Abstract
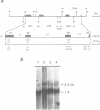
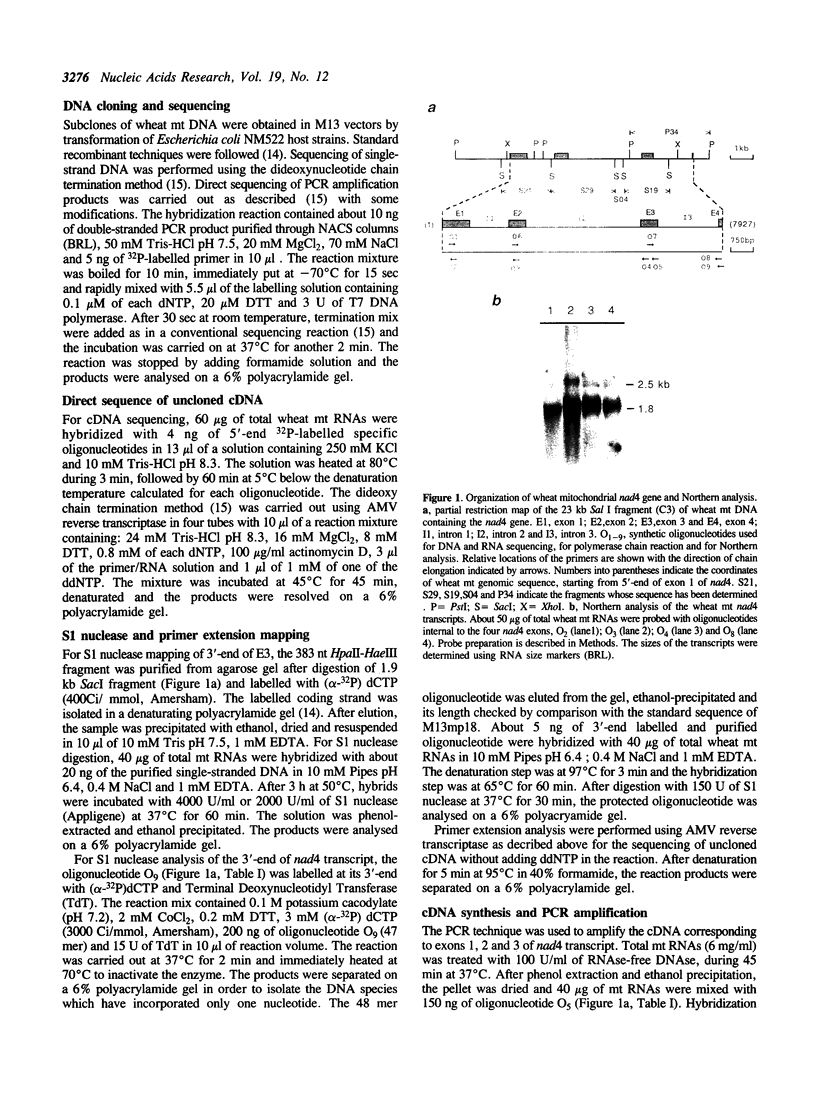
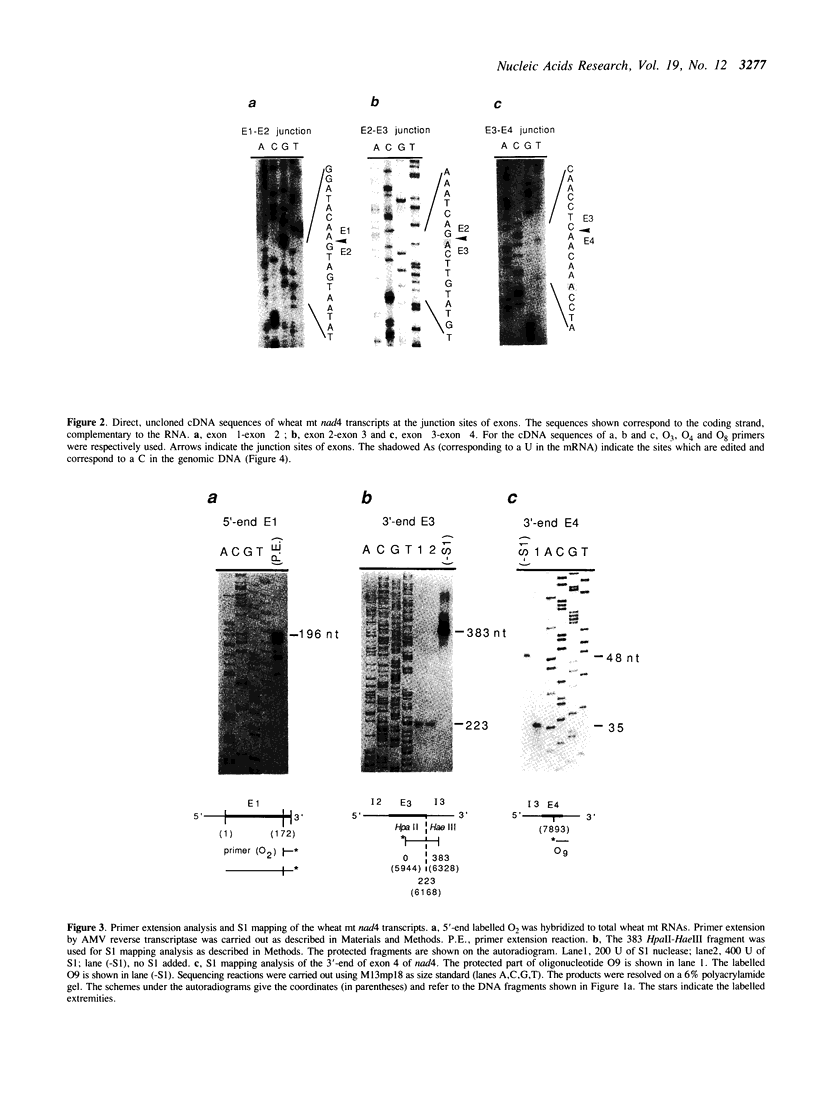
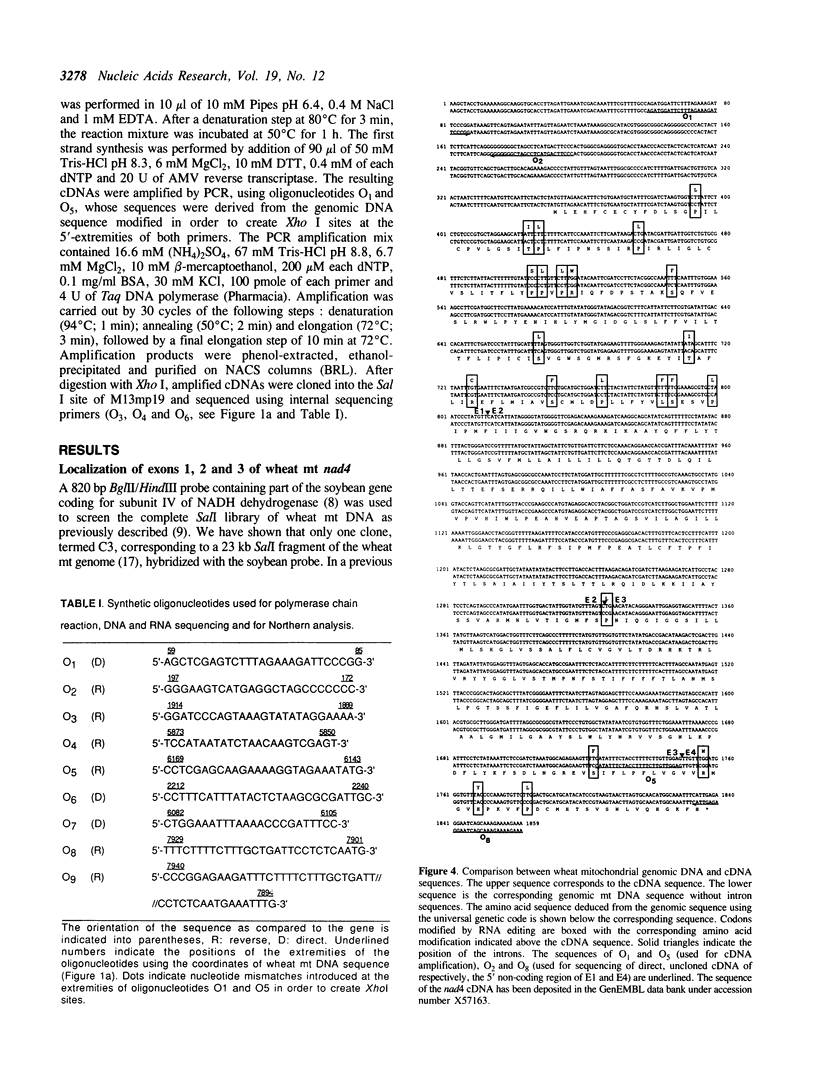
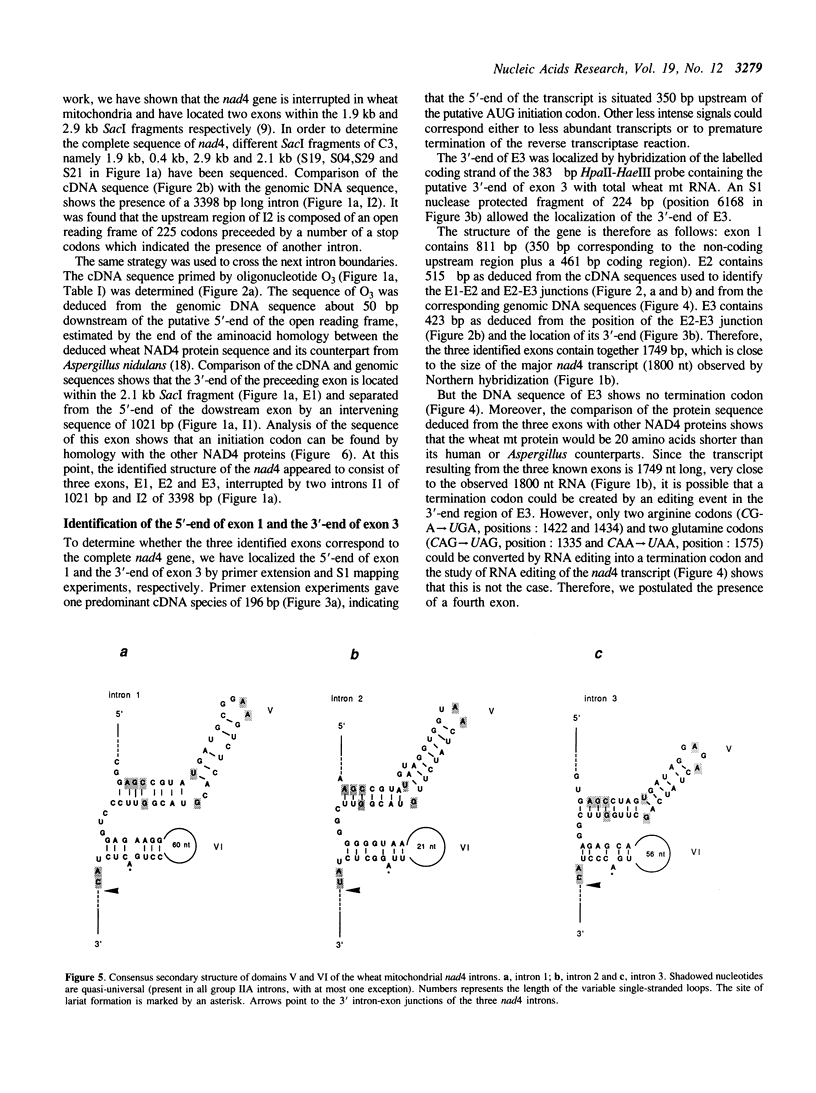
The wheat mitochondrial (mt) NADH dehydrogenase subunit 4 gene (nad4) has been localized and sequenced. This gene, about 8 kb long, is composed of four exons separated by three class II introns. The nad4 gene exists as a single copy in the wheat mitochondrial genome and it is transcribed into one abundant mRNA of 1.8 kb, whose extremities have been mapped. The complete cDNA sequence corresponding to the nad4 transcript has been determined by combining the direct sequencing of uncloned cDNA and a method involving cDNA synthesis and PCR amplification using specific oligonucleotides as primers, followed by cloning and sequencing of the amplification product. Comparison of the genomic sequence with that of the cDNA shows that all nad4 transcripts are fully edited at 23 positions, with an uneven distribution of the editing sites between the different exons: While exon 1 and exon 4 are extensively edited (with a change of 11% of the amino acid sequence), exon 2 is not edited at all and exon 3 is 0.5% edited. This uneven distribution is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Benne R. RNA editing in trypanosomes: is there a message? Trends Genet. 1990 Jun;6(6):177–181. doi: 10.1016/0168-9525(90)90173-4. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Blake C. Exons--present from the beginning? Nature. 1983 Dec 8;306(5943):535–537. doi: 10.1038/306535a0. [DOI] [PubMed] [Google Scholar]
- Brown T. A., Davies R. W., Ray J. A., Waring R. B., Scazzocchio C. The mitochondnal genome of Aspergillus nidulans contains reading frames homologous to the human URFs 1 and 4. EMBO J. 1983;2(3):427–435. doi: 10.1002/j.1460-2075.1983.tb01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. L., Pannetier C., Jaulin C., Kourilsky P. Optimal conditions for directly sequencing double-stranded PCR products with sequenase. Nucleic Acids Res. 1990 Jul 11;18(13):4028–4028. doi: 10.1093/nar/18.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A., Mariottini P., Cleeter M. W., Ragan C. I., Matsuno-Yagi A., Hatefi Y., Doolittle R. F., Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature. 1985 Apr 18;314(6012):592–597. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Dorit R. L., Schoenbach L., Gilbert W. How big is the universe of exons? Science. 1990 Dec 7;250(4986):1377–1382. doi: 10.1126/science.2255907. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Wintz H., Weil J. H., Grienenberger J. M. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988 Dec;215(1):118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Lamattina L., Weil J. H., Grienenberger J. M. RNA editing at a splicing site of NADH dehydrogenase subunit IV gene transcript in wheat mitochondria. FEBS Lett. 1989 Nov 20;258(1):79–83. doi: 10.1016/0014-5793(89)81620-5. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989 Oct 15;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Müller M. W. Self-splicing of group II introns in vitro: lariat formation and 3' splice site selection in mutant RNAs. Cell. 1987 Dec 4;51(5):753–762. doi: 10.1016/0092-8674(87)90098-5. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Bang A. G., Thompson W. F. The watermelon mitochondrial URF-1 gene: evidence for a complex structure. Curr Genet. 1986;10(11):857–869. doi: 10.1007/BF00418532. [DOI] [PubMed] [Google Scholar]
- Wahleithner J. A., MacFarlane J. L., Wolstenholme D. R. A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci U S A. 1990 Jan;87(2):548–552. doi: 10.1073/pnas.87.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H., Chen H. C., Pillay D. T. Partial characterization of the gene coding for subunit IV of soybean mitochondrial NADH dehydrogenase. Curr Genet. 1989 Feb;15(2):155–160. doi: 10.1007/BF00435463. [DOI] [PubMed] [Google Scholar]
- Wissinger B., Hiesel R., Schuster W., Brennicke A. The NADH-dehydrogenase subunit 5 gene in Oenothera mitochondria contains two introns and is co-transcribed with the 5 S rRNA gene. Mol Gen Genet. 1988 Apr;212(1):56–65. doi: 10.1007/BF00322444. [DOI] [PubMed] [Google Scholar]
- Xue Y. B., Davies D. R., Thomas C. M. Sugarbeet mitochondria contain an open reading frame showing extensive sequence homology to the subunit 2 gene of the NADH: ubiquinone reductase complex. Mol Gen Genet. 1990 Apr;221(2):195–198. doi: 10.1007/BF00261720. [DOI] [PubMed] [Google Scholar]