of genes for Charcot-Marie-Tooth disease. Clin Chem. 2007;53:349–352. doi: 10.1373/clinchem.2006.080010. [DOI] [PubMed] [Google Scholar]
- Kaur M, DeScipio C, McCallum J, et al. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Wagenstaller J, Spranger S, Lorenz-Depiereux B, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Redon R, Sanlaville D, et al. NIPBL mutations and genetic heterogeneity in Cornelia de Lange syndrome. J Med Genet. 2004;41:e128. doi: 10.1136/jmg.2004.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J, Wincent J, Barbaro M, et al. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet. 2007;15:143–149. doi: 10.1038/sj.ejhg.5201737. [DOI] [PubMed] [Google Scholar]
- Barisic I, Tokic V, Loane M, et al. Descriptive epidemiology of Cornelia de Lange syndrome in Europe. Am J Med Genet A. 2008;146A:51–59. doi: 10.1002/ajmg.a.32016. [DOI] [PubMed] [Google Scholar]
- Castronovo P, Gervasini C, Cereda A, et al. Premature chromatid separation is not a useful diagnostic marker for Cornelia de Lange syndrome. Chromosome Res. 2009;17:763–771. doi: 10.1007/s10577-009-9066-6. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann NY Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.