Abstract
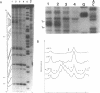
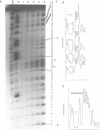
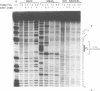
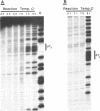
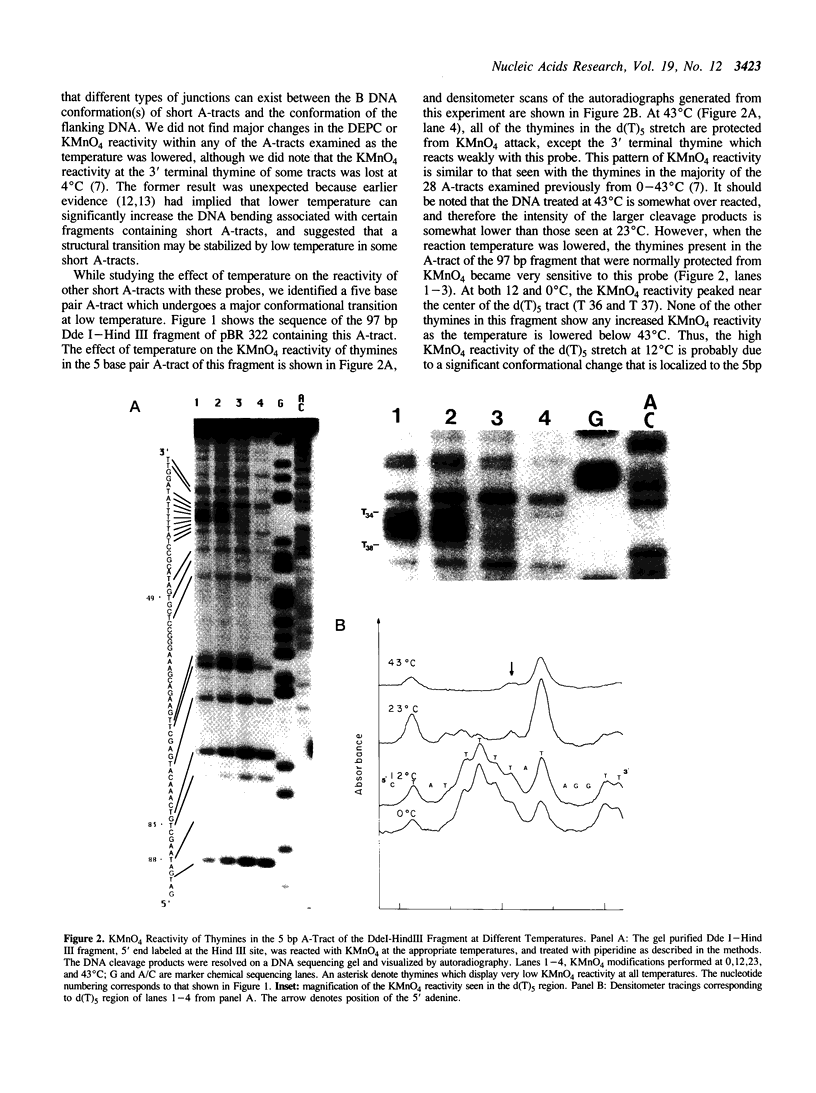
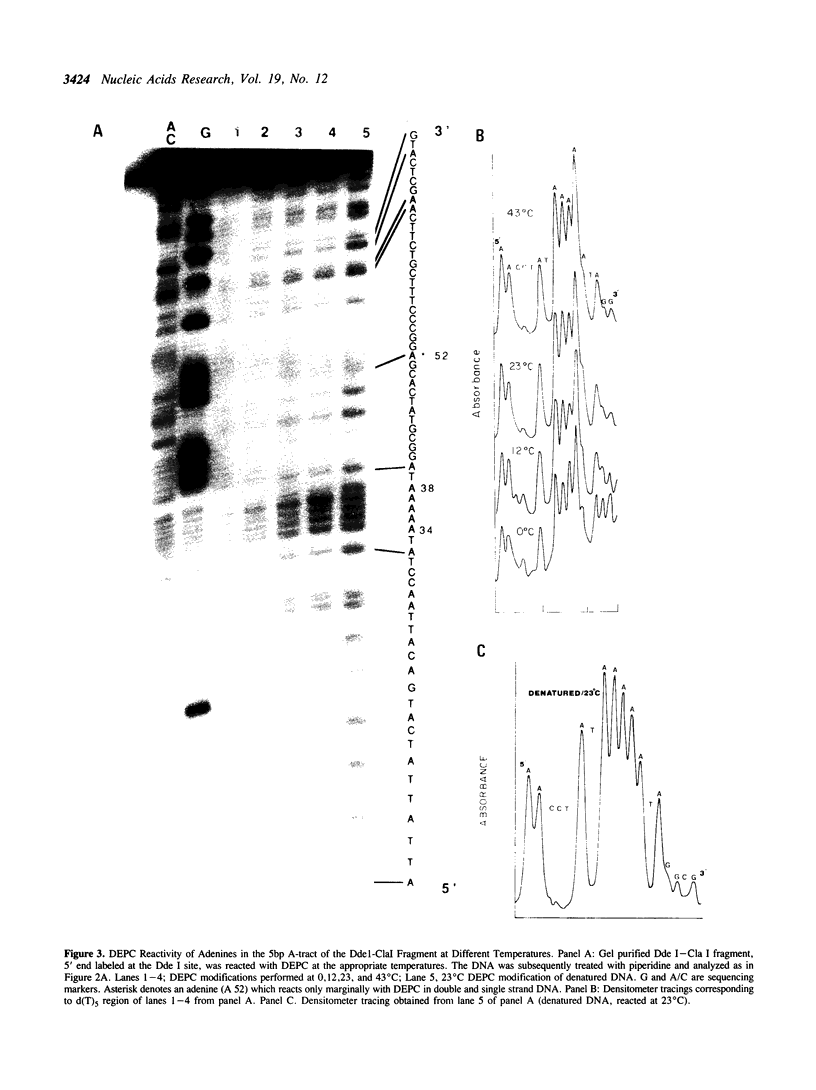
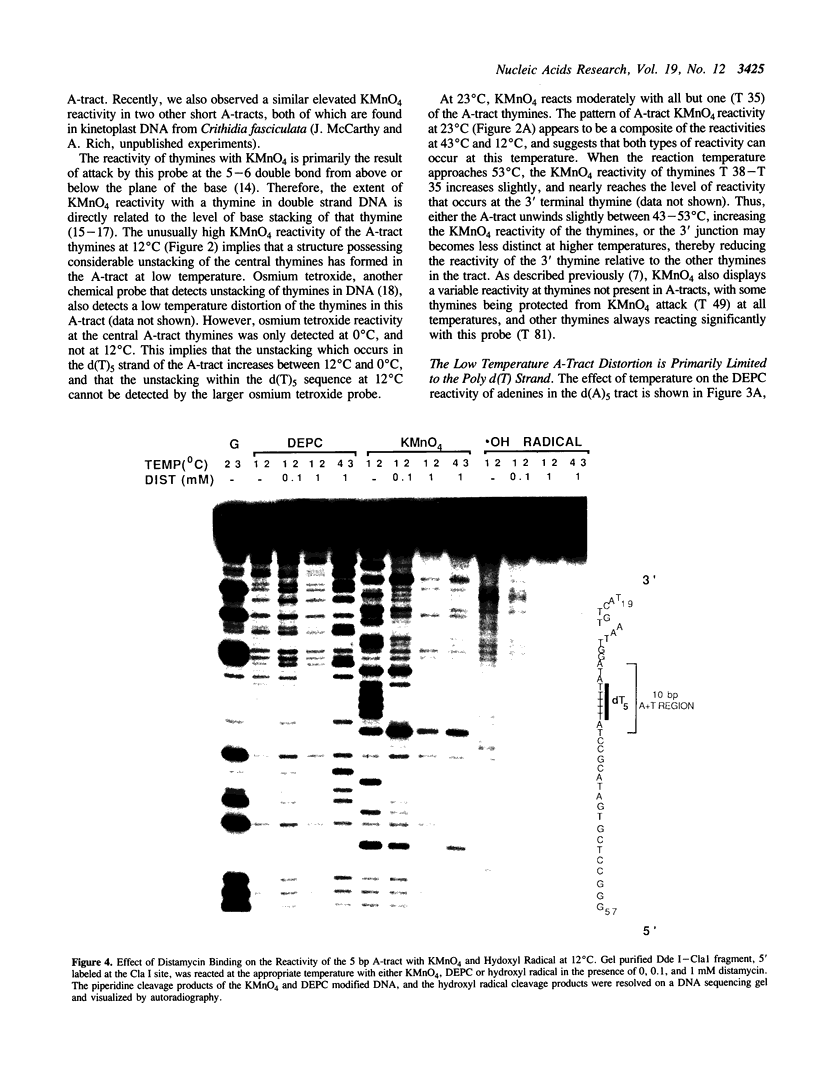
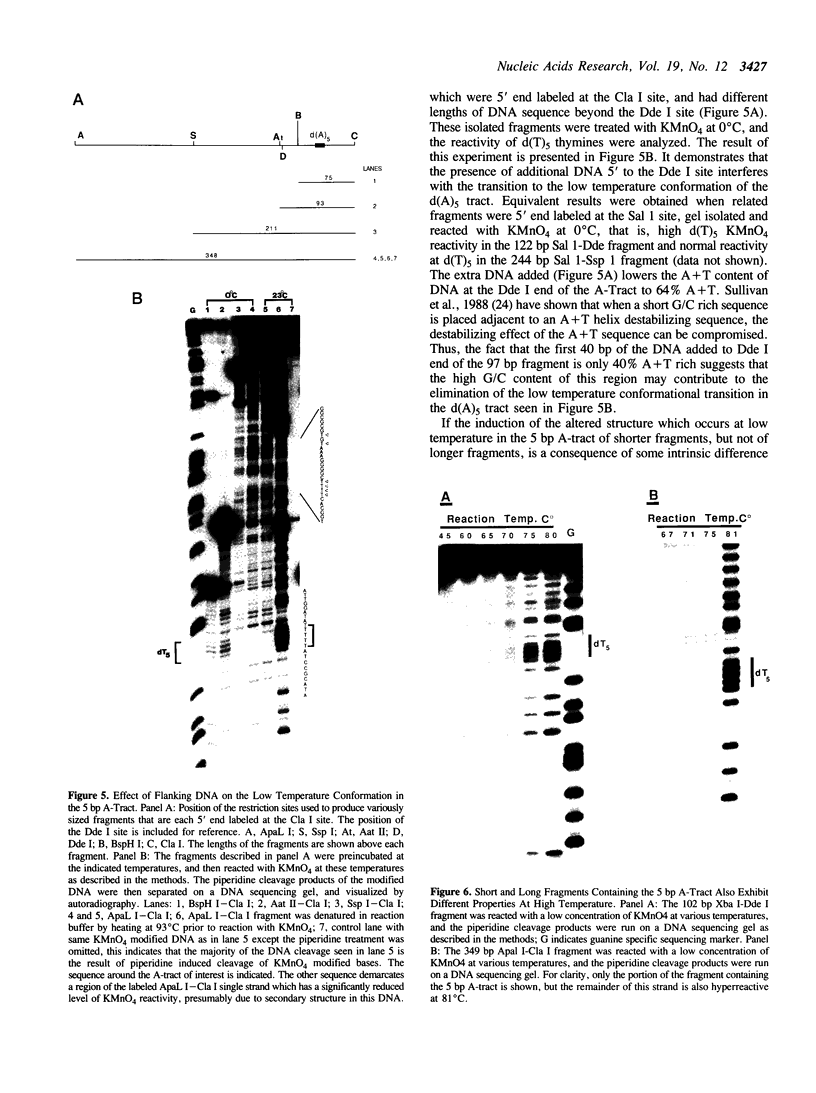
The chemical probes potassium permanganate (KMnO4) and diethylpyrocarbonate (DEPC) can be used to study the conformational flexibility of short tracts of adenine (A-tracts) present in DNA. With these probes, we demonstrate that a novel distortion is induced in a 5 base pair A-tract at low temperature. Formation of this distorted A-tract structure, which occurs in a DNA fragment from the promoter region of the plasmid pBR322, is distinguished by a dramatic increase in the KMnO4 reactivity of the central thymines in this tract at 12 degrees C. This alteration occurs in the absence of any detectable rearrangement in the conformation of the adenines in the complementary strand. Induction of this low temperature A-tract structure is blocked by the minor groove binding drug distamycin. Hydroxyl radical footprinting of distamycin binding to the fragment containing the d(A)5 tract at 12 degrees C suggests that this drug has two different modes of binding to DNA in agreement with recent NMR data. These experiments show that short A-tracts are capable of forming more than one structural variant of B DNA in solution. The possible relationship between the intrinsic bending of DNA containing short phased A-tracts and the low temperature A-tract conformation is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. Structural details of an adenine tract that does not cause DNA to bend. Nature. 1988 Feb 4;331(6155):455–457. doi: 10.1038/331455a0. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Nielsen P. E. Detection of intercalation-induced changes in DNA structure by reaction with diethyl pyrocarbonate or potassium permanganate. Evidence against the induction of Hoogsteen base pairing by echinomycin. FEBS Lett. 1988 Apr 11;231(1):172–176. doi: 10.1016/0014-5793(88)80725-7. [DOI] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y., Fujii S., Fujisawa R., Tomita K. One- and two-dimensional NMR studies on the conformation of DNA containing the oligo(dA)oligo(dT) tract. Nucleic Acids Res. 1988 Sep 12;16(17):8619–8632. doi: 10.1093/nar/16.17.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintanar A., Klevit R. E., Reid B. R. Two-dimensional NMR investigation of a bent DNA fragment: assignment of the proton resonances and preliminary structure analysis. Nucleic Acids Res. 1987 Jul 24;15(14):5845–5862. doi: 10.1093/nar/15.14.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G. An improvement in thymine specific chemical DNA sequencing. Nucleic Acids Res. 1989 Sep 25;17(18):7541–7541. doi: 10.1093/nar/17.18.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. G., Williams L. D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990 Jun 26;29(25):6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5723–5727. doi: 10.1073/pnas.86.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton J. G., Wemmer D. E. Structural modeling of the distamycin A-d(CGCGAATTCGCG)2 complex using 2D NMR and molecular mechanics. Biochemistry. 1988 Oct 18;27(21):8088–8096. doi: 10.1021/bi00421a018. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Hydroxyl radical footprinting of the sequence-selective binding of netropsin and distamycin to DNA. FEBS Lett. 1987 Dec 10;225(1-2):195–200. doi: 10.1016/0014-5793(87)81156-0. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz P. G., Dervan P. B. Distamycin and penta-N-methylpyrrolecarboxamide binding sites on native DNA. A comparison of methidiumpropyl-EDTA-Fe(II) footprinting and DNA affinity cleaving. J Biomol Struct Dyn. 1984 Mar;1(5):1133–1147. doi: 10.1080/07391102.1984.10507508. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Murchie A. I., Lilley D. M. Long range structural communication between sequences in supercoiled DNA. Sequence dependence of contextual influence on cruciform extrusion mechanism. J Biol Chem. 1988 Sep 15;263(26):13074–13082. [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]