Background: Lipins are phosphatidic acid phosphatases. In yeast, lipin is activated by the Nem1p-Spo7p complex. There is controversy as to whether a mammalian Spo7p ortholog exists.
Results: The metazoan Spo7p ortholog is now identified and shown to interact with lipins in yeast, nematodes, and mammalian cells.
Conclusion: NEP1-R1 is the metazoan Spo7p ortholog.
Significance: The lipin activation system is conserved in evolution.
Keywords: Diacylglycerol, Endoplasmic Reticulum (ER), Nucleus, Phosphatidic Acid, Protein Phosphatase, NEM1, SPO7, TMEM188, dullard, Lipin
Abstract
Lipin-1 catalyzes the formation of diacylglycerol from phosphatidic acid. Lipin-1 mutations cause lipodystrophy in mice and acute myopathy in humans. It is heavily phosphorylated, and the yeast ortholog Pah1p becomes membrane-associated and active upon dephosphorylation by the Nem1p-Spo7p membrane complex. A mammalian ortholog of Nem1p is the C-terminal domain nuclear envelope phosphatase 1 (CTDNEP1, formerly “dullard”), but its Spo7p-like partner is unknown, and the need for its existence is debated. Here, we identify the metazoan ortholog of Spo7p, TMEM188, renamed nuclear envelope phosphatase 1-regulatory subunit 1 (NEP1-R1). CTDNEP1 and NEP1-R1 together complement a nem1Δspo7Δ strain to block endoplasmic reticulum proliferation and restore triacylglycerol levels and lipid droplet number. The two human orthologs are in a complex in cells, and the amount of CTDNEP1 is increased in the presence of NEP1-R1. In the Caenorhabditis elegans embryo, expression of nematode CTDNEP1 and NEP1-R1, as well as lipin-1, is required for normal nuclear membrane breakdown after zygote formation. The expression pattern of NEP1-R1 and CTDNEP1 in human and mouse tissues closely mirrors that of lipin-1. CTDNEP1 can dephosphorylate lipins-1a, -1b, and -2 in human cells only in the presence of NEP1-R1. The nuclear fraction of lipin-1b is increased when CTDNEP1 and NEP1-R1 are co-expressed. Therefore, NEP1-R1 is functionally conserved from yeast to humans and functions in the lipin activation pathway.
Introduction
Phosphatidic acid (PA)4 and diacylglycerol (DAG) are lipid second messengers involved in several essential signaling and trafficking functions. For example, PA activates mammalian target of rapamycin, S6 kinase, and other mitogenic signaling molecules, recruits PKCs and RhoGDIs to membranes for cytoskeletal rearrangements and cell migration, and is involved in vesicle budding and fusion (1–5). DAG activates several signaling molecules such as PKCs, PKDs, and ion channels through their C-1 domains (6–8). These two lipids are also at the cross-roads of phospholipid synthesis and neutral lipid storage (9, 10). PA serves as a precursor of phospholipids through the CDP-DAG pathway and in yeast can negatively regulate phospholipid synthesis through sequestration of a transcription factor (11, 12). DAG is the precursor of phospholipids through the de novo (Kennedy) pathway and also is required for the formation of triacylglycerols.
Phosphatidic acid and diacylglycerol can be interconverted by phosphatidic acid phosphatases and DAG kinases, and thus these enzymes can control cell signaling and movement, protein trafficking, and biosynthesis of bulk membrane and storage lipids (13, 14). In mammals, Mg2+-dependent phosphatidic acid phosphatase activity is encoded by three lipin genes encoding lipin-1, lipin-2, and lipin-3 and related splice variants (15–18). Lpin1 was originally identified by characterization of a spontaneous mutation in the mouse that resulted in a lipodystrophy-like syndrome characterized by loss of body fat, fatty liver, hypertriglyceridemia, and insulin resistance (19, 20). Lipin genes were found to be highly conserved from mammals to yeast (19). In Saccharomyces cerevisiae, the sole known lipin ortholog is Pah1p and was the first lipin protein shown to act as a Mg2+-dependent phosphatidic acid phosphatase enzyme (16, 21, 22).
Lipins, including Pah1p, are regulated by multiple phosphorylations (23–25). For example, mouse lipin-1 is phosphorylated through the mammalian target of rapamycin pathway in response to insulin (26). In budding yeast, Pah1p is inactivated by phosphorylation by Cdc28p and likely by other kinases (21, 27) and activated by dephosphorylation by a nuclear envelope membrane complex consisting of Nem1p and Spo7p, with Nem1p being the catalytic subunit (21). In the absence of Pah1p or its activators in yeast, there is a severe decrease in TAG levels and lipid droplets and an expansion of the nuclear envelope, consistent with a decrease in DAG and an increase in phospholipid synthesis caused by the sequestration of the transcriptional regulator Opi1p by PA in the ER membrane (11, 16, 28, 29). Lipin also plays a role in nuclear envelope breakdown during mitosis in the Caenorhabditis elegans embryo. Lowering lipin-1 levels inhibits nuclear envelope breakdown, suggesting a role for DAG in this process (30, 31).
A study in 2007 (32) demonstrated that human C- terminal Domain Nuclear Envelope Phosphatase 1 (CTDNEP1, originally termed “dullard” for its effects on Xenopus neural development (33)) is the ortholog of Nem1p and could functionally replace Nem1p in yeast. CTDNEP1 had no lipin dephosphorylation activity in two of three cultured cell lines tested, suggesting it required a hypothetical human Spo7p as a binding partner. However, a soluble fragment of CTDNEP1 was able to dephosphorylate peptides derived from the lipin-1 sequence with good kinetics, leading the authors to postulate that there was no need for a human Spo7p (34).
Although S. cerevisiae Spo7p was characterized over a decade ago, identification of a Spo7p homolog in higher eukaryotes has proven challenging even with recent advances in bioinformatic search methods. We now report that TMEM188 is the metazoan Spo7p ortholog and have renamed it NEP1-R1 (Nuclear Envelope Phosphatase 1-Regulatory subunit 1). NEP1-R1 and CTDNEP1 are shown to physically and functionally interact. Knockdown of the two corresponding genes have similar phenotypes in C. elegans and share nearly identical expression profiles with lipin-1 in mouse and human tissues. CTDNEP1 can dephosphorylate lipin-1 isoforms and lipin-2 in cells only in the presence of NEP1-R1. Finally, in the presence of CTDNEP1 and NEP1-R1 together, there is an accumulation of lipin in the nucleus.
EXPERIMENTAL PROCEDURES
Homology Searching
Servers hosting PROCAIN and HHPRED used to identify animal SPO7 homologs can be found on line.
Experiments in Yeast
Strains and Growth Conditions
All yeast strains used in this study are listed in supplemental Table S1 (top section). The BY4742 strain was used as wild-type control unless indicated. Cells were grown in SD (yeast nitrogen base + 2% dextrose) medium with appropriate amino acid and base supplements for auxotrophies in a 30 °C shaker-incubator and harvested in late log phase (OD600 1.2–2.0) for observation and analysis, because spo7Δ and nem1Δspo7Δ show the most prominent lipid-related phenotype in late log phase. To generate spo7Δ or nem1Δspo7Δ, the SPO7 allele was replaced with the HIS3 auxotrophic marker by homologous recombination. Yeast strains were transformed by the lithium acetate method (35). Correct integration was verified by PCR of genomic DNA. For the sporulation assay, diploid strains were sporulated (36) and observed by light microscopy after 5 days at 26 °C. Asci containing at least two spores were counted as sporulated.
Plasmid Vectors for Yeast Transformation
All plasmids and oligonucleotides used in yeast are listed in supplemental Tables S1 (bottom section) and S2, respectively. Standard recombinant DNA techniques were used. Bacterial strain DH10β (Invitrogen) was used for all bacterial transformations. NEP1-R1 (TMEM188, ID 5266084) and CTDNEP1 (dullard, ID 4123279) cDNA were purchased from Open Biosystems. The open reading frames encoding NEP1-R1 and CTDNEP1 were amplified by PCR and subcloned into pRS315-PGK1 and pRS316-PGK1, respectively (37). For the C-terminally tagged versions, pRS315-PGK1-HA-ProtA was derived from plasmid BG1805 (38), and pRS315-PGK1–2×FLAG was generated by inserting oligonucleotides encoding tandem repeated FLAG sequence (KLDYKDDDDKDYKDDDDK, additional amino acid sequence is underlined) to the HindIII/SalI site of pRS316-PGK1. Subsequently, NEP1-R1 and CTDNEP1 ORFs were subcloned into pRS315-PGK1-HA-ProtA and pRS316-PGK1–2×FLAG, yielding pRS315 PGK1-NEP1-R1-HA-ProtA and pRS316-PGK1-CTDNEP1–2×FLAG, respectively. The catalytic inactive version CTDNEP1 D67N-2×FLAG was generated from pRS316-PGK1-CTDNEP1–2×FLAG by rapid PCR site-directed mutagenesis (39). The open reading frames of SPO7 and NEM1 were PCR-amplified from genomic DNA and subcloned into pRS315-PGK1 and pRS316-PGK1, respectively. For an endoplasmic reticulum marker, a Sec63p-CFP fusion construct (40) was generated as follows. First, pFA6-CFP hygroMX vector was derived from pRS315-PGK-CFP-HDEL (41) and pFA6 hph (kindly provided by B. Tu laboratory, University of Texas Southwestern) (42, 43). Then, using this vector as a PCR template, SEC63 ORF was C-terminally tagged with the CFP-encoding sequence in wild-type yeast as described before (43). From that strain, Sec63p-CFP fusion gene was amplified by PCR and subcloned first into pRS315-PGK1 and then into the pRS317 vector, yielding pRS317-PGK1-SEC63-CFP. DNA sequences were verified by The McDermott Center for Human Growth and Development (University of Texas Southwestern).
Fluorescence Microscopy
To visualize lipid droplets, yeast cells in early or late log phase were stained with BODIPY 493/503 (Invitrogen) as described previously (41). All images were acquired with Slidebook software (Intelligent Imaging Innovations, version 4.1.0.3) using a microscope (Axiovert 200 M; Carl Zeiss Microimaging, Inc) with a 100 × 1.3 NA oil immersion objective lens (plan-NeoFluar) equipped with a digital camera (Sensican, Cooke). BODIPY images were acquired using the fluorescein isothiocyanate (FITC) filter set (excitation 490/20 and emission 528/38), and CFP was imaged with the CFP filter set (excitation 430/25; emission 470/30) (Chroma Technology Corp). Z-stacks were obtained at 0.3-μm spacing, and Slidebook commands of deconvolution (nearest neighbor option) were used to build images through cells. Transmission images were acquired using bright field optics. Micrograph figures were prepared with Adobe Photoshop and Illustrator.
Lipid Analysis
Lipid extraction and one-dimensional thin layer chromatography were performed as described previously (37) with a few modifications. Yeast cell pellets were lysed with acid-washed glass beads (Sigma, G8772, 425–600 μm) in lysis buffer (50 mm Tris-Cl, pH 8.0, 1 mm EDTA, 150 mm NaCl, 10% glycerol, 0.5 mm dithiothreitol) containing 0.1% (v/v) IGEPAL CA-630 (Sigma, I-3021, equivalent to Nonidet P-40) in the cold room for 30 min. Extracts were treated with 70 °C hot isopropyl alcohol for 30 min and then extracted as before. Lipid bands were digitally scanned, and their intensities were determined by ImageJ (version 1.41, National Institutes of Health). Concentrations were calculated relative to serial dilutions of lipid standards (Nu-Chek Prep, TLC 18–5C).
Immunoblotting and Co-immunoprecipitation
For examining protein expression, whole lysates were prepared from yeast as described previously (44) and subjected to SDS-PAGE followed by immunoblotting. For the co-immunoprecipitation experiment, yeast cell pellets were lysed with glass beads in lysis buffer containing 1% (v/v) protease mixture inhibitors (Sigma, P-8215) as described under “Lipid Analysis.” About 1.5 mg of clarified lysates (equal protein in each experiment; protein was determined with protein assay kit (Bio-Rad)) of each sample was subjected to immunoprecipitation with 25 μl of IgG-Sepharose 6 Fast flow bead s(GE Healthcare, 17-0969-01) in 1.5-ml tubes for 2 h at 4 °C and separated into bound or unbound fractions by centrifugation. The beads (bound fraction) were washed at least three times with lysis buffer. Equivalent amounts were saved for total lysates. Subsequently, the equivalent amounts of fractions (corresponding to 10–30 μg of total lysate) were resolved by SDS-PAGE and immunoblotted following standard protocols. Hemagglutinin (HA) and FLAG epitopes were probed with mouse monoclonal anti-HA (Roche Applied Science, 12CA5) and mouse monoclonal anti-FLAG M2 (Sigma, F3165) antibody, respectively. Anti-glucose-6-phosphate dehydrogenase (Zwf1p) antibody (Invitrogen, A9521) was used for a loading control. Horseradish peroxidase-conjugated goat anti-mouse IgG (Invitrogen, G21040) or rabbit anti-mouse IgG (Invitrogen, G21234) was used as secondary antibody. Bound antibody was detected using ECL Western blotting detection reagents (GE Healthcare).
Experiments in C. elegans
For C. elegans RNA-mediated interference and embryo imaging, double-stranded RNA (dsRNA) was prepared as described previously (45) from templates by using primers for scpl-2 (5′-AATTAACCCTCACTAAAGGATGACAACAATCGCACAGTCT-3′; 5′-TAATACGACTCACTATAGGGAGACGGAGAAGGTGTAGAAC-3′), T19A6.3 (5′-AATTAACCCTCACTAAAGGGATTCAGGCCAGAAGAATGC-3′; 5′-TAATACGACTCACTATAGGTGGAGCAGAATTACGAACTGC-3′), and lpin-1(5′-AATTAACCCTCACTAAAGGGGCCATTTGTCCACTCTCAT-3′; 5′-TAATACGACTCACTATAGGCTTACACACTCGGCGGTTTT-3′) to amplify N2 genomic DNA or N2 cDNA. For depletion of scpl-2 and T19A6.3, L4 hermaphrodites were injected with dsRNA and incubated at 20 °C for 48 h before analysis. For depletion of lpin-1, young adult hermaphrodites were injected with dsRNA and incubated for 12–16 h at 25 °C. For each condition, two different C. elegans strains expressing fluorescent markers had twinned nuclei in the two-cell stage. Embryos were mounted as described previously (45), and live imaging was conducted at 20 °C using a spinning-disc confocal microscopy head (CSU-10) mounted on either a Nikon TE2000-E inverted microscope or an Andor Revolution spinning disc confocal microscope system controlled by the Andor iQ software (Andor Technology). In all cases a ×60, 1.4 NA Plan Apochromat lens with 2 × 2 binning was used with a charge-coupled device camera (Orca-ER; Hamamatsu). Images were processed using ImageJ software.
Experiments in Mice and Mammalian Cultured Cells
Gene Expression in Mouse and Human Tissues
BALB/cByJ-Lpin1+/fld mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and bred to generate homozygous wild-type and fld/fld mice. Mouse tissue RNA was isolated with TRIzol, reverse-transcribed (OmniscriptTM, Qiagen, Valencia, CA), and quantitated by real time PCR using the iCycler (Bio-Rad) and SensiMix SYBR kit (Bioline USA Inc., Tauton, MA). Mouse gene expression levels were normalized to TATA box-binding protein mRNA. Gene expression studies in human tissues were performed with a panel of normalized first strand cDNA preparations pooled from multiple individuals (Human MTC Panel I and Digestive System MTC Panel, Clontech) and normalized to levels of β2-microglobulin and hypoxanthine phosphoribosyltransferase mRNA. PCR primer sequences are provided in supplemental Table S3
Gene Knockdown in Mouse Cells
Mouse myoblast C2C12 cells were cultured in DMEM (Invitrogen) containing 10% FBS supplemented with 100 μg/ml penicillin/streptomycin (Invitrogen). All gene knockdown experiments were carried out in 12-well plates using the BioT transfection reagent (Bioland Scientific LLC, La Palma, CA) according to the manufacturer's instructions. Briefly, C2C12 myoblasts were plated to 50–90% confluence and transfected with 200 nm lipin-1 antisense oligonucleotide (Isis Pharmaceuticals, Inc., Carlsbad, CA) or siTMEM188 (ON-TARGETplus SMART pool, Dharmacon Inc., Chicago) (see supplemental Table S3). After 24 h, the medium was replaced with DMEM containing 2% horse serum and then cultured for 48 h. The cells were harvested in 0.6 ml of TRIzol reagent. RNA isolation and real time PCR was then performed as described above. Expression levels were normalized to TATA box-binding protein, β2-microglobulin, and hypoxanthine-guanine phosphoribosyltransferase. Data were expressed as fold induction or repression by normalizing the data to the control condition.
DNA Constructs for Transfection
Construction and description of CTDNEP1- and NEP1-R1-encoding plasmids for mammalian expression are listed in supplemental Table S4.
Transfections and Cell Manipulations
HEK293T or HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For biochemistry experiments, cells were transfected with DNA using Lipofectamine 2000 (Invitrogen). For immunofluorescence experiments, cells were transfected with DNA using X-tremeGENE HP (Roche Applied Science). For stability experiments, cells were transfected with 6 μg of CTDNEP1-V5His and increasing amounts (0, 1, 3, and 6 μg) of NEP1-R1-HA-ProtA keeping total DNA amounts constant at 12 μg. Cells were lysed with 1% SDS with 2 mm EDTA in 10 mm Tris-HCl, pH 7.2, and total protein amounts were measured using the BCA protein assay kit (Thermo Scientific) for equal loading. For co-immunoprecipitation, cells were harvested in immunoprecipitation buffer (32) and lysed by passing through a 25–5/8-gauge needle 10 times before centrifuging at 10,000 × g for 5 min. Cleared lysates were further spun down at 14,000 × g for 10 min at 4 °C twice to isolate fully detergent-soluble lysates. About 3.5 mg of lysates were applied to IgG-Sepharose. The subsequent steps were the same as described under “Experiments in Yeast.”
Antibodies
For detection of FLAG- and HA-tagged proteins, these epitopes were probed as described for yeast. For detection of mammalian cell lysates by immunoblot, the following antibodies were used: anti-V5 (Invitrogen, R960-25) at 1:5000; anti-HA (Roche Applied Science, 12CA5) at 1:1000; and anti-tubulin (Sigma, DM1α) at 1:5000. Primary antibodies for immunofluorescence were as follows: anti-HA.11 at 1:5000 (Sigma) and anti-FLAG at 1:1000 (Invitrogen). Secondary antibodies anti-mouse Cy2 and anti-rabbit Cy3 (Jackson ImmunoResearch) were each used at 1:250.
Indirect Immunofluorescence and Fixed Cell Analysis
Cells were fixed immediately after aspiration of media with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature (RT), then permeabilized with 0.1% Triton X-100 in PBS for 5 min, rinsed three times with PBS, and incubated with 0.5% BSA for 30 min at RT. Fixed cells were incubated with primary antibodies and secondary antibodies diluted in 0.5% BSA for 1 h and 30 min, respectively, at RT. Between antibody incubations, cells were rinsed three times with PBS. Cells were mounted with ProLong Gold mounting media containing DAPI (Invitrogen). Images were recorded on a DeltaVision microscope at 1 × 1 binning with a 100× NA 1.3 U-PlanApo objective (Olympus). Z-stacks (0.5 μm between sections) were deconvolved using SoftWoRx (Applied Precision) and processed in Photoshop. All images shown are single sections that were processed similarly between different conditions. For quantification of nuclear accumulation of Lipin-1b, images using a ×40 NA 1.3 U-PlanApo objective (Olympus) were analyzed using ImageJ. Using the DAPI channel as a reference, the nuclear region was traced to measure the mean nuclear fluorescence. Cell boundaries were traced using the lipin-1b channel using an area at least 20 pixels larger than the nuclear area to measure mean cell fluorescence (nuclear and cytoplasmic). To calculate total fluorescence in each region, mean fluorescence was multiplied by area and corrected by subtracting the total camera background for that same area. The nuclear fraction is equal to total nuclear fluorescence divided by total cell (nuclear and cytoplasmic) fluorescence.
In Vivo Dephosphorylation of Lipins in HEK293 Cells
Cells were transfected in 10-cm dishes with 6 μg each of CTDNEP1–2×FLAG or CTDNEP1-V5His and NEP1-R1-HA-ProtA or NEP1-R1-FLAG together with 2 μg of mouse lipin-1a, -1b, or -2 fused to V5His or HA epitopes. Twenty four hours after transfection, cells were lysed with 1 ml of cold lysis buffer (50 mm NaF, 1 mm EDTA, 1 mm EGTA, 10 mm Na2HPO4, 50 mm β-glycerophosphate, pH 7.4, 0.1% Triton X-100, and a protease inhibitor tablet). Lysates were resuspended in SDS loading buffer, and 1% of total lysate from each sample was loaded and resolved on a SDS-polyacrylamide gel. For λ-phosphatase treatment, 50 μg of total lysate was added to 400 units of λ-phosphatase in 1× PMP buffer (New England Biolabs, P0753), supplemented with 1 mm MnCl2, and was incubated at 30 °C for 30 min. Reactions were stopped by addition of SDS sample buffer, and ∼20% of the reaction was loaded and resolved on an SDS-polyacrylamide gel. For immunoprecipitations, precleared cell lysates were incubated with 2 μg of anti-mid-lipin-1b (32) for 2 h at 4 °C. After washing with lysis buffer, immune complexes were washed twice with phosphatase wash buffer (50 mm HEPES, 100 mm NaCl, 2 mm DTT, and 0.01% Brij). After the last wash, the supernatant was removed, and beads were incubated with the reaction buffer ± phosphatase, as above.
RESULTS
TMEM188 Is a Candidate for the Human Spo7p Ortholog
In yeast, Nem1p associates with Spo7p in a phosphatase complex to dephosphorylate the yeast lipin ortholog, Pah1p (21). The Nem1p mammalian ortholog, CTDNEP1, was unable to dephosphorylate lipin-1 in HeLa or HEK293 cells, suggesting that these cells are missing a Spo7p-like partner (32).
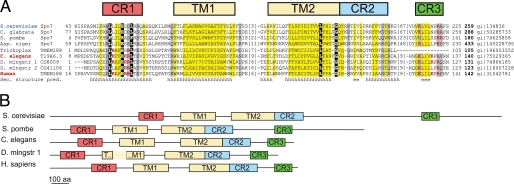
Spo7p orthologs have been found in several fungal species, including Schizosaccharomyces pombe. However, identification of nonfungal homologs has been hindered by very low sequence similarity. Standard approaches such as BLAST and PSI-BLAST failed to find statistically significant hits outside fungi, and analysis of marginal hits below default threshold (E-value above 0.005) did not suggest useful leads. This difficulty is largely caused by the small size of the fungal family and the transmembrane nature of the protein. The first hint of an Spo7p mammalian homolog was given by the PROCAIN program (47), which found TMEM188 (GI 31542781) as the first hit encoded by the human genome. TMEM188 is a hypothetical membrane protein of unknown function or significance that has been identified in vertebrates (fish and tetrapods). Visual inspection of the alignment of Spo7p with TMEM188 revealed a few conserved positions concentrating in several motifs. Some of the invariant positions were occupied by nonhydrophobic amino acids, which increased our confidence in homology for these transmembrane proteins (Fig. 1). Homology was further supported by the HHpred server (48). Although S. cerevisiae Spo7p sequence used as HHpred query did not find TMEM188 proteins with statistical significance, querying the human proteome with the S. pombe Spo7p sequence returned TMEM188 with HHpred probability of 95%. Additional homologs were found in Drosophila, C. elegans, and Trichoplax. Their multiple alignment (Fig. 1A) looked convincing enough to warrant experimental investigation into their functions.
FIGURE 1.
Spo7p is conserved from fungi through mammals. A, multiple sequence alignment of representative Spo7p homologs. Sequences are labeled with organism names and gene IDs. GenBankTM gi numbers are at the end of each sequence. PSI-PRED consensus secondary structure prediction (pred.) is shown in the last line (h, α-helix; e, β-strand). Alignment columns are colored according to conserved residue type as follows: mainly hydrophobic positions are highlighted yellow and mainly hydrophilic positions are highlighted gray. Positions invariant in this set of sequences are white on black. Positively charged amino acids at positions with high content of positive charges are shown in red. Residue numbers are indicated to the right and to the left of each sequence, and removed segments are accounted for with the number of omitted residues in brackets. The number in boldface at the end of each line is sequence length. Abbreviations used are as follows: C. glabrata, Candida glabrata strain 138; Trichoplax, Trichoplax adherens; D. mlngstr, Drosophila melanogaster; human, Homo sapiens; TM, transmembrane domain; CR, conserved region. B, comparison of selected Spo7p homologs showing the placement of transmembrane domains (TM1 and TM2) and conserved regions (CR) along their lengths. aa, amino acids.
All candidate proteins have a very similar topology with a few invariant amino acids. Similar to the yeast Spo7p, the animal forms are predicted to have two membrane spans (TM1 and TM2 in Fig. 1) and three other regions of homology (CR1, CR2, and CR3). The Spo7p forms in yeast were longer than the vertebrate proteins (Fig. 1B). In S. cerevisiae, Candida glabrata, and Aspergillus niger, the SPO7-encoded proteins were predicted to have 259, 288, and 433 amino acids, respectively; the protein in the fission yeast S. pombe was predicted to have 180 amino acids, which corresponded more closely to the length of the animal proteins, 123–141 amino acids (excluding the Trichoplax sequence of 105 amino acids). The difference in length between budding yeast and animal forms is a longer N-terminal extension, an additional internal region between CR2 and CR3, and an extended C terminus compared with the animal homologs.
NEP1-R1 (TMEM188) in Combination with CTDNEP1 Can Rescue the spo7Δnem1Δ Lipid Phenotype in Yeast
To determine whether human TMEM188, which we rename nuclear envelope phosphatase 1-regulatory subunit 1 (NEP1-R1), is a Spo7p ortholog, we first tested whether it could replace Spo7p in a spo7Δ strain. spo7Δ cells have an elongated and often stacked perinuclear endoplasmic reticulum resulting in aberrant nuclear shape (21, 29). This is, at least in part, a result of sequestering the phospholipid synthesis repressor Opi1p by its binding to excess phosphatidate in membranes lowering its nuclear concentration (11). In addition, we have recently shown that the BODIPY 493/503, a neutral lipid dye that usually is specific for lipid droplets (49), stains the nuclear envelope in the spo7Δ strain (50). The strain also has fewer lipid droplets, presumably a result of lower DAG levels because Pah1p is not activated (28); however, besides being a precursor for TAG, DAG itself plays a role in droplet formation (50, 51).
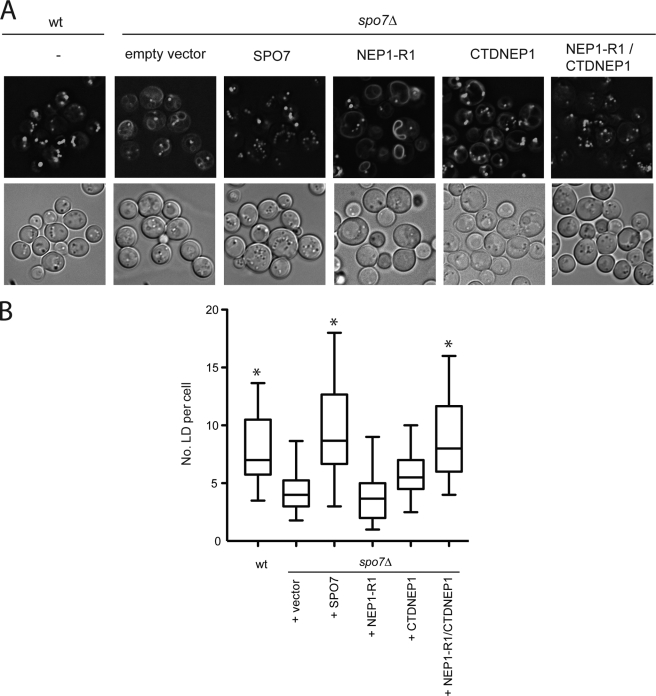
Expression in spo7Δ cells of human NEP1-R1, driven by the PGK1 promoter on a low copy plasmid, did not reverse the BODIPY staining of perinuclear rings or increase the number of lipid droplets (Fig. 2). Expression of CTDNEP1 alone resulted in a small decrease in nuclear ring staining with BODIPY but no significant increase in droplet number. However, when both human CTDNEP1 and NEP1-R1 were co-expressed in the spo7Δ strain, droplet numbers were greatly increased, and BODIPY staining of the ER was eliminated (Fig. 2). Our interpretation is that NEP1-R1 increased the effectiveness of CTDNEP1 to dephosphorylate Pah1p. These data also suggest that, although NEP1-R1 is conserved between fungi and mammals, the mammalian form interacts more effectively with the mammalian Nem1p ortholog, CTDNEP1 than the yeast form.
FIGURE 2.
NEP1-R1 requires CTDNEP1 for complementing spo7Δ phenotype. spo7Δ cells were transformed with empty vector or vector containing open reading frames for SPO7, NEP1-R1, CTDNEP1, or NEP1-R1 plus CTDNEP1, all under the PGK1 promoter. A, cells were stained with BODIPY 493/503 (top panels) and bright field images shown below. B, quantification of lipid droplet number. At least 100 cells from each strain are shown. Vertical sides of boxes in this box-and-whisker graph represent the 25 and 75% range of distributions of droplets per cell; the bar is median. Vertical lines represent 5–95% of distribution. Asterisks signify p < 0.01 compared with spo7Δ with empty vector. Data were collected from at least two independent experiments.
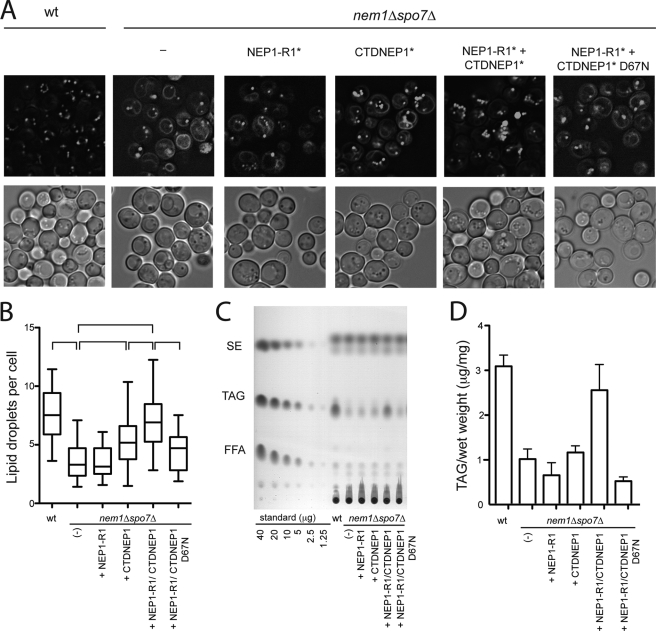
We next tested whether CTDNEP1 in combination with NEP1-R1 can functionally complement the nem1Δspo7Δ double deletion strain. The nem1Δspo7Δ cells had a similar phenotype to spo7Δ as follows: proliferated ER that stained with BODIPY, fewer lipid droplets, and low TAG levels (Fig. 3). Unlike the case for spo7Δ cells, where CTDNEP1 had no activity (Fig. 2), CTDNEP1 partially complemented the double deletion mutant to increase droplet number and lower BODIPY staining of membranes. The difference could reflect titration of factors (such as Pah1p) by Nem1p in spo7Δ cells. The presence of both human proteins together in the double deletion mutant resulted in virtually complete loss of BODIPY membrane staining, in a droplet number comparable with wild type, and in a nearly normal level of TAG; neither human protein alone was able to raise TAG levels (Fig. 3, C and D). This complementation was dependent on active CTDNEP1 phosphatase, because a CTDNEP1 inactive mutant, D67N (32), could not substitute for wild type in lowering BODIPY staining intensity of ER rings or increasing droplet number or TAG level (Fig. 3).
FIGURE 3.
NEP1-R1 and CTPNEP1 in combination complement nem1Δspo7Δ phenotypes. nem1Δspo7Δ cells were transformed with empty vector or vector expressing NEP1-R1-HA-ProtA, CTDNEP1–2×FLAG, both, or both but with inactive CTDNEP1 (D67N). A, cells grown to late log in SD medium were stained with BODIPY. Bright field images are shown below. The asterisks signify that tagged versions were used, as just described, in A and B; the tags were omitted from the labels because they were not used for detection. B, quantification of lipid droplets. Vertical sides of boxes in this box-and-whisker graph represent the 25 and 75% range of distributions of droplets per cell; the bar is median. Vertical lines represent 5–95% of distribution. Brackets above represent significant differences (p ≤ 0.01) according to one-way analysis of variance. Data were collected from at least two independent experiments. C, total lipids were extracted from each strain and subjected to thin layer chromatography to separate neutral lipids in this representative experiment. SE, steryl ester; FFA, free fatty acid. D, quantification of triacylglycerol from TLC analysis (mean ± S.E. from three experiments).
It is not clear why CTDNEP1 alone increased droplet number but had no effect on TAG levels, but it may point to the dual roles of DAG in droplet formation and TAG synthesis (50). The former effect may be more sensitive to small changes in DAG concentration. Regardless, these results indicate that human CTDNEP1 and NEP1-R1 can functionally replace the Nem1p-Spo7 complex in yeast with respect to droplet number and lipids.
Although CTDNEP1 and NEP1-R1 complemented the low droplet and low TAG phenotype of the nem1Δspo7Δ strain, NEP1-R1 or the two human proteins in combination could not rescue the sporulation defect of spo7Δ (52), indicating a yeast-specific function for SPO7 (data not shown). There was partial complementation of the ER proliferation seen in the double deletion mutant (data not shown) (21).
NEP1-R1 Stabilizes and Physically Interacts with CTDNEP1 in Yeast
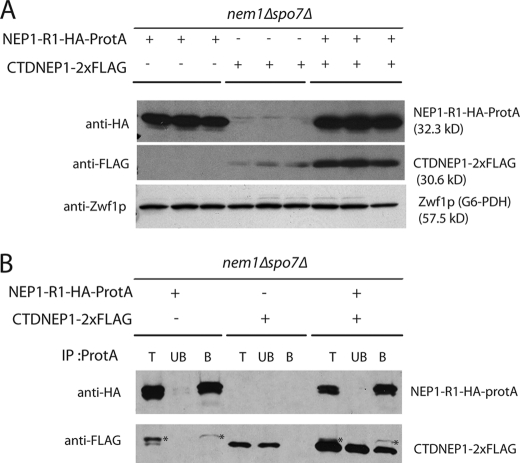
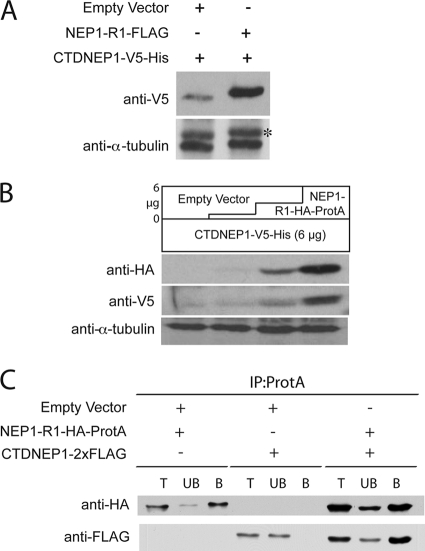
Nem1p and Spo7p form a complex in the ER (29). To test whether human CTDNEP1 and NEP1-R1 can physically interact in yeast, we generated FLAG-tagged CTDNEP1 and hemagglutinin (HA)-protein A-tagged NEP1-R1. Both proteins could be visualized on immunoblots, but the level of FLAG-tagged CTDNEP1 was low when expressed alone (Fig. 4A). In combination with NEP1-R1, however, the steady-state level of CTDNEP1 was increased 6-fold (average from three experiments), suggesting that NEP1-R1 stabilizes CTDNEP1. To determine whether the human proteins can physically interact in yeast, we tested for co-precipitation of CTDNEP1 with NEP1-R1 from cell extracts. About 45% (average from two experiments) of CTDNEP1 could be co-immunoprecipitated with NEP1-R1, although none was precipitated in the absence of NEP1-R1 (Fig. 4B). This experiment indicates that, at least when expressed in yeast, NEP1-R1 is in a complex with CTDNEP1, as is the case for Spo7p and Nem1p.
FIGURE 4.
NEP1-R1 interacts with CTDNEP1. A, NEP1-R1 stabilizes CTDNEP1. nem1Δspo7Δ cells were transformed with NEP1-R1-HA-ProtA, CTDNEP1–2×FLAG, or both. Extracts from triplicate cultures were subjected to SDS-PAGE and immunoblotting. NEP1-R1 and CTPNEP1 were detected by anti-HA and anti-FLAG antibody, respectively. B, CTDNEP1 is co-immunoprecipitated with NEP1-R1. Cell lysates from cells expressing NEP1-R1-HA-ProtA, CTDNEP1–2×FLAG, or both were immunoprecipitated (IP) with IgG-Sepharose beads and analyzed by immunoblot, in which NEP1-R1 and CTPNEP1 were probed by anti-HA and anti-FLAG antibody, respectively. T, total lysate; UB, unbound; B, bound fraction to IgG-Sepharose beads. Asterisk indicates weak interacting product of protein A and anti-FLAG antibody.
Knockdown of NEP1-R1 and CTDNEP1 Share a Similar Phenotype in C. elegans
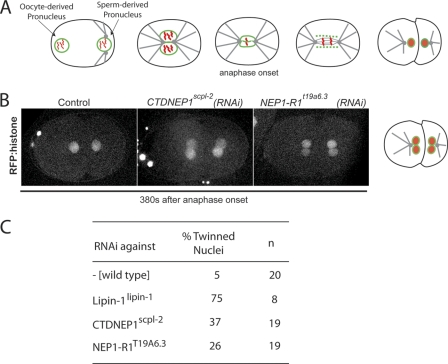
It has been demonstrated previously that the deficiency of the C. elegans lipin ortholog, LPIN-1, leads to impaired embryonic development associated with abnormal nuclear dynamics (30, 31). We hypothesized that if CTDNEP1 and NEP1-R1 have conserved roles in activation of LPIN-1 in C. elegans, their depletion might produce a similar phenotype. Following fertilization of C. elegans embryos, the expanded maternal and paternal pronuclei meet, nuclear membranes disassemble, and the maternal and paternal haploid genomes are mixed. After chromosome segregation, a single nuclear envelope forms around the chromatin in each daughter cell (Fig. 5A) (53). In embryos depleted of LPIN-1, two nuclei are formed in each daughter cell, rather than a single nucleus per cell (30, 31). The “twinned nuclei” phenotype was also observed when either C. elegans CTDNEP1 (scpl-2) or NEP1-R1 (T19A6.3) was depleted (Fig. 5, B and C). Despite extensive functional genomic analyses of phenotypes in the first C. elegans embryonic division (54), few genes have been reported to result in twinned nuclei suggesting that CTDNEP1scpl-2, NEP1-R1T19A6.3, and lipinlpin-1 are in the same pathway and that these proteins are functionally conserved in C. elegans.
FIGURE 5.
C. elegans NEP1-R1 knockdown in C. elegans embryos results in twinned nuclei similar to depletion of C. elegans CTDNEP1 and lipin. A, diagram highlighting the dynamics of the oocyte- and sperm-derived pronuclear envelopes (green) and chromosomes (55) during the first division of the C. elegans embryo. B, spinning disk confocal optics were used to obtain four-dimensional images of embryos expressing mCherry-histone in control and the indicated RNAi knockdown strains. Representative central z-sections 380 s after anaphase onset are shown. C, extent of “twinning” of nuclei in the various strains; n, number of embryos separately subjected to time-lapse imaging.
Expression of CTDNEP1 and NEP1-R1 Parallels That of lipin-1 in Human and Mouse Tissues
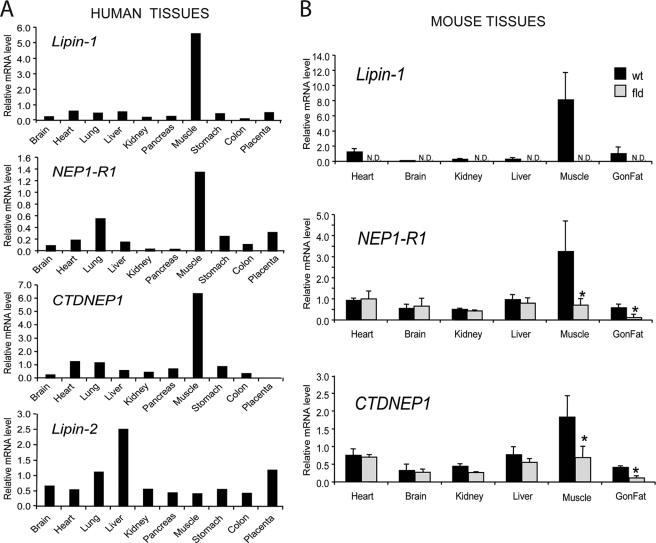
If lipin-1, CTDNEP1, and NEP1-R1 function in the same pathway, they should have similar tissue expression patterns. We compared mRNA expression patterns for lipin-1 and the two membrane phosphatase complex proteins in both mouse and human tissue panels. In agreement with previous studies (15), lipin-1 is expressed most prominently in skeletal muscle in the human (Fig. 6A) and mouse (Fig. 6B, black bars), with lower levels in other metabolic tissues such as fat, heart, and liver. Consistent with a role in the regulation of lipin-1 dephosphorylation, there was a striking concordance among expression of lipin-1, NEP1-R1, and CTDNEP1, with very high levels in skeletal muscle and lower levels in other metabolic tissues (Fig. 6). Interestingly, in the fatty liver dystrophy fld mouse model, which carries a null mutation in Lpin1, both NEP1-R1 and CTDNEP1 mRNA levels in muscle and fat were reduced compared with wild-type mice (Fig. 6B, gray bars). These results suggest that there is coordinate regulation between lipin-1 and the two phosphatase components. The expression patterns of mRNAs encoding NEP1-R1 and CTDNEP1 are consistent with a critical function in muscle, where lipin-1 deficiency results in severe muscle disease (55, 56). This pattern was distinct from that of lipin-2 (Fig. 6A), another member of the phosphatidate phosphatase family, which causes a nonmuscle-related syndrome when mutated in humans (57).
FIGURE 6.
Expression of lipin-1 and its activators in human and mouse tissues. A, quantitative PCR analysis of pooled human tissue samples shows coordinate mRNA expression patterns for the proteins human lipin-1, NEP1-R1, and CTDNEP1. Lipin-2 mRNA exhibits a distinct expression pattern. B, both CTDNEP1 and NEP1-R1 expressions are reduced in muscle and adipose tissue from the fld mouse. Coordinate expression of mRNA for lipin-1, NEP1-R1, and CTDNEP1 in tissues from wild-type mice (black bars). In Lpin1-deficient fld mice, lipin-1 mRNA expression is undetectable (N.D.), and levels of NEP1-R1 and CTDNEP1 mRNA in muscle and gonadal fat (Gon Fat) are significantly reduced. Values represent the mean ± S.D. for tissues from three mice. *, p < 0.05 in wild type versus fld.
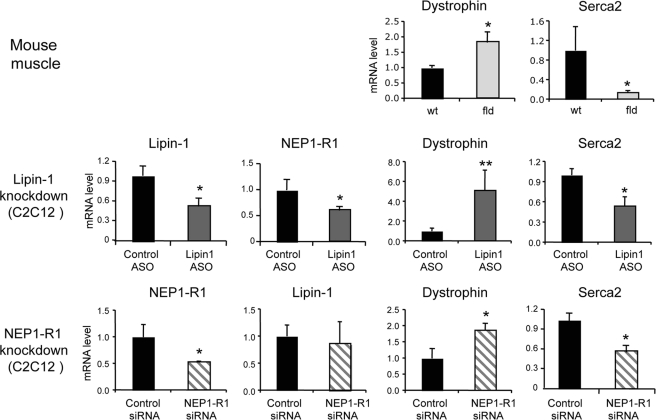
Skeletal muscle from lipin-1-deficient fld mice exhibits altered expression levels for key genes that are important for muscle function (data not shown). These include elevated levels of dystrophin and reduced levels of Serca2, a sarcoplasmic protein involved in regulation of calcium levels. We could replicate the pattern of gene expression observed in fld muscle by knockdown of lipin-1 in C2C12 myocytes (Fig. 7). Consistent with a role for NEP1-R1 in lipin-1 function, knockdown of NEP1-R1 also resulted in elevated dystrophin and reduced Serca2 levels. Interestingly, lipin-1 knockdown led to reduced NEP1-R1 expression, although NEP1-R1 knockdown did not influence lipin-1 levels. These results indicate that expression of lipin-1, CTDNEP1, and NEP1-R1 is closely linked in mammals.
FIGURE 7.
Knockdown of NEP1-R1 and lipin-1 in C2C12 cells has similar effects on expression of dystrophin and Serca2. Top row, gene expression changes in muscle from lipin-1-deficient fld mice. Lack of lipin-1 in muscle leads to enhanced expression of dystrophin and reduced expression of Serca2. Middle row, knockdown of lipin-1 expression in C2C12 myocytes using a lipin-1-specific antisense oligonucleotide (ASO) recapitulates the effects on dystrophin and Serca2 gene expression observed in fld muscle and leads to reduced expression of NEP1-R1. Bottom row, knockdown of NEP1-R1 expression in C2C12 myocytes using a specific siRNA leads to similar changes in dystrophin and Serca2 expression as observed in fld muscle. NEP1-R1 knockdown does not affect lipin-1 mRNA levels. Relative mRNA levels represent the mean ± S.D. for three separate cultures. *, p < 0.05 or **, p < 0.01 versus relevant wild type or control sample.
CTDNEP1 and NEP1-R1 Form a Complex in Human Cells
Although we showed that CTDNEP1 and NEP1-R1 can interact in yeast cells, the more physiological milieu is within human cells. Therefore, we expressed both proteins in HEK293 cells. CTDNEP1 tagged with V5His was visible on immunoblots when expressed alone, but expression was increased severalfold when co-expressed with NEP1-R1 (Fig. 8A), similar to our findings in yeast (Fig. 4A). To examine this more carefully, we transfected cells with increasing amounts of NEP1-R1 in the presence of a constant amount of CTDNEP1. Levels of expressed phosphatase increased in proportion to expression of NEP1-R1 (Fig. 8B). Thus, CTDNEP1 is stabilized by NEP1-R1 both in yeast and human cells.
FIGURE 8.
NEP1-R1 binds to and stabilizes CTDNEP1 in human cells. A, immunoblot of whole cell lysates of HEK293 cells transfected with 6 μg of CTDNEP1-V5His DNA and either 6 μg of empty vector or NEP1-R1-FLAG DNA. CTDNEP1 is stabilized in the presence of NEP1-R1 (anti-V5). Blot was stripped and reprobed with anti-tubulin as loading control. *, nonspecific cross-reacting band. B, immunoblot of whole cell lysates of HEK293 cells transfected with 6 μg of CTDNEP1 and increasing amounts of NEP1-R1 DNA as shown with total transfected DNA kept constant at 12 μg by transfecting with empty vector. C, CTDNEP1 is co-immunoprecipitated with NEP1-R1. HEK293 cell lysates expressing empty vector, NEP1-R1-HA-ProtA DNA, CTDNEP1–2×FLAG DNA, or both were immunoprecipitated (IP) with IgG-Sepharose beads and analyzed by immunoblot with HA or FLAG antibodies, as shown. T, total lysate; UB, unbound fraction; B, IgG-Sepharose-bound fraction. Equivalent amounts of all fractions are shown.
To determine whether these two proteins physically interact in HEK293 cells, we tested for co-precipitation of FLAG-tagged CTDNEP1 with HA-Protein A-tagged NEP1-R1. Most of the phosphatase could be pulled down with NEP1-R1, indicating that the two proteins interact within human cells (Fig. 8C).
CTDNEP1 Requires NEP1-R1 to Dephosphorylate Lipins-1 and -2
CTDNEP1 was originally identified as the Nem1p ortholog and activator of lipin-1 by demonstrating that expression in BHK cells led to dephosphorylation of lipin-1b, a splice variant of lipin-1. However, this activity was absent in HeLa and HEK293T cells, suggesting that an Spo7p ortholog was missing in these lines (32). To test whether NEP1-R1 can function as the missing partner, HEK293 cells expressing mouse HA-lipin-1b were transfected with CTDNEP1-V5His and NEP1-R1-FLAG, or the two in combination (Fig. 9A). As shown previously, CTDNEP1 was not sufficient for lipin-1b dephosphorylation. However, in combination with NEP1-R1, most of the lipin-1 migrated faster on an SDS gel, suggesting that it was dephosphorylated (Fig. 9A). This was confirmed after immunoprecipitation of lipin-1b and treatment with λ phosphatase, which increased lipin-1b gel migration to the same extent (Fig. 9B). Thus, NEP1-R1 can act as a partner to CTDNEP1 in dephosphorylating lipin-1b in HEK293 cells.
FIGURE 9.
CTDNEP1 requires NEP1-R1 to dephosphorylate lipins. A, HEK293 cells were transfected with DNA for HA-lipin-1b, CTDNEP1-V5His, and NEP1-R1-FLAG, and the electrophoretic mobility of HA-lipin-1b was assessed by SDS-PAGE, followed by immunoblot (IB) for HA-lipin-1b. An increased electrophoretic mobility of HA-lipin-1b is only seen in the presence of both CTDNEP1 and NEP1-R1. B, total cell lysates expressing HA-lipin-1b or immunoprecipitated (IP) HA-lipin-1b were treated with λ phosphatase in the presence of both CTDNEP1 and NEP1-R1 or either construct alone showing that the increased electrophoretic mobility was due to dephosphorylation. C, CTDNEP1 can dephosphorylate lipin-1a and lipin-2 in the presence of the NEP1-R1. Total cell lysate expressing lipin-1a or lipin-1b-V5 in the presence of CTDNEP1–2×FLAG alone or in combination with NEP1-R1-HA-ProtA ± λ phosphatase were analyzed by immunoblot against V5.
To determine whether dephosphorylation is specific to lipin-1b, we substituted lipin-1a and lipin-2 in this assay. Neither lipin species was dephosphorylated by CTDNEP1 alone but could be dephosphorylated if both CTDNEP1 and NEP1-R1 were present (Fig. 9C). Thus, the human phosphatase complex is capable of dephosphorylating at least these three lipins. We could not show a reproducible band shift with lipin-3 (data not shown). Whether this reaction occurs with equal efficiency with the endogenously expressed proteins must await future experiments.
Lipin-1b Accumulates in the Nucleus When CTDNEP1 and NEP1R1 Are Expressed
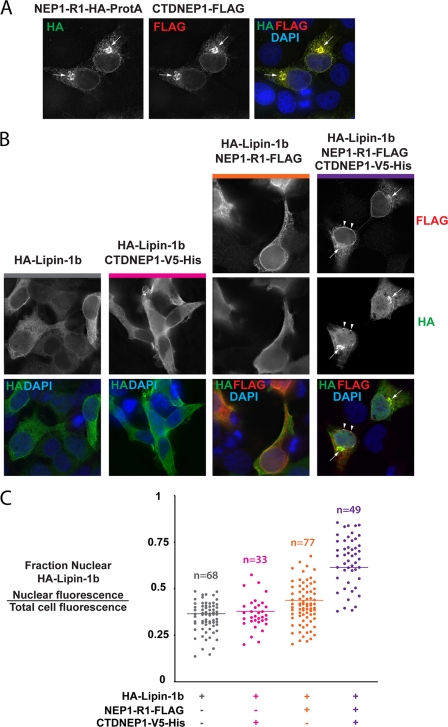
It has been shown recently that the mammalian target of rapamycin kinase affects the localization of lipin-1 between the cytoplasm and nucleus (58). To determine whether the mammalian lipin phosphatase complex can also control localization of lipin-1, we co-expressed these proteins in human cells. We first confirmed co-localization of tagged CTDNEP1 and NEP1-R1. As expected from the co-immunoprecipitation results (Fig. 8C), CTDNEP1 and NEP1-R1 co-localized in cells, both appearing in a filamentous pattern in the cytoplasm, a perinuclear accumulation (arrows), and along the nuclear rim (Fig. 10A). Next, we determined the extent of nuclear localization of HA-tagged lipin-1b in the absence and presence of CTDNEP1 or NEP1-R1. Lipin-1b, when expressed alone, showed diffuse localization in cells (Fig. 10B), predominantly in the cytoplasm (Fig. 10C). Although expression of CTDNEP1 alone did not affect the nuclear localization of lipin-1b as compared with control, expression of NEP1-R1 alone resulted in a slight increase in nuclear lipin, possibly due to its ability to function with endogenous CTDNEP1 (Fig. 10, B and C). When both CTDNEP1 and NEP1-R1 were expressed, some focal co-localization of lipin-1b at the nuclear rim with NEP1-R1 was apparent in 25% (n = 123 cells) of HEK293 cells (arrowheads) but not in HeLa cells (Fig. 10, B and C; data not shown). Lipin also tended to accumulate in the perinuclear region (arrows) with NEP1-R1 (Fig. 10B). Importantly, the fraction of lipin within the nucleus increased significantly in both cell types (Fig. 10C and data not shown). Thus, the combination of CTDNEP1 and NEP1-R1 can change the phosphorylation status of lipin-1b and affect its localization, and likely function, in human cells.
FIGURE 10.
Lipin-1b accumulates in the nucleus when co-expressed with NEP1-R1 and CTDNEP1. A, co-localization of tagged CTDNEP1 with NEP1-R1 by immunostaining. HeLa cells were transfected with plasmids encoding NEP1-R1-HA-ProtA and CTDNEP1–2×FLAG. Antibodies against HA and FLAG were used to localize the corresponding proteins. Proteins show co-localization at the nuclear rim, perinuclear accumulation (arrows), and a filamentous pattern in the cytoplasm that is likely the ER. B, immunostaining in HEK293 cells of HA-lipin-1b (HA) when expressed alone (1st column), with CTDNEP1-V5His (2nd column), NEP1-R1-FLAG (3rd column), and a combination of CTDNEP1-V5His and NEP1-R1-FLAG (4th column). Note that HA-lipin-1b is mostly diffuse with a slight nuclear accumulation when co-expressed with NEP1-R1 and a significant nuclear and perinuclear (arrows) accumulation when co-expressed with both CTDNEP1 and NEP1-R1. Arrowheads show foci of HA-lipin-1b that co-localize with NEP1-R1-FLAG at the nuclear rim. C, quantification of the nuclear fraction of HA-lipin-1b in individual HEK293 cells co-stained with HA and FLAG in the four conditions in B is shown. The nuclear fraction of lipin-1b is not significantly different when expressed alone or with CTDNEP1-V5His (mean ± S.E., 95% confidence interval is 0.35 ± 0.02 and 0.36 ± 0.03, respectively). To be certain that the nuclear fraction of lipin-1b in co-transfected cells was measured, measurements were also made in cells co-expressing HA-lipin-1b with CTDNEP1–2×FLAG. A similar nuclear fraction of HA-lipin-1b (0.31 ± 0.03; n = 29) was found in these cells (data not shown). The HA-lipin-1b nuclear fraction increased to 0.42 ± 0.03 in the presence of NEP1-R1 and 0.59 ± 0.03 when both NEP1-R1 and CTDNEP1 are expressed (p < 0.001 by Student's t test compared with cells expressing HA-lipin-1b alone).
DISCUSSION
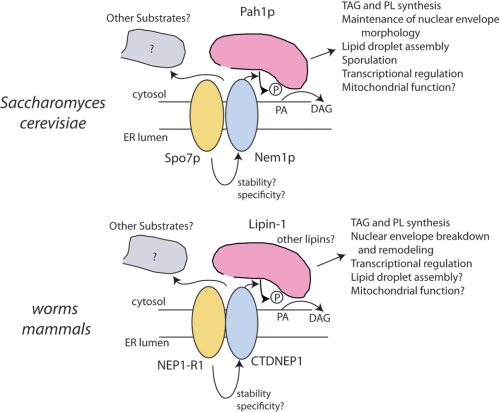
We have found the metazoan ortholog for the lipin-1/Pah1p coactivator Spo7p, TMEM188, here renamed NEP1-R1. Addition of CTDNEP1 and NEP1-R1 to nem1Δspo7Δ cells is able to rescue the neutral lipid synthesis defect in yeast. NEP1-R1 stabilizes and forms a physical complex with CTDNEP1. The expression patterns of NEP1-R1 are nearly identical to CTDNEP1 and lipin-1 in mouse and human tissues. Human CTDNEP1·NEP1-R1 is able to dephosphorylate lipin-1 in HEK293 cells in a manner similar to the function of the Nem1p-Spo7p complex acting on Pah1p in yeast. The conservation of structure and function between yeast and metazoans is depicted in Fig. 11.
FIGURE 11.
Conservation of function in Pah1p/lipin-1 activation among yeast, nematodes, and mammals. The proteins and their functions are compared between yeast and metazoan systems. Spo7p/NEP1-R1 is likely to stabilize the Nem1p/CTDNEP1 phosphatase. Spo7p/NEP1-R1 may also influence assembly of the complex or specificity for substrates. The gray shape indicates that there may be other unknown substrates for the phosphatase complex.
What exactly is Spo7p/NEP1-R1 doing in facilitating the action of Nem1p/CTDNEP1? In some contexts the phosphatase subunit appears to function alone; in vitro, a soluble form of human CTDNEP1 expressed in bacteria can dephosphorylate synthetic peptides corresponding to sequences within lipin-1; the authors postulated that there was no need for a Spo7p-like adapter protein (34). In vivo, CTDNEP1 can complement the nem1Δ strain in yeast, and the human protein when expressed in BHK cells can dephosphorylate lipin-1 (32). However, the contribution of the endogenous Spo7p/NEP1-R1 to CTDNEP1 activity was unknown in this study. Neither Nem1p nor CTDNEP1 in vitro can by itself dephosphorylate Pah1p or lipin-1, respectively (21, 32), suggesting an essential contribution of endogenous binding partners in vivo. We observe that CTDNEP1 expressed from the strong PGK1 promoter in nem1Δspo7Δ cells can moderately increase lipid droplet number and decrease nuclear envelope proliferation (Fig. 3 and data not shown), suggesting that a partner is not absolutely required for lipin-1 dephosphorylation.
Our data show that protein concentration of CTDNEP1 is much higher when co-expressed with NEP1-R1 both in yeast and mammalian cells. Because the corresponding yeast proteins stably associate in a 1:1 molar ratio (29), it is reasonable to hypothesize that the human proteins associate similarly and that CTDNEP1 is stabilized by NEP1-R1. It remains to be seen whether CTDNEP1 alone is degraded at a faster rate than when associated with its binding partner, accounting for the contrast in expression levels. NEP1-R1 could also influence the initial interaction of CTDNEP1 with the ER membrane. Full-length yeast Nem1p can assemble into membranes without Spo7p, but deletions in Nem1p confer Spo7p dependence for membrane association (29), suggesting that NEP1-R1 might recruit CTDNEP1 to the membrane. However, human CTDNEP1 is predicted (with 91% probability) to have an N-terminal signal sequence, so it is likely to insert into the ER through the classical Sec61 channel independent of NEP1-R1. Whether NEP1-R1 has other effects, such as to increase the affinity of CTDNEP1 for lipin-1, the specificity of CTDNEP1 for lipin-1, or whether the complex can dephosphorylate other substrates, is unknown (Fig. 11).
Unlike the yeast proteins, human CTDNEP1 and NEP1-R1 in combination were not able to reverse the sporulation defect, suggesting that the requirement of Spo7p for sporulation may extend beyond the role of activating Pah1p. This function may reside in the large regions of yeast Spo7p not found in the mammalian proteins (Fig. 1A). However, the Spo7p homolog in the fission yeast S. pombe, which sporulates, does not have the large inserts of the budding yeast homologs, although it has not yet been shown that the S. pombe SPO7 is required for sporulation.
In the worm, the knockdown of NEP1-R1T19A6.3 or CTDNEP1scpl-2 phenocopies the knockdown of lpin-1, causing the appearance of twinned nuclei in the second embryonic division. This study suggests that these three proteins work in the same pathway. Similar to the nuclear envelope expansion phenotype in yeast, reducing NEP1-R1T19A6.3 or CTDNEP1scpl-2 levels results in a less severe nuclear phenotype than depleting lipinlpin-1 itself suggesting that, as in yeast, dephosphorylation of lipin by the complex is only partially responsible for determining the activity of lipin-1. However, we cannot exclude the possibility of different knockdown efficiencies. Unlike fungi, in metazoans a filamentous lamin network underlies the nuclear envelope, and it has been shown that LPIN-1 functions in lamin disassembly (31), although the precise mechanism is not known.
Lipin-1 in mammals is most highly expressed in skeletal muscle, and we find that this is also true for CTDNEP1 and NEP1-R1. These proteins have virtually identical expression profiles in human and mouse tissues, both are down-regulated in the fld mouse, and the knockdown of either results in similar changes in expression of the dystrophin and SERCA1 genes in muscle, strongly suggesting they work in unison. The importance of lipin-1 for normal muscle function is evident by the occurrence of severe myopathy in patients lacking lipin-1 function (55, 56). Because muscles have a high respiratory demand, it is tempting to speculate that lipin-1, activated by the CTDNEP1·NEP1-R1 complex, is involved in mitochondrial homeostasis; for example, phosphatidate is the precursor of cardiolipin, the major phospholipid in mitochondria (59–61). The DAG produced by lipin-1 may be used in mitochondrial phospholipid biosynthesis and provided through ER-mitochondrial junctions (62). The respiratory defect seen in deletions of SPO7 or PAH1 in yeast (46, 63) is consistent with a mitochondrial connection to these proteins. Whether the action of Pah1p on normal mitochondria is mediated at ER-mitochondrial junctions by direct binding of Pah1p to mitochondria or by a more indirect mechanism is unknown.
In a previous study, CTDNEP1 alone was able to dephosphorylate lipin-1b in BHK cells but unable in HEK293A or HeLa cells (32). Although the most likely explanation is that the incompetent cells lacked NEP1-R1, we find that mRNA levels for this protein are similar in all three lines (data not shown). At this time, we do not know if the corresponding NEP1-R1 protein levels are similar (we lack an antibody against the endogenous protein). Therefore, we do not know whether protein levels are very different in the lines, whether NEP1-R1 undergoes a critical modification essential for its function only in BHK cells, or whether an unknown factor in BHK is responsible for its phosphatase activity. We also do not know the physiological significance of the ability of overexpressed CTDNEP1·NEP1-R1 to dephosphorylate lipin-2. Although we have shown that this can happen, there is relatively little mRNA of either protein in liver cells, the main source of lipin-2 (Fig. 6). There may be related but yet unknown paralogs in this tissue that act upon lipin-2.
The accumulation of mouse lipin-1b to the nucleus in the presence of the phosphatase complex in HEK293 (Fig. 10) and HeLa cells (data not shown) is intriguing given recent work showing that mTORC1 can negatively control nuclear accumulation of lipin through phosphorylation and that mutation of 17 of the phosphorylation sites in lipin results in its constitutive accumulation in the nucleus (58). The likely possibility is that the phosphatase complex can positively regulate this process by directing lipin toward the nucleus. Further work with endogenous proteins will be necessary to determine the conditions under which the phosphatase complex is necessary to regulate lipin phosphorylation status and localization.
Although we report here that NEP1-R1 is an essential component of the CTDNEP1 complex, two other important questions of specificity remain. Although the complex can dephosphorylate more than one lipin, is this broad specificity physiological? Moreover, considering other signaling cascades, are lipins the only substrates for CTDNEP1·NEP1-R1? We shall attempt to probe for other substrates of this fascinating complex.
Supplementary Material
Acknowledgments
We thank the Goodman laboratory for useful discussions. We are very grateful to Barbara Barylko and Joe Albanesi for helping S. Han with experiments using mammalian cell cultures.
This work was supported, in whole or in part, by National Institutes of Health Grants GM094575, HL90553 (to K. R.), DK18849 and DK18024 (to J. E. D.), and GM084210 (to J. M. G.). This work was also supported by The Jan and Bob Bullock Endowment (to S. H.) and The Welch Foundation I-1506 (to N. G.),

This article contains supplemental Tables S1–S4.
- PA
- phosphatidic acid
- ER
- endoplasmic reticulum
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- CFP
- cyan fluorescent protein
- BHK
- baby hamster kidney.
REFERENCES
- 1. Chianale F., Rainero E., Cianflone C., Bettio V., Pighini A., Porporato P. E., Filigheddu N., Serini G., Sinigaglia F., Baldanzi G., Graziani A. (2010) Diacylglycerol kinase α mediates HGF-induced Rac activation and membrane ruffling by regulating atypical PKC and RhoGDI. Proc. Natl. Acad. Sci. U.S.A. 107, 4182–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knapek K., Frondorf K., Post J., Short S., Cox D., Gomez-Cambronero J. (2010) The molecular basis of phospholipase D2-induced chemotaxis. Elucidation of differential pathways in macrophages and fibroblasts. Mol. Cell. Biol. 30, 4492–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roth M. G. (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 4. Sun Y., Chen J. (2008) mTOR signaling. PLD takes center stage. Cell Cycle 7, 3118–3123 [DOI] [PubMed] [Google Scholar]
- 5. Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 6. Zhong X. P., Guo R., Zhou H., Liu C., Wan C. K. (2008) Diacylglycerol kinases in immune cell function and self-tolerance. Immunol. Rev. 224, 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goto K., Hozumi Y., Nakano T., Saino-Saito S., Martelli A. M. (2008) Lipid messenger, diacylglycerol, and its regulator, diacylglycerol kinase, in cells, organs, and animals. History and perspective. Tohoku J. Exp. Med. 214, 199–212 [DOI] [PubMed] [Google Scholar]
- 8. Topham M. K. (2006) Signaling roles of diacylglycerol kinases. J. Cell. Biochem. 97, 474–484 [DOI] [PubMed] [Google Scholar]
- 9. Carman G. M., Han G. S. (2009) Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50, S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coleman R. A., Lee D. P. (2004) Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176 [DOI] [PubMed] [Google Scholar]
- 11. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 12. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai J., Abramovici H., Gee S. H., Topham M. K. (2009) Diacylglycerol kinases as sources of phosphatidic acid. Biochim. Biophys. Acta 1791, 942–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Csaki L. S., Reue K. (2010) Lipins. Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 30, 257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 [DOI] [PubMed] [Google Scholar]
- 16. Han G. S., Wu W. I., Carman G. M. (2006) The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han G. S., Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reue K. (2009) The lipin family: mutations and metabolism. Curr. Opin. Lipidol. 20, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Péterfy M., Phan J., Xu P., Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 [DOI] [PubMed] [Google Scholar]
- 20. Reue K., Xu P., Wang X. P., Slavin B. G. (2000) Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41, 1067–1076 [PubMed] [Google Scholar]
- 21. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carman G. M. (1997) Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim. Biophys. Acta 1348, 45–55 [DOI] [PubMed] [Google Scholar]
- 23. O'Hara L., Han G. S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han G. S., Siniossoglou S., Carman G. M. (2007) The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 [DOI] [PubMed] [Google Scholar]
- 25. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286 [DOI] [PubMed] [Google Scholar]
- 26. Huffman T. A., Mothe-Satney I., Lawrence J. C., Jr. (2002) Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. U.S.A. 99, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi H. S., Su W. M., Morgan J. M., Han G. S., Xu Z., Karanasios E., Siniossoglou S., Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae. Identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., Yang H. (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Golden A., Liu J., Cohen-Fix O. (2009) Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122, 1970–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gorjánácz M., Mattaj I. W. (2009) Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J. Cell Sci. 122, 1963–1969 [DOI] [PubMed] [Google Scholar]
- 32. Kim Y., Gentry M. S., Harris T. E., Wiley S. E., Lawrence J. C., Jr., Dixon J. E. (2007) A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6596–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Satow R., Chan T. C., Asashima M. (2002) Molecular cloning and characterization of dullard. A novel gene required for neural development. Biochem. Biophys. Res. Commun. 295, 85–91 [DOI] [PubMed] [Google Scholar]
- 34. Wu R., Garland M., Dunaway-Mariano D., Allen K. N. (2011) Homo sapiens dullard protein phosphatase shows a preference for the insulin-dependent phosphorylation site of lipin1. Biochemistry 50, 3045–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 37. Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. (2006) An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., Dumont M. E., Phizicky E. M., Snyder M., Grayhack E. J. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiner M. P., Costa G. L. (1994) Rapid PCR site-directed mutagenesis. PCR Methods Appl. 4, S131–S136 [DOI] [PubMed] [Google Scholar]
- 40. Prinz W. A., Grzyb L., Veenhuis M., Kahana J. A., Silver P. A., Rapoport T. A. (2000) Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W. P., Agarwal A. K., Garg A., Anderson R. G., Goodman J. M. (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldstein A. L., McCusker J. H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 43. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 44. Kushnirov V. V. (2000) Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 45. Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Merz S., Westermann B. (2009) Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance, and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Sadreyev R. I., Grishin N. V. (2009) PROCAIN. Protein profile comparison with assisting information. Nucleic Acids Res. 37, 3522–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gocze P. M., Freeman D. A. (1994) Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry 17, 151–158 [DOI] [PubMed] [Google Scholar]
- 50. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skinner J. R., Shew T. M., Schwartz D. M., Tzekov A., Lepus C. M., Abumrad N. A., Wolins N. E. (2009) Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284, 30941–30948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Esposito M. S., Esposito R. E. (1969) The genetic control of sporulation in Saccharomyces. I. The isolation of temperature-sensitive sporulation-deficient mutants. Genetics 61, 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oegema K., Hyman A. A. (Jan. 19, 2005) WormBook, The C. elegans Research Community, doi/ 10.1895/wormbook.1.72.1, www.wormbook.org [DOI]
- 54. Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., Hewitson M., Holz C., Khan M., Lazik S., Martin C., Nitzsche B., Ruer M., Stamford J., Winzi M., Heinkel R., Röder M., Finell J., Häntsch H., Jones S. J., Jones M., Piano F., Gunsalus K. C., Oegema K., Gönczy P., Coulson A., Hyman A. A., Echeverri C. J. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 [DOI] [PubMed] [Google Scholar]
- 55. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., Vaz F. M., Pines O., Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Michot C., Hubert L., Brivet M., De Meirleir L., Valayannopoulos V., Müller-Felber W., Venkateswaran R., Ogier H., Desguerre I., Altuzarra C., Thompson E., Smitka M., Huebner A., Husson M., Horvath R., Chinnery P., Vaz F. M., Munnich A., Elpeleg O., Delahodde A., de Keyzer Y., de Lonlay P. (2010) LPIN1 gene mutations. A major cause of severe rhabdomyolysis in early childhood. Hum. Mutat. 31, E1564–E1573 [DOI] [PubMed] [Google Scholar]
- 57. Ferguson P. J., Chen S., Tayeh M. K., Ochoa L., Leal S. M., Pelet A., Munnich A., Lyonnet S., Majeed H. A., El-Shanti H. (2005) Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J. Med. Genet. 42, 551–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., Sabatini D. M. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schlame M., Brody S., Hostetler K. Y. (1993) Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur. J. Biochem. 212, 727–735 [DOI] [PubMed] [Google Scholar]
- 61. Schlame M., Haldar D. (1993) Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 268, 74–79 [PubMed] [Google Scholar]
- 62. Kornmann B., Walter P. (2010) ERMES-mediated ER-mitochondria contacts. Molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 123, 1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Irie K., Takase M., Araki H., Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.