Abstract
A Mediterranean diet rich in olive oil has been associated with health benefits in humans. It is unclear if and to what extent olive oil phenolics may mediate these health benefits. In this study, we fed senescence-accelerated mouse-prone 8 (SAMP8, n=11 per group) semisynthetic diets with 10% olive oil containing either high (HP) or low amounts of olive oil phenolics (LP) for 4.5 months. Mice consuming the HP diet had significantly lower concentrations of the oxidative damage markers thiobarbituric acid–reactive substances and protein carbonyls in the heart, whereas proteasomal activity was similar in both groups. Nrf2-dependent gene expression may be impaired during the aging process. Therefore, we measured Nrf2 and its target genes glutathione-S-transferase (GST), γ-glutamyl cysteine synthetase (γ-GCS), nicotinamide adenine dinucleotide phosphate [NAD(P)H]:quinone oxidoreductase (NQO1), and paraoxonase-2 (PON2) in the hearts of these mice. Nrf2 as well as GST, γ-GCS, NQO1, and PON2 mRNA levels were significantly higher in heart tissue of the HP as compared to the LP group. The HP-fed mice had significantly higher PON1 activity in serum compared to those receiving the LP diet. Furthermore, HP feeding increased relative SIRT1 mRNA levels. Additional mechanistic cell culture experiments were performed, and they suggest that the olive oil phenolic hydroxytyrosol present in the HP oil may be responsible for the induction of Nrf2-dependent gene expression and the increase in PON activity. In conclusion, a diet rich in olive oil phenolics may prevent oxidative stress in the heart of SAMP8 mice by modulating Nrf2-dependent gene expression.
Introduction
Epidemiological studies suggest an inverse relationship between coronary heart disease and the consumption of a so-called Mediterranean diet.1–3 Despite the scarcity of clinical studies investigating the underlying mechanisms, beneficial effects of the Mediterranean diet have partly been attributed to the use of olive oil.4–7 Olive oil is a rich source of oleic acid.8 However, it has been suggested that the beneficial effects of its consumption in the prevention of chronic diseases may not only be attributed to its fatty acid composition but also to its high content of phenolics.9–11 Studies in cultured cells12–19 and laboratory animals20–23 have demonstrated that the olive oil phenolics tyrosol, hydroxytyrosol, and oleuropein exert antioxidant, antiinflammatory and gene regulatory activities.
Rodent models may be used to understand the cellular and molecular mechanisms of age-dependent degeneration and to develop dietary interventions for healthy aging.24 It has been previously shown that biomarkers of oxidative stress are elevated in the heart tissue of the senescence-accelerated mouse-prone 8 (SAMP8) mouse as compared to the senescence-accelerated mouse-resistant strain.25,26 Furthermore, SAMP8 mice may exhibit elevated biomarkers of lipid27–29 and protein oxidation,30–32 mitochondrial dysfunction,29,33 and early onset of atherogenesis,34 all of which ultimately lead to a decreased life span in this mouse strain.35 These properties render SAMP8 mice a suitable rodent model for experimental aging research.35
The transcription factor Nrf2 is an important molecular switch that orchestrates the gene expression of antioxidant and phase 2 enzymes.36,37 Nrf2 is a basic leucine zipper transcription factor that binds to the antioxidant response element (ARE) in the promoter region of many adaptive genes, such glutathione-S-transferase (GST, EC 2.5.1.18), γ-glutamyl cysteine synthetase (γ-GCS; EC 6.3.2.2), and NAD(P)H:quinone oxidoreductase 1 (NQO1; EC 1.6.99.2).38 Under basal conditions, Nrf2 is bound to Keap1 in the cytosol. Upon activation, Nrf2 is released and then translocates into the nucleus, where it heterodimerizes, binds to the ARE, and subsequently increases Nrf2 target gene expression.39
The PON1 gene is mainly expressed in the liver and, albeit to a lower extent, also in the heart.40,41 Paraoxonase-1 (PON1) circulates in the blood bound to high-density lipoprotein (HDL) and prevents or delays the oxidation of low-density lipoprotein (LDL). High PON1 activity may be associated with a reduced cardiovascular disease risk. Paraoxonase-2 (PON2) is expressed ubiquitously42 and PON2 is primarily localized in the plasma membrane, where it prevents cell-mediated lipid peroxidation.43 PON1 has been suggested as a longevity gene due to its modulation of cardiovascular disease risk.44 However, in a recently conducted meta-analysis, no effects or only population-specific effects of PON1 on human longevity were found.45
It has been shown that hydroxytyrosol induces antioxidant/detoxificant enzymes and Nrf2 translocation in hepatocytes.46 Furthermore, hydroxytyrosol-induced cytoprotection against oxidative injury in vascular endothelial cells via Nrf2-dependent signal transduction pathway.23 Little is known about the role of olive oil phenolics on Nrf2-dependent gene expression in vivo. Furthermore, activation of SIRT1 by polyphenols may be beneficial for controlling of oxidative stress, cellular senescence, and metabolism.47
Overall, aging seems to be associated with changes in the oxidant/antioxidant equilibrium, impaired Nrf2 signaling and phase 2 response, and decreased SIRT1 activity. Therefore, the aim of the present feeding study in SAMP8 mice was to answer the question of whether a diet rich in olive oil phenolics may affect age-related changes in heart oxidant/antioxidant status, Nrf2-dependent gene expression, and SIRT1 status.
Materials and Methods
Chemicals and reagents
High-performance liquid chromatography (HPLC)-grade methanol was obtained from J.T. Baker (Deventer, The Netherlands), and acetonitrile was obtained from Sigma Aldrich (Steinheim, Germany). Standards of tyrosol (CAS no. 501-94-0), vanillic acid (CAS no. 121-34-6), caffeic acid (CAS no. 331-39-5), p-coumaric acid (CAS no. 501-98-4), and ferulic acid (CAS no. 1135-24-6) were purchased from Sigma Aldrich (Steinheim, Germany). Oleuropein (CAS no. 32619-42-4) and hydroxytyrosol (CAS no. 10597-60-1) were supplied by Extrasynthese (Genay Cedex, France) and pinoresinol by Separation Research (Turku, Finland). Reference standards of all analyzed compounds were HPLC grade, with purity higher than 98% (except oleuropein, >90%). Stock solutions were prepared in methanol and stored at −20°C. HPLC grade 60% perchloric acid was obtained from Fisher Scientific (Leicestershire, UK).
For the protein carbonyl quantification, potassium chloride, monopotassium phosphate, sodium chloride, sodium phosphate dibasic dihydrate, disodium phosphate, monosodium phosphate and guanidine hydrochloride, citric acid, sulfuric acid, hydrogen chloride, sodium hydroxide, bovine serum albumin, and Roti Quant® were purchased from Carl Roth GmbH (Karlsruhe, Germany). Tween 20, 2,4-dinitrophenylhydrazine, biotin-conjugated rabbit immunoglobulin G (IgG) polyclonal antibody raised against a dinitrophenol (DNP) conjugate of keyhole limpet hemocyanin (anti-DNP), streptavidin, biotinylated horseradish peroxidase, and o-phenylenediamine were from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). Hydrogen peroxide (H2O2) was obtained from Merck KGaA (Darmstadt, Germany).
Spectrophotometric determination of total phenols and HPLC analysis of olive oil phenolics
The total phenolic content of the extracts was determined according to the Folin–Ciocalteu spectrophotometric method.48 Diluted Folin–Ciocalteu reagent and 20% sodium carbonate solution were added to the phenolic extract. The solution was left in the dark for 45 min. The absorbance of each sample as well as of gallic acid standards was determined at 750 nm. The results were expressed as mg of gallic acid/kg of oil. HPLC analyses of olive oil phenolics were carried out on a Jasco system (Jasco GmbH Deutschland, Gross-Umstadt, Germany) consisting of a pump (PU-2085) and autosampler (XLC-3059AS), and detection was carried out on an eight-channel ESA 5600A CoulArray Detector with integrated column oven (ESA Inc., Chelmsford, MA). Separation was achieved by gradient elution with water (pH 3.1), methanol, and acetonitrile (all containing 60 mM LiClO4) as mobile phases on a Kinetex Luna C18 column (100×4.6 mm, 2.6 μm, Phenomenex, USA), and analytes were quantified against authentic compounds as external standards.
Ferric reducing ability of plasma and Trolox equivalent antioxidant capacity assays
The ferric reducing ability of plasma (FRAP) assay was conducted according to Benzie and Strain,49 using ascorbic acid as a reference. The Trolox equivalent antioxidant capacity (TEAC) assay was conducted as described by Re et al.,50 using Trolox as a reference.
Animals and study design
The animal experiment was performed according to German animal welfare laws and regulations and with permission of the Ministry of Agriculture, Environment and Rural Areas of the state of Schleswig-Holstein (Germany). Twenty-two female SAMP8 mice, aged 9–10 weeks, were obtained from Harlan Winkelmann GmbH (Borchen, Germany). The mice were housed in groups (3–5 animals per cage) in type II polypropylene cages equipped with soft wood bedding, a water bottle, a mouse house, and a table tennis ball in a climate-controlled room (temperature, 22±2°C; humidity, 55%±5%) with a 12-hr light/dark cycle.
Female mice were used because they can be housed in groups, which we considered superior to individual housing for an experimental period of 4.5 months. One of the SAMP8 mice died of natural causes during the experiment. Mice were randomly divided into two groups of 11 animals each and fed with a pelletized Western-type diet (Altromin Spezialfutter GmbH & Co. KG, Lage, Germany) with 0.15% cholesterol and 20% fat, in which 10% of fat was from olive oil containing either low (44 mg gallic acid/kg oil.) or high (532 mg gallic acid/kg oil; Fratelli Ferrara s.a.s, Italy) amounts of phenolics. Both oils contained 74%–75% oleic acid and 11% palmitic acid as determined by gas chromatography (LUFA-ITL GmbH, Kiel, Germany, DGF C VI 10a/11a). All animals had free access to feed and water. Feed intake was controlled daily, and body weights were measured weekly during the 4.5-month feeding trial. At the end of the experiment, the mice were starved overnight before anesthesia by CO2 and decapitation. Blood samples were collected in tubes and serum was separated by centrifugation (Eppendorf 5804 R, Rotor F34-6-38, Wesseling-Berzdorf, Germany). Tissues were excised, snap-frozen in liquid nitrogen, and stored at −80°C until analyzed.
Preparation of heart tissue homogenates
Heart tissue homogenates were prepared from ≈20 mg of tissue in 250 μL of ice-cold phosphate-buffered saline (PBS; pH 7.4) at 26,000 rpm with a Miccra D-8 homogenizer (ART Prozess- & Labortechnik GmbH & Co. KG, Mullheim, Germany). Homogenates were centrifuged at 4,800×g at 4°C for 10 min (Eppendorf 5804 R, Rotor F34-6-38, Wesseling-Berzdorf, Germany), and the supernatant was stored at −80°C until further use.
Lipid peroxidation
Lipid peroxidation in heart homogenates was assayed fluorometrically as thiobarbituric acid-reactive substances (TBARS) with a Tecan Infinite 200 microplate reader (Tecan Group Ltd., Crailsheim, Germany) after protein precipitation with trichloroacetic acid (TCA) and extraction in 1-butanol. Excitation and emission wavelengths were 520 nm and 560 nm, respectively. A calibration curve was prepared with 1,1,3,3-tetraethoxypropane (TEP) as an external standard.51
Quantification of protein carbonyls
Protein carbonyl content was determined in the homogenized heart tissue supernatant according to the enzyme-linked immunosorbent assay (ELISA) method of Buss et al.52 with required modifications. The detection system used an anti-dinitrophenyl rabbit IgG-antiserum (Sigma Aldrich, Steinheim, Germany) as the primary antibody and a monoclonal antirabbit IgG antibody peroxidase conjugate (Sigma Aldrich, Steinheim, Germany) as the secondary antibody. Color change was induced with o-phenylenediamine and H2O2.
Proteasomal activity
Tissue (20–40 mg) was homogenized in lysis buffer (250 mM sucrose, 25 mM HEPES, 10 mM magnesium chloride, 1 mmol/L EDTA, and 1.7 mmol/L dithiothreitol [DTT]) using a homogenizer (Ultra-Turrax®) and then centrifuged at 14,000×g for 30 min. The supernatant was used for determination of protein content using the Bradford assay and for measurement of the proteasomal activity. For proteasomal activity, samples were incubated in 225 mmol/L Tris buffer (pH 7.8), 45 mmol/L potassium chloride, 7.5 mmol/L magnesium acetate, 7.5 mmol/L magnesium chloride, and 1 mmol/L DTT. For the peptidyl-glutamyl like-(β1), trypsin like-(β2), and chymotrypsin like-(β5) activity the substrates Z-Leu-Leu-Glu-MCA (Biochem, Boston, MA), Ac-Arg-Leu-Arg-MCA (Biochem, Boston, MA), and N-succinyl-Leu-Leu-Val-Tyr-MCA (Sigma Aldrich, Steinheim, Germany) were used, respectively. MCA liberation of the substrates was measured with a fluorescence reader at 360 nm excitation and 460 nm emmission. Free MCA was used as a standard.53
PON1 arylesterase activity in serum
Arylesterase activity was measured by using phenylacetate as an artificial substrate for PON1. Initial rates of hydrolysis were determined spectrophotometrically at 270 nm. The assay mixture included 4 mmol/L of phenylacetate and 1 mmol/L of CaCl2 in 20 mmol/L of Tris HCl, pH 8.0. Nonenzymatic hydrolysis of phenylacetate was subtracted from the total rate of hydrolysis. One unit of arylesterase activity is equal to 1 μmol of phenylacetate hydrolyzed per minute per milliliter.54
RNA isolation and real-time quantitative RT-PCR
RNA was isolated from heart samples (20–30 mg) using TRIsure lysis reagent (Bioline, Luckenwalde, Germany). Real-time quantitative PCR was performed as a one-step procedure (SensiMix One-step Kit; Quantace, Berlin, Germany) with SYBR Green detection using a Rotorgene cycler (Corbett Life Science, Sydney, Australia). Relative messenger RNA (mRNA) concentrations of genes were quantified by the use of a standard curve. Target gene mRNA concentration was normalized to the mRNA concentration of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers were designed by standard tools (Spidey, Primer3 and National Center for Biotechnology Information [NCBI Blast]) and purchased from MWG (Ebersberg, Germany). Primer sequences for analyzed genes are summarized in Table 1.
Table 1.
Primers Used in Real-Time PCR Experiments
| Primers | Sequence | Annealing temperature (°C) |
|---|---|---|
| GAPDH | F: GACAGGATGCAGAAGAGATTACT R: TGATCCACATCTGCTGGAAGGT |
55 |
| γ-GCS | F: AACACAGACCCAACCCAGAG R: GTCTGGCTGAGAAGCCTTTG |
59 |
| GST | F: TACTTTGATGGCAGGGGAAG R: TCATCCCGTCGATCTCTACC |
58 |
| NQO1 | F: TTCTTCTGGCCGATTCAGAGT R: TCCAGACGTTTCTTCCATCC |
55 |
| Nrf2 | F: GGGGACAGAATCACCATTTG R: GATGCAGGCTGACATTCTGA |
57 |
| PON2 | F: ATGGTGGCTCTGAGTTTGCT R: TCCTCAGCTCCAGTTTCGAT |
57 |
| SIRT1 | F: GTCTCCTGTGGGATTCCTGA R: ACACAGAGACGGCTGGAACT |
57 |
GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; γ-GCS, γ-glutamyl cysteine synthetase; GST, glutathione-S-transferase; NQO1, nicotinamide adenine dinucleotide phosphate [NAD(P)H]:quinone oxidoreductase; PON2, paraoxonase-2; F, forward; R, reverse.
Cell culture
NIH 3T3 fibroblasts (German collection of microorganisms and cell cultures, Braunschweig, Germany) were maintained in Dulbecco modified Eagle medium (DMEM) containing 4.5 g/L glucose, 4 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, 10% fetal calf serum (FCS), 100 U/mL penicillin and 100 μg/mL streptomycin (PAA, Coelbe, Germany). Cells were grown in 5% CO2 at 37°C under a humidified atmosphere. NIH 3T3 cells comprise a nontransformed standard fibroblast cell line that exhibits robust growth patterns and are easily transfected, thereby delivering reliable reporter gene data. Furthermore, we have used this cell line because of its murine origin to be consistent with our mouse study. Although NIH 3T3 cells do not fully reflect the cellular metabolism of cardiomyocytes, they comprise a valuable experimental tool to screen for Nrf2-inducing activity of plant bioactives.55
All cell culture plasticware was purchased from Sarstedt (Nuembrecht, Germany) unless otherwise stated. For cell culture experiments, the olive oil phenolics were dissolved in dimethylsulfoxide (DMSO; Carl Roth, Karlsruhe, Germany), and stock solutions were stored at −80°C until usage. Vehicle controls have been performed, and the solvents did not affect any of the parameters measured.
Cytotoxicity measurement
Cytotoxicity was determined via the Neutral Red assay.56 The Neutral Red assay is based on the pH-dependent accumulation of Neutral Red in the lysosomes of viable cells.56 NIH 3T3 cells were seeded in 24-well plates (Fisher Scientific, Schwerte, Germany) at a density of 50,000 cells/well, precultured for 24 hr, and treated with the olive oil phenolics for 24 hr, respectively. In brief, the culture medium containing the olive oil phenolics was replaced with fresh serum-containing medium including 50 μg/mL of Neutral Red (Carl Roth). After incubation for 3 hr, the medium was removed and the cells were extracted using a solution comprising 50:49:1 (vol/vol/vol) ethanol, water, and glacial acetic acid. The absorbance was measured in a plate reader (Labsystems, Helsinki, Finland) at 540 nm. For all subsequent experiments, noncytotoxic concentrations were used.
Nrf2 reporter gene assay
NIH 3T3 cells were grown to 60%–80% confluence in 24-well plates for 24 hr. The cells were transiently transfected with an expression vector containing a 25-bp oligonucleotide derived from the promoter region of the gastrointestinal glutathione peroxidase 2 comprising the conserved ARE-motif and the reporter gene firefly luciferase (pARE_GIGPx) kindly provided by A. Banning and R. Brigelius-Flohé (DIfE, Potsdam-Rehbruecke, Germany)57 and a normalization vector phRL-TK (Promega, Mannheim, Germany) containing the Renilla reniformis luciferase gene. Transfection was performed using JetPEI transfection reagent (Polyplus transfection, Illkirch Cedex, France) according to manufacturer's instructions. Following 24 hr of transfection, cells were incubated with the test compounds for 24 hr in serum-containing medium. Subsequently, cells were lysed, and luciferase activity was measured using the Dual-Luciferase reporter gene assay system (Promega) according to the manufacturer's protocol in a Tecan Infinite 200 microplate reader (Tecan Group Ltd, Crailsheim, Germany).
Nrf2 western blot
For Nrf2 detection in nucleic fractions, NIH 3T3 cells were treated with test compounds for 6 hr. Subsequently, cells were washed with ice-cold PBS, scraped off, and centrifuged. For whole-cell extracts, the remaining cell pellet was stored at −80°C, whereas nuclear extracts were prepared subsequently. Samples for western blotting were prepared as described in Wagner et al.58 A quantity of 30 μg of protein of each sample were mixed with loading buffer, incubated at 95°C for 5 min and separated on a 12% sodium dodecyl sulfate polyacrylamide gel. Subsequently, the samples were transferred onto a polyvinylidene fluoride membrane and blocked with 3% (wt/vol) skim milk dissolved in Tris-buffered saline plus 0.05% (vol/vol) Tween 20 for at least 2 hr and probed with the respective antibody against Nrf2 (Santa Cruz; 1:200) or TATA-binding protein (TBP; 1:200, Santa Cruz) at 4°C overnight. Subsequently, the membranes were incubated with a secondary antibody (1:4,000) anti-rabbit (Bio-Rad, Munich, Germany) for 1 hr, and the bands were visualized by using enhanced chemilumescent reagent (Thermo Scientific) in a ChemiDoc XRS system (BioRad). Molecular weight of the protein bands was estimated using a western C protein standard (Bio-Rad).
PON1 reporter gene assay
We selected HuH7 liver hepatoma cells for the PON1 promoter activity studies because PON1 is mainly synthesized in the liver and, to a lower extent, in the heart.40,41 HuH7 liver hepatoma cells of human origin stably transfected with a 1,000-bp fragment of the human PON1 promoter (PON1-HuH7; originating from X. Coumoul, INSERM, France) were cultivated in DMEM with 10% heat-inactivated FCS, 100 U/mL streptomycin, and 100 mg/mL penicillin (all from PAA, Coelbe, Germany). PON1-HuH7 cells were seeded at an initial density of 150,000 cells per well (24-well plate) and incubated with the olive oil phenolics hydroxytyrosol, tyrosol, oleuropein, pinoresinol, caffeic acid, p-coumaric acid, and vanillic acid and resveratrol (used as a positive control), for 48 hr as described recently.59 Afterward, the cells were washed with PBS, lysed, and subjected to luciferase activity measurement as described above.
Statistical analysis
Results are expressed as mean values with standard error of the mean (SEM). Statistical analysis was performed using PASW Statistics 18 (IBM, Chicago, IL). Data were analyzed for normality of distribution (Kolmogorow–Smirnov and Shapiro–Wik tests) and equality of variance (Levene test) before the t-test for independent samples and, in the case of nonparametric data, Mann–Whitney U-test. Differences were considered significant when the p value was ≤0.05.
Results
The two olive oils used in this study differed notably in their content of the major olive oil phenolics hydroxytyrosol, tyrosol, oleuropein, and pinoresionol, and this was reflected in the corresponding FRAP and TEAC values of the two oils (Table 2). Importantly, the HP olive oil contained 12-fold higher concentrations of total phenolics than the LP olive oil. However, α-tocopherol concentrations were similar in both oils (Table 2).
Table 2.
Concentrations of Phenols and TEAC and FRAP Values of the High-Polyphenol and Low-Polyphenol Olive Oils
| High-polyphenol olive oil | Low-polyphenol olive oil | |
|---|---|---|
| Phenolic compounds (mg/kg oil) | ||
| Tyrosol | 20.78 | 7.03 |
| Hydroxytyrosol | 18.87 | 0.54 |
| Pinoresinol | 3.56 | 1.85 |
| Oleuropein | 1.33 | 1.19 |
| Caffeic acid | 0.56 | 0.91 |
| Vanillic acid | 0.34 | 0.16 |
| p-Coumaric acid | 0.13 | 0.20 |
| Ferulic acid | 0.09 | 0.08 |
| α-Tocopherol | 217 | 197 |
| Total phenolics (mg GA equivalent/kg oil) | 532 | 44 |
| FRAP (mmol AA equivalent/kg oil) | 1.27 | 0.17 |
| TEAC (mmol trolox equivalent/kg oil) | 1.01 | 0.11 |
TEAC, Trolox equivalent antioxidant capacity; FRAP, ferric reducing ability of plasma; GA, .
Mean daily feed intake was similar in mice fed the LP (3.13±0.06 g/day) and HP (3.26±0.47 g/day) diets. At the end of the 4.5-month feeding trial, no differences in the final body weight of HP mice (27.2±1.4 g) compared to LP (29.1±1.3 g) mice were observed.
To determine whether olive oil phenolics may affect lipid and protein oxidation, TBARS and protein carbonyl concentrations were determined in heart tissue (Table 3). Feeding the HP diet resulted in significantly lower TBARS and protein carbonyl concentrations in heart tissue relative to the LP diet. Because mice fed the HP diet exhibited reduced heart protein carbonyl concentrations, we determined whether this was related to differences in proteasomal activity. However, proteasomal activities of the subunits β-1, β-2, and β-5 in heart tissue remained unchanged by the different dietary treatments (Table 3).
Table 3.
Concentrations of TBARS and Protein Carbonyls As Well As Proteasomal Activities in Heart Homogenates from SAMP8 Mice Fed for 4.5 Months Diets with 10% High-Polyphenol or Low-Polyphenol Olive Oil
| Low-polyphenol diet | High-polyphenol diet | p | |
|---|---|---|---|
| TBARS (nmol/g tissue) | 428±28 | 274±22a | 0.0002 |
| Protein carbonyls (nmol/g protein) | 958±70 | 664±37 | 0.004 |
| Proteasomal activity (μmol/mg·min) | |||
| β-1 subunit | 26.1±1.1 | 31.7±3.4 | 0.173 |
| β-2 subunit | 8.2±0.4 | 6.9±0.4 | 0.060 |
| β-5 subunit | 90.3±9.1 | 79.3±11.5 | 0.357 |
Values are expressed as mean±standard error of the mean (SEM) (n=10).
Means are significantly different at the given p value.
TBARS, Thiobarbituric acid-reactive substances; SAMP8, senescence-accelerated mouse-prone 8.
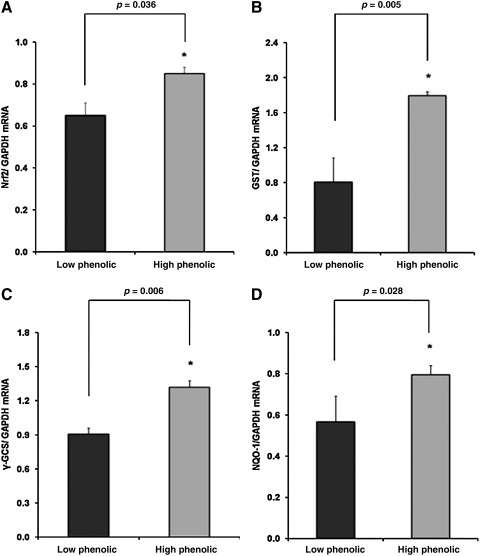
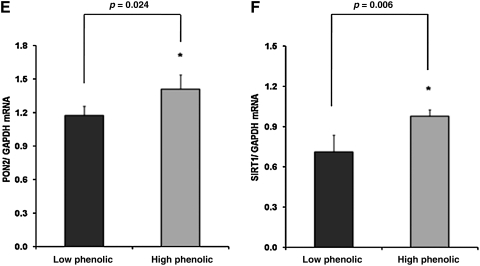
The transcription factor Nrf2 plays a pivotal role in antioxidant and phase 2 defense mechanisms. Impaired Nrf2 signaling may be associated with increased oxidative stress. Because Nrf2 controls its own gene expression, Nrf2 mRNA levels were measured. SAMP8 mice receiving the LP diet exhibited significantly lower Nrf2 mRNA levels as compared to mice receiving the HP diet (Fig. 1A). Nrf2 controls the gene expression of GST, γ-GCS, NQO1, and PON2. Interestingly, differences in Nrf2 expression between the two groups were also reflected in terms of differences in GST (Fig. 1B), γ-GCS (Fig. 1C), NQO1 (Fig. 1D), and PON2 (Fig. 1E) mRNA. Thus, feeding the HP diet resulted in significantly elevated mRNA levels of genes encoding for antioxidant and phase 2 enzymes in the heart of our mice. Recently, it has been shown that there is crosstalk between SIRT1 and Nrf2 because SIRT1 was reduced in Nrf2−/− murine fibroblasts.60 In the present study, the increase in Nrf2 and Nrf2-dependent gene expression due to the HP diet was associated with a significant increase in heart SIRT1 mRNA levels (Fig. 1F).
FIG. 1.
Relative messenger RNA (mRNA) expression (normalized for glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) of Nrf2 (A), glutathione-S-transferase (GST) (B), γ-glutamyl cysteine synthetase (γ-GCS) (C), nicotinamide adenine dinucleotide phosphate [NAD(P)H]:quinone oxidoreductase 1 (NQO1) (D), paraoxonase-2 (PON2) (E), and SIRT-1 (F) in hearts of senescence-accelerated mouse-prone 8 (SAMP8) mice fed for 4.5 months a Western type diet with 0.15% cholesterol and 20% fat, in which 10% of fat was from olive oil containing either low or high amount of phenolics. Mice were killed at 7 months of age. Values are expressed as mean±standard error of the mean (SEM) (n=10). * indicates statistical significant differences between groups.
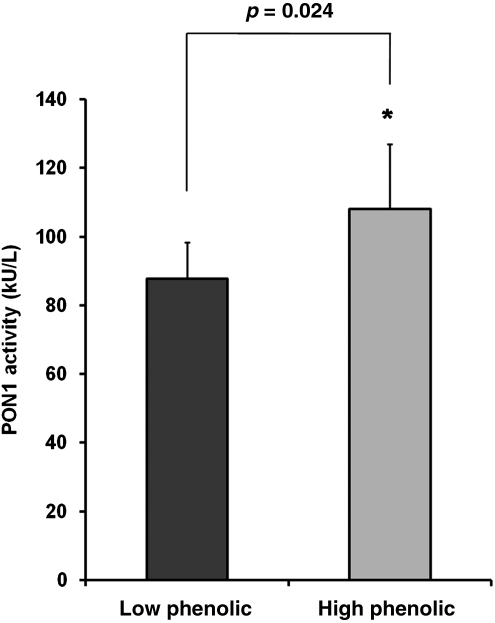
Apart from PON2 gene expression in the heart, we looked also into serum PON1 activity levels in our mice in response to dietary supplementation with LP and HP olive oil. Mice receiving the LP diet exhibited significantly lower PON1 activity levels in serum as compared to those receiving the HP diet (Fig. 2).
FIG. 2.
Mean (±standard error of the mean [SEM]) paraoxonase-1 (PON1) activity in serum of senescence-accelerated mouse-prone 8 (SAMP8) mice fed a Western type diet with 0.15% cholesterol and 20% fat, in which 10% of fat was from olive oil containing either low or high amount of phenolics (n=9). *indicates statistical significant differences between groups.
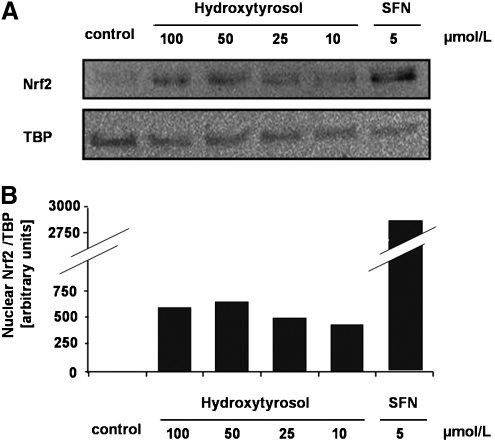
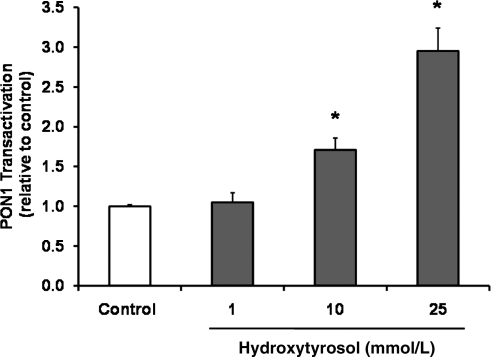
To determine which phenol present in olive oil may have induced Nrf2-dependent gene expression and the increase in PON status, additional cell culture studies in NIH 3T3 and HuH7 cells were performed. In total, we tested seven different phenolic compounds present in olive oil, including hydroxytyrosol, tyrosol, oleuropein, pinoresinol, caffeic acid, p-coumaric acid, and vanillic acid, for their ability to affect Nrf2 and PON1 transactivation. Under the conditions investigated, only hydroxytyrosol, but not tyrosol, oleuropein, pinoresinol, caffeic acid, p-coumaric acid, or vanillic acid, increased Nrf2 transactivation (Table 4). Furthermore, we determined nuclear Nrf2 protein levels in NIH 3T3 cells and found an increase in nuclear Nrf2 in hydroxytyrosol-treated cells (Fig. 3). Also, treatment of HuH7 cells with hydroxytyrosol resulted in a dose-dependent increase in PON1 transactivation (Fig. 4).
Table 4.
Nrf2 Transactivation in NIH 3T3 Cells Incubated with 25 μmol/L of the Respective Olive Oil Phenolics
| Test component | Fold Nrf2 transactivation |
|---|---|
| Control | 1.00±0.12a |
| Tyrosol | 0.97±0.10a |
| Hydroxytyrosol | 1.48±0.10b |
| Pinoresinol | 1.03±0.06a |
| Oleuropein | 1.05±0.05a |
| Caffeic acid | 1.20±0.03a |
| Vanillic acid | 1.03±0.07a |
| p-Coumaric acid | 0.96±0.07a |
Mean values (±SEM) with different superscript letters are significantly different (p<0.05).
FIG. 3.
Representative western blot (including densitometry) of Nrf2 in NIH 3T3 nuclear cell extracts in response to hydroxytyrosol following 6 hr of incubation with increasing concentrations of the test compound. Sulforaphane (SFN) was used as positive and TATA-binding protein (TBP) as a loading control. In densitometric analysis, the control equals one arbitrary unit.
FIG. 4.
Induction of PON1 transactivation by hydroxytyrosol in stably transfected HuH7 liver cells. Values are expressed as mean±standard error of the mean (SEM) of three independent experiments performed in triplicate. * indicates statistical significant differences between treatment and control (p<0.05).
Discussion
In the present study, feeding a HP diet reduced biomarkers of oxidative stress, induced Nrf2, and increased SIRT1 gene expression in the heart of SAMP8 mice compared to animals fed a LP diet. Mice fed the HP diet had lower concentrations of TBARS and protein carbonyls in the heart than mice on the LP diet (Table 3). However, proteasomal activity was not changed. Thus, differences in heart protein carbonyl levels were independent of proteasomal activity. In a comparable study with SAMP8 mice, neither curcumin nor Ginkgo biloba extract, when fed for 5 months, reduced protein carbonyl concentrations in the heart.61
On the basis of the present data, it is suggested that differences in the oxidant/antioxidant status between the LP and HP groups may be partly due to differences in Nrf2-dependent gene expression. The latter drives the expression of genes encoding phase 2 and antioxidant enzymes. However, it is unclear which phenols present in olive oil may have improved antioxidant status and Nrf2-dependent gene expression in our mice. Because we found an induction of GST, γ-GCS, NQO1, and PON in response to the HP diet, we performed additional cell culture studies with purified compounds to investigate which particular olive polyphenol may have induced Nrf2-dependent gene expression. Our cell culture data in NIH 3T3 and HuH7 cells suggest that hydroxytyrosol may have mediated the induction of Nrf2-dependent gene expression and the increase in PON status in our mice. Our data in NIH 3T3 cells are in accordance with a previous study by Martín et al.,46 demonstrating an induction of Nrf2 activity by hydroxtyrosol in HepG2 cells. Interestingly, in the current study, hydroxytyrosol, but not tyrosol, resulted in an induction of Nrf2 transactivation and PON1 activity. Thus, the presence of the 3-hydroxyl group may be an important structural determinant as far as Nrf2 and PON1 induction are concerned. In our HuH7 cell culture studies, we used hydroxytyrosol concentration up to 25 μmol/L. This hydroxytyrosol concentration is similar to those reported in human plasma after the consumption of 40 mL of olive oil62 or 20 olives.63 However, in other human studies, hydroxytyrosol concentrations in plasma following olive oil consumption were in the lower micromolar range.64
In the present cell culture and mouse studies, we also observed an induction of PON1 and PON2 due to hydroxytyrosol and a HP diet, respectively. PON2 prevents cell-mediated lipid peroxidation in the heart.65 Thus, the decreased lipid peroxidation levels in the heart of our mice in response to the HP diet may be partly mediated by PON2-induction.
Mice fed the HP diet exhibited an improved PON1 status. In a human study, plasma oxidized LDL concentrations were negatively correlated with the phenol content of LDL in men who ingested different olive oils.66 Because PON1 prevents oxidation of LDL, lower oxidized LDL concentrations might be due to PON1 induction.
In a previous study, SIRT gene expression and protein levels were decreased in SAMP8 mice compared to normal aging mice.67 Furthermore, a moderate induction of SIRT retarded aging of the heart and induced resistance to oxidative stress.68 Thus, the decreased concentrations of biomarkers of oxidative stress in mice fed the HP diet might be related to the moderate SIRT1 induction observed in these animals.
Our results suggest that a diet rich in olive oil phenolics reduces oxidative stress in the heart of SAMP8 mice, most likely by induction of Nrf2-dependent gene expression. The content of hydroxytyrosol seems to be of importance for the potential health benefits of olive oils. The present data from studies with cultured cells and mice need to be confirmed in humans. Ultimately, studies in other model organisms are warranted to determine whether the induction of Nrf2-dependent gene expression by hydroxytyrosol-rich olive oil may also affect life span.
Overall, a Mediterranean diet rich in olive oil may be an important cornerstone in the prevention of age-dependent chronic diseases such as coronary heart disease. Therefore, dietary recommendations, as far as healthy aging is concerned, should promote an increased consumption of olive oil.69 Health benefits of olive oil may be partly attributed to its content of olive oil phenolics. Thus, olive producers as well as the food industry are encouraged to establish means by which they may increase the phenolic content of olives and derived products.
Acknowledgments
B.B. is supported by TUBITAK (The Scientific and Technological Research Council of Turkey). J.F. is supported by grant no. FR 2478/4-1 from the German Research Foundation (DFG) and grant no. 0315679A from the German Federal Ministry of Education and Research (BMBF). A.E.W. is supported by the DFG Clusters of Excellence “Inflammation of Interfaces.” T.G. and G.R. are supported by the Federal Ministry of Education and Research (BMBF) and the DFG. C.S. is supported by a grant of the Christian-Albrechts University of Kiel.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Fung TT. Rexrode KM. Mantzoros CS. Manson JE. Willett WC. Hu FB. Mediterranean diet and incidence and mortality of coronary heart disease and stroke in women. Circulation. 2009;119:1093–1100. doi: 10.1161/CIRCULATIONAHA.108.816736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perona JS. Covas MI. Fitó M. Cabello-Moruno R. Aros F. Corella D. Rose E. Garcia M. Estruch R. Martinez-Gonzalez MA. Ruiz-Gutierrez V. Reduction in systemic and VLDL triacylglycerol concentration after a 3-month Mediterranean-style diet in high-cardiovascular-risk subjects. J Nutr Biochem. 2010;21:892–898. doi: 10.1016/j.jnutbio.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Martínez-González MA. García-López M. Bes-Rastrollo M. Toledo E. Martínez-Lapiscina EH. Delgado-Rodriguez M. Vazquez Z. Benito S. Beunza JJ. Mediterranean diet and the incidence of cardiovascular disease: A Spanish cohort. Nutr Metab Cardiovas Dis. 2011;21:237–244. doi: 10.1016/j.numecd.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Alonso A. Martínez-González MA. Olive oil consumption and reduced incidence of hypertension: The SUN study. Lipids. 2004;39:1233–1238. doi: 10.1007/s11745-004-1352-x. [DOI] [PubMed] [Google Scholar]
- 5.Esposito K. Marfella R. Ciotola M. Di Palo C. Giugliano F. Giugliano G. D'Armiento M. D'Andrea F. Giugliano D. Effect of a Mediterranean-style diet on endothelial dysfunction and markers of vascular inflammation in the metabolic syndrome: A randomized trial. JAMA. 2004;292:1440–1446. doi: 10.1001/jama.292.12.1440. [DOI] [PubMed] [Google Scholar]
- 6.Estruch R. Martínez-González MA. Corella D. Salas-Salvadó J. Ruiz-Gutiérrez V. Covas MI. Fiol M. Gómez-Gracia E. López-Sabater MC. Vinyoles E. Arós F. Conde M. Lahoz C. Lapetra J. Sáez G. Ros E. Effects of a Mediterranean style diet on cardiovascular risk factors: A randomized trial. Ann Intern Med. 2006;145:1–11. doi: 10.7326/0003-4819-145-1-200607040-00004. [DOI] [PubMed] [Google Scholar]
- 7.Kontogianni MD. Panagiotakos DB. Chrysohoou C. Pitsavos C. Zampelas A. Stefanadis C. The impact of olive oil consumption pattern on the risk of acute coronary syndromes: The CARDIO2000 case-control study. Clin Cardiol. 2007;30:125–129. doi: 10.1002/clc.20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covas MI. Olive oil and cardiovascular health. J Cardiovasc Pharmacol. 2009;54:477–482. doi: 10.1097/FJC.0b013e3181c5e7fd. [DOI] [PubMed] [Google Scholar]
- 9.Visioli F. Galli C. Olive oil: more than just oleic acid. Am J Clin Nutr. 2000;72:853–856. doi: 10.1093/ajcn/72.3.853. [DOI] [PubMed] [Google Scholar]
- 10.Fitó M. Cladellas M. de la Torre R. Martí J. Alcántara M. Pujadas-Bastardes M. Marrugat J. Bruguera J. López-Sabater MC. Vila J. Covas MI. Antioxidant effect of virgin olive oil in patients with stable coronary heart disease: A randomized, crossover, controlled, clinical trial. Atherosclerosis. 2005;181:149–158. doi: 10.1016/j.atherosclerosis.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Bogani P. Galli C. Villa M. Visioli F. Postprandial anti-inflammatory and antioxidant effects of extra virgin olive oil. Atherosclerosis. 2007;190:181–186. doi: 10.1016/j.atherosclerosis.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Visioli F. Bellosta S. Galli C. Oleuropein, the bitter principle of olives, enhances nitric oxide production by mouse macrophages. Life Sci. 1998;62:541–546. doi: 10.1016/s0024-3205(97)01150-8. [DOI] [PubMed] [Google Scholar]
- 13.Manna C. Galletti P. Cucciolla V. Montedoro G. Zappia V. Olive oil hydroxytyrosol protects human erythrocytes against oxidative damages. J Nutr Biochem. 1999;10:159–165. doi: 10.1016/s0955-2863(98)00085-0. [DOI] [PubMed] [Google Scholar]
- 14.Manna C. D'angelo S. Migliardi V. Loffredi E. Mazzoni O. Morrica P. Galletti P. Zappia V. Protective effect of the phenolic fraction from virgin olive oils against oxidative stress in human cells. J Agric Food Chem. 2002;50:6521–6526. doi: 10.1021/jf020565+. [DOI] [PubMed] [Google Scholar]
- 15.Turner R. Etiene N. Garcia-Alonso M. De Pascual-Teresa S. Minihane AM. Weinberg PD. Rimbach G. Antioxidant and anti-atherogenic activities of olive oil phenolics. Int J Vit Nutr Res. 2005;75:61–70. doi: 10.1024/0300-9831.75.1.61. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X. Cao J. Zhon L. Hydroxytyrosol inhibits pro-inflammatory cytokines, iNOS, and COX-2 expression in human monocytic cells. Naunyn-Schmied Arch Pharmacol. 2009;379:581–586. doi: 10.1007/s00210-009-0399-7. [DOI] [PubMed] [Google Scholar]
- 17.Deiana M. Corona G. Incani A. Loru D. Rosa A. Atzeri A. Melis MP. Dessì MA. Protective effect of simple phenols from extra virgin olive oil against lipid peroxidation in intestinal Caco-2 cells. Food Chem Toxicol. 2010;48:3008–3016. doi: 10.1016/j.fct.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 18.Abe R. Beckett J. Abe R. Nixon A. Rochier A. Yamashita N. Sumpio B. Olive oil polyphenol oleuropein inhibits smooth muscle cell proliferation. Eur J Vasc Endovasc Surg. 2011;41:814–820. doi: 10.1016/j.ejvs.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Cumaoglu A. Ari N. Kartal M. Karasu C. Polyphenolic extracts from Olea europea L. protect against cytokine induced β-cell damage through maintenance of redox homeostasis. Rejuvenation Res. 2011;14:325–334. doi: 10.1089/rej.2010.1111. [DOI] [PubMed] [Google Scholar]
- 20.Coni E. Di Benedetto R. Di Pasquale M. Masella R. Modesti D. Mattei R. Carlini EA. Protective effect of oleuropein, an olive oil biophenol, on low density lipoprotein oxidizability in rabbits. Lipids. 2000;35:45–54. doi: 10.1007/s11745-000-0493-2. [DOI] [PubMed] [Google Scholar]
- 21.González-Santiago M. Martín-Bautista E. Carrero JJ. Fonollá J. Baró L. Bartolomé MV. Gil-Loyzaga P. López-Huertas E. One-month administration of hydroxytyrosol, a phenolic antioxidant present in olive oil, to hyperlipemic rabbits improves blood lipid profile, antioxidant status and reduces atherosclerosis development. Atherosclerosis. 2006;188:35–42. doi: 10.1016/j.atherosclerosis.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Jacomelli M. Pitozzi V. Zaid M. Larrosa M. Tonini G. Martini A. Urbani S. Taicchi A. Servili M. Dolara P. Giovannelli L. Dietary extra-virgin olive oil rich in phenolic antioxidants and the aging process: Long-term effects in the rat. J Nutr Biochem. 2010;21:290–296. doi: 10.1016/j.jnutbio.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Zrelli H. Matsuoka M. Kitazaki S. Araki M. Kusunoki M. Zarrouk M. Miyazaki H. Hydroxytyrosol induces proliferation and cytoprotection against oxidative injury in vascular endothelial cells: Role of Nrf2 activation and HO-1 induction. J Agric Food Chem. 2011;59:4473–4482. doi: 10.1021/jf104151d. [DOI] [PubMed] [Google Scholar]
- 24.Chiba Y. Shimada A. Kumagai N. Yoshikawa K. Ishii S. Furukawa A. Takei S. Sakura M. Kawamura N. Hosokawa M. The senescence-accelerated mouse (SAM): A higher oxidative stress and age-dependent degenerative diseases model. Neurochem Res. 2009;34:679–687. doi: 10.1007/s11064-008-9812-8. [DOI] [PubMed] [Google Scholar]
- 25.Rebrin I. Zicker S. Wedekind KJ. Paetau-Robinson I. Packer L. Sohal RS. Effect of antioxidant-enriched diets on glutathione redox status in tissue homogenates and mitochondria of the senescence-accelerated mouse. Free Radic Biol Med. 2005;39:549–557. doi: 10.1016/j.freeradbiomed.2005.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forman K. Vara E. Garcia C. Ariznavarreta C. Escames G. Tresguerres JAF. Cardiological aging in SAM model: Effect of chronic treatment with growth hormone. Biogerontology. 2010;11:275–286. doi: 10.1007/s10522-009-9245-z. [DOI] [PubMed] [Google Scholar]
- 27.Matsugo S. Kitagawa T. Minami S. Esashi Y. Oomura Y. Tokumaru S. Kojo S. Matsushima K. Sasaki K. Age-dependent changes in lipid peroxide levels in peripheral organs, but not in brain, in senescence-accelerated mice. Neurosci Lett. 2000;278:105–108. doi: 10.1016/s0304-3940(99)00907-6. [DOI] [PubMed] [Google Scholar]
- 28.Yasui F. Ishibashi M. Matsugo S. Kojo S. Oomura Y. Sasaki K. Brain lipid hydroperoxide level increases in senescence-accelerated mice at an early age. Neurosci Lett. 2003;350:66–68. doi: 10.1016/s0304-3940(03)00827-9. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez MI. Escames G. López LC. López A. García JA. Ortiz F. Sánchez V. Romeu M. Acuña-Castroviejo D. Improved mitochondrial function and increased lifespanafter chronic melatonin treatment in senescent prone mice. Exp Gerontol. 2008;43:749–756. doi: 10.1016/j.exger.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Okatani Y. Wakatsuki A. Reiter RJ. Miyahara Y. Melatonin reduces oxidative damage of neural lipids and proteins in senescence-accelerated mouse. Neurobiol Aging. 2002;23:639–644. doi: 10.1016/s0197-4580(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 31.Nabeshi H. Oikawa S. Inoue S. Nishino K. Kawanishi S. Proteomic analysis for protein carbonyl as an indicator of oxidative damage in senescence-accelerated mice. Free Radic Res. 2006;40:1173–1181. doi: 10.1080/10715760600847580. [DOI] [PubMed] [Google Scholar]
- 32.Caballero B. Vega-Naredo I. Sierra V. Huidobro-Fernández C. Soria-Valles C. De Gonzalo-Calvo D. Tolivia D. Gutierrez-Cuesta J. Pallas M. Camins A. Rodríguez-Colunga MJ. Coto-Montes A. Favorable effects of a prolonged treatment with melatonin on the level of oxidative damage and neurodegeneration in senescence-accelerated mice. J Pineal Res. 2008;45:302–311. doi: 10.1111/j.1600-079X.2008.00591.x. [DOI] [PubMed] [Google Scholar]
- 33.Carretero M. Escames G. López LC. Venegas C. Dayoub JC. García L. Acuňa-Castroviejo D. Long-term melatonin administration protects brain mitochondria from aging. J Pineal Res. 2009;47:192–200. doi: 10.1111/j.1600-079X.2009.00700.x. [DOI] [PubMed] [Google Scholar]
- 34.Fenton M. Huang HL. Hong Y. Hawe E. Kurz DJ. Erusalimsky JD. Early atherogenesis in senescence-accelerated mice. Exp Gerontol. 2004;39:115–122. doi: 10.1016/j.exger.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Takeda T. Hosokawa M. Higuchi K. Senescence-accelerated mouse (SAM): A novel murine model of senescence. Exp Gerontol. 1997;32:105–109. doi: 10.1016/s0531-5565(96)00036-8. [DOI] [PubMed] [Google Scholar]
- 36.Lewis KN. Mele J. Hayes JD. Buffenstein R. Nrf2, a guardian of health span and gatekeeper of species longevity. Integr Comp Bio. 2010;50:829–843. doi: 10.1093/icb/icq034. : [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sykioti GP. Habeos IG. Samuelson AW. Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr Opin Clin Nutr Metab Care. 2011;14:41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaspar JW. Niture SK. Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 40.Parker-Katiraee L. Bousiaki E. Monk D. Moore GE. Nakabayashi K. Scherer SW. Dynamic variation in allele-specific gene expression of Paraoxonase-1 in murine and human tissues. Hum Mol Genet. 2008;17:3263–3270. doi: 10.1093/hmg/ddn222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackness B. Beltran-Debon R. Aragones G. Joven J. Camps J. Mackness M. Human tissue distribution of paraoxonases 1 and 2 mRNA. IUBMB Life. 2010;62:480–482. doi: 10.1002/iub.347. [DOI] [PubMed] [Google Scholar]
- 42.Hegele RA. Paraoxonase genes and disease. Ann Med. 1999;31:217–224. doi: 10.3109/07853899909115981. [DOI] [PubMed] [Google Scholar]
- 43.Precourt LP. Amre D. Denis MC. Lavoie JC. Delvin E. Seidman E. Levy E. The three-gene paraoxonase family: Physiologic roles, actions and regulation. Atherosclerosis. 2011;214:20–36. doi: 10.1016/j.atherosclerosis.2010.08.076. [DOI] [PubMed] [Google Scholar]
- 44.Lescai F. Marchegiani F. Franceschi C. PON1 is a longevity gene: Results of a meta-analysis. Ageing Res Rev. 2009;8:277–284. doi: 10.1016/j.arr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Caliebe A. Kleindorp R. Blanché H. Christiansen L. Puca AA. Rea IM. Slagboom E. Flachsbart F. Christensen K. Rimbach G. Schreiber S. Nebel A. No or only population-specific effect of PON1 on human longevity: A comprehensive meta-analysis. Ageing Res Rev. 2010;9:238–244. doi: 10.1016/j.arr.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Martín MA. Ramos S. Granado-Serrano AB. Rodríguez-Ramiro I. Trujillo M. Bravo L. Goya L. Hydroxytyrosol induces antioxidant/detoxificant enzymes and Nrf2 translocation via extracellular regulated kinases and phosphatidylinositol-3-kinase/protein kinase B pathways in HepG2 cells. Mol Nutr Food Res. 2010;54:956–966. doi: 10.1002/mnfr.200900159. [DOI] [PubMed] [Google Scholar]
- 47.Chung S. Yao H. Caito S. Hwang JW. Arunachalam G. Rahman I. Regulation of SIRT1 in cellular functions: Role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singleton VL. Orthofer R. Lamuela-Raventós RM. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin ciocalteu reagent. Methods Enzymol. 1999;299:152–178. [Google Scholar]
- 49.Benzie IF. Strain JJ. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- 50.Re R. Pellegrini N. Proteggente A. Pannala A. Yang M. Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 51.Morel DW. Hessler JR. Chisolm GM. Low density lipoprotein cytotoxicity induced by free radical peroxidation of lipid. J Lipid Res. 1983;24:1070–1076. [PubMed] [Google Scholar]
- 52.Buss H. Chan T P. Sluis K B. Domigan NM. Winterbourn CC. Protein carbonyl measurement by a sensitive ELISA method. Free Radic Biol Med. 1997;23:361–366. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 53.Breusing N. Grune T. 2008 Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 54.Fuhrman B. Volkova N. Aviram A. Postprandial serum triacylglycerols and oxidative stress in mice after consumption of fish oil, soy oil or olive oil: Possible role for paraoxonase-1 triacylglycerol lipase-like activity. Nutrition. 2006;22:922–930. doi: 10.1016/j.nut.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 55.Ernst IM. Wagner AE. Schuemann C. Storm N. Höppner W. Döring F. Stocker A. Rimbach G. Allyl-, butyl- and phenylethyl-isothiocyanate activate Nrf2 in cultured fibroblasts. Pharmacol Res. 2011;63:233–240. doi: 10.1016/j.phrs.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Borenfreund E. Puerner JA. Toxicity determined in vitro by morphological alterations and neutral red absorption. Toxicol Lett. 1985;24:119–124. doi: 10.1016/0378-4274(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 57.Banning A. Deubel S. Kluth D. Zhou Z. Brigelius-Flohe R. The GI-GPx gene is a target for Nrf2. Mol Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner AE. Ernst I. Iori R. Desel C. Rimbach G. Sulforaphane but not ascorbigen, indole-3-carbinole and ascorbic acid activate the transcription factor Nrf2 and induce phase-2 and antioxidant enzymes in human keratinocytes in culture. Exp Dermatol. 2010;19:137–144. doi: 10.1111/j.1600-0625.2009.00928.x. [DOI] [PubMed] [Google Scholar]
- 59.Gouedard C. Barouki R. Morel Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol Cell Biol. 2004;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jódar L. Mercken EM. Ariza J. Younts C. González-Reyes JA. Alcaín FJ. Burón I. de Cabo R. Villalba JM. Genetic deletion of Nrf2 promotes immortalization and decreases lifespanof murine embryonic fibroblasts. J Gerontol A Biol Sci Med Sci. 2011;66:247–256. doi: 10.1093/gerona/glq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiborr C. Eckert GP. Weissenberger J. Müller WE. Schwamm D. Grune T. Rimbach G. Frank J. Cardiac oxidative stress and inflammation are similar in SAMP8 and SAMR1 mice and unaltered by curcumin and Ginkgo biloba extract intake. Curr Pharma Biotech. 2010;11:861–867. doi: 10.2174/138920110793262006. [DOI] [PubMed] [Google Scholar]
- 62.Covas MI. de la Torre K. Farré-Albaladejo M. Kaikkonen J. Fitó M. López-Sabater C. Pujadas-Bastardes MA. Joglar J. Weinbrenner T. Lamuela-Raventós RM. de la Torre R. Postprandial LDL phenolic content and LDL oxidation are modulated by olive oil phenolic compounds in humans. Free Radic Biol Med. 2006;40:608–616. doi: 10.1016/j.freeradbiomed.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 63.Kountouri AM. Mylona A. Kaliora AC. Andrikopoulos NK. Bioavailability of the phenolic compounds of the fruits (drupes) of Olea europaea (olives): Impact on plasma antioxidant status in humans. Phytomed. 2007;14:659–667. doi: 10.1016/j.phymed.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Suárez M. Valls RM. Romero MP. Macià A. Fernández S. Giralt M. Solà R. Motilva MJ. Bioavailability of phenols from a phenol-enriched olive oil. Br J Nutr. 2011 doi: 10.1017/S0007114511002200. [DOI] [PubMed] [Google Scholar]
- 65.Mackness B. Durrington PN. Mackness MI. The paraoxonase gene family and coronary heart disease. Curr Opin Lipid. 2002;13:357–362. doi: 10.1097/00041433-200208000-00002. [DOI] [PubMed] [Google Scholar]
- 66.de la Torre-Carbot K. Chávez-Servín JL. Jaregui O. Castellote AI. Lamuela-Raventós RM. Nurmi T. Poulsen HE. Gaddi AV. Kaikkonen J. Zunft JF. Kiesewetter H. Fitó M. Covas MI. López-Sabater MC. Elevated circulating LDL phenol levels in men who consumed virgin rather than refined olive oil are associated with less oxidation of plasma LDL. J Nutr. 2010;140:501–508. doi: 10.3945/jn.109.112912. [DOI] [PubMed] [Google Scholar]
- 67.Tajes M. Gutierrez-Cuesta J. Folch J. Ortuño-Sahagun D. Verdaguer E. Jiménez A. Junyent F. Lau A. Camins A. Pallàs M. Neuroprotective role of intermittent fasting in senescence-accelerated mice P8 (SAMP8) Exp Gerontol. 2010;45:702–710. doi: 10.1016/j.exger.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 68.Alcendor RR. Gao S. Zhai P. Zablocki D. Holle E. Yu X. Tian B. Wagner T. Vatner SF. Sadoshima J. SIRT1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 69.Masala G. Ceroti M. Pala V. Krogh V. Vineis P. Sacerdote C. Saieva C. Salvini S. Sieri S. Berrino F. Panico S. Mattiello A. Tumino R. Giurdanella MC. Bamia C. Trichopoulou A. Riboli E. Palli D. A dietary pattern rich in olive oil and raw vegetables is associated with lower mortality in Italian elderly subjects. Br J Nutr. 2007;98:406–415. doi: 10.1017/S0007114507704981. [DOI] [PubMed] [Google Scholar]