Abstract
Neuritogenesis, or neurite outgrowth, is a critical process for neuronal differentiation and maturation in which growth cones are formed from highly dynamic actin structures. Gas7 (growth arrest-specific gene 7), a new member of the PCH (Pombe Cdc15 homology) protein family, is predominantly expressed in neurons and is required for the maturation of primary cultured Purkinje neurons as well as the neuron-like differentiation of PC12 cells upon nerve growth factor stimulation. We report that Gas7 co-localizes and physically interacts with N-WASP, a key regulator of Arp2/3 complex-mediated actin polymerization, in the cortical region of Gas7-transfected Neuro-2a cells and growth cones of hippocampal neurons. The interaction between Gas7 and N-WASP is mediated by WW-Pro domains, which is unique in the PCH protein family, where most interactions are of the SH3-Pro kind. The interaction contributes to the formation of membrane protrusions and processes by recruiting the Arp2/3 complex in a Cdc42-independent manner. Importantly, specific interaction between Gas7 and N-WASP is required for regular neurite outgrowth of hippocampal neurons. The data demonstrate an essential role of Gas7 through its interaction with N-WASP during neuronal maturation/differentiation.
Keywords: Cell/Neuron, Cytoskeleton/Actin, Membrane, Neuroblastoma, Neurodifferentiation, Gas7, N-WASP, PCH (Pombe Cdc15 Homology), Neurite Outgrowth
Introduction
Neurite outgrowth, which begins with the extension of membrane protrusions of neurons, is an important characteristic of neuronal differentiation that largely depends on actin dynamics and/or cortical cytoskeleton rearrangement. The significance of actin polymerization and dynamics during neuronal development is well known, as neurite formation and growth cone motility are closely associated with actin reorganization (1).
Growth arrest-specific (gas) genes were primarily identified from cultured cells that had entered a quiescent state induced by serum starvation or confluence (2–4). Apart from their expression in response to growth arrest, these gas genes show no sequence homology. However, gas genes have been implicated in the regulation of a variety of biological processes, including microfilament organization (5), neuronal differentiation (6), and apoptosis (7, 8). Gas7, isolated from growth-arrested NIH3T3 cells, is abundantly expressed in early embryonic cells, testis, and neurons of mouse brain, including cerebral cortex, hippocampus, and cerebellum (4, 9–12). Its overexpression can induce formation of membrane protrusions on the surface of Neuro-2a cells and potentiate nerve growth factor-stimulated differentiation of PC12 cells (9, 13, 14). A requirement for Gas7 during neuronal differentiation has also been demonstrated in cultured Purkinje neurons (9). In addition, Gas7 directly interacts with actin, promoting its assembly, cross-linking F-actin into bundles, and forming oligomers via its C-terminal (160–421 residue) region (15, 16).
Recently, Gas7 was categorized as a member of the PCH (Pombe Cdc15 homology) family and belongs to the PSTPIP subfamily (12, 17, 18). PCH proteins are a group of adaptor proteins that coordinate and regulate cytokinesis, the actin cytoskeleton, and membrane dynamics (19). Most PCH proteins share a similar domain architecture; that is, an N-terminal Fes/CIP4 homology (FCH)2 domain, a coiled-coil (CC) region, and one or two C-terminal Src homology domains (17). Both the mouse and human Gas7 isoform c (gi:41406079) possess a similar domain structure, except that the SH3 domain is absent in mouse Gas7. FCH-containing proteins have been implicated in the regulation of cytoskeletal rearrangement, vesicular transport, and endocytosis (20). Furthermore, the FCH domain and the CC region have also been represented as a single extended domain, the F-BAR domain, which mediates oligomer formation and interactions with the submembrane region (18, 21–24).
Neural-Wiskott Aldrich syndrome protein (N-WASP), a critical regulator of actin dynamics, was originally isolated as an Ash/Grb2-interacting protein that regulates the actin-related protein 2 and 3 (Arp2/3) complex (25–28). It consists of a WH1 domain at the N terminus followed by the basic region (BR), a GTPase-binding domain/Cdc42-Rac interactive binding domain (GBD/CRIB), a proline-rich domain, and a VCA domain at the C terminus (26, 29, 30). Expression of N-WASP is ubiquitous; however, it is more abundant in the brain (26, 29), where it is essential for the differentiation of hippocampal neurons (31). Previous studies have shown that, when inactive N-WASP becomes activated after an appropriate stimulus, it is unwound by active Cdc42 and phosphoinositol 4,5-bisphosphate into its active form, exposing the VCA domain to recruit the Arp2/3 complex and initiate actin polymerization. The formation of actin filaments in the submembrane region is followed by formation of spike-like cell protrusions called filopodia (30, 32–34).
Filopodia are dynamic actin-based structures present in the growth cones of neurons that drive neurite outgrowth in the process of neuronal differentiation (1). There is considerable evidence that several SH3-containing proteins, such as WISH, Abil, Nck, Grb2, and Toca-1, activate quiescent N-WASP by binding to its Pro region (35–39). These N-WASP-associated proteins participate in various cellular processes that require N-WASP-Arp2/3-mediated cytoskeleton reorganization. Some of them cooperate with GTP-Cdc42/phosphoinositol 4,5-bisphosphate to fully activate N-WASP, whereas others work in a Cdc42/phosphoinositol 4,5-bisphosphate-independent manner.
The importance of Gas7 during neuronal differentiation has recently emerged; however, it remains unclear how Gas7 functions in neurons. A Gas7-interacting protein, WISH (WASP interacting SH3 protein), has been isolated in our laboratory by yeast two-hybrid screens from a cDNA library of mouse brain.3 Previous work elsewhere showed that WISH is an N-WASP-interacting protein that binds and activates N-WASP to recruit the Arp2/3 complex in a Cdc42-independent manner (36). Our preliminary studies on the relationship between Gas7, WISH, and N-WASP found that Gas7 directly interacts with N-WASP and that they are co-localized to the membrane protrusions induced by Gas7 overexpression. This prompted us to look further into the interaction between Gas7 and N-WASP and the mechanism of their effects on membrane deformation or cytoskeleton rearrangement. Neuro-2a cells and hippocampal primary cultured neurons, as in vitro and semi-in vivo models, were used to investigate the role of Gas7-N-WASP in Arp2/3-mediated cytoskeleton rearrangement and to address the physiological significance of Gas7-N-WASP interaction in neuronal differentiation.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Recombinant Proteins
Gas7 cDNA was isolated as previously described (9). The various glutathione S-transferase (GST)-tagged Gas7 constructs, GST/Cdc42 (gi:20379097) and HA/Cdc42N17, were generated by a GST gene fusion system in the pGEX-4T vector (Amersham Biosciences) and the pcDNA3 (Clontech) vector by subcloning or by PCR cloning. All GST fusion proteins were produced in Escherichia coli BL21 and affinity-purified on glutathione-agarose beads. Gas7 internal deletion mutants (Gas7ΔWW, Gas7ΔFCH, and Gas7Δcoiled) were amplified by PCR with specific primers. The recombinant Gas7 genes, Gas7WW2 (residues 1–151) or Pro domain of N-WASP (residues 234–401), were subcloned into pcDNA3.1/Myc-His C (Invitrogen) and pEGFP systems (Clontech), respectively. Rat N-WASP in pcDL-SRα was a gift from Dr. Takenawa. Recombinant constructs for wild-type HA/N-WASP, the HA/N-WASPΔcof mutant, and the HA/N-WASP H208D mutant were prepared according to a published protocol with modifications (40). ECFP-CA was cloned as described by Rohatgi et al. (27) with minor modifications. The CA region of N-WASP was amplified with specific primers and subcloned into pECFP-C1 (Clontech). His-tagged N-WASP was cloned in pFastBac-HTa and produced by the Bac-to-Bac baculovirus expression system (Invitrogen) in Sf21 cells. His-tagged WH1, GBD, Pro, and VCA of N-WASP were subcloned by PCR with specific primers into His/pET32c vector (Novagen), and proteins were produced in E. coli BL21. All His-tagged proteins were purified on His*bind Fractogel resin, as described by the manufacturer (Novagen).
For the RNAi assay, GFP, shGas7/GFP, and shN-WASP/GFP were gifts from the National RNAi Core Facility of Academia Sinica. shGas7/GFP and shN-WASP/GFP specifically targets the sequence 5′-GTGGAAATGATCCGACAACAT-3′ and 5′-TCAGGAAACAAAGCAGCTCTT-3′ located on the open reading frame of gas7 and N-WASP, respectively, to knock down Gas7 and N-WASP expression.
Antibodies
Polyclonal antibodies against Gas7 were generated as previously described (9). Antibodies against rat N-WASP were produced as follows. Partial cDNA fragments encoding amino acids 392–500 (VCA) were amplified with the primer set 5′-ATG AAT TCT GAC CAT CAA GTT CCA GCT-3′ and 5′-ATG CGT CGA CGT CTT CCC ACT CAT CCT G-3′, which were ligated into the EcoRI and SalI sites (shown in bold) of the His/pET32c expression vector (Novagen). His-tagged VCA recombinant protein was expressed in E. coli BL21 (Novagen) and purified on His*bind Fractogel resin. The eluted recombinant proteins were injected into rabbits to produce a polyclonal antiserum. Other specific antibodies were purchased from standard suppliers: MAPII and HA monoclonal from Upstate, Myc monoclonal from Santa Cruz Biotechnology, α-tubulin monoclonal from Chemicon, and His polyclonal from Bioman. Hoechst 33258, secondary antibodies conjugated with Alexa Fluor (488, 555, or 633), or Alexa Fluor 633-conjugated phalloidin for immunofluorescence and secondary antibodies conjugated with horseradish peroxidase for Western blots were purchased from Molecular Probes and Amersham Biosciences.
Cell Culture, Primary Culture, and Transfection
Neuro-2a, COS-7, and NIH3T3 cells from ATCC were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 ng/ml streptomycin, 100 units/ml penicillin, and 1 mm sodium pyruvate under humid air with 5% CO2 in air at 37 °C. Neuro-2a or arrest-induced NIH3T3 cells were seeded at about 3 × 105 on poly-l-lysine-coated glass coverslips (22 × 22 mm) for transfection, immunofluorescence, and/or knockdown assay. COS-7 cells were seeded on 10-cm round dishes at 80–90% confluence for transfection and co-immunoprecipitation.
Primary dissected hippocampal cells from C57BL/6 mice at embryonic day 16.5 were kept in Hanks' balanced salt buffer with 1% glucose. The tissues were digested in 0.05% trypsin/EDTA for 3 min and triturated through a flame-polished Pasteur pipette to disrupt cell-cell connections. Dispersed cells were centrifuged and cultured in Neural Basal medium (Invitrogen) with B27 complex and 1 mm l-glutamine. The primary neurons were seeded on 4-well (1 × 105 cells per well) chamber slides or 6-well plates (3 × 105 cells/well) coated with poly-l-lysine for immunostaining and Western blots, respectively. The cells were cultured in vitro and collected for immunostaining or Western blots as indicated. Transient expression was brought about by transfection with Lipofectamine 2000 for cell lines and Lipofectamine LTX (Invitrogen) for cultured hippocampal neurons, according to the manufacturer's instructions with minor modifications.
Immunohistochemistry, Immunofluorescence Staining, and Morphological Characterization
Adult C57BL/6 mice were perfused with PBS and fixed with 4% paraformaldehyde. Whole brains were dissected out and kept in the fixative overnight before 50-μm sections were taken with a Vibratome. The slices were washed in PBS 3 times and blocked with blocking buffer (5% bovine serum albumin, 3% normal goat serum, 0.2% Triton X-100 in PBS) overnight. After 3 washes in PBS, the slices were incubated with the first primary antibody, anti-MAPII monoclonal, for 1 h at room temperature and washed before incubation with another primary antibody, anti-N-WASP polyclonal antibody, or antigen-preabsorbed anti-N-WASP polyclonal antibody (negative control) overnight at 4 °C. After washing, the slices were incubated with Hoechst 33258 and secondary fluorescent-linked antibodies for 1 h. Finally, the slices were incubated with anti-Gas7 polyclonal antibody (previously linked to Alexa Fluor 488 with a Zenon Alexa Fluor 488 Rabbit IgG labeling kit, Z25302; Invitrogen) or antigen preabsorbed anti-Gas7 polyclonal antibody (negative control) for 1 h at room temperature and mounted in 30% glycerol for observation.
Transfected Neuro-2a cells were washed 3 times with PBS and fixed with 4% paraformaldehyde in PBS for 10 min. After fixation, the cells were washed 3 times with PBS and permeabilized with 0.2% Triton X-100 in PBS for 10 min. Cells were blocked with 1% bovine serum albumin, 1% normal goat serum in PBS for 1 h and washed 3 times with PBS. Specimens were incubated with primary antibodies as indicated in PBS buffer for 1 h at room temperature. After washing, they were incubated with secondary antibodies linked to a fluorescent agent for 1 h, and nuclei were then stained with Hoechst 33258 for 20 min. For primary neurons, cells were fixed with 4% paraformaldehyde followed by methanol. Specimens were stained as described above (except for blocking with Image Signal Enhancer (Invitrogen) for 30 min) and incubated overnight at 4 °C with anti-Gas7 polyclonal antibody linked to Alexa Fluor 488. Finally, slides were mounted in 30% glycerol and imaged by confocal microscopy (LSM 510 META; Zeiss).
For morphological characterization of Neuro-2a cells, confocal images were used to count the number of cells with membrane protrusions in 3 independent determinations, using 50–150 cells each time. For primary neurons, the cells were collected from 2 or 3 independent transfections, and images were captured with a CoolPix digital camera (AxioImager-Z1; Zeiss) and analyzed with Neuro J (a plugin for ImageJ freeware, rsbweb.nih.gov) to determine neurite length and cell body diameter. All data sets were compared by paired or unpaired (2-sample) Student's t tests using GraphPad Prism 5.
In Vitro Binding Assay
Several GST fusion proteins were immobilized on glutathione-agarose beads and mixed with purified His-tagged proteins in NETN buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm EDTA, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride) at 4 °C for 1 h. After 3 washes in NETN buffer, the beads were suspended in Laemmli sample buffer, separated by SDS-PAGE, and analyzed by Western blot.
Immunoprecipitation
Mouse whole brain lysates were homogenized with a Dounce homogenizer and lysed with ice-cold radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol) containing a protease inhibitor mixture (1.04 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.8 μm aprotinin, 20 μm leupeptin, 40 μm bestatin, 15 μm pepstatin A, 14 μm E-64; Sigma) on ice for 20 min. For transfected COS-7 cells, cells were also lysed with ice-cold radioimmune precipitation assay buffer on ice for 20 min after 24 h of transfection.
Brain or cell lysates were centrifuged at 12,000 × g at 4 °C for 15 min. Supernatants containing about 1000 μg of protein were incubated with rabbit anti-Gas7 polyclonal antibody and/or preimmune serum (as negative control) for 1 h at 4 °C and subsequently incubated with 45 μl of protein A-Sepharose (Amersham Biosciences) slurry for another 1 h. The co-immunoprecipitated complexes were washed with ice-cold radioimmune precipitation assay buffer three times and released in Laemmli sample buffer by heating for 5 min at 95–100 °C. For co-immunoprecipitation of HA/N-WASP or derivatives, anti-HA monoclonal antibody was cross-linked, and the complex released as previously described (41). Finally, the samples were separated by SDS-PAGE and immunoblotted with specific antibodies as indicated.
In Vitro Actin Polymerization Assay
Purified pyrene actin and Arp2/3 complex from Cytoskeleton Co. were resuspended as per the manufacturer instructions. Pyrene actin was depolymerized with G-buffer (5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2, 0.2 mm ATP, 0.2 mm dithiothreitol) on ice for 1 h and centrifuged at 14,000 × g for 30 min to remove residual F-actin. Other purified proteins (GST-Cdc42, His-N-WASP, GST, and GST-Gas7) were concentrated and dialyzed in preservation buffer (20 mm Tris, pH 7.5, 25 mm KCl, 1 mm MgCl2, 1 mm dithiothreitol). Activated GST-Cdc42 was charged with GTPγS as described previously (42). These proteins (Fig. 6D) were premixed in the preservation buffer and incubated at room temperature for 10 min. Finally, they were added to the actin polymerization buffer (10 mm Tris-HCl, pH 7.5, 50 mm KCl, 2 mm MgCl2, and 1 mm ATP), and polymerization was started by pyrene G-actin addition. The process was monitored spectroscopically using a SpectraMax M5 (Molecular Devices).
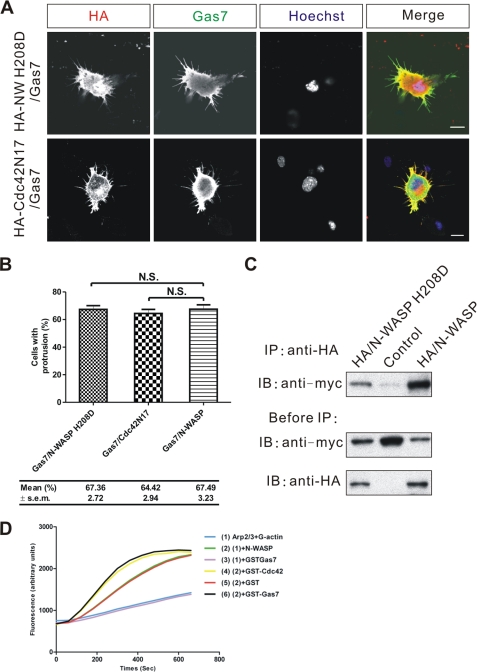
FIGURE 6.
Functional interaction between Gas7 and N-WASP is Cdc42-independent. A, shown is immunostaining of Neuro-2a cells co-transfected with Gas7/Myc and either loss-of-function N-WASP (HA/N-WASP H208D) or Cdc42 dominant negative (HA/Cdc42N17) mutants to examine the role of Cdc42 in Gas7-induced membrane protrusion. Scale bar, 10 μm. B, shown are the fractions of cells in A extending membrane protrusions. Data are presented as the mean ± S.E. of three independent determinations, each counting at least 100 cells. Data sets were statistically compared with the control group (Gas7 only) by paired Student's t test; N.S., no significant differences were seen. C, extracts from COS-7 cells transfected with Gas7/Myc alone (negative control) or in combination with either wild-type HA/N-WASP (positive control) or HA/N-WASP H208D were analyzed by co-IP to assay the interaction of HA-N-WASP H208D with Gas7. Gas7 was detected in complex with both N-WASP H208D and N-WASP. D, the effect of Gas7 in the N-WASP/Arp2/3-mediated actin polymerization process was shown. The actin polymerization process was modeled in vitro with pyrene actin (2.6 μm), Arp2/3 complex (60 nm), and His-NWASP (200 nm) in actin polymerization buffer. GST-Cdc42 (500 nm) charged with GTPγS was a positive control. Experiments were repeated three times, and each independent trial was done in duplicate. Gas7 (200 nm) activated N-WASP directly to induce actin polymerization without the Cdc42 molecule, which was not seen in control experiment (GST, 200 nm) or without N-WASP.
RESULTS
Gas7 Induces Actin Reorganization, Is Predominantly Localized in the Submembrane Region, and Directly Interacts with N-WASP
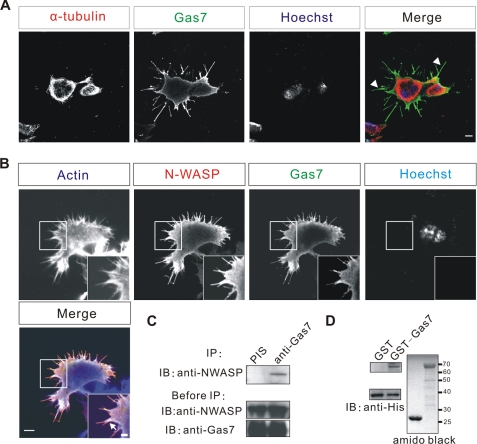
Ju et al. (9) reported that overexpression of Gas7 in neuroblastoma cells induced membrane protrusions. To investigate the cytoskeleton reorganization underlying these protrusions, we overexpressed Gas7 in Neuro-2a cells (which do not normally express Gas7). Immunostaining analysis (Fig. 1B) showed that the membrane protrusions induced by Gas7 contain actin, where both Gas7 and actin are co-localized (arrow), but not microtubules (arrowhead, Fig. 1A). Thus, the membrane protrusions result from actin reorganization. Because N-WASP regulates actin dynamics, we looked for a physiological correlation between Gas7 and N-WASP, initially examining the spatial relationship of the two proteins in Neuro-2a cells. Neuro-2a cells were cotransfected with Gas7 and N-WASP tagged with HA (HA-NWASP) peptide, and the subsequent distributions of Gas7, N-WASP, and F-actin were analyzed. N-WASP overexpression alone did not affect or induce any membrane protrusions without an activator (supplemental Fig. S1); in contrast, Gas7-transfected cells had membrane protrusions, as previously reported (9). As well as actin, Gas7 was also co-localized with N-WASP in the membrane protrusions and the submembrane region. To test whether Gas7 and N-WASP form an intracellular complex, we co-expressed these two proteins in COS-7 cells for a co-immunoprecipitation assay with rabbit polyclonal anti-Gas7 antibodies (Fig. 1C). N-WASP was only detected in the Gas7-associated complex and not in the preimmune serum sample, suggesting that Gas7 and N-WASP form a complex in cells. In an in vitro binding assay, recombinant GST-Gas7, but not the control GST protein, directly pulled down purified N-WASP (Fig. 1D), thereby confirming that Gas7 and N-WASP physically interact with each other.
FIGURE 1.
Co-localization and direct interaction between Gas7 and N-WASP in vitro. The microtubule organization (A), actin reorganization, and co-localization of Gas7 and N-WASP (B) in transfected neuroblastoma cells is shown. Neuro-2a cells were transfected only with Gas7/Myc or co-transfected with HA/N-WASP to examine the distribution of microtubule, actin, Gas7, and N-WASP. Gas7, N-WASP, and actin filaments are co-localized at the submembrane region as well as in protrusions (arrows). Scale bar, 5 μm (2-μm inset). C, interaction of Gas7 and N-WASP in cells is shown. Extracts from COS-7 cells co-transfected with Gas7/Myc and HA/N-WASP were analyzed by co-immunoprecipitation (IP). N-WASP was only detected in association with Gas7. Antibodies against Gas7 were raised in rabbits, and preimmune serum (PIS) was used as a negative control. D, direct interaction between Gas7 and N-WASP in vitro is shown. Recombinant GST-Gas7 and His-NWASP were used in a GST pulldown assay to examine the interaction between Gas7 and N-WASP. His-N-WASP was directly pulled down by immobilized Gas7 but not by GST alone (negative control).
Co-localization and Interaction of Gas7 and N-WASP in Hippocampal Neurons
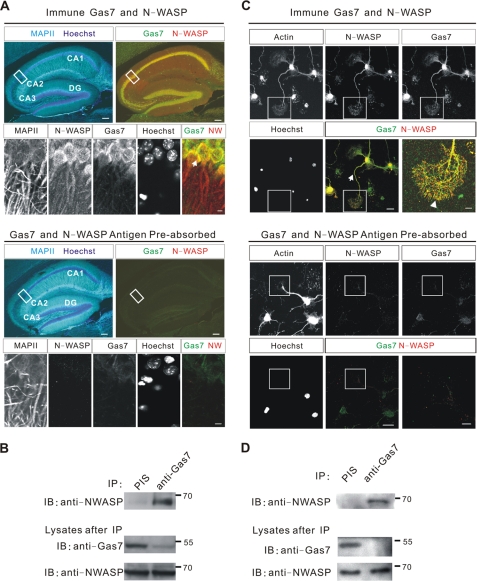
Gas7 and N-WASP are strongly expressed in the hippocampus (9, 10, 26, 29). To confirm the correlation between Gas7 and N-WASP in vivo, the localization of Gas7 and N-WASP in the mouse hippocampus was examined. Adult mouse brains were sliced at 50 μm and stained with anti-Gas7/anti-N-WASP or antigen preabsorbed antibodies (negative control). Anti-MAPII was used to indicate the position of neurons. Both Gas7 and N-WASP were distributed in a neuron-specific pattern (Fig. 2A) throughout the hippocampus from CA1 to CA3 and the dentate gyrus. N-WASP occurred in the cytoplasm and neurites and in a punctate manner in the nucleus. Gas7 was localized throughout the cytoplasm with small amounts seen in neurites and the nucleus. This co-localization was confirmed by co-immunoprecipitation assay of brain lysates (Fig. 2B), which showed clearly that Gas7 and N-WASP form a complex in neurons of the hippocampus.
FIGURE 2.
Co-localization and interaction between Gas7 and N-WASP in vivo. A, distribution of Gas7 and N-WASP in brain of 3-month mice is shown. Brain tissue sections were stained with various antibodies to visualize Gas7 and N-WASP; MAPII staining represented the neurons. Gas7 and N-WASP expression was seen in hippocampal pyramidal neurons, from CA1 to CA3, and in the dentate gyrus (DG) but not with antigen preabsorbed antibodies (negative control, lower panels). In superimposed images, areas with co-localized Gas7 (green) and N-WASP (red) are yellow. Scale bar, 100 μm (5 μm in magnified area). B, interaction of Gas7 and N-WASP in adult mice brain is shown. Adult whole brain lysates were analyzed by co-IP to examine the in vivo interaction of Gas7 and N-WASP. N-WASP was detected in the anti-Gas7 polyclonal antibody sample. PIS, preimmune serum. C, the co-localization of Gas7 and N-WASP in primary culture of embryonic E16.5 mouse hippocampal neurons at 5 DIV. Gas7 (green) and N-WASP (red) co-localized in the neurites (arrows) and in the tip of growth cone filopodia (arrowhead, inset). Antigen preabsorbed antibodies (lower panels) demonstrate the specificity of the interaction, with only very faint staining for Gas7 was still evident in nuclei. Hoechst dye: nuclei. Scale bar, 20 μm (5 μm in magnified area, seen enlarged in the final panel). D, interaction of Gas7 and N-WASP in cultured hippocampal neurons is shown. Lysates from 2 × 106 cultured hippocampal neurons at DIV6 were used in co-IP to determine the interaction between Gas7 and N-WASP. Significant amounts of N-WASP were detected in the anti-Gas7 polyclonal antibody sample but not in control.
In the rest of the cortex, Gas7 was located predominantly in the cytoplasm of cortical neurons; N-WASP was present in a punctate distribution in both the cytoplasm and the nucleus of cortical neurons (supplemental Fig. S2A). However, in cerebellar neurons, Gas7 was present in large amounts in the cytoplasm, whereas N-WASP was present in only small amounts in the Purkinje neurons (supplemental Fig. S2B), with no obvious co-localization of Gas7 and N-WASP in Purkinje neurons.
We also examined the correlation of Gas7 and N-WASP in primary cultures of embryonic hippocampal neurons by staining dissociated neurons at 5 days in vitro (DIV) with specific antibodies. Gas7 was present throughout the neurons, including the cytoplasm, nucleus, and neurites (Fig. 2C, upper panel); however, only minimal nuclear signal remained after an antigen competition assay (Fig. 2C, bottom panel). Gas7 was predominantly localized in the cytoplasm and neurites of hippocampal neurons, with a small amount in the nucleus. N-WASP signal was present throughout the hippocampal neurons but was barely detectable after an antigen competition assay (Fig. 2C, bottom panel), from which it can be concluded that N-WASP was present in the nucleus, cytoplasm, and neurites. Both Gas7 and N-WASP were also co-localized in the filopodia of growth cones of cultured hippocampal neurons (arrowhead, Fig. 2C). In conclusion, Gas7 and N-WASP were mainly co-localized in the cytoplasm, neurites, and growth cones of hippocampal neurons. This association was confirmed by co-immunoprecipitation from lysates of cultured hippocampal neurons (Fig. 2D) in which N-WASP was only present in the Gas7-associated complex and not in the controls.
Gas7 Interacts with N-WASP Directly via the WW-Pro Domain
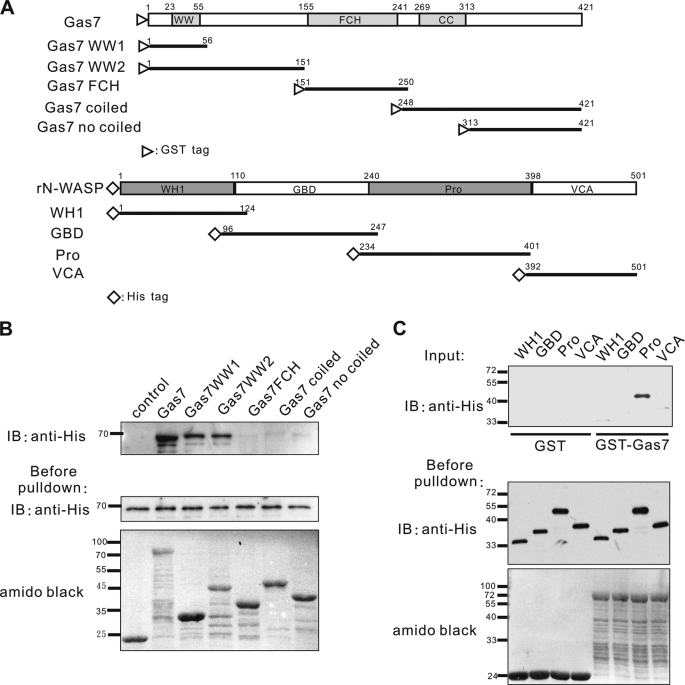
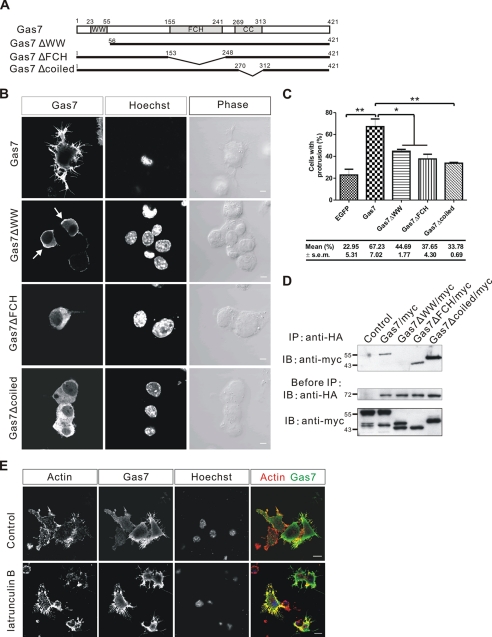
A series of functional domain truncates of Gas7 with a GST tag on the N terminus was generated (Fig. 3A, upper panel). These were immobilized on glutathione-agarose beads and mixed with full-length His-tagged N-WASP protein for an in vitro binding assay followed by Western blot analysis. N-WASP was pulled down by intact Gas7 and by WW-containing Gas7 truncates (GST-Gas7WW1 and GST-Gas7WW2)but not by the N-truncated Gas7 derivatives (GST-Gas7FCH, GST-Gas7 coiled, and GST-Gas7 no coiled, Fig. 3B).
FIGURE 3.
Mapping of interacting domains between Gas7 and N-WASP. A, schematic representation of various Gas7 (upper panel) and N-WASP truncates (lower panel) according to functional domains. Open triangles and diamonds indicate GST and His tags, respectively. B, Gas7 derivatives (A, upper panel) were produced in vitro, purified, and used in a pulldown assay with purified His-tagged N-WASP. His-N-WASP was pulled down by WW domain-containing proteins (GST-Gas7, GST-Gas7WW1, and GST-Gas7WW2). C, His-N-WASP derivatives (A, lower panel) were generated in vitro, purified, and used in a pulldown assay with immobilized Gas7. The Pro region of N-WASP was pulled down by GST-Gas7 but not by GST (negative control). WW, WW domain; WH1, WASP homology domain 1; Pro, proline-rich region.
The region responsible for N-WASP interaction with Gas7 was also determined. In a similar fashion, a series of His N-terminal tagged N-WASP truncates (Fig. 3A, lower panel) was assayed against immobilized GST or full-length GST-Gas7 (Fig. 3C). The Pro region of N-WASP was required for interaction with Gas7. Thus, the interaction between Gas7 and N-WASP requires both the WW domain and the Pro region.
Intact Gas7 Is Required for Membrane Process Formation in Neuro-2a Cells
Previous studies have shown that overexpression of Gas7 in neuroblastoma cells results in the formation of membrane protrusions (9). To determine whether the interaction between Gas7 and N-WASP is involved in the formation of these structures, Gas7 truncates lacking different functional domains (Fig. 4A) were transiently expressed in Neuro-2a cells. Formation of membrane protrusions (Fig. 4B) was reduced in Gas7ΔWW-transfected Neuro-2a cells (44.7% had processes) after transfection for 12–16 h, compared with the intact Gas7-transfected cells (67.2%, p < 0.05). Unexpectedly, formation of membrane protrusions was also significantly inhibited in both Gas7ΔFCH- and Gas7Δcoiled-transfected Neuro-2a cells, where the number of cells with processes fell to 37.6% (p < 0.05) and 33.8% (p < 0.01), respectively. Actin and microtubule distributions were also examined in the transfected cells (supplemental Figs. S3 and S4). Furthermore, the localization of Gas7 truncates also changed. Gas7ΔWW was localized predominantly in the submembrane region (Fig. 4B, arrow), like the wild-type Gas7 protein. However, Gas7ΔFCH and Gas7Δcoiled were evenly distributed in the cells. Although the loss of the FCH or coiled-coil domains had no effect on Gas7 interaction with endogenous N-WASP (Fig. 4D), they did disturb its submembrane localization (Fig. 4B). Thus, this specific interaction between Gas7 and N-WASP is required for the formation of membrane protrusions as well as for the submembrane localization of Gas7. The latter is dependent on an intact F-BAR domain of Gas7 (both the FCH and the coiled coil regions, Fig. 4B).
FIGURE 4.
Requirement of intact Gas7 for Gas7-induced membrane protrusion in Neuro-2a. A, a schematic representation of Gas7 truncates with various internal deletions is shown. Gas7, Gas7ΔWW, Gas7ΔFCH, and Gas7Δcoiled were tagged with His-Myc peptides at the C terminus. B, Gas7 truncates were transiently expressed in Neuro-2a cells, and cell morphology was examined. Neuro-2a cells transfected with either Gas7ΔWW/Myc, Gas7ΔFCH/Myc, or Gas7Δcoiled/Myc failed to form processes. Note that Gas7 and Gas7ΔWW predominantly localized in the submembrane region, whereas Gas7ΔFCH and Gas7Δcoiled were evenly distributed in the cytosol. Scale bar, 5 μm. C, shown is the fraction of Neuro-2a cells transfected with either Gas7/Myc, Gas7ΔWW/Myc, Gas7ΔFCH/Myc, or Gas7Δcoiled/Myc successfully extending membrane protrusions. Data are the means ± S.E. of 3 independent determinations using 100–130 cells. Significant differences (paired Student's t test) in group means compared with the control group (Gas7 transfection) are indicated; *, p < 0.05; **, p < 0.01. D, extracts from COS-7 cells co-transfected with HA/N-WASP and either Gas7/Myc (positive control), Gas7ΔWW/Myc, Gas7ΔFCH/Myc or Gas7Δcoiled/Myc were subjected to co-IP to examine in vivo interactions of HA-N-WASP with various Gas7 internal deletions. Only Gas7ΔWW failed to interact with N-WASP. E, the effect on Gas7-induced membrane protrusions of treatment with an actin-depolymerizing reagent is shown. Gas7/Myc-transfected Neuro-2a cells were treated with 2 μm latrunculin B for 30 min and immunostained for actin and Gas7. Gas7-expressing Neuro-2a cells had similar membrane protrusions after latrunculin B treatment as before. Scale bar, 10 μm.
To test whether Gas7-induced membrane protrusions in the Neuro-2a cells are stable in the absence of actin filaments, we treated Gas7-expressing Neuro-2a cells with the actin depolymerizing reagent, latrunculin B, and used immunostaining to observe the resultant cell morphology. Membrane protrusions induced by Gas7 remained stable in cells after latrunculin B treatment, as in control cells (Fig. 4E). This result agrees with Suetsugu et al. (43) for the RCB domain (N-terminal Rac binding domain).
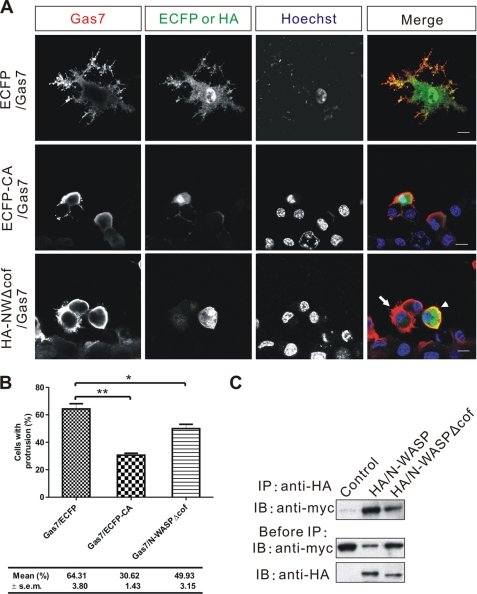
Involvement of N-WASP in the Formation of Gas7-induced Membrane Protrusions
To address the role of N-WASP in the formation of Gas7-induced membrane protrusions, two N-WASP mutants were co-expressed with Gas7 in Neuro-2a cells. The C-terminal region of N-WASP protein interacts directly with Arp2/3, resulting in a loss of function of the Arp2/3 complex in vitro (27) due to its aberrant subcellular localization. An ECFP fusion with the C-terminal region of N-WASP protein (ECFP-CA) was expressed simultaneously with Gas7 in Neuro-2a cells to see whether the Gas7-induced formation of membrane protrusions was affected. Overexpressed ECFP-CA competed against endogenous N-WASP for the interaction with Arp2/3 complex (Fig. 5A) and resulted in decreased formation of Gas7-induced membrane protrusions in Neuro-2a cells (30.6%, versus Gas7/ECFP control 64.3%; p < 0.01; Fig. 5B). Another dominant-negative mutant, N-WASPΔcof, was used to confirm that the interaction of N-WASP and Arp2/3 is essential for the formation of Gas7-induced membrane protrusions. The cofilin-homology region of N-WASP is an accessory domain that facilitates binding of the Arp2/3 complex to the acidic domain of N-WASP (27, 31, 42, 44). When N-WASPΔcof and Gas7 were co-expressed in Neuro-2a cells, the cells were unable to form membrane protrusions (Fig. 5A, arrowhead) compared with normal Gas7-expressing cells (Fig. 5A, arrow). The fraction with protrusions was reduced in cells that co-expressed Gas7 and N-WASPΔcof (49.9% versus control 64.3%; p < 0.05; Fig. 5B). These results were confirmed by an in vitro co-precipitation assay with HA-tagged intact N-WASP or N-WASPΔcof proteins. N-WASPΔcof physically interacted with Gas7 in the same way as intact N-WASP (Fig. 5C), suggesting that N-WASP works with the Arp2/3 complex in the formation of Gas7-induced membrane protrusions in Neuro-2a cells.
FIGURE 5.
N-WASP is involved in Gas7-induced membrane protrusions in Neuro-2a cells. A, Neuro-2a cells co-transfected with Gas7 (red) and either ECFP (control, green) or N-WASP mutants (ECFP-CA or HA/N-WASPΔcof) (green) were immunostained, and nuclei were counterstained with Hoechst 33258 (cyan). In the lower right panel, the arrow shows successful extension of membrane protrusion; the arrowhead shows a lack of extension. Scale bar, 10 μm. B, shown is the fractions of cells in A with membrane protrusions. Data are presented as the mean ± S.E. of three independent determinations, each counting at least 100 cells. Statistically significant decreases of the fraction of cells that extend processes compared with the control group (ECFP/Gas7 co-transfection) are indicated (paired Student's t test; *, p < 0.05; **, p < 0.01). C, extracts from COS-7 cells transfected with Gas7/Myc alone (negative control) or in combination with either wild-type HA/N-WASP (positive control) or HA/N-WASPΔcof were subjected to co-IP to assay the interaction between HA-N-WASPΔcof and Gas7. Gas7 was detected in complex with both N-WASPΔcof and N-WASP.
Formation of Gas7-induced Membrane Protrusions and N-WASP-mediated Actin Polymerization Are Cdc42-independent
During N-WASP-mediated cytoskeleton rearrangement, simultaneous binding of Cdc42 and phosphoinositol 4,5-bisphosphate are required for full activation of N-WASP (29, 30). However, some N-WASP activators such as WISH activate N-WASP in a Cdc42-independent manner (36). To clarify whether or not the involvement of N-WASP in Gas7-induced membrane protrusions is Cdc42-dependent in Neuro-2a cells, a N-WASP mutant (N-WASP H208D) that failed to bind Cdc42 (40) was used to compete against the interaction of endogenous N-WASP with Gas7, and membrane protrusion formation was examined. Neuro-2a cells that co-expressed Gas7 and N-WASP H208D did form membrane protrusions (Fig. 6A). No significant difference was observed in membrane protrusion formation between cells that co-expressed Gas7 and N-WASP H208D (67.4%) or Gas7 and normal N-WASP (67.5%; Fig. 6B). In addition, the dominant-negative mutant of Cdc42, HA/Cdc42N17, which has a higher affinity for GDP and is, thus, a loss-of-function Cdc42, was used to confirm that Cdc42 activity is not required in Gas7-induced formation of membrane protrusions (Fig. 6A). The same extent of membrane protrusion formation was seen in Gas7 and Cdc42N17 co-expressing cells as in Gas7 and N-WASP co-expressing cells (Fig. 6B). In vitro co-precipitation confirmed that N-WASP H208D physically interacted with Gas7 equally well as intact N-WASP (Fig. 6C). We also used an in vitro reconstituted system to assess whether Gas7 acts as an effector, like Cdc42, in activating N-WASP activity, resulting in actin polymerization. N-WASP alone could not significantly enhance the Arp2/3-mediated actin polymerization process without any effector or activator (sample 2; Fig. 6D). On the contrary, the GTPγS-charged Cdc42 did do so, enhancing N-WASP activity to induce actin polymerization as mentioned above (sample 4). In parallel, we found that Gas7 in the reconstituted system accelerated actin polymerization to a similar extent as Cdc42 (sample 6 versus 4). In contrast, this was not detected in the GST negative control (sample 5) or without N-WASP (sample 3). These data indicate that Gas7 indeed functions as an effector, interacting with N-WASP directly to induce actin polymerization in a Cdc42-independent manner.
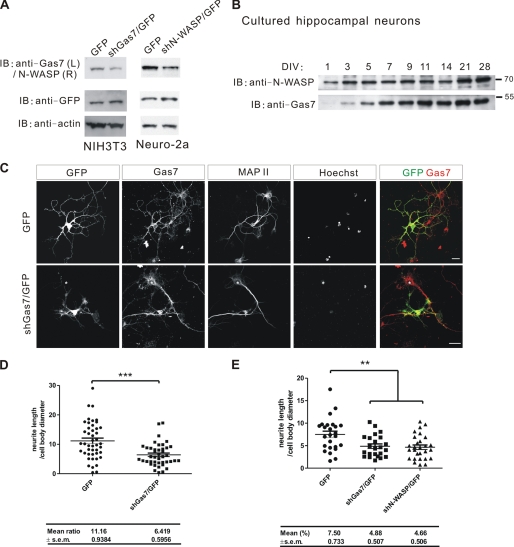
Requirement of Gas7 in the Neurite Outgrowth of Cultured Hippocampal Neurons
Banzai et al. (31) have shown that N-WASP is indispensable in the neurite outgrowth of hippocampal neurons. We found that Gas7 was co-localized with N-WASP in growth cones of cultured hippocampal neurons (Fig. 2C). The dynamic assembly of actin in growth cones is a driving force for neurite outgrowth during neuronal differentiation (1, 45). To investigate whether Gas7 is required for neurite outgrowth of hippocampal neurons as for Purkinje neurons (9), endogenous Gas7 in cultured hippocampal neurons was knocked out by transient transfection with small hairpin Gas7/GFP (shGas7/GFP). Knockdown efficiency of shGas7/GFP in arrest-induced NIH3T3 cells (Fig. 7A, left panel) was first examined. Expression of endogenous Gas7 was significantly reduced in shGas7/GFP-transfected NIH3T3 cells. In parallel, we also generated a shN-WASP/GFP construct to knock down N-WASP expression. The performance of shN-WASP was validated in Neuro-2a cells (Fig. 7A, right panel), with less endogenous N-WASP protein detected in the shN-WASP/GFP-transfected cells than in the control cells.
FIGURE 7.
The requirement for Gas7 in the neurite outgrowth of hippocampal neurons. A, knockdown of the endogenous Gas7 in NIH3T3 cells with shGas7 and of N-WASP in Neuro-2a cells with shN-WASP is shown. Cell extracts were prepared from shGas7/GFP-transfected NIH3T3 cells or shN-WASP/GFP-transfected Neuro-2a cells after 48 h of transfection and assayed by Western blot with GFP as a negative control. Both endogenous Gas7 and N-WASP expression were significantly reduced by small hairpin transfection. B, shown is the time-course of Gas7 and N-WASP expression in cultured hippocampal neurons. Lysates were prepared from cultured embryonic hippocampal neurons at the indicated days in vitro (DIV) and assayed by Western blot. N-WASP is expressed constantly from DIV 1–28, peaking after 2 weeks. In contrast, Gas7 expression begins during DIV 3–5 and then continues, with one peak in the second week and a second peak between weeks 3 and 4. C, immunostaining of Gas7-knockdown hippocampal neurons is shown. Cultured hippocampal neurons were transfected with GFP or shGas7/GFP at DIV2 and immunostained at DIV5. Neurite outgrowth was aberrant in Gas7-knockdown neurons as compared with neurons expressing Gas7 normally (GFP panel or the star in shGas7/GFP-transfected panel). Scale bar, 20 μm. D, quantification of morphological changes in neurite outgrowth from C. The ratio of longest neurite length to cell body diameter decreased significantly (shGas7/GFP (n = 42) versus control GFP (n = 45); unpaired Student's t test; ***, p < 0.001). E, quantification of morphological changes in neurite outgrowth is shown. Cultured hippocampal neurons were transfected with GFP, shGas7/GFP, or shN-WASP/GFP at DIV2 and immunostained at DIV4. The ratio of longest neurite length to cell body diameter decreased significantly (shGas7/GFP (n = 28) or shN-WASP/GFP (n = 23) versus control GFP (n = 25); unpaired Student's t test; **, p < 0.01).
The time-course of Gas7 expression in cultured embryonic hippocampal neurons was determined by Western blot analysis of cells collected at different DIV. Gas7 expression began between DIV3 and DIV5 and continued strongly until DIV11, with another peak of expression between DIV14 and DIV21 (Fig. 7B). N-WASP was expressed continually from DIV1 to DIV14 with another peak after DIV14, consistent with a report by Tomasevic et al. (46). Accordingly, hippocampal neurons were transfected at DIV2 with shGas7/GFP plasmid, carrying a separate GFP operon as a transfection reporter, and immunostained at DIV5 to examine the morphological changes in hippocampal neurons. Gas7 expression was reduced significantly, and neurite outgrowth was abnormal in the shGas7/GFP-transfected hippocampal neurons compared with GFP alone-transfected cells (star in shGas7/GFP panels; Fig. 7C). Neurites in the Gas7-silenced hippocampal neurons were also much shorter; the ratio of longest neurite length to cell body diameter was almost half that of control cells (Fig. 7D; p < 0.001). The shN-WASP/GFP-transfected cells showed a similar defect of neurite outgrowth to the shGas7/GFP-transfected neurons (Fig. 7E). These results indicate that the presence of both Gas7 and N-WASP is required for regular neurite outgrowth of hippocampal neurons.
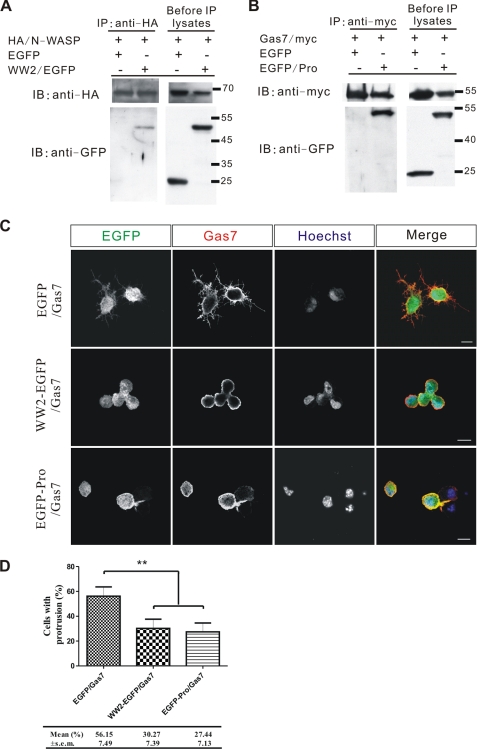
Interaction between Gas7 and N-WASP Is Required for Regular Neurite Outgrowth of Hippocampal Neurons
As shown above, both Gas7 and N-WASP are indispensable for regular neurite outgrowth of hippocampal neurons. To determine whether a specific interaction between Gas7 and N-WASP in neurons is necessary for their function, we generated a truncated Gas7 derivative that contained Gas7 residues 1–151 (WW2) fused with EGFP at the N terminus to compete with endogenous Gas7 and also to reduce the specific interaction between Gas7 and N-WASP in cultured hippocampal neurons. Likewise, an N-WASP-derived truncated protein, EGFP-Pro domain fusion peptide, was generated to compete for endogenous N-WASP. In transfected COS-7 cells, we found that WW2-EGFP fusion protein but not EGFP alone interacted with N-WASP (Fig. 8A); likewise, the EGFP-Pro specifically interacted with Gas7 (Fig. 8B). Unlike Gas7, the production of WW2-EGFP was unable to induce the extension of membrane protrusions in Neuro-2a cells (supplemental Fig. S5), and this fusion protein is likely to be a Gas7 dominant negative mutant. This result echoed our previous result that only intact Gas7 produced membrane protrusion in Neuro-2a cells (Fig. 4B).
FIGURE 8.
WW2-EGFP and EGFP-Pro function as dominant negative mutants in the Gas7-induced membrane protrusion. A, extracts from COS-7 cells co-transfected with HA/N-WASP, and either EGFP (negative control) or WW2/EGFP was analyzed by co-IP to assay the interaction between N-WASP and WW2-EGFP. A specific interaction between N-WASP and WW2-EGFP was found. B, extracts from COS-7 cells co-transfected with Gas7/Myc and either EGFP (negative control) or Pro/EGFP were analyzed by co-IP to assay the interaction between Gas7 and Pro-EGFP. A strong interaction between Gas7 and Pro-EGFP was detected. C, immunostaining of Neuro-2a cells co-transfected with Gas7/Myc and either Gas7 dominant negative (WW2/EGFP) or N-WASP dominant negative (EGFP/Pro) mutants to examine competition performance in Gas7-induced membrane protrusion. Unlike control (Gas7/EGFP) cells, Gas7/WW2-EGFP and Gas7/EGFP-Pro co-transfected Neuro-2a cells did not show any membrane protrusions. Scale bar, 10 μm. D, fractions of cells in C with membrane protrusions are shown. Data are presented as the mean ± S.E. of three independent determinations, each counting at least 50 cells. Statistically significant decreases of the fraction of cells that extended processes compared with the control group (EGFP/Gas7 co-transfection) are indicated (paired Student's t test; **, p < 0.01).
To test their efficiency as competition agents, we looked at both constructs in the cell line system shown in Fig. 8C. Co-transfection with Gas7/Myc and WW2/EGFP did not produce any membrane protrusions in Neuro-2a cells, unlike the control experiment (EGFP/Gas7). The same outcome was also observed in cells co-transfected with EGFP/Pro and Gas7/Myc. Both results were significant (Fig. 8D); the fractions of cells with membrane protrusions in the Gas7/WW2-EGFP (30.27%) and Gas7/EGFP-Pro (27.44%) co-transfection cells were much smaller than control (Gas7/EGFP, 56.15%; p < 0.01). These results confirm that both Gas7 and N-WASP constructs functioned as dominant negative mutants.
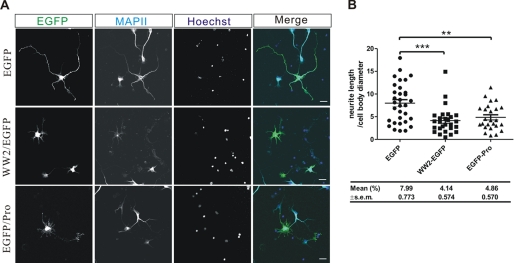
Cultured hippocampal neurons were transfected with either WW2-EGFP or EGFP-Pro plasmids at DIV2 and immunostained at DIV4 (Fig. 9A). When WW2-EGFP or EGFP-Pro was successfully expressed in hippocampal neurons, the neurite outgrowth was abnormal, with significantly shorter neurites as in the case of Gas7 or N-WASP-deleted hippocampal neurons (Fig. 9B). Collectively, our data confirm that a specific interaction of Gas7 with N-WASP directly contributes to regular neurite outgrowth of cultured hippocampal neurons.
FIGURE 9.
A specific interaction between Gas7 and N-WASP regulates the neurite outgrowth of hippocampal neurons. A, immunostaining of cultured hippocampal neurons transfected with EGFP, WW2/EGFP, or EGFP/Pro is shown. Cultured hippocampal neurons were transfected with EGFP, WW2/EGFP, or EGFP/Pro at DIV2 and immunostained at DIV4. Neurite outgrowth was aberrant in WW2/EGFP and EGFP/Pro-overexpressing cultured hippocampal neurons as compared with control EGFP-overexpressing cells. Scale bar, 20 μm. B, shown is quantification of morphological changes in neurite outgrowth from A. The ratio of longest neurite length to cell body diameter decreased significantly with mutant transfection (WW2-EGFP, n = 27; EGFP-Pro, n = 25; control EGFP, n = 32; unpaired Student's t test; **, p < 0.01; ***, p < 0.001).
DISCUSSION
Neurite outgrowth during the initial stages of neuronal differentiation depends largely on the formation of the growth cone, a specialized structure of highly dynamic actin (1, 45). She et al. (15) have demonstrated that association of Gas7 with actin can induce the reorganization of microfilaments and promote actin polymerization in vitro. Morphological changes directly correlated with Gas7 expression in Neuro-2a cells were reported by Ju et al. (9). To explore the molecular machinery that underlies Gas7-induced morphological changes, we looked for Gas7-interacting proteins. Among several candidates was WISH, a protein known to bind and activate N-WASP, leading to recruitment of the Arp2/3 complex in a Cdc42-independent manner (36). Unexpectedly, preliminary studies of the interaction between Gas7, WISH, and N-WASP by GST pulldown assays showed that Gas7 itself directly interacts with N-WASP. We have now demonstrated that Gas7 directly interacts with N-WASP in a Cdc42-independent manner to induce membrane protrusions.
Ju et al. (9) found that Gas7 is predominantly expressed in brain, and its loss impedes neurite outgrowth in primary Purkinje cell cultures. Following up on this, we found highly co-localized expression of Gas7 and N-WASP in the cytoplasm and neurites of hippocampal neurons (Fig. 2, A and C) but disjoint or only slightly overlapping distributions in other cortical neurons or cerebellar Purkinje cells (supplemental Fig. S2). This indicates that the role of Gas7 in the maturation of Purkinje cells may be in some other aspect of morphological differentiation rather than the actin-based morphological changes associated with the N-WASP-mediated pathway. More specifically, Gas7 is co-localized with N-WASP in the growth cones of hippocampal neurons. This strongly suggests an involvement of Gas7 in neurite outgrowth through N-WASP-mediated actin organization in hippocampal neurons.
Compared with other N-WASP-associated proteins such as WISH or Toca-1, which contain the SH3 domain and activate N-WASP through the interaction of SH3 and Pro domains (36, 37), mouse Gas7, which lacks the SH3 domain, interacts directly with N-WASP via the WW domain (Fig. 3) as well as FBP11 (47). The WW domain structurally resembles the SH3 domain and also binds specifically to proline-rich regions (48). This is the first indication that the WW-Pro interaction functionally activates N-WASP in the PCH family. Furthermore, human Gas7 isoform c contains both the SH3 and the WW domains at its N terminus (18, 49). The comparable domain architecture suggests that human Gas7-c functions similarly to mGas7 in N-WASP-mediated cytoskeleton rearrangement.
The two mouse Gas7 isoforms, Gas7 and Gas7-cb, possess the same C-terminal region of 320 amino acid residues, including the FCH and coiled-coil domains. Endogenous Gas7-cb is abundant in the nuclei of cerebellar neurons (10). Ectopic expression of Gas7-cb in Neuro-2a cells showed that Gas7-cb is also localized mainly in the nucleus and has no effect on morphological events (supplemental Fig. S6). It is suggested that both the WW domain and the correct spatial expression of Gas7 are required for the formation of membrane protrusions in Neuro-2a cells. This idea is supported by the co-localization of Gas7 and N-WASP in the submembrane region (Figs. 1B and 4B).
We found that loss of the interaction between Gas7 and N-WASP abolished the formation of Gas7-induced membrane protrusions. The C-terminal domains of Gas7 are essential for the submembrane localization that is required for Gas7-induced membrane protrusion formation (Fig. 4). The C-terminal region of Gas7 contains the coiled-coil domain, which has been demonstrated to mediate the oligomerization of Gas7 (16). The functional deficit of Gas7 after loss of the coiled-coil domain suggests that oligomerization of Gas7 may be essential for the interaction of Gas7 and N-WASP. Moreover, the FCH and coiled-coil domains are represented as an extended domain, the F-BAR domain, which mediates interaction with membrane phospholipids (18). The loss of either the FCH or the coiled-coil domains renders Gas7 unable to induce the formation of membrane protrusions. As seen by the localization patterns of the various Gas7 truncates in neuroblastoma cells (Fig. 4B), those with incomplete C termini may lead to a failure of Gas7 targeting to the submembrane region and an inability to activate N-WASP, which suggests an interaction of Gas7 with membrane phospholipids could be required for the induction of membrane protrusions. Interestingly, we observed the formation of tubular structures in Gas7-EGFP-expressing COS-7 cells. Supplemental Fig. S7A shows a video of one such event. A tubule started to form at the cell margin as a concentrated dot signal (indicated by an arrow at time 0 min, 00 s, supplemental Fig. S7A). The dot signal elongated and propagated continually toward the nucleus until 13 min, 06 s. Finally, it formed a stable structure in the cell. Immunostaining showed the tubular structures were co-localized with the membrane (supplemental Fig. S7B, lower panel) but not with the cellular cytoskeleton (supplemental Fig. S7B, upper and middle panels). Immunostaining also showed (preliminary results as yet) that Gas7 induces membrane invagination like other proteins containing an F-BAR domain such as FBP17 (50). However, how the Gas7 induces membrane deformation and coordinates with actin remains unclear.
N-WASP is activated by Cdc42 and phosphoinositol 4,5-bisphosphate after the recruitment of Arp2/3 complex to initiate actin polymerization (30, 32–34). Two dominant-negative mutants of N-WASP were used to analyze the involvement of N-WASP and Arp2/3 in Gas7-induced formation of membrane protrusions. Both mutants have been shown to interfere with the function of Arp2/3 as mentioned above. Our data clearly demonstrate that Gas7-induced membrane protrusions are required for N-WASP and Arp2/3 in the cells (Fig. 5). The in vitro actin polymerization assay showed clearly that Gas7 induces actin polymerization, and therefore, the subsequent extension of membrane protrusions through an N-WASP-Arp2/3-mediated pathway.
During neuronal differentiation, filopodia and lamellipodia always appear simultaneously in the growth cone of neurons and participate in neurite outgrowth. Cdc42-dependent N-WASP/Arp2/3-mediated actin polymerization is essential for the formation of filopodia (40). We used a dominant-negative mutant of N-WASP, which failed to bind Cdc42, to determine the Cdc42 dependence of the Gas7-induced formation of membrane protrusions. Gas7-induced morphological changes were unaffected irrespective of the lack of interaction between mutant N-WASP and Cdc42 (Fig. 1A versus Fig. 6A, upper panel). We also used a dominant-negative mutant of Cdc42, which has abnormal Cdc42 function because of a higher affinity for GDP, to examine the Cdc42 dependence of the Gas7-induced membrane protrusion formation. The induction by Gas7 of membrane protrusions was not significantly affected by the absence of Cdc42 activity (Fig. 6B), which indicates that Gas7-induced formation of membrane protrusions is Cdc42-independent. This data were also validated in the in vitro actin polymerization assay (Fig. 6D).
To elucidate the role that the interaction of Gas7 with N-WASP plays in hippocampal neurons, we inhibited the expression of gas7 in cultured hippocampal neurons by shGas7/GFP (Fig. 7) or blocked the specific association of Gas7 and N-WASP with the truncate construct WW2-EGFP (Fig. 9). Neurites in both Gas7-silenced (Fig. 7) and WW2-EGFP-transfected (Fig. 9) hippocampal neurons were shorter than in the wild type (GFP- or EGFP-transfected controls). A similar phenomenon was also observed in N-WASP-deficient and EGFP-Pro-transfected hippocampal neurons. This demonstrated that, in addition to N-WASP (31), Gas7 is as critical for regular neurite outgrowth in cultured hippocampal neurons as N-WASP and implied that Gas7 functions with N-WASP in the neurite extension of differentiating hippocampal neurons.
Dendritic spines, which are the postsynaptic receptive regions of most excitatory synapses, are an important feature of mature neurons and have been characterized as emerging from many thin headless protrusions called filopodia. Studies of dendritic spines have demonstrated correlations between abnormal spine morphology and brain dysfunction. Morphological events including the size, shape, and number of spines are believed to be associated with the formation and maintenance of memory and learning (51–54). Recently, Wegner et al. (55) reported N-WASP is also predominant in the dendritic spine and is required for spinogenesis. As Gas7 induces striking morphological changes in Gas7-transfected Neuro-2a cells, which lead to the extension of membrane protrusions similar to dendritic spines, Gas7 may be also implicated in the formation of dendritic spines in hippocampal neurons. Indeed, we also found here that Gas7 expression peaked during spinogenesis (after DIV14, Fig. 7B) and that Gas7 co-localizes with N-WASP in the dendritic spines (supplemental Fig. S8A) and in the synaptic vesicle (supplemental Fig. S8B) of cultured hippocampal neurons. These findings implicate Gas7 in the regulation of spinogenesis and synaptogenesis in hippocampal neurons via N-WASP-mediated actin dynamics. Whether Gas7 in the presence or absence of N-WASP plays a direct role in spinogenesis or synaptogenesis in hippocampal neurons to regulate neuronal plasticity is another intriguing question to be answered.
In summary, Gas7 and N-WASP participate in a direct interaction mediated through their WW (Gas7) and proline-rich (N-WASP) domains. This interaction changes the conformation of N-WASP and activates it. After exposure of the VCA domains in the C-terminal region of N-WASP, the Arp2/3 complex can subsequently bind and switch on cytoskeletal rearrangement (33, 56). Another C-terminal Gas7 (residues 160–421) participates in de novo actin polymerization and the bundling of the filament (15) in the cells, which also suggested contribution to the residual process in the Gas7ΔWW-transfected Neuro-2a cells as compared with EGFP-transfected cells (p < 0.05) (Fig. 4C). A combination of these processes initiates the formation of membrane protrusions, and their continuation results in the normal neurite outgrowth of hippocampal neurons. Gas7 interacts with N-WASP at submembrane regions to coordinate cortical cytoskeleton reorganization and induce membrane protrusions in neuroblastoma cells. This specific interaction is also required for regular neurite outgrowth of hippocampal neurons. Whether the interaction of Gas7 and N-WASP regulates spinogenesis/synaptogenesis and precisely how Gas7 coordinates actin and membrane dynamics in spinogenesis remain to be determined.
Supplementary Material
Acknowledgments
RNAi reagents were obtained from the National RNAi Core Facility located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica, supported by the National Research Program for Genomic Medicine Grants of the National Science Council (NSC 97–3112-B-001-016). We are grateful to Dr. Tadaomi Takenawa for the gift of rat N-WASP plasmid, Dr. Wenlynn B. Su for comments on the manuscript, Dr. Harry Wilson of Academia Sinica for manuscript editing, and Sue-Ping Lee of IMB Confocal Microscope Facility for technical assistance.
This work was supported by an intramural fund and Program Project Grant AS-95-TP-B04 from Academia Sinica and by grants from the National Science Council of Taiwan. This work fulfilled in part the requirements for the Ph.D. thesis of J.-J. You, Graduate Institute of Molecular Medicine, College of Medicine, National Taiwan University, Taiwan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
S.-L. Chen and S. Lin-Chao, unpublished data.
- FCH
- Fes/CIP4 homology
- CC
- coiled-coil
- GBD
- GTPase-binding domain
- CRIB
- Cdc42-Rac interactive binding
- VCA
- verproline homology, cofilin homology, acidic domain
- WISH
- WASP interacting SH3 protein
- GST
- glutathione S-transferase
- shGas7
- small hairpin Gas7
- DIV
- days in vitro
- IP
- immunoprecipitation
- IB
- immunoblot
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- ECFP
- enhanced cyan fluorescent protein
- GFP
- green fluorescent protein.
REFERENCES
- 1. Dent E. W., Gertler F. B. (2003) Neuron 40, 209–227 [DOI] [PubMed] [Google Scholar]
- 2. Schneider C., King R. M., Philipson L. (1988) Cell 54, 787–793 [DOI] [PubMed] [Google Scholar]
- 3. Lih C. J., Cohen S. N., Wang C., Lin-Chao S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenner D. G., Lin-Chao S., Cohen S. N. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5517–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brancolini C., Bottega S., Schneider C. (1992) J. Cell Biol. 117, 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adlkofer K., Martini R., Aguzzi A., Zielasek J., Toyka K. V., Suter U. (1995) Nat. Genet. 11, 274–280 [DOI] [PubMed] [Google Scholar]
- 7. Fabbretti E., Edomi P., Brancolini C., Schneider C. (1995) Genes Dev. 9, 1846–1856 [DOI] [PubMed] [Google Scholar]
- 8. Lee K. K., Tang M. K., Yew D. T., Chow P. H., Yee S. P., Schneider C., Brancolini C. (1999) Dev. Biol. 207, 14–25 [DOI] [PubMed] [Google Scholar]
- 9. Ju Y. T., Chang A. C., She B. R., Tsaur M. L., Hwang H. M., Chao C. C., Cohen S. N., Lin-Chao S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11423–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lazakovitch E. M., She B. R., Lien C. L., Woo W. M., Ju Y. T., Lin-Chao S. (1999) Genomics 61, 298–306 [DOI] [PubMed] [Google Scholar]
- 11. Moorthy P. P., Kumar A. A., Devaraj H. (2005) Stem Cells Dev. 14, 664–670 [DOI] [PubMed] [Google Scholar]
- 12. Su W. B., You J.-J., Huang B.-T., Sivakumar V., Yeh S.-D., Lin-Chao S. (2009) in The Pombe Cdc15 Homology Proteins, Madame Curie Biosience Database, Landes Bioscience, Austin, TX [Google Scholar]
- 13. Lortie K., Huang D., Chakravarthy B., Comas T., Hou S. T., Lin-Chao S., Morley P. ( 2005) Brain Res. 1036, 27– 34 [DOI] [PubMed] [Google Scholar]
- 14. Chao C. C., Su L. J., Sun N. K., Ju Y. T., Lih J. C., Lin-Chao S. (2003) J. Neurosci. Res. 74, 248–254 [DOI] [PubMed] [Google Scholar]
- 15. She B. R., Liou G. G., Lin-Chao S. (2002) Exp. Cell Res. 273, 34–44 [DOI] [PubMed] [Google Scholar]
- 16. So C. W., Lin M., Ayton P. M., Chen E. H., Cleary M. L. (2003) Cancer Cell 4, 99–110 [DOI] [PubMed] [Google Scholar]
- 17. Chitu V., Stanley E. R. (2007) Trends Cell Biol. 17, 145–156 [DOI] [PubMed] [Google Scholar]
- 18. Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006) J. Cell Biol. 172, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lippincott J., Li R. (2000) Microsc. Res. Tech. 49, 168–172 [DOI] [PubMed] [Google Scholar]
- 20. Aspenström P. (1997) Curr. Biol. 7, 479–487 [DOI] [PubMed] [Google Scholar]
- 21. Henne W. M., Kent H. M., Ford M. G., Hegde B. G., Daumke O., Butler P. J., Mittal R., Langen R., Evans P. R., McMahon H. T. (2007) Structure 15, 839–852 [DOI] [PubMed] [Google Scholar]
- 22. Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 23. Shimada A., Niwa H., Tsujita K., Suetsugu S., Nitta K., Hanawa-Suetsugu K., Akasaka R., Nishino Y., Toyama M., Chen L., Liu Z. J., Wang B. C., Yamamoto M., Terada T., Miyazawa A., Tanaka A., Sugano S., Shirouzu M., Nagayama K., Takenawa T., Yokoyama S. (2007) Cell 129, 761–772 [DOI] [PubMed] [Google Scholar]
- 24. So C. W., Karsunky H., Passegué E., Cozzio A., Weissman I. L., Cleary M. L. (2003) Cancer Cell 3, 161–171 [DOI] [PubMed] [Google Scholar]
- 25. Miki H., Miura K., Matuoka K., Nakata T., Hirokawa N., Orita S., Kaibuchi K., Takai Y., Takenawa T. (1994) J. Biol. Chem. 269, 5489–5492 [PubMed] [Google Scholar]
- 26. Miura K., Miki H., Shimazaki K., Kawai N., Takenawa T. (1996) Biochem. J. 316, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) Cell 97, 221–231 [DOI] [PubMed] [Google Scholar]
- 28. Yamaguchi H., Miki H., Suetsugu S., Ma L., Kirschner M. W., Takenawa T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miki H., Miura K., Takenawa T. (1996) EMBO J. 15, 5326–5335 [PMC free article] [PubMed] [Google Scholar]
- 30. Rohatgi R., Ho H. Y., Kirschner M. W. (2000) J. Cell Biol. 150, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Banzai Y., Miki H., Yamaguchi H., Takenawa T. (2000) J. Biol. Chem. 275, 11987–11992 [DOI] [PubMed] [Google Scholar]
- 32. Takenawa T., Suetsugu S. (2007) Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 33. Bompard G., Caron E. (2004) J. Cell Biol. 166, 957–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. (2000) Nature 404, 151–158 [DOI] [PubMed] [Google Scholar]
- 35. Carlier M. F., Nioche P., Broutin-L'Hermite I., Boujemaa R., Le Clainche C., Egile C., Garbay C., Ducruix A., Sansonetti P., Pantaloni D. (2000) J. Biol. Chem. 275, 21946–21952 [DOI] [PubMed] [Google Scholar]
- 36. Fukuoka M., Suetsugu S., Miki H., Fukami K., Endo T., Takenawa T. (2001) J. Cell Biol. 152, 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho H. Y., Rohatgi R., Lebensohn A. M., Le Ma., Li J., Gygi S. P., Kirschner M. W. (2004) Cell 118, 203–216 [DOI] [PubMed] [Google Scholar]
- 38. Innocenti M., Gerboth S., Rottner K., Lai F. P., Hertzog M., Stradal T. E., Frittoli E., Didry D., Polo S., Disanza A., Benesch S., Di Fiore P. P., Carlier M. F., Scita G. (2005) Nat. Cell Biol. 7, 969–976 [DOI] [PubMed] [Google Scholar]
- 39. Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., Mayer B. J. (2001) J. Biol. Chem. 276, 26448–26452 [DOI] [PubMed] [Google Scholar]
- 40. Miki H., Sasaki T., Takai Y., Takenawa T. (1998) Nature 391, 93–96 [DOI] [PubMed] [Google Scholar]
- 41. Vanzo N. F., Li Y. S., Py B., Blum E., Higgins C. F., Raynal L. C., Krisch H. M., Carpousis A. J. (1998) Genes Dev. 12, 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abe T., Kato M., Miki H., Takenawa T., Endo T. (2003) J. Cell Sci. 116, 155–168 [DOI] [PubMed] [Google Scholar]
- 43. Suetsugu S., Murayama K., Sakamoto A., Hanawa-Suetsugu K., Seto A., Oikawa T., Mishima C., Shirouzu M., Takenawa T., Yokoyama S. (2006) J. Biol. Chem. 281, 35347–35358 [DOI] [PubMed] [Google Scholar]
- 44. Zalevsky J., Lempert L., Kranitz H., Mullins R. D. (2001) Curr. Biol. 11, 1903–1913 [DOI] [PubMed] [Google Scholar]
- 45. Dent E. W., Tang F., Kalil K. (2003) Neuroscientist 9, 343–353 [DOI] [PubMed] [Google Scholar]
- 46. Tomasevic N., Jia Z., Russell A., Fujii T., Hartman J. J., Clancy S., Wang M., Beraud C., Wood K. W., Sakowicz R. (2007) Biochemistry 46, 3494–3502 [DOI] [PubMed] [Google Scholar]
- 47. Mizutani K., Suetsugu S., Takenawa T. (2004) Biochem. Biophys Res. Commun. 313, 468–474 [DOI] [PubMed] [Google Scholar]
- 48. Macias M. J., Wiesner S., Sudol M. (2002) FEBS Lett. 513, 30–37 [DOI] [PubMed] [Google Scholar]
- 49. Chao C. C., Chang P. Y., Lu H. H. (2005) J. Neurosci. Res. 81, 153–162 [DOI] [PubMed] [Google Scholar]
- 50. Kamioka Y., Fukuhara S., Sawa H., Nagashima K., Masuda M., Matsuda M., Mochizuki N. (2004) J. Biol. Chem. 279, 40091–40099 [DOI] [PubMed] [Google Scholar]
- 51. Irwin S. A., Galvez R., Greenough W. T. (2000) Cereb. Cortex 10, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 52. Purpura D. P., Bodick N., Suzuki K., Rapin I., Wurzelmann S. (1982) Brain Res. 281, 287–297 [DOI] [PubMed] [Google Scholar]
- 53. Sekino Y., Kojima N., Shirao T. (2007) Neurochem. Int. 51, 92–104 [DOI] [PubMed] [Google Scholar]
- 54. Wisniewski K. E., Segan S. M., Miezejeski C. M., Sersen E. A., Rudelli R. D. (1991) Am. J. Med. Genet. 38, 476–480 [DOI] [PubMed] [Google Scholar]
- 55. Wegner A. M., Nebhan C. A., Hu L., Majumdar D., Meier K. M., Weaver A. M., Webb D. J. (2008) J. Biol. Chem. 283, 15912–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takenawa T. (2005) Novartis Found. Symp. 269, 3–10;discussion 10–15, 30–34 passim [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.