Abstract
Under a shortage of oxygen, bacterial growth can be faced mainly by two ATP-generating mechanisms: (i) by synthesis of specific high-affinity terminal oxidases that allow bacteria to use traces of oxygen or (ii) by utilizing other substrates as final electron acceptors such as nitrate, which can be reduced to dinitrogen gas through denitrification or to ammonium. This bacterial respiratory shift from oxic to microoxic and anoxic conditions requires a regulatory strategy which ensures that cells can sense and respond to changes in oxygen tension and to the availability of other electron acceptors. Bacteria can sense oxygen by direct interaction of this molecule with a membrane protein receptor (e.g., FixL) or by interaction with a cytoplasmic transcriptional factor (e.g., Fnr). A third type of oxygen perception is based on sensing changes in redox state of molecules within the cell. Redox-responsive regulatory systems (e.g., ArcBA, RegBA/PrrBA, RoxSR, RegSR, ActSR, ResDE, and Rex) integrate the response to multiple signals (e.g., ubiquinone, menaquinone, redox active cysteine, electron transport to terminal oxidases, and NAD/NADH) and activate or repress target genes to coordinate the adaptation of bacterial respiration from oxic to anoxic conditions. Here, we provide a compilation of the current knowledge about proteins and regulatory networks involved in the redox control of the respiratory adaptation of different bacterial species to microxic and anoxic environments. Antioxid. Redox Signal. 16, 819–852.
I. Introduction
Respiration is a fundamental process to all living cells in which electrons produced from oxidation of low-redox-potential electron donors such as NADH are transferred sequentially through a series of membrane-bound or membrane-associated protein carriers, the electron transport chain (ETC), which terminates in the reduction of the high-redox-potential electron acceptor, oxygen (Fig. 1a). The free energy released during this electron transfer process is used to drive the translocation of protons across the membrane to generate an electrochemical gradient that can be used for a variety of purposes, such as ATP synthesis and active transport (Fig. 1a). In contrast to the respiratory systems found in the mitochondria of many higher eukaryotic organisms, prokaryotic cells can induce branched-respiratory chains terminating in multiple oxidases with different affinities for the oxygen or use alternative electron acceptors, which contributes to their ability to colonize many microoxic and anoxic environments [reviewed in (61, 183, 186, 197)] (Fig. 1b).
FIG. 1.
Respiratory chain(s) in mammalian mitochondrion and bacteria. (a) A summary of the topology and bioenergetics of a basic aerobic respiratory electron transport system of a mammalian mitochondrion is shown. This figure is adapted from ref. (197). (b) Schematic representation of aerobic and anaerobic nitrate respiration pathways in bacteria. MK, menaquinone; UQ, ubiquinone; Cyt, cytochrome; SDH, succinate dehydrogenase; NDH, NADH dehydrogenase; cd1Nir, cd1-type nitrite reductase; CuNir, Cu-type nitrite reductase; Nor, nitric oxide reductase; Nos, nitrous oxide reductase; Nar, membrane-bound nitrate reductase; Nap, periplasmic nitrate reductase; NrfA; cytochrome c nitrite reductase; UQH2, ubihydroquinone; MKH2, menahydroquinone.
The oxidation of organic molecules in cell respiration releases electrons that are transferred to membrane mobile quinones which constitute the link between the electron donating enzymes and the electron accepting enzymes (Fig. 1b). Quinones are small, freely diffusible, lipophilic, membrane-entrapped organic molecules that can carry two electrons and two protons when fully reduced, that is, in the quinol state [for a review, see (221)]. Different kinds of quinones have different electrochemical potentials, and many bacteria can synthesize more than one type of quinone. Escherichia coli synthesize three types of quinones, a benzoquinone (UQ-8), and two naphthoquinones, menaquinone (MK) and demethylmenaquinone. Ubiquinone (UQ) predominates under aerobic conditions, and MK predominates under anaerobic conditions when the cellular state is more reduced (221). From the lipophilic hydroquinones, electrons can be carried to two different types of terminal oxidases: cytochrome c oxidases or quinol oxidases, where dioxygen is reduced to water (Fig. 1b). When oxygen is not present in the medium, electrons can be transmitted to alternative reductases that reduce substrates such as nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O), dimethyl-sulphoxide (DMSO), trimethylamine N-oxide, sulfate, sulfite, and fumarate as final electron acceptors. Among them, nitrate is one of the essential environmental components in the biosphere. It serves as a nutrient for plants and microorganisms, and is used as an electron acceptor by many bacteria, archaea, and also by several eukaryotes (109, 125, 266). During anaerobic respiration, nitrate can be reduced to dinitrogen gas (N2) or to ammonium. The anaerobic reduction of nitrate to N2 gas is called denitrification, and it constitutes one of the more important processes in the N-cycle (141). This reductive process occurs in four stages, reduction of NO3− to NO2− and via the gaseous intermediates NO and N2O to N2. The enzymes involved in denitrification are nitrate-, nitrite-, nitric oxide-, and nitrous oxide reductase, encoded by nar/nap, nir, nor, and nos genes, respectively [reviewed in (125, 247, 248)]. In agriculture and wastewater treatment, denitrification by microorganisms is an important issue due to the economical, environmental, and public health implications of this process (10, 105).
The bacterial respiratory flexibility requires a regulatory strategy which ensures that prokaryotic cells sustain life under different environments in response to changing oxygen tension. To date, three main modes to sense O2 have been described in bacteria: two types by direct interaction of O2 with a membrane protein receptor (as in the heme-based sensor kinase FixL in rhizobia [reviewed in (83, 101, 205)] or by interaction with a regulatory protein such as the Fe-S-based fumarate and nitrate reductase regulatory protein (Fnr) in E. coli [reviewed in (65, 68, 84, 53, 101, 261)]. In addition, a third type of O2 perception is based on monitoring environmental oxygen concentration by sensing changes in the redox state of molecules or pools of molecules within the cell. These changes are detected by various protein sensors that convert the redox signals into regulatory output at the transcriptional or post-transcriptional level. Redox signals are many and diverse, and can involve redox-active cofactors such as heme, flavins, pyridine nucleotides, iron-sulfur clusters, and redox-sensitive amino-acids chains such as cysteine thiols among others [reviewed in (3, 14, 102)]. This article focuses on a comparative study about the regulatory mechanisms used by different bacterial groups to undergo respiratory shifts from oxic to microoxic and anoxic environments.
II. Aerobic Respiration Under Microoxic Conditions
The branched ETC present in many bacteria often contains several terminal oxidases (Fig. 1b), which can be grouped into two main types based on the substrates used as electron donors [reviewed in (61, 177, 183)]. Reduced c-type cytochromes donate electrons to cytochrome oxidases, whereas hydroquinones deliver electrons to quinol oxidases (Fig. 1b). Both types of oxidases belong to the extensively studied family of heme-copper oxidases (HCOs). The common denominator in HCOs is a membrane-integral subunit I that carries as cofactors a low-spin heme and a high-spin heme-copper binuclear center (CuB site) where reduction of O2 to H2O takes place (Fig. 2). Among this family, quinol oxidases possess a cofactor-free subunit II, whereas cytochrome c oxidases have cofactors bound to subunit II. In most cases, this is a binuclear Cu–Cu center (CuA site) that is liganded by six highly conserved amino acids (87, 176). The HCO superfamily is very versatile due to the wide range of electron donors they can use, their subunit composition, and the types of heme groups that they contain. Studies of evolutionary relationships led to classifying the HCO enzymes in three different types (176, 177) (Fig. 2): (i) type A oxidases (oxidases aa3), which are structurally and functionally close to mitochondrial oxidases, (ii) type B oxidases, grouped as cytochrome oxidases bo3, and (iii) type C oxidases, which include the cytochrome c oxidases cbb3. To gain new insights into the oxygen respiration process, C-family HCOs from pathogenic bacteria have been recently structurally characterized, and it has been shown that C-HCOs differs from the two other HCO families, A and B (40). The X-ray structure of the C-family cbb3 oxidase from Pseudomonas stutzeri shows an electron supply system different from families A and B.
FIG. 2.
Summary of the main types of heme–copper oxygen reductases belonging to the A, B, and C families and the bd-type quinol oxidase.
Under O2 limitation (microoxic conditions), many bacteria induce high-affinity oxidases to respire traces of molecular oxygen. The cbb3-type cytochrome oxidase is known to have a high affinity for oxygen. Thus far, the enzyme from Bradyrhizobium japonicum is the only cytochrome cbb3 oxidase in which substrate affinity has been measured showing a Km for dioxygen in the order of 7 nM (185). As a consequence, it allows respiration under microoxic conditions, such as those encountered by rhizobia in the legume nodule ([O2] estimated to be 30 nM) (61). This oxidase has a subunit II (CcoO or FixO) that is a monoheme c-type cytochrome instead of the CuA-containing protein (67, 176). Subunit III in cbb3-type oxidases, which is a diheme cytochrome c (CcoP or FixP), is thought to relay the electrons from the cytochrome bc1 complex via CcoO to the redox centers of subunit I (126, 265). Subunit III of all other HCOs is cofactor-free, just like the nonconserved small subunit IV (176). Homology modelling and recent studies on the thermodynamic properties of the redox centers in cbb3 oxidase from Rhodobacter sphaeroides using a combination of optical and electron paramagnetic resonance redox titrations have allowed the characterization of the cbb3 active site (191). Furthermore, it has been recently shown that the redox-coupled proton movement in the proximal cavity of cbb3-enzymes contributes to the low redox potential of heme b3, and suggests its potential implications for the high oxygen affinity of these enzymes (219).
In addition to C-family cbb3 oxidases, another family of high-affinity oxidases comprises cytochrome bd-type oxidases (29). Among them, the bd oxidase of E. coli is known to have a high affinity for oxygen showing the Kd(O2) value of 0.28 μM (19). The bd-type oxidases are quinol oxidases that show no sequence homology to any subunit of heme-cooper family members. They do not contain any copper atoms in the catalytic center, but probably replace the CuB atom with a second heme (Fig. 2). In contrast to the heme-copper enzymes that are found at most branches of the tree of life, bd-type oxidases are only found in bacteria and archaea (183). The bd-type cytochromes do not pump protons and, therefore, have a lower total energetic efficiency compared with the heme-copper type oxidases (29).
A. Oxygen control of cbb3 oxidase expression
As just mentioned, cbb3 oxidase is quite distinct from other bacterial HCOs in terms of its strategy for receiving electrons, the heme prosthetic group present in the active site, and its affinity for oxygen. Cytochrome cbb3 oxidases have been purified from several organisms, including Paracoccus denitrificans, Rho. sphaeroides, Rhodobacter capsulatus, and Br. japonicum [reviewed in (179)]. The biogenesis of this multisubunit enzyme, encoded by the ccoNOQP operon, depends on the ccoGHIS gene products, which are proposed to be specifically required for cofactor insertion and maturation of cbb3-type cytochrome c oxidases (185). In the facultative photosynthetic model organism Rho. capsulatus, CcoN, CcoO, and CcoQ assemble first into an inactive 210 kDa sub-complex, which is stabilized via its interaction with CcoH and CcoS. Binding of CcoP, and probably subsequent dissociation of CcoH and CcoS, generates the active 230 kDa complex (126). Recent results have proposed that CcoH behaves more similar to a subunit of the cbb3 oxidase rather than a transient assembly factor per se (173). The insertion of the heme cofactors into the c-type cytochromes CcoP and CcoO precedes sub-complex formation, while the cofactor insertion into CcoN could occur either before or after the 210 kDa sub-complex formation during the assembly of the cbb3-type oxidase (126). CcoQ is required for optimal cbb3-type oxidase activity, because it stabilizes the interaction of CcoP with the CcoNO core complex, leading subsequently to the formation of the active 230-kDa cbb3-type oxidase complex (178).
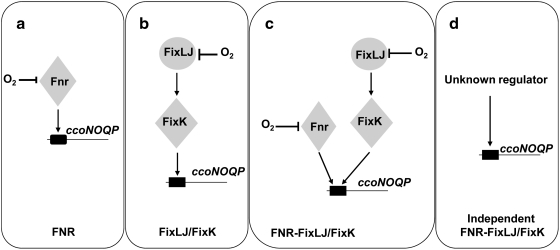
Genes encoding the cbb3 complex were initially isolated from rhizobia and named fixNOQP due to its requirement for symbiotic nitrogen fixation (185). Since then, orthologous genes called ccoNOQP were identified in other Proteobacteria including photosynthetic and pathogenic bacteria, suggesting that this oxidase is not only required for diazotrophs to sustain N2 fixation, but also for the successful colonization of anoxic tissues by human pathogens [reviewed in (50, 61, 179)]. Regulation of ccoNOQP expression has been investigated in many bacteria with oxygen being the key effector that controls its expression. To date, four different strategies that activate the ccoNOQP operon have been described (Fig. 3): (a) in Rho. sphaeroides, Pa. denitrificans, Pseudomonas aeruginosa, Rhizobium leguminosarum bv. viciae (UPM791), and Rho. capsulatus, Fnr acts as oxygen sensor and transcriptional factor; (b) in rhizobial species, such as Sinorhizobium meliloti, Br. japonicum, and Azorhizobium caulinodans, FixLJ/FixK is the system implicated in the activation of ccoNOQP under oxygen deprivation; (c) in Rhi. leguminosarum (VF39) and Rhizobium etli CFN42, a mixed regulation strategy operates that involves both Fnr and FixK proteins; (d) and finally, in the human pathogenic bacteria Helicobacter pylori and Campylobacter jejuni (ɛ-Proteobacteria), which lack fnr and fixLJ-fixK genes, it has been proposed that cbb3 may be constitutively expressed allowing the growth under microoxic conditions in its hosts. In addition to the positive control of Fnr on fixNOQP expression, it has been shown in Rhi. etli that two novel Fnr-type transcriptional regulators (StoRd and StoRf, symbiotic terminal oxidase regulators) have a negative control on microoxic expression of fixNOQP genes (100).
FIG. 3.
Regulation strategies of ccoNOQP gene transcription. Four regulation strategies have been described in several bacterial groups (see text for details) that involve (a) Fnr, (b) FixLJ-FixK, (c) Fnr and FixLJ-FixK, or (d) an unknown regulator. Gene transcriptional activation is indicated by arrows. Protein inactivation by O2 is indicated by perpendicular lines. This figure is adapted from ref. (50).
1. Involvement of Fnr
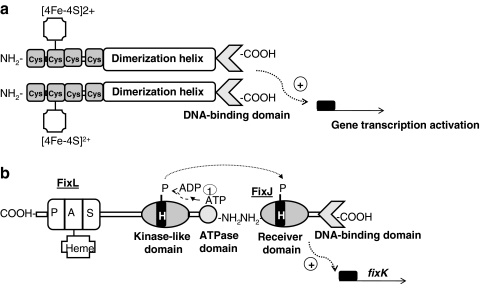
The O2-sensitive protein Fnr belongs to the cyclic AMP (cAMP) receptor protein (Crp) superfamily of transcription factors that trigger physiological changes in response to a variety of metabolic and environmental cues [reviewed in (53, 65, 68, 84, 101, 103, 124, 261)]. All members of this family are predicted to be structurally related to Crp. They consist of four functionally distinct domains: (i) a sensor domain, (ii) a series of β-strands (β-roll structure) that form a loop-like structure making contact with the RNA polymerase holoenzyme (RNAP), (iii) a long α-helix acting as a dimerization interface, and (iv) a C-terminal helix-turn-helix motif (H-T-H) involved in DNA binding (Fig. 4a). The best-characterized Fnr protein is that of E. coli. In E. coli, Fnr acts as a direct oxygen sensor and is the primary transcriptional regulator of the switch between aerobic and anaerobic growth. Thus, under anoxic conditions, Fnr is in its active state and is able to bind specific palindromic sequences of DNA (Fnr box). Once bound to DNA, Fnr activates target gene expression by recruiting RNAP or represses transcription by preventing the formation of productive RNAP:DNA complexes (53). Active Fnr regulates genes involved in the adaptation of E. coli for anaerobic growth and controls one of the best-studied gene regulatory networks in the cell. Very recently, the Fnr regulatory network has been reviewed by Tolla and Savageau (238). Generally, Fnr activates genes encoding products involved in anaerobic metabolism, such as the nar operon (nitrate reductase), the dms operon (DMSO reductase), and the frd operon (fumarate reductase), and represses genes encoding products involved in aerobic metabolism, such as the sdh operon (succinate dehydrogenase) and ndh (NADH dehydrogenase II). Fnr is activated under anoxic conditions by the acquisition of one [4Fe-4S]2+ cluster per monomer (designated 4Fe-Fnr). Each [4Fe-4S]2+ cluster is ligated by four cysteine residues (Cys20, Cys23, Cys29, and Cys122), and the presence of the cluster promotes Fnr dimerization, increasing its capacity to bind specifically to DNA (52, 54, 132) (Fig. 4a). The [4Fe-4S]2+ cluster is susceptible to be attacked by O2, resulting in rapid and reversible conversion to a [2Fe-2S]2+ and finally to apoFnr on exposure to high oxygen concentrations. In addition to its reaction with oxygen, the Fnr [4Fe-4S]2+ cluster is also sensitive to NO. On exposure to NO, [4Fe-4S]2+ becomes nitrosylated, forming a combination of monomeric and dimeric dinitrosyl-iron-cysteine complexes, again abolishing its ability to bind DNA (53, 57). It has been reported that Fnr represses genes involved in NO detoxification (e.g., hmp, which encodes the flavohemoglobin) (182). When cultures of E. coli are exposed to NO, Fnr repression of hmp is relieved, suggesting that the reaction between Fnr and NO is physiologically significant. This suggestion is supported by transcript profiling experiments which reveal that the abundances of many Fnr-activated genes are lower, and many Fnr-repressed genes are higher, when E. coli is exposed to NO (119, 187).
FIG. 4.
Functional domains and model of Fnr and FixLJ mediated oxygen activation. (a) Fnr senses oxygen via an iron-sulfur center, which is coordinated with four cysteine residues. Under anoxic conditions, Fnr is in its active state and it promotes DNA-binding by formation of a homodimer leading to activation of gene transcription. (b) FixL is an oxygen receptor in which oxygen binds directly to a heme group that is coordinated to a histidine residue within a PAS domain. Detection of low oxygen tension changes the conformation of the input PAS domain, rendering increased autophosphorylation activity of the transmitter domain in His residue and repressing the phosphatase activity of FixL. FixL activates FixJ transcriptional activity by transferring the phosphoryl group to the N-terminal domain of FixJ (+) gene transcription activation.
2. Involvement of FixLJ/K
In some rhizobial species, the fixNOQP operon is under the control of one Fnr-type transcriptional regulator named FixK, which is also able to recognize Fnr boxes but lacks [4Fe–4S]2+ clusters. FixK is thereby unable to sense oxygen directly. In this case, it is the two-component regulatory system FixLJ that ensures oxygen sensing under low-oxygen conditions and activates the transcription of the fixK gene. This regulatory system FixLJ/FixK has been studied in rhizobial species where it controls the expression of genes for microoxic respiration, as well as various other functions, in response to low oxygen (28, 56, 83, 145, 160). Interestingly, it has been shown that Rhodopseudomonas palustris FixK is not only required for optimal microaerobic growth, but is also required for phototrophic and autotrophic growth, and for growth on aromatic compounds (194). These authors also propose that FixK helps the control of the transition of R. palustris to anaerobic growth by activating the expression of the oxygen-sensing Fnr homolog, AadR.
The biochemistry of the FixLJ two-component system has been characterized in Si. meliloti and Br. japonicum (96, 211, 223). FixL is a histidine kinase that binds heme at an amino-terminal PAS domain (Fig. 4b). PAS domains are present in many proteins involved in the sensing of oxygen, redox, or light. They were first found in eukaryotes, and were named after homology to the Drosophila period protein (PER), the aryl hydrocarbon receptor nuclear translocator protein (ARNT), and the Drosophila single-minded protein (SIM). The binding of oxygen to heme inactivates FixL kinase activity. FixL, and the response regulator and transcription factor FixJ form a complex in which FixJ allosterically stimulates autophosphorylation of FixL. At very low oxygen concentrations, FixL transphosphorylates FixJ, which, in turn, activates expression of the transcription factor gene fixK (Fig. 4b). In Br. japonicum, the nitrogen-fixing root-nodule endosymbiont of soybean (Glycine max), a FixK-like protein called FixK2, acts as the key distributor of the low-oxygen signal perceived at the level of the FixLJ two-component regulatory system. FixK2 is one of the 16 (Crp/Fnr)-type transcriptional regulators whose genes are present in the Br. japonicum genome [reviewed in (146)]. FixK2 recognizes a palindromic sequence motif (TTG-N8-CAA, termed the FixK2 box) that is located around position–41 upstream of the transcription start site in the regulated promoters. Microoxically induced targets of FixK2 include the operons fixNOQP and fixGHIS, both essential for microoxic respiration, several heme biosynthesis genes (hemA, hemB, hemN1, and hemN2), denitrification genes, and some hydrogen oxidation genes (hup genes) (16, 160). Recently, the Br. japonicum FixK2 regulon was unraveled by using a transcriptomics approach (145). DNA binding site predictions, together with a FixK2-dependent in vitro transcription assay, demonstrated that the fixNOQP operon is a direct target for FixK2. The latter studies, carried out with purified FixK2 protein, showed that FixK2 is sufficient to activate transcription in vitro without any identified effector (148). This is puzzling in view of the fact that all Crp/Fnr-like proteins can be positively or negatively modulated in their activity through bound cofactors or intrinsic, reactive amino acids (101). However, recent findings have reported that posttranslational control occurs at FixK2, whereby a critical cysteine at position 183 in the polypeptide chain is a target for oxidation. This provides a second, important means of affecting FixK2 activity, in addition to the regulation of its expression by FixLJ (147).
III. Redox Control of the Adaptation of Aerobic Respiration from Oxic to Microoxic Conditions
A. E. coli ArcBA
E. coli has three terminal quinol oxidases, of which one is a heme-copper bo3 oxidase and two are bd-type oxidases [reviewed in (183, 186)]. The quinol bo3 oxidase contains a heme-Cu cofactor (CuB) and is essential when oxygen is present at high concentrations. The protein complex is encoded by the cyoABCDE operon (186). Under conditions of low aeration, the high-affinity bd oxidase is the preferred terminal reductase in E. coli (29). The structural components are encoded by the genes cydAB, which form a heterodimer. CydC and D are encoded in the same operon as CydA and B and thought to form a heme transporter necessary for cytochrome bd oxidase biogenesis (29). There is a third terminal oxidase present in E. coli encoded by the genes cyxAB. It is a bd type oxidase but its function is unclear. It appears to be expressed under microoxic conditions (29).
Cytochrome bd oxidase prevails under oxygen-limiting conditions. Consistent with these properties, transcription of the cydAB operon is activated when oxygen becomes limiting. Under fully anoxic conditions, cydAB expression is subsequently repressed to an intermediate level relative to microoxic and oxygen-rich conditions (241). It is now solidly established that this oxygen control is achieved through the combined action of the oxygen sensing transcription factor Fnr and the aerobic respiration control (Arc)B/ArcA two-component system (217). Under anoxic conditions, Fnr acts as a repressor of the cydAB operon (Fig. 5). ArcBA is a two-component regulatory system involved in the transcriptional regulatory network that allows facultative aerobic bacteria to sense and respond to various respiratory conditions. While the Fnr protein is directly implicated in sensing oxygen, the ArcBA system activity is proposed to be modulated by the redox state of the quinones in the membrane (137, 138). The ArcBA system comprises the ArcB protein, a tripartite membrane-bound sensor kinase, and the cognate response regulator ArcA (Fig. 6a). ArcB belongs to the subclass of hybrid sensor kinases that also includes BarA, BvgS, EvgS, LemA (GacS), RteA, and TorS (2, 138). Under limiting oxygen conditions, ArcB autophosphorylates residue His292 and, through a phosphorelay system involving Asp576 and His717, transphosphorylates ArcA (Fig. 6a). It has been previously reported that histidine kinases act as homodimers and that they autophosphorylate by an intermolecular reaction (229). That is, the gamma-phosphoryl group of ATP, which is bound to one monomer in the homodimer, is transferred to the other monomer. On the contrary, it has been recently proposed that ArcB autophosphorylates through an intramolecular and not through an intermolecular reaction as previously proposed (175).
FIG. 5.
Regulation of bd and bo3 terminal oxidase expression in Escherichia coli. Fnr and ArcB provide negative or positive transcriptional control in response to oxygen availability. The genes cydAB and cyoABCDE encode bd and bo3 oxidase polypeptides, respectively. Positive regulation (+) is denoted by arrows, and negative regulation (−) is indicated by perpendicular lines.
FIG. 6.
Functional domains and model of ArcBA-mediated redox control. (a) ArcB is attached to the membrane by two transmembrane regions (TM). A linker region containing a putative leucine-zipper and a PAS domain connects TM2 with the catalytic domains. The two cysteine-residues (Cys180 and Cys241) in the linker region are able to undergo oxidation and to form intermolecular disulfide bonds between both ArcB monomers. The transmitter domain (H1) contains the conserved His292 together with the G1 and G2 nucleotide-binding motifs. The receiver domain (D1) harbors the conserved Asp576, and the histidine phosphotransfer domain (Hpt/H2) contains the conserved His717. The ArcA component is represented with its N-terminal receiver domain carrying the conserved Asp54 and its C-terminal H-T-H) domain. This figure is adapted from ref. (138). (b) MKH2, which predominates under anaerobic conditions, would be oxidized in the presence of low levels of oxygen (∼20% aerobiosis), leading to increased levels of MKs and inactivation of ArcB kinase activity. UQH2 predominate under microoxic conditions (∼80% aerobiosis), giving rise to activation of ArcB kinase activity. In fully aerobic conditions (100%), the UQH2 pool is subjected to oxidation, which results in increasing levels of UQs and oxidation of the key cysteines, leading to inactivation of the ArcB kinase activity. This figure is adapted from ref. (18).
On a shift to aerobic conditions, ArcB can form intermolecular disulphide bonds via Cys180 and Cys241 located in the PAS domain (Fig. 6a). The kinase activity of ArcB is highly dependent on this covalent linkage (137). The regulatory mechanisms that underlie the function of the ArcBA two-component system have been the subject of numerous studies. ArcB does not sense oxygen directly but is thought to sense the redox state of the UQ-ubiquinol pool in the aerobic respiratory chain, because UQ inhibits ArcB autokinase activity (137). However, this regulatory network becomes complex, as recent observations could not demonstrate a correlation between ArcB activation and oxygen availability (18). Bekker and colleagues (18) proposed that the in vivo activity of ArcB in E. coli is also modulated by the redox state of the MK pool and that the UQ/ubiquinol ratio is not the only determinant of ArcB activity (Fig. 6b). This combined regulation by the redox state of the UQ pool and MK pool provides a mechanism to explain the observed complex regulation of ArcB activation at variable rates of oxygen supply (Fig. 6b). Menaquinols would be the dominant activators under anoxic conditions, where the size of the UQ pool is approximately five times less than the size of the MK pool (18). Recent work has used controlled chemostat cultures subject to gradually decrease oxygen concentration to study the effects of oxygen availability on the transcriptome, redox state of the UQ-ubiquinol, outer membrane protein profiles, and terminal oxidase proteins of E. coli K-12 (208). A probabilistic model was used to predict the activity of ArcBA across the aerobiosis range. The model implied that the activity of the regulator ArcA correlated with aerobiosis, but not with the redox state of the UQ pool. Rolfe and colleagues (208) have proposed that, at least under the conditions used in this work, the rate of fermentation product synthesis (qacetate) exerts a greater influence on ArcA activity than the redox state of the quinone pools. It is perhaps not surprising that, as a regulator of genes that have fundamental roles in different processes of the bacterial metabolism (aerobic respiration, anaerobic respiration, and fermentation), ArcB should integrate the response to multiple signals (e.g., UQ, MK, acetate, and other fermentation products) in response to different growth conditions.
Phosphorylated ArcA activates genes or operons needed to use traces of oxygen in the medium, for example, those for terminal oxidase bd, and represses expression of the aerobic cytochrome bo3 terminal oxidase (136). Regulation of bd and cytochrome bo3 terminal oxidases in response to oxygen concentration variation is also controlled by Fnr as just described (Fig. 5b). Fnr inhibits cyd and cyo gene activation from 0% to 10% oxygen tension. However, ArcA represses cyo expression and activates cyd transcription when O2 concentration is between 10% and 20% (241) (Fig. 5). These authors proposed that when the oxygen tension drops, ArcA is phosphorylated by ArcB and activates cydAB transcription reaching a maximum at <2% O2 tension. When oxygen is decreased further, Fnr becomes active and represses cydAB transcription. Fnr repression of cydAB requires the presence of a functional ArcA protein (51, 99). According to this observation, Fnr has been proposed to serve as an antiactivator by counteracting ArcA-mediated activation, rather than directly repressing transcription. This indirect regulation of cydAB repression by Fnr through ArcA is supported by gene expression profiling analyses where nearly two-thirds of the genes for which expression is affected by Fnr are also affected by ArcA (209, 210). In addition to controlling genes encoding respiratory oxidases, ArcA∼P represses the expression of genes encoding dehydrogenases of the tricarboxylic acid cycle and enzymes of the glyoxylate shunt. Recent studies have demonstrated that ArcAB exerts an important influence on carbon and redox balances in E. coli growing under carbon-limited conditions with a restricted oxygen supply (163).
B. Rhodobacter species RegBA/PrrBA
Photosynthetic bacteria such as Rho. Capsulatus and Rho. sphaeroides conserve energy under oxic or microoxic conditions by driving electrons through branched electron transfer chains to terminal oxidases with different affinity for oxygen (Fig. 7). One ubiquinol oxidase is found in Rho. capsulatus (cydAB), while two are found in Rho. sphaeroides (qoxBA and qxtAB). The cytochrome c oxidases depend on the cytochrome bc1 complex (petABC in Rho. capsulatus and fbcFBC in Rho. sphaeroides), and cytochromes c2 (cycA) or cy (cycY). While Rho. capsulatus contains only one cbb3-type oxidase (ccoNOQP), an aa3- (ctaDC) and a cbb3-type oxidase (ccoNOQP) are found in Rho. sphaeroides [reviewed in (72, 120, 166, 233, 258)]. Gene expression experiments using the lacZ as reporter gene revealed that the cbb3 oxidase from Rho. capsulatus and aa3 oxidase from Rho. sphaeroides are considered low-oxygen-affinity oxidases which are induced under oxic conditions (233). However, the Rho. capsulatus ubiquinol bd oxidase and Rho. sphaeroides cbb3 oxidase are classified as high-affinity terminal oxidases and are highly induced under oxygen-limiting conditions (233).
FIG. 7.
Aerobic respiratory chains in Rhodobacter capsulatus and Rhodobacter sphaeroides. The quinone-reducing (left) and quinol-oxidizing (right) branches with terminal oxidases induced under oxic conditions (gray boxes), and microoxic conditions (white boxes), are shown. Black arrows indicate the influx of reducing equivalents.
RegBA/PrrBA are members of the family of the two-component regulatory systems present in a large number of Proteobacteria. These proteins are named RegBA in Rho. capsulatus, Rhodovulum sulfidophilum, and Roseobacter denitrificans [reviewed in (72, 258)], PrrBA in Rho. sphaeroides (106, 167), ActSR in Si. meliloti (73) and Agrobacterium tumefaciens (8), RegSR in Br. japonicum (15), and RoxSR in Ps. aeruginosa (46). In Rhodobacter species, the RegBA/PrrBA regulon encodes proteins involved in numerous energy-generating and energy-utilizing processes such as photosynthesis, carbon fixation, nitrogen fixation, hydrogen utilization, aerobic and anaerobic respiration, and aerotaxis [reviewed in (72, 233, 258)]. Recent analyses of the transcriptome and the proteome of Rho. sphaeroides prrA mutant revealed that, in addition to the numerous PrrA gene targets already known, genes encoding proteins whose functions are involved in intermediary metabolism, repair of DNA and protein damage, cell motility and secretion, and translation constituted new targets for PrrA regulation (77).
The RegBA/PrrBA two-component systems comprise the membrane-associated RegB/PrrB histidine protein kinase, sensing changes in redox state and its cognate PrrA/RegA response regulador. Under conditions where the redox state of the cell is altered due to generation of an excess of reducing potential, produced by either an increase in the input of reductants into the system (e.g., presence of reduced carbon source) or a shortage of the terminal respiratory electron acceptor (e.g., oxygen deprivation), the kinase activity of RegB/PrrB is stimulated relative to its phosphatase activity (72). This increases phosphorylation of the partner response regulators RegA/PrrA, which are transcription factors that bind DNA and activate or repress gene expression. The membrane-bound sensor kinase proteins RegB/PrrB contain an H-box site of autophosphorylation (His225), a highly conserved quinone binding site (the heptapeptide consensus sequence GGXXNPF, which is totally conserved among all known RegB homologues), and a conserved redox-active cysteine (Cys265, located in a “redox box”). The mechanism by which RegB controls kinase activity in response to redox changes has been an active area of investigation. A previous study demonstrated that RegB Cys265 is partially responsible for redox control of kinase activity. Under oxidizing growth conditions, Cys265 can form an intermolecular disulfide bond to convert active RegB dimers into inactive tetramers (235) (Fig. 8a). The highly conserved sequence, GGXXNPF, located in a short periplasmic loop of the RegB transmembrane domain has also been implicated in redox sensing by interacting with the UQ pool [Swem et al. (234)] (Fig. 8a). Recently, kinase activity assays together with isothermal titration calorimetry (ITC) measurements indicated that RegB with a substitution in the cytosolic cysteine by serine in position 265 (RegB C265S) binds both oxidized and reduced UQ with almost equal affinity. However, only the oxidized UQ inhibits RegB kinase activity (259). The observation that the RegB C265S mutant is still redox responsive suggests that UQ binding is a signal input able of functioning independently from Cys265. However, the contribution of each redox sensing inputs is unknown.
FIG. 8.
Proposed models for redox-mediated sensing by RegBA/PrrBA. (a) In Rho. capsulatus, the conserved cysteine C265 of RegB functions as a redox switch that controls RegB kinase activity through a metal-dependent formation of a disulfide bond in response to redox changes (235). Binding of UQ to the RegB transmembrane domain has also been proposed to inhibit RegB kinase activity, whereas UQH2 does not affect RegB kinase activity (234). (b) In Rho. sphaeroides, the cbb3 oxidase generates an “inhibitory” signal that can directly stimulates the phosphatase activity of PrrB. Under low oxygen conditions when the electron flow is reduced, the inhibitory signal from cbb3 is weakened, and PrrB would retain its kinase activity (165). PrrC has been proposed as a signal mediator between the cbb3 oxidase and the sensor kinases PrrB (7).
In Rho. sphaeroides, the PrrB histidine kinase is a bifunctional enzyme that possesses both kinase and phosphatase activities (168). Several reports proposed that the cbb3 oxidase transduced an inhibitory signal to the PrrBA under oxic conditions to prevent gene expression (165, 166). The dual function of the cbb3 oxidase as both terminal oxidase and O2/redox sensor and modulator of PrrB kinase/phosphatase activity represents a new model of redox sensing. In this model (Fig. 8b), the UQ binding site within the PrrB transmembrane domain is not required for monitoring the PrrB kinase activity. Instead, a control based in direct interaction between components of the terminal oxidase cbb3 and PrrB is strengthened (122).
The photosynthetic regulatory response protein (PrrC) is a Sco homolog present in Rho. sphaeroides (75). Sco is thought to be involved in donating copper to the CuA center and, thus, it has a central role in cytochrome oxidase synthesis (12). Rho. sphaeroides PrrC, which reduces Cu2+ to Cu+, and possesses disulfide reductase activity, is required for the correct functioning of the sensor kinase/phosphatase PrrB (7). Similarly, the Rho. capsulatus SenC protein, homologous to PrrC, which is required for synthesis of a functional cytochrome c oxidase (232), might act as a signal mediator between the Q-pool and the sensor kinase RegB. However, at present, there is no direct evidence that SenC or cbb3 oxidase directly modulates the activity of the RegBA regulatory system.
RegA/PrrA contain conserved domains that are typical in two component response regulators such as a phosphate accepting aspartate, an “acid box” containing two highly conserved aspartate residues and a H-T-H DNA-binding motif (72). The phosphorylated form of RegA/PrrA has increased DNA binding capacity (129, 189). Under oxidizing conditions, RegB/PrrB shifts the relative equilibrium from the kinase to the phosphatase mode resulting in a dephosphorylated inactive RegA/PrrA form. Despite this evidence, it has been reported that inactivation of the regA gene affects expression of many different genes under oxidizing (aerobic) conditions, suggesting that both phosphorylated and unphosphorylated RegA/PrrA may be active transcriptional regulators (72, 233). In this context, it has been shown that both phosphorylated and unphosphorylated forms of RegA/PrrA are capable of binding DNA in vitro and activating transcription (189). Based on recent findings, there are two types of PrrA binding sites within the promoter of Rho. sphaeroides hemA, which codes for one of two isoenzymes catalyzing 5-aminolevulinate synthesis: (i) one site for which unphosphorylated PrrA has greater affinity, and (ii) another for which phosphorylated PrrA has greater affinity (190). Several consensus DNA binding sequences have been previously reported for PrrA (128, 139) and its homologs RegA in Rho. capsulatus (233, 253) and RegR in Br. japonicum (73, 134). While there is a measure of agreement in terms of the assignment of two imperfect quasisymmetrical inverted repeats, or half-sites separated by a spacer region possessing variable length, the DNA sequence per se shows a high degree of degeneracy. Very recently, it has been demonstrated that PrrA can bind in vitro to DNA sequences with different lengths in the spacer regions between the half sites (76).
In Rho. capsulatus, RegA has been reported to activate the expression of both the bd and cytochrome cbb3 oxidases (Fig. 9a). An interesting paradox is that RegA activates expression of cytochrome cbb3 oxidase under oxic conditions, but represses expression of this oxidase under anoxic conditions (233). In many respects, this is similar to what occurs in E. coli where ArcA acts as an oxic activator, as well as an anoxic repressor of cytochrome bo3 oxidase (see section III.A; Fig. 5b).
FIG. 9.
Regulation of bd and cbb3 terminal oxidases expression in Rho. capsulatus (a) and Rho. sphaeroides (b). In Rho. capsulatus, RegA regulates gene transcription in response to both oxic and anoxic conditions, FnrL, HvrA, in response to anaerobiosis, and AerR and CrtJ in response to aerobiosis. In Rho. sphaeroides, FnrL, PrrA, and PspR regulate gene transcription in response to anoxic conditions. Genes cydAB and ccoNOQP encode bd and cytochrome cbb3 oxidase polypeptides, respectively. Positive regulation (+) is denoted by arrows, and negative regulation (−) is indicated by perpendicular lines.
Terminal oxidase expression in Rho. capsulatus involves a complex set of regulators beyond that of RegBA. In response to oxygen availability, these regulators provide negative or positive transcriptional control to coordinate enzyme synthesis for optimal growth. A regulatory scheme for control of terminal oxidases in response to oxygen availability is shown in Figure 9a. Specifically, in Rho. capsulatus cytochrome cbb3, oxidase expression is regulated by RegA and FnrL, as well as moderately regulated by HvrA, an activator of various photosynthetic components under low-light conditions [(Swem et al. (231)]. Ubiquinol bd oxidase expression was found to be regulated by RegA and HvrA as well as by AerR and CrtJ, which are aerobic repressors of photosystem gene expression (231). In Rho. sphaeroides, microarray analyses and quantitative reverse transcriptase–polymerase chain reaction showed that both PrrA and the repressor of genes involved in photosynthesis, PspR, affect the transcription of the ccoNOQP operon (Fig. 9b). The absence of PrrA prevented the expression of the cco genes, and a prrA-pspR double mutant leads to derepresion of ccoNOQP operon (34). The fact that PpsR binds directly the cco promoter indicates that PrrA/PspR could act as an antirepressor/repressor system in which PspR acts as the direct repressor of the cco operon and PrrA is the PspR antirepressor regulator (34). Thus, RegBA/PrrBA appears to be just one component of a more complex regulatory network that controls many cellular processes. In this context, recent comparative genomics analyses and characterization of Rho. sphaeroides FnrL regulon have revealed that ccoNOQP is also positively controlled by this protein (68).
C. Pseudomonas species RoxSR
Pseudomonads are opportunistic pathogens that inhabit a wide variety of environments, including soil, water, and animal, human,- and plant roots. They are endowed with versatile respiratory chains that can be adapted to changing oxygen concentrations. Although they preferentially obtain energy from aerobic respiration, some of them, such as Ps. aeruginosa, can also grow anaerobically using nitrate as a final electron acceptor, or can even ferment arginine and pyruvate (254). The genome sequence and some biochemical studies have revealed that Pseudomonads have at least five terminal oxidases for aerobic respiration [reviewed in (183)] (Fig. 10a). In Ps. aeruginosa, UQs can be oxidized by either a bo3-type oxidase, a cyanide-insensitive oxidase (named CIO), or a bc1 complex that transfers electrons to a cytochrome c, which, in turn, can be oxidized by an aa3 oxidase or by two related cbb3 oxidases, named cbb3-1 and cbb3-2 (46, 47, 49). The Pseudomonas putida genome contains genes coding for all these terminal oxidases (161). Each oxidase is expected to have a specific affinity for oxygen, efficiency of proton translocation, and redox potential. Ps. aeruginosa has three predicted high-affinity oxidases; CIO, cbb3-1, and cbb3-2, which are believed to be important in the adaptation to oxygen-limiting conditions during growth under microoxic conditions. In this context, Alvarez-Ortega and Harwood (1) reported that any one of CIO, cbb3-1 and cbb3-2, could support growth under 2% oxygen concentrations and that either of cbb3-1 and cbb3-2 was indispensable for the growth under 0.4% oxygen.
FIG. 10.
Aerobic respiratory chains (a) and regulation of CIO, cbb3, and aa3 terminal oxidase expression (b) in Pseudomonas aeruginosa. In (a), the quinone-reducing (left) and quinol-oxidizing (right) branches with terminal oxidases induced under oxic conditions (gray boxes), and microoxic conditions (white boxes), are shown. In (b), Anr and RoxSR regulators provide negative or positive transcriptional control, in response to oxygen availability. Genes cox, cioAB, and ccoNOQP encode aa3, CIO, and cbb3 oxidase polypeptides, respectively. Positive regulation (+) is denoted by arrows, and negative regulation (−) is indicated by perpendicular lines. CIO, cyanide insensitive oxidase.
In Ps. aeruginosa, the Anr global transcriptional regulator plays a pivotal role in adaptation to microoxic or anoxic conditions. Anr is a homolog of E. coli Fnr and regulates expression of the enzymes required for nitrate respiration (4, 213), and of at least some of the aerobic terminal oxidases, such as the CIO oxidase and cytochrome cbb3-2 oxidase (47, 49). Recently, it has been shown that under low-oxygen conditions, Anr is involved in the repression of the CIO genes and induction of the cbb3-2 genes, while the other three terminal oxidases are not significantly regulated by Anr (Fig. 10b) (121). Similarly, in Ps. Putida, it has been shown that, under oxygen limitation, inactivation of anr led to an increased expression of the bo3-type and the CIO terminal oxidases, and to a much lower expression of cbb3-1 (equivalent to Ps. aeruginosa cbb3-2), suggesting that Anr is a transcriptional activator of cbb3-1 and a repressor of CIO genes (244).
In addition to Anr, another redox-responsive regulatory system has been proposed to be important for the regulation of terminal oxidases in Pseudomonas species, the RoxSR system. RoxSR belongs to the family of RegBA and PrrBA regulatory systems. It has been postulated in Ps. aeruginosa that the activation signal perceived by RoxS might depend on electron transport signals emerging from the oxidase cbb3-1 (47). Microarray analyses performed in Ps. putida (152) showed that a cyo mutation leads to a significant change in the transcriptome profile. They showed that the absence of bo3-type oxidase was compensated by upregulation of CIO and cbb3-1 (corresponding to cbb3-2 of Ps. aeruginosa). A regulatory system monitoring the electron flow through the bo3-type oxidase, which is similar to PrrBA activity modulation by the cbb3 oxidase, might be operative in Ps. putida.
Microarray studies have reported that RoxRS is involved in the regulation of the terminal oxidases genes in Ps. putida (78) and Ps. aeruginosa (121). In Ps. aeruginosa, RoxSR functions as a positive regulator of the putative high-affinity oxidases CIO, cbb3-1 and cbb3-2, and at the stationary phase where the dissolved oxygen concentration is low due to the high cell density, as a negative regulator of the putative low-affinity oxidase aa3 (121) (Fig. 10b). The RoxSR regulon of Ps. putida includes genes for respiratory function and maintenance of the redox balance, genes involved in sugar and amino acid metabolism, and the sulfur starvation response (78). These authors also showed that RoxSR participates in cell density signal transduction in Ps. putida. In Ps. aeruginosa, however, only a few genes related to sugar and amino-acid metabolism were identified as the members of the RoxSR regulon (121). The number of the putative RoxSR-regulated genes in Ps. aeruginosa seemed to be low compared with that in Ps. putida. These differences are presumably due to the different growth conditions used for the microarray analysis in the two Pseudomonas species.
D. Br. japonicum RegSR
The genera Allorhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium (Ensifer), collectively referred to as rhizobia, are members, along with other genera, of the bacterial order Rhizobiales of the α-Proteobacteria. Rhizobia are soil bacteria with the unique ability to establish a N2-fixing symbiosis on legume roots and on the stems of some aquatic legumes. During this interaction, bacteroids, as rhizobia are called in the symbiotic state, are contained in intracellular compartments called symbiosomes within a specialized organ, the nodule, where they fix N2 [reviewed in (116, 169)]. In the nodule, maintenance of nitrogenase activity is subject to a delicate equilibrium. A high rate of O2 respiration is necessary to supply the energy demands of the N2-reduction process; but, on the other hand, O2 also irreversibly inactivates the nitrogenase complex. In order to keep the steady-state concentration of free-O2 low, the cortex of nodules acts as a diffusion barrier, which greatly limits permeability to O2 [reviewed in (150)]. Oxygen is delivered to the symbiosomes by the plant O2-carrier leghemoglobin (Lb), which transports O2 at a low, but stable, concentration, allowing for the simultaneous operation of nitrogenase activity and bacteroid respiration [(66) and references therein]. N2-fixing bacteroids deal with the low levels of free O2 by inducing a high-affinity cytochrome cbb3-type oxidase [reviewed in (61)].
Similar to many other anaerobic facultative bacteria, Br. japonicum adapts to different environmental O2 concentrations by inducing multiple terminal oxidases with different affinities for O2. Br. japonicum has eight terminal oxidases, of which two are bd-type oxidases, and six are heme-copper oxidases, the latter being further divided into two quinol oxidases and four cytochrome oxidases [reviewed in (38)] (Fig. 11). Of particular interest is the fixNOQP-encoded cbb3-type oxidase, because it supports Br. japonicum aerobic respiration under free-living microoxic conditions and enables endosymbiotic Br. japonicum cells (bacteroids) to support the nitrogen fixation process. Cytochrome cbb3 oxidase has been purified from Br. japonicum (185), where the experimentally determined Km for O2 is in the order of 7 nM, a value that is consistent with its function in the bacteroid.
FIG. 11.
Aerobic respiratory chains in Bradyrhizobium japonicum. The quinone-reducing (left) and quinol-oxidizing (right) branches with terminal oxidases are shown. The coxBACF-encoded cytochrome aa3 is the predominant heme-copper oxidase for aerobic growth (gray box), and the fixNOQP-encoded cbb3-type oxidase (white box) supports respiration under free-living microoxic and under symbiotic conditions. aGene number according to RhizoBase (http://genome.kazusa.or.jp/rhizobase).
The contribution of Br. japonicum CoxG and ScoI to the biogenesis of the cbb3 oxidase has been recently investigated (38). These proteins are the respective paralogs of the mitochondrial chaperones Cox11 and Sco1 involved in the formation of CuB and CuA centers of cytochrome aa3 (12). Analyses of Br. japonicum coxG and scoI mutants revealed that disparate pathways are used for aa3- and cbb3-type oxidases biogenesis [(Bühler et al. (38)].
Oxygen concentration is the primary effector of fixNOQP expression in Br. japonicum. Under O2 limitation, the FixJ protein of the FixLJ system is phosphorylated by the oxygen-inhibitable, heme-based sensor kinase FixL (see section II.A Fig. 4b). The only known target of FixJ in Br. japonicum is fixK2, whose product, FixK2, has been shown to activate genes involved in anoxic or microoxic energy metabolism, including the fixNOQP operon (161). By using a transcriptomics approach (145), and FixK2-dependent in vitro transcription assay (148), the involvement of FixK2 as a direct transcriptional regulator of cbb3 has been demonstrated.
RegSR are members of the family of two-component regulatory redox-responsive proteins. In Br. japonicum, RegSR induces expression of the fixR-nifA operon, which encodes the NifA regulatory protein (15) (see Fig. 15). Targets of NifA include nif and fix genes, which are directly involved in nitrogen fixation, and also genes that are indirectly related to nitrogen fixation or have a unknown function in this process (83, 108, 162). Although fixNOQP genes are involved in nitrogen fixation, these genes were not identified as targets of NifA in microarray-based experiments (108). Transcription of fixR-nifA is dependent on NifA and on RegSR, respectively (Fig. 15). RegR induces expression of the fixR-nifA operon in both oxic and anoxic conditions, suggesting that RegSR responds to the overall redox state of the cell rather than to oxygen directly (15). Br. japonicum RegS possesses a highly conserved quinone binding site and a conserved redox-active cysteine, which suggests that the redox state of the membrane-localized quinone pool or the redox-active cysteine might be involved in redox sensing in Br. japonicum. However, the precise nature of the signal that is transduced by the Br. japonicum RegSR is unknown.
FIG. 15.
Schematic representation of the regulatory network of denitrification in Br. japonicum. FixLJ regulates fixK2 gene transcription in response to a shortage of oxygen, and FixK2 provides control of transcription on nap and nirK denitrification genes as well as the nnrR gene, whose product, in turn, activates transcription of nor genes. RegSR activates expression of fixRnifA operon presumably by sensing changes in the redox state of the cell. The NifA regulatory protein activates gene expression at very low oxygen concentrations. Control of denitrification genes by NifA has been demonstrated by our group (36). Furthermore, recent results have shown that NorC expression is significantly reduced in a Br. japonicum regR mutant (239). Sigma54 links both regulatory cascades. Positive regulation is denoted by arrows, and negative regulation is indicated by perpendicular lines.
Null mutations in the regR gene led to a dramatically decreased nitrogen fixation activity (15). Surprisingly, the phenotypic properties of regS mutants were largely indistinguishable from the wild type. Although it has been demonstrated, in vitro, that RegS is both a functional autokinase and a kinase (74), the in vivo role of RegS in RegR-mediated transcriptional activation is still unclear. In addition to fixR-nifA, a large number of novel members of the RegR regulon have been identified by transcriptomic analyses (134). Among them, a putative operon that encodes a predicted multidrug efflux system required for an efficient symbiosis specifically with soybean has been recently characterized (133).
In addition to oxygen respiration, Br. japonicum is able to obtain energy and grow from nitrate reduction to N2 through denitrification when cultured under oxygen-limiting conditions with nitrate as the terminal electron acceptor. In Br. japonicum, denitrification reactions depend on napEDABC, nirK, norCBQD, and nosRZDYFLX genes encoding the nitrate-, nitrite-, nitric oxide-, and nitrous oxide reductases, respectively (see section V.C). Inactivation of Br. japonicum cycA or napC, which encode cytochromes c involved in the electron transfer through denitrification pathway, decreases the expression of Br. japonicum cbb3 oxidase during nitrate-dependent anaerobic growth (35, 37). These results suggest that a change in the electron flow through the denitrification pathway may affect the quinone redox state, leading to alterations in cbb3 expression. To further explore the possibility of redox-dependent regulation of Br. japonicum cbb3 oxidase, the effect of reduced and oxidized carbon substrates on β-galactosidase activity from a fixP’–’lacZ fusion was investigated. Levels of fixP’–’lacZ expression were largely dependent on the oxidized or reduced nature of the carbon source in the medium (37). In order to study the involvement of Br. japonicum RegR in the redox-dependent regulation of fixNOQP genes, Bueno and colleagues (37) analyzed the expression of the fixP’–’lacZ fusion in a Br. japonicum regR mutant. When cells were grown under denitrifying conditions with an oxidized carbon source, there was a significant decrease in β-galactosidase activity in the regR mutant compared with the expression levels detected in the wild-type strain. These observations suggest that Br. japonicum fixNOQP operon might be under the control of RegR. However, a whole-genome transcription-profiling analysis of the Br. japonicum regR mutant grown under microoxic conditions demonstrated that transcript levels of fixNOQP operon were not RegR-dependent (134). In recent studies from our group (Maria J. Torres et al., unpublished results), fixNOQP genes were not identified as RegR targets after transcriptomic analyses of the regR mutant grown under denitrifying conditions with succinate (an oxidized carbon source).
E. Bacilus subtilis ResDE and Rex
The endospore-forming Gram-positive bacterium Bacilus subtilis utilizes a branched ETC under aerobic conditions (250). To date, three terminal oxidases have been identified in Ba. subtilis (Fig. 12a). Both cytochromes aa3 and caa3 have been identified as heme-copper oxidases. The third oxidase has been shown to be a member of the cytochrome bd family encoded by the cydABCD operon (255). In addition, the presence of a fourth terminal oxidase of bd type, YthAB, can be predicted from the genome sequence (6).
FIG. 12.
Aerobic respiratory chains (a) and regulation of bd, aa3, and caa3 terminal oxidase expression (b) in Bacilus subtilis. In (a), the quinone-reducing and quinol-oxidizing branches with terminal oxidases induced under oxic conditions (gray boxes), and microoxic conditions (white boxes), are shown. In (b), ResDE and Fnr provide positive transcriptional control in response to oxygen availability. ResD, but not ResE, is required for transcription of cyd operon (this complex regulation is indicated with a question mark in the figure). Redox-dependent repression of cydABCD (bd oxidase) by Rex is controlled by NAD+/NADH ratio in the cells. Operons qox and cta encode the aerobic terminal oxidases aa3 and caa3. Positive regulation (+) is denoted by arrows, and negative regulation (−) is indicated by perpendicular lines.
Cytochrome bd is produced under conditions of low-oxygen tension and in cells grown in the presence of glucose. A perfect 16-bp palindromic sequence, upstream of the translation start site for cydA, was proposed as a potential operator binding site for a regulatory protein (255). Gel-shift and DNase I footprinting analyses identified YdiH as a negative regulator of cyd genes (212). YdiH is a redox sensor protein whose activity is regulated by the levels of NAD+ and NADH in the cell (104). YdiH has been renamed Rex, as it is an ortholog of Rex in Streptomyces coelicolor (31) where this redox-sensitive transcription regulator was first described. In St. coelicolor, the Rex-DNA target region (Rex operator, ROP) is an 8-bp inverted repeat located upstream of genes encoding respiratory proteins, such as heme biosynthetic enzymes, NADH dehydrogenase, Rex itself, and cytochrome bd oxidase. In Staphylococcus aureus, it has been recently demonstrated, by combining various protein–DNA interaction studies with transcriptional analyses, that Rex acts on a multitude of anaerobically induced genes (172).
Brekasis and Paget (31) showed by electrophoretic mobility shift assay (EMSA) experiments that St. coelicolor Rex DNA-binding properties are influenced by the NADH/NAD+ ratio. Under aerobic conditions, raised NAD+/NADH ratio results in an increased binding of NAD+ to Rex, leading to its activation (Fig. 12b). Activated Rex binds to ROP, and inhibits the expression of several respiratory genes. However, under oxygen-limiting conditions, NADH replaces NAD+ bound to Rex, triggering a conformational change that dissociates Rex from the cyd-ROP sequence. Derepression of the cyd operon enables the cell to respire micromolar concentration of oxygen (31) (Fig. 12b). DNA binding studies of Ba. subtilis Rex with NADH or NAD+ showed that NAD+ boosted the binding activity of Rex, but that NADH seemed to have a negligible effect or a partial negative effect on DNA-binding activity (104). By contrast, in St. coelicolor, NADH completely inhibits DNA-binding activity. These data suggest that DNA-binding determinants of Ba. subtilis Rex are distinct from those of St. coelicolor Rex (31). Gyan and colleagues (104) proposed that Rex and NADH dehydrogenase together form a regulatory loop which functions to prevent a large fluctuation in the NADH/NAD+ratio in Ba. subtilis.
In order to understand the conformational modifications involved in the redox sensing by Rex, Sickmier and colleagues (220) performed X-ray studies of the Rex homolog in the thermophile Thermus aquaticus (T-Rex). Its structure comprises two main domains, an N-terminal domain that adopts a winged H-T-H fold and which most likely is the region that interacts with DNA and a C-terminal NADH binding Rossmann fold domain (220). These studies revealed that T-Rex possesses the same functional characteristics as the St. coelicolor homolog Rex, which lead to speculatation that the mechanism of repression of gene activation in response to the NADH/NAD+ redox pair is conserved among Rex family members. Small-angle X-ray scattering has been recently applied to obtain solution structures for Ba. subtilis apo B-Rex, B-Rex:NADH, and B-Rex:NAD+ showing that B-Rex presents rigid conformations for the complexes with NADH and NAD+ (251). Results from these studies indicated that the domain movements of B-Rex from the DNA bound form to the NADH (DNA-free) form could be different from the reported mechanism for T-Rex which is based on crystal structures (220).
In addition to Rex, transcription of the Ba. subtilis cytochrome bd oxidase during the transition between aerobic to anaerobic lifestyle is subjected to the action of other regulatory proteins such as Fnr, CcpA (carbon catabolite regulator protein), and the two-component system ResDE (188).
The ResD/ResE two-component signal transduction system has an important role in the regulation of both aerobic and anaerobic respiration [reviewed in (92)]. ResE is a membrane-bound sensor histidine kinase that, by responding to a redox signal(s), which have been postulated to be the redox state of intracellular MK, autophosphorylates and subsequently donates a phosphate to its cognate response regulator, ResD. It has been demonstrated that ResD activates transcription by interaction with the C-terminal domain of the alpha subunit of RNAP (91). The ResD/ResE system activates transcription of genes involved in the respiratory pathway that transfers electrons to oxygen (under oxic conditions) or to nitrate (under anoxic conditions) as well as genes encoding transcriptional factors such as Fnr (91, 92) (Fig. 12b). Since ResDE is essential for both ctaA and ctaB expression, which are required for heme A biosynthesis, resDE mutant strains lack cytochromes aa3 and caa3. ResD also activates the genes ctaCDEF, encoding the structural proteins for cytochrome caa3 (264). By using a cydA:lacZ transcriptional fusion in resD and resE mutant backgrounds, it was shown that ResD, but not ResE, is required for the transcription of the cydA promoter, suggesting that another sensor might be involved in cydA activation by ResD. The direct interaction of ResD with the cydA promoter was found around −58 and −108 nucleotides upstream from the transcription start site by using a DNase I footprinting assay (188).
IV. Nitrate Respiration: Denitrification and Ammonification
When faced with a shortage of oxygen, many bacterial species are able to use nitrate as an electron acceptor of the respiratory chain. This switch from oxygen to nitrate respiration leads to a reduction in the ATP yield rates, but allows bacteria to survive and multiply (221). Denitrification has been defined as the dissimilatory reduction of nitrate or nitrite to a gaseous N-oxide concomitant with free energy transduction (266). The process requires four separate enzymatically catalyzed reactions:
 |
For many years, it was believed that denitrification is performed exclusively by eubacteria. However, there are indications that some fungi (e.g., the pathogenic species Fusarium oxysporum) and archaea are also able to denitrify (236, 266). It has also been shown that nitrifiers also have genes involved in denitrification (44). Some bacteria such as E. coli or Ba. subtilis are able to perform nitrate respiration and release gaseous nitrogen oxides, but they do not denitrify with dinitrogen as a product. Instead, they reduce nitrate to ammonium, so-called nitrate-ammonification (45). In many species of nitrate ammonifying bacteria, there are two biochemically distinct nitrate reductases, one membrane-bound with the active site located in the cytoplasm and the other in the periplasm. These are coupled to two nitrite reductases (Nir) that provide independent pathways for nitrate reduction to ammonium (nitrate-ammonification) in the two cellular compartments. In the cytoplasm, nitrate is reduced to nitrite by a membrane-bound respiratory nitrate reductase system (NarGHI):
 |
The nitrite produced can then be further reduced to ammonium by a siroheme-containing Nir (NirB):
 |
In the periplasm, the process involves two different enzymes, a periplasmic nitrate reductase (NapA) that reduces nitrate to nitrite and a periplasmic cytochrome c Nir that further reduces nitrite to ammonium. This process of nitrate ammonification produces nitric oxide and nitrous oxide as side-products (95, 149, 222, 225).
Products of denitrification and nitrate ammonification have manifold, mainly adverse, effects on the atmosphere, soils, and waters and thus, have both an agronomic and environmental impact (10, 105). When nitrate is converted to gaseous nitrogen by denitrifying bacteria in agricultural soils, nitrogen is lost as an essential nutrient for the growth of plants. In contrast to ammonium, which is tightly bound in soil, nitrate is easily washed out and flows to the groundwater where it (and its reduction product nitrite) adversely affects water quality. In addition, nitrogenous oxides released from soils and waters are in part responsible for the depletion of the ozone layer above the Antarctic, and in part for the initiation of acid rain and global warming (192). Among them, N2O, commonly known as laughing gas, has received special attention in the last years, because it is a powerful greenhouse gas that can persist for up to 150 years while it is slowly broken down in the stratosphere. Although N2O only accounts for around 0.03% of total greenhouse gas emissions, it has a 300-fold greater potential for global warming effects, based on its radiative capacity compared with that of carbon dioxide (10, 13, 192, 195).
A. Respiratory nitrate reductases
The first reaction of denitrification and ammonification (reactions 1 and 5), the conversion of nitrate to nitrite, is catalyzed by a membrane-bound nitrate reductase (Nar), or a Nap [reviewed in (98, 153, 154, 184, 198, 199, 201, 247)].
The Nar enzyme employs a redox loop to couple quinol oxidation with proton translocation and energy conservation, which permits cell growth under oxygen-limiting conditions (23, 221). Nap is also linked to quinol oxidation, but does not transduce the free energy in the QH2-NO3− couple into proton motive force (PMF) to synthesize ATP (221). NO3—reduction via Nap can only be coupled to free-energy transduction if the primary quinone reductase, for example NADH dehydrogenase or formate dehydrogenase, generates an H+-electrochemical gradient (221). In contrast to Nar, which has a respiratory function, Nap systems have a range of physiological functions that include the disposal of reducing equivalents during aerobic growth on reduced carbon substrates and anaerobic nitrate respiration as a part of bacterial ammonification or denitrification pathways (184).
1. Membrane-bound respiratory nitrate reductase
The E. coli and Paracoccus Nar enzymes have been the focus of the most biochemical and genetic studies (reviewed in references just mentioned). Nar is a structurally defined 3-subunit enzyme composed of NarGHI (Fig. 13a) (24, 118). NarG is the catalytic subunit of about 140 kDa that contains a bis-molybdopterin guanine dinucleotide (bis-MGD) cofactor and a [4Fe-4S] cluster. NarH, of about 60 kDa, contains four additional iron-sulfur centers: one [3Fe-4S] and three [4Fe-4S]. NarG and NarH are located in the cytoplasm and associate with NarI. NarI is an integral membrane protein of about 25 kDa with five transmembrane helices and the N-terminus facing the periplasm (Fig. 13a). Nar proteins are encoded by genes of a narGHJI operon. The organization of this operon is conserved in most species that express Nar. The narGHI genes encode the structural subunits, and narJ encodes a dedicated chaperone required for the proper maturation and membrane insertion of Nar (27). E. coli has a functional duplicate of the narGHJI operon named narZYWV. The subunits of the two enzymes encoded by both operons are interexchangeable, but physiologically, NarZYWV has a function during stress response rather than anaerobic respiration per se (26, 224). A number of nar gene clusters in denitrifying bacteria also have a gene encoding a nitrogen ox anion transporter NarK that can transport nitrate into the cell and nitrite out of the cell (97, 257), a homolog of which is also found in many nitrate-ammonifying bacteria (45). A variation of the Nar system occurs in some archaea and bacteria where the NarGH subunits are on the outside, rather than the inside of the cytoplasmic membrane. These systems have thus far been less well studied biochemically, but a nitrate transporter is not needed for these systems (142).
FIG. 13.
Denitrification pathway (a) and regulation (b) in bacteria. In (a), the topological organization of denitrification enzymes is shown. The membrane-bound (NarGHI), and periplasmic, (NapABC) nitrate reductases as well as the nitrite reductases (Cu-type or cd1-type), nitric oxide reductases (cNor, qNor, and qCuANor), and nitrous oxide reductase (NosZ) are shown. (b) Regulatory network of denitrification genes in response to O2 concentration, nitrate/nitrite, and nitric oxide (NO). Positive regulation is denoted by arrows, and negative regulation is indicated by perpendicular lines. Further details are given in the text.
2. Periplasmic nitrate reductase
Nap is widespread in all classes of denitrifying and non-denitrifying proteobacteria [reviewed in (98, 153, 154, 184, 198, 199, 201, 247)]. The best-studied Nap enzymes were isolated from Paracoccus pantotrophus E. coli, Rho. sphaeroides, and Desulfovibrio desfuromonas (5, 64, 115). Nap is commonly a 2-subunit enzyme composed of the NapAB complex located in the periplasm and associates with a transmembrane NapC component (Fig. 13a). The catalytic subunit NapA contains the bis-MGD cofactor at its active site and an FeS center. NapB is diheme cytochrome c552, and NapC is a c-type tetra-heme membrane-anchored protein that is involved in the electron transfer from the quinol pool to NapAB (43, 207). Mono-subunit forms of NapA also occur, which may interact with small FeS proteins as well as cytochromes (114, 115, 140).
In contrast to the nar operon, the nap operons present considerable heterogeneity in gene composition as well as in ordering. Eight different genes have been identified as components for operons that encode Naps in different organisms (199). Except for Shewanella oneidensis, Wollinella succinogenes, Desulfobacterum hafniensi, and C. jejuni, which lack napB or napC, all operons studied thus far have the napABC genes in common. The remaining napDEFKL genes encode for different proteins that are not directly involved in the nitrate reduction. NapD is a cytoplasmic protein that acts as a chaperone. NapF is a cytoplasmic iron–sulfur containing protein with four loosely bound [4Fe–4S] clusters, and is thought to participate in the assembling of the iron–sulfur cluster of NapA (164, 170). The napEKL genes encode for proteins with so far unknown functions. In E. coli, the nap operon includes napGH genes encoding a periplasmic and an integral membrane protein with [4Fe–4S] clusters. NapH and NapG interact, making an electron transfer supercomplex that can channel electrons from both menaquinol and ubiquinol to NapA (32, 33).
B. Other enzymes in bacterial denitrification
The enzymes involved in denitrification are nitrate-, nitrite-, nitric oxide-, and nitrous oxide reductase, encoded by nar/nap, nir, nor, and nos genes, respectively. A scheme of the denitrification reactions is shown in Figure 13a. Comprehensive reviews covering the physiology, biochemistry, and molecular genetics of denitrification have been published elsewhere (11, 16, 110, 125, 199, 247, 248, 266).
1. Nitrite reductases
In denitrifying bacteria, two types of respiratory Nir have been described: the NirS cd1 nitrite reductase, a homodimeric enzyme with hemes c and d1, and the NirK, a copper-containing Nir [reviewed in (202, 203, 246, 247)]. Both are located in the periplasmic space, and receive electrons from cytochrome c and/or a blue copper protein, pseudoazurin via the cytochrome bc1 complex (Fig. 13a). They catalyze the one-electron reduction of nitrite to nitric oxide. Neither of the enzymes is electrogenic.
The cd1-type nitrite reductase (cd1Nir) is a homodimer, and each monomer carries a small heme c binding domain and a larger heme d1-binding domain. Electrons are transferred from the electron donor via the heme c of NirS to heme d1, where nitrite binds and is reduced to nitric oxide (202). The nirS gene encoding the cd1-Nir polypeptide is part of a nir gene cluster. The best-characterized clusters are those from the denitrifying species Ps. aeruginosa (nirSMCFDLGHJEN), Pa. denitrificans (nirXISECFDLGHJN), and Ps. stutzeri (nirSTBMCFDLGH and nirJEN, the two clusters being separated by a part of the nor gene cluster encoding nitric oxide reductase [Nor]).
Cu-type nitrite reductases (CuNir) are homotrimeric complexes harboring three type I copper centers, and three type II copper centers, which form the active site. Nitrite binds to the copper ion in the type II center, replacing an exogenous ligand (water or chloride), and by electron transfer from the type I copper site, nitrite is reduced to nitric oxide. The nirK gene encodes the CuNir (203).
2. Nitric oxide reductases
To date, three types of Nor have been characterized, and they are classified according to the nature of their electron donor as c-type nitric oxide-reductase (cNor), qNor, and qCuANor [reviewed in (59, 247, 248, 267)]. As a unique case, the Nor of Ros. denitrificans is similar to cNor, but differs in that it contains copper (143). The best-studied Nor is the cNor that uses membrane or soluble c-type cytochromes or small soluble blue copper proteins (azurin, pseudoazurin) as physiological electron donors. The qNor uses quinol or menaquinol as electron donors and it is found not only in denitrifying archaea and soil bacteria, but also in pathogenic microorganisms that do not denitrify (60). The qCuANor has thus far been found only in the Gram-positive bacterium Bacillus azotoformans (230). This enzyme is bifunctional using both menahydroquinone (MKH2) and a specific c-type cytochrome c551 as electron donor. It was suggested that the MKH2-linked activity of qCuANor serves detoxification and the c551 pathway has a bioenergetic function.
The cNor is an integral membrane enzyme harboring two subunits, NorC with a heme c group and NorB containing hemes b and a non-heme iron (Fig. 13a). Electron transfer from donor molecules to cNor is mediated by the cytochrome bc1 complex and a soluble cytochrome c or pseudoazurin (174). Electrons are transferred to the heme c in subunit II and then via the heme b to the active site. There, two molecules of nitric oxide are reduced. The molecular mechanism of the NO reduction by Nor has been extensively studied through chemical, biochemical, and physicochemical techniques (80, 81, 252). The crystal structure of Nor from Ps. aeruginosa has been recently solved (112). Although the overall structure of Nor is closely related to the heme copper oxidases, neither the D- nor K-proton pathway, which connect the HCO active center to the intracellular space, was observed. This confirmed early studies with NorCB in intact cells of Rho. capsulatus which showed that it was not a proton pump or electrogenic (196). Site-specific mutagenesis has suggested that protons required for the Nor reaction are probably provided from the periplasmic side of the membrane via a channel that involves conserved glutamate residues (41, 85, 86, 237).
cNOR is encoded by the norCBQD operon. The norC and norB genes encode subunit II and subunit I, respectively. The norQ and norD genes encode proteins essential for activation of cNor. Some more specialized denitrifiers have additional norEF genes, the products of which are involved in maturation and/or stability of Nor activity (107).
3. Nitrous oxide reductase
The final step in denitrification consists of the two-electron reduction of nitrous oxide to dinitrogen gas. This reaction is performed by a copper containing homodimeric soluble protein located in the periplasmic space, the nitrous oxide reductase (Nos) [reviewed in (246–248, 268)] (Fig. 13a). The enzyme has been purified from a large number of denitrifying strains, including Pa. denitrificans Pa. Pantotrophus, and Ps. stutzeri. Nos is a homo-dimer of a 65 kDa copper-containing subunit. Each monomer is made up of two domains: the “CuA domain” and the “‘Cuz domain.” The recently reported structure of purple N2OR from Ps. stutzeri has revealed that N2O bound side-on at CuZ, in close proximity to CuA (181). Electron input into CuA is usually via c-type cytochromes or cupredoxins (20, 22). The nos gene clusters often comprise the nosRZDFYLX genes. The nosZ gene encodes the monomers of Nos. The nosDFYL genes encode proteins that are apparently required for copper assemblage into Nos, although their specific role still remains unknown. The NosRX proteins have roles in transcription regulation, activation, and Cu assemblage of Nos (268).
C. Oxygen and nitrogen oxides control of nitrate respiration
The key molecules that act as signals for the regulation of nitrate respiration through denitrification or ammonification pathways are oxygen, nitrate, nitrite, and NO. Sensing of each of these molecules involves several types of sensor proteins (see Fig. 13b).
1. Oxygen sensors
There are two types of oxygen sensors involved in regulation of nitrate respiration, Fnr and FixL (reviewed in section II.A; see Fig. 4). Fnr controls the expression of genes involved in anaerobic respiration when O2 is absent. For example, the nar and nap operons in E. coli and Ba. subtilis (193, 228, 238, 241) are activated by Fnr under anaerobic conditions (Fig. 13b). Pa. denitrificans FnrP controls expression of the nar gene cluster and the cco-gene cluster encoding the cbb3-type oxidase (30, 249). Oxygen tension is sensed in Ps. aeruginosa by the Anr regulator, which carries a [4Fe-4S]2+ cluster (262).
2. Nitrate and nitrite sensors
In denitrifying species and species that anaerobically reduce nitrate to amonium, there are three types of nitrate/nitrite sensing systems: NarXL, NarQP, and NarR. NarXL and NarQP are members of two-component regulatory systems (Fig. 13b). The NarX and NarQ proteins are the signal sensors, and NarL and NarP proteins are their cognate response regulators, respectively (227). In E. coli NarL and NarP bind DNA to control induction of the nar and nap operons and repression of genes encoding alternate anaerobic respiratory enzymes (58, 227, 228). The effects of nitrate and nitrite on the E. coli transcriptome during anaerobic growth have been investigated, resulting in a new list of operons that are regulated by Fnr, NarL, and NarP (48). To date, narXL and narQP genes are confined to species classified in the γ and β subdivisions of the proteobacteria such as Escherichia, Salmonella, Klebsiella,Yersinia, Burkholderia, Ralstonia, Neisseria, and Pseudomonas species among others. In Ps. aeruginosa, the regulators Anr, Dnr, and NarL in concert with integration host factor (IHF) activate transcription of the narK1K2GHJI operon encoding nitrate reductase and two transporters in response to oxygen limitation, nitrate, and N-oxides (213). Recently, it has been shown that during anaerobic growth of Ps. aeruginosa PAO1, NarL directly represses expression of Nap, while inducing maximal expression of membrane nitrate reductase (245).
NarR is a member of the Fnr family of transcription activators, but it lacks a [4Fe-4S] cluster. NarR of Pa. pantotrophus and Pa. denitrificans is specifically required for transcription of the narKGHJI genes in the presence of nitrite (249, 256) (Fig. 13b). Genes encoding NarR are found in the α-proteobacteria Brucella suis, Brucella melitensis, Pa. Denitrificans, and Pa. pantotrophus. There are no indications that they have counterparts of narXL. It, therefore, seems that NarR substitutes the NarXL system in the α-proteobacteria. A recent proteomic analysis of Pa. denitrificans fnrP, nnr, and narR mutant strains grown oxically, microoxically, and microoxically in the presence of nitrate has revealed new proteins involved in the FnrP, Nnr, and NarR regulons (30). Data from this work are in full agreement with the current view on the FnrP protein being a sensor for oxygen and on the Nnr and NarR proteins being sensors for intermediates of nitrate respiration. This study has also revealed that FnrP, Nnr, and NarR control transcription of target genes having both activating and repressing functions (30).
3. NO sensors
NO is a toxic, free radical gas with diverse biological roles in eukaryotes and bacteria. Several bacterial transcriptional regulators sense this molecule and regulate the expression of genes involved in both denitrification and NO detoxification [reviewed in (206, 226, 243, 267)]. Bacterial NO regulatory proteins can be roughly divided into (i) proteins whose primary role is not to sense NO, but which contain metal centers or cysteines that are nitrosylated by NO and (ii) those that appear to be solely dedicated to sensing NO (i.e., they regulate genes involved in NO detoxification). An example of the first class is Fnr, whose primary role is to sense oxygen but that has also been shown to sense and respond to NO in E. coli (see section II.A).
The second class of NO sensors includes nitrite and nitric oxide reductase regulator (NnrR), NorR, and NsrR. NnrR also belongs to the Fnr family of transcription activators, but, similar to NarR, it lacks the cysteines to incorporate a [4Fe-4S] cluster. NnrR orthologs, sometimes named as Nnr, DnrR, or FnrD, are found in nearly all species containing the reductases Nir and Nor. In the presence of NO, these regulators orchestrate the expression of the nir and nor gene clusters. In many species, nnrR gene and its target genes are situated in close proximity on the chromosome, suggesting a local specificity. The promoters of these operons contain NnrR binding sites that resemble the consensus Fnr-box to a large extent. Nnr homologs, including NnrR in Rho. sphaeroides (131) and Dnr in Ps. aeruginosa, most likely sense NO via a ligated heme (93, 94). In denitrifying bacteria, knockout mutagenesis has shown that the Dnr and NnrR regulators are needed for the transcriptional control of the nir, nor, and nos operons.
NorR is another NO-responsive protein that was first identified in Ralstonia eutropha (180). This bacterium has two copies of the norR gene, both of which are located upstream of their norAB gene clusters where norB encodes a single-subunit Nor of the qNor type. NorR is a member of the NtrC family of response regulators. The absence of possible phosphorylation sites as well as the presence of a conserved GAF domain indicative for signal perception suggest that the protein belongs to a sub-family of response regulators which sense their signal themselves rather than via a cognate signal sensor (55). In response to anaerobiosis and the presence of NO, the NorR proteins specifically activate transcription of the σ54-dependent norAB promoters and repress their own synthesis via negative autoregulation (39). NorR has also been found in E. Coli, which senses NO directly through a mononuclear nonheme iron center and responds by switching on expression of the flavorubredoxin NorVW to detoxify NO (242).
NsrR is an iron–sulfur-containing protein that senses NO directly via a [2Fe–2S] cluster. Nitrosylation of this cluster leads to a loss of DNA binding activity and, hence, derepression of NsrR target genes. In E. coli, this transcription repressor was shown to sense reactive nitrogen species and to switch on a regulon of at least 60 genes, including genes involved in nitrate respiration (82, 226, 243). In denitrifying bacteria, NsrR appears to have a specific role in coordinating production of the nitrite and NO reductase enzymes to prevent the build up of NO (243). Intriguingly, the same role is performed by Nnr homologs in denitrifying bacteria that do not contain NsrR. Two components of the denitrification pathway are transcriptionally regulated by NsrR in Neisseria meningitidis: the membrane-bound nitrite reductase AniA and the respiratory NO reductase NorB (111). In the closely related Neisseria gonorrhoeae, NsrR represses both norB and aniA expression in the absence of NO and, most notably, also exerts an autoregulatory effect on its own expression (113, 171).
4. Other regulators in denitrifying bacteria
In addition to the regulatory proteins that can monitor oxygen, nitrate, nitrite, and NO, regulation of the on-set and fine-tuning of nitrate respiration in some bacteria involves copper responsive regulators, redox sensing mechanisms and the NosR and NirI proteins (Van Spanning, 2011). The NosR is a membrane-bound protein containing six transmembrane helices, a large periplasmic domain, and cysteine-rich cytoplasmic domains that resemble the binding sites of [4Fe-4S] clusters. The periplasmic domain of NosR has a structural similarity with the FMN-binding domain of the NqrC subunit of the Na+-translocating NADH:quinone oxidoreductase and may play a role in activating or repairing oxygen damaged Nos (260). Although NosR proteins do not have properties of transcription factors, they are required for transcription activation of the nos genes in denitrifying bacteria. However, the regulatory function of NosR is not well understood (268). In Pa. Denitrificans, a homolog of nosR, nirI, representing thus far a singular case among the denitrifying bacteria, has been studied for its role in the transcription of nirS, encoding the cytochrome cd1Nir.
V. Redox Control of Nitrate Respiration
Nitrate reduction by the membrane-bound Nar generates an H+-motive force (PMF) across the cytoplasmic membrane of the cell, allowing ATP synthesis (197, 221). During denitrification, the periplasmic Nir and Nos cannot themselves be involved in H+-movement across the membrane. Although Nor is an integral membrane protein, several lines of evidence show that H+ required for reduction of NO are taken from the periplasm and thus, no net charge movement occurs (see section IV.B). However, e-transfer from ubiquinol to NO2−, NO or N2O is coupled to H+-translocation across the membrane, because this involves the cytochrome bc1 complex (197). Quinol oxidation by the periplasmic Nap is not directly coupled to the generation of a PMF and is independent of the cytochrome bc1 complex. In many bacteria, Nap has a role in redox balancing under aerobic conditions (20, 42); indeed it seems to be biochemically tuned for this function (88). The need to dissipate reductants is likely to be most acute during the metabolism of a reduced carbon substrate under conditions that are both oxygen and energy sufficient. Accordingly, expression of the Nap is significantly higher in Paracoccus species grown aerobically with butyrate or caproate to that when grown with malate or succinate (69, 71, 196, 215, 216). Taken together, these results suggest that Nap constitutes an example of redox-dependent regulation of the respiratory network in Paracoccus species. Two crucial regulatory sequences that control expression of the nap operon in response to the redox status of the cell were identified in Pa. pantotrophus, but the redox responsive transcriptional regulator has yet to be described (70). The use of Nap in redox homoeostasis has been also shown in photosynthetic bacteria where it regulates the redox poise of the cyclic photosynthetic electron transport system (117), particularly during photoheterotrophic growth on reduced carbon substrates (89, 200).
Many bacteria can also recruit Nap for anaerobic nitrate respiration as a part of bacterial ammonification (Enterobacteriaceae) or to promote denitrification (Rhizobiaceae). In fact, some rhizobia species (e.g., Pseudomonas sp. G179 [actually Rhizobium galegae], Br. japonicum) can express nap genes under anaerobic conditions, and disruption of the nap genes is lethal for growth under denitrifying conditions (17, 62). In addition, in Rho. sphaeroides f. sp. denitrificans, which can express both Nar and Nap, inactivation of Nap is lethal for anaerobic denitrification (135). Thus, in these organisms, the physiological role of Nap is in anaerobic denitrification. When considered as single enzymes, the energy coupling of Nar and Nap appear markedly different: q+/2e−=6 (Nar) and 4 (Nap) with NADH as electron donor and 2 (Nar) and 0 (Nap) with succinate as electron donor. However, when considered in the context of the entire denitrification pathway, the q+/2e− ratio is 24 (Nar) or 22 (Nap) with NADH and 8 (Nar) or 6 (Nap) with succinate. Thus, the energetic loss of using Nap rather than Nar is only 8% when NADH is the electron donor to the respiratory system (197).
To date, the knowledge about the redox-dependent control of anaerobic nitrate respiration as well as the identification of the regulators and the signal transduction mechanism involved in this response is limited. In this section, we will focus on recent advances in understanding nitrate respiration redox-dependent control in Rho. sphaeroides, Ag. tumefaciens, Br. japonicum, and Ba. subtilis.
A. Rho. sphaeroides
Rho. sphaeroides strains are photosynthetic bacteria able to induce the denitrification pathway by expressing the enzymes Nap, Nir, and cNor, which are encoded by the nap, nirK, and nor genes, respectively, when oxygen concentration is limited and nitrate or nitrite is present in the medium. In Rho. sphaeroides 2.4.3., Nap is the enzyme responsible for denitrification as judged by the fact that strains with mutations in the nap gene cluster lost nitrate reductase activity as well as the ability to grow with nitrate under anaerobic-dark conditions (135). In the strain IL106 of Rho. sphaeroides, nap genes showed the same level of expression under both oxic and denitrifying conditions with nitrate. Expression increases in the presence of the reduced carbon compound butyrate, indicating that this protein probably plays a role in redox homeostasis [reviewed in (218)]. A slightly different nap expression pattern was observed in Rho. sphaeroides strain 2.4.1, where expression of the nap operon increases significantly when cells are shifted from anoxic photosynthetic conditions to oxic conditions and do not require nitrate [reviewed in (218)].
Although the expression of nap has been shown to be variable, the expression of the genes encoding Nir and Nor only occurs under microoxic conditions. In Rho. sphaeroides 2.4.3, nir and nor expression requires low oxygen and also requires the presence of a nitrogen oxide. It has been shown that NnrR is necessary for the expression of nirK and nor during denitrification (240). Studies on regulation by NnrR demonstrated that the presence of NO is required for transcriptional activation of nirK and nor, yet the mechanism whereby NnrR senses NO is not known. Analysis of the Rho. sphaeroides 2.4.3 genome and expression studies demonstrated that genes designated paz and norEF are also members of the NnrR regulon involved in denitrification (107).
Genes for anaerobic growth in many photosynthetic bacteria are also regulated by the two-component PrrBA system. In Rho. sphaeroides strain 2.4.1 where nap genes are maximally expressed under oxic conditions, the phosphorylated form of the response regulator PrrA is responsible for repressing nap expression under low oxygen [reviewed in (218)], see Fig. 14a). By contrast, in Rho. sphaeroides 2.4.3, inactivation of prrA eliminated the ability to grow both photosynthetically and anaerobically in the dark on nitrite-amended medium (130). The PrrA-deficient strain exhibited a severe decrease in both Nir activity and expression of a nirK-lacZ fusion (130) when environmental oxygen tension was limited. This regulation is not mediated by NnrR, as nnrR is fully expressed in a PrrA mutant background (Fig. 14a). By using truncated versions of the nirK promoter fused to the lacZ maker gene, Laratta and colleagues (130) revealed that the DNA region expanded upstream to the putative NnrR binding site is required for full activation of nirK. These results suggested that this DNA region could act as a binding site of PrrA or additional regulatory proteins in order to control nirK expression. In fact, the presence upstream of the nirK start site of 4 imperfect repeats proposed as binding sites for RegR (a PrrA homolog in Br. japonicum) suggests again that PrrA could bind directly to such sites. However, binding assays using purified PrrA would be required to confirm this. Unlike the nirK promoter region, the nor promoter has no sites with similarity to the imperfect repeats proposed as binding sites for PrrA homologs, suggesting that PrrA may not directly regulate norB expression (130). The model proposed by Laratta and colleagues (130) to understand the mechanism of nirK control by PrrBA is that under low oxygen tensions, the kinase activity of PrrB increases relative to its phosphatase activity, resulting in an increase in the concentration of PrrA-P. Under microoxic conditions and in the presence of NO, NnrR together with PrrA-P activates transcription of nirK (Fig. 14a). As mentioned in section III.B (Fig. 8c), inactivation of genes encoding the cbb3 oxidase in Rho. sphaeroides has been shown to lead to high levels of PrrA-P under both oxic and microoxic conditions (190). Unexpectedly, loss of the cbb3 oxidase resulted in a significant decrease in nirK expression and Nir activity (130). Recent results have demonstrated that the cbb3 mutant retains Nir activity at very low oxygen concentrations (107). The simplest explanation for this result is that, under transition conditions, loss of the cbb3 oxidase results in high residual oxygen levels in the culture, which prevents expression of nirK and nor.
FIG. 14.
Schematic representation of denitrification control in Rho. sphaeroides (a) and Agrobacterium tumefaciens (b). (a) In Rho. Sphaeroides, PrrBA provides control of nirK expression in response to external redox variations, and NnrR provides control of expression of nirK and nor genes in response to NO. This figure is adapted from ref. (218). (b) In Ag. tumefaciens, ActSR provides control of nirK expression presumably in response to external redox variations, and FnrN and NnrR provide control of nirK and nor expression in response to low oxygen tension and presence of NO, respectively. Positive regulation is denoted by arrows, and negative regulation is indicated by perpendicular lines.
B. Ag. tumefaciens
Ag. tumefaciens is a plant pathogen that can grow under free-living conditions in the absence of oxygen via denitrification. The ability to respire nitrogen oxides could be significant for the survival and growth of free-living and plant-associated Ag. tumefaciens cells. The ability to respire nitrate has been shown to be advantageous to bacteria in the rhizosphere, as nitrate provides an alternative oxidant when oxygen concentrations are low (25). In addition, the ability to reduce nitrogen oxides may be useful for countering some of the plant defenses against plant pathogens (63, 79). Ag. tumefaciens is considered a partial denitrifier, as it lacks the genes encoding Nos (9). Consequently, nitrous oxide is the final product of denitrification.
Ag. tumefaciens has a nap cluster that is maximally expressed under denitrifying conditions, but is not influenced by nitrate (218). The regulator(s) responsible for expression of nap in Ag. tumefaciens are uncertain, as there are no conserved sequence motifs in the promoter region that might indicate the involvement of a particular family of regulators. Similar to Rho. sphaeroides, activation of nirK and norCBQD expression requires low oxygen concentration and the presence of NO (8, 9, 21). By using the lacZ marker gene fused to the nirK and nor promoter and by in vitro binding assay experiments, it has been demonstrated that activation of nirK and nor requires NnrR. This protein is directly activated by FnrN when oxygen tension in the environment becomes depleted (Fig. 14b). However, activation of nirK and nor is raised when NO, as a reduction product of nitrate or nitrite, is present in the medium, due to the maximal NO-dependent activation of NnrR. In Ag. tumefaciens, electron transfer through the denitrification pathway is facilitated by soluble small cytochromes such as the copper-containing pseudoazurin, encoded by the paz gene, whose expression also depends on FnrN and NnrR. In addition to FnrN and NnrR, the ActRS system is involved in denitrification (Fig. 14b). As observed in Rho. sphaeroides, insertional inactivation of the response regulator ActR significantly reduced nirK expression and Nir activity but not nnrR expression. In Ag. tumefaciens, a putative ActR binding site was identified in the nirK promoter region using mutational analysis and an in vitro binding assay (8). These studies also show that purified ActR bound to the nirK promoter but not to the nor or nnrR promoter. Additional experiments revealed that expression of paz was highly reduced in the actR or fnrN mutants and that ActR binds to the paz promoter (8). Due to the similarity to the PrrBA/RegBA systems, the ActSR system is believed to mediate gene activation on sensing redox changes originated from the ETC through cbb3-oxidase (PrrB) or through the redox state of the UQ pool (RegB). However, there is no experimental evidence as yet to confirm these hypotheses.
With all of this experimental evidence, we can see that activation of denitrification in Ag. tumefaciens depends on regulatory proteins involved in sensing different environmental signals. While the Fe-S-containing FnrN protein is an oxygen sensor, NnrR activates denitrification genes in response to NO. In this complex regulatory process, the Prr/Reg orthologs protein ActSR would adjust the denitrification machinery in response to redox status as an indirect measure of external oxygen tension (Fig. 14b).
C. Br. japonicum
Similar to Ag. tumefaciens, the nitrogen-fixing root nodule symbiont of soybean Br. japonicum has the ability to sustain cell growth by using nitrate as respiratory substrate when oxygen concentrations are very low. In contrast to Ag. tumefaciens, Br. japonicum contains all the denitrification enzymatic machinery necessary to reduce sequentially nitrate to molecular nitrogen.
In Br. japonicum, denitrification is dependent on the napEDABC, nirK, norCBQD, and nosRZDYFLX genes that encode an Nap, a Cu-containing Nir (NirK), a cNor, and an Nos, respectively (16). In addition, accessory cytochromes are necessary to support electron transport during denitrification. For example, the cytochrome c550, encoded by cycA, is essential for the electron delivery to the NirK reductase (35). Very recently, it has also been found that the cytochrome c6, encoded by the cycS gene, has a role in denitrification, although its precise biochemical function still remains unknown (145). Neither azurin- nor pseudoazurin-like copper proteins has been annotated in the genome sequence of Br. japonicum (www.kazusa.jp/rhizobase/).
In Br. japonicum, maximal expression of denitrification genes requires both the absence of oxygen and the presence of nitrate or a derived N-oxide. Microoxic induction of transcriptional fusions from the nap, nir, nor, and nos promoter regions to the lacZ reporter gene depends on the FixLJ/FixK2 regulatory cascade (Fig. 15) (16, 204). Sequences homologous to the consensus DNA binding sites of the Fnr and FixK proteins are present in the promoter region of each denitrification gene. Regulatory studies indicated that, in addition to low oxygen, nitrate, or an N-oxide derived from it, presumably either NO2− or NO or both, are required for maximal induction of Br. japonicum denitrification genes. This N oxide-mediated induction of nap, nir, and nor genes depended on NnrR (144, 204). Thus, NnrR expands the FixLJ/FixK2 regulatory cascade by an additional control level that integrates the N oxide signal required for maximal induction of the denitrification genes (Fig. 15). Induction of the norCBQD promoter is completely abolished in the absence of a functional nnrR gene. By contrast, microoxic induction of the nap or nirK promoters is retained in a nnrR mutant background, implying that the nap or nirK and the norCBQD promoters exhibit slight differences with regard to their dependence on FixK2. In this context, recent results from our group have demonstrated that purified FixK2 activates transcription from nap- or nirK-dependent promoters but not from nor-dependent promoter (E. Bueno, unpublished work). By contrast, ITC allowed us to demonstrate that NnrR bound to a specific DNA fragment from the promoter region of the nor genes, but not to those from the nap and nirK genes and that this interaction requires anoxic conditions but not the presence of an N oxide (unpublished results from our laboratory). Supporting these observations, a genome-wide transcription profiling of Br. japonicum fixJ and fixK2 mutant strains grown in free-living microoxic conditions have shown that nap, nirK, and nnrR but not nor are targets of fixK2 regulatory gene (145).
In addition to FixLJ/FixK2-NnrR, in Br. Japonicum, a second oxygen responsive regulatory cascade has been described, the RegSR/NifA cascade (Fig. 15). NifA activates gene expression in concert with the RNAP containing the alternative sigma factor (σ54), which, in Br. japonicum, is encoded by the two highly similar and functionally equivalent genes (rpoN1 and rpoN2) (127). Since rpoN1 is under the control of FixK2, this gene represents the link between the two regulatory cascades.
While the FixLJ/FixK2-NnrR cascade activates gene transcription at a moderately low oxygen concentration (≤5%), activation of the RegSR/NifA system requires oxygen concentration ≤0.5% (214). Recent results from our group showed that NifA is required for maximal expression of nap, nirK, and nor genes, suggesting a role for the RegSR/NifA regulatory cascade in the control of the denitrification process in Br. japonicum (36). In that study, it was shown that disruption of nifA caused a growth defect in Br. japonicum cells when grown under denitrifying conditions, as well as decreased activity of Nap and Nir enzymes. Furthermore, expression of napE–lacZ, nirK–lacZ, or norC–lacZ transcriptional fusions, as well as levels of nirK transcripts, were significantly reduced in the nifA mutant after incubation under nitrate-respiring conditions. These results suggest that maximal induction of denitrification genes is only triggered once the cellular oxygen falls to a sufficiently low level, that is, within the sensory range of the NifA-dependent regulatory cascade. Similarly, in Ag. tumefaciens, a study of expression and activity of Nir and Nor as a function of oxygen has shown that nirK and nor are expressed once oxygen reaches <0.5 μM or approximately 0.2% of air-saturating oxygen (21). These oxygen concentrations are low enough to allow formation of ActR-P necessary for nirK activation in Ag. tumefaciens.
Concerning the involvement of Br. japonicum RegSR in denitrification, it has been shown that free-living growth of regR mutants under anoxic conditions with nitrate as the terminal electron acceptor was severely impaired [Bauer et al. (15)]. Furthermore, expression of NorC is significantly lower in membranes from the regR mutant (239). As just mentioned, it has been shown that disruption of Rho. sphaeroides prrA or prrB causes a significant decrease in both nirK expression and Nir activity. Similarly, in Ag. tumefaciens, it has been shown that purified ActR binds to the nirK promoter, but not to the nor or nnrR promoters. In Br. japonicum, the involvement of RegR in nirK expression is at the moment unknown, and further investigations are needed to demonstrate the involvement of RegR in norCBQD gene expression and to establish whether these genes are direct, or indirect, targets of RegR.
D. Ba. subtilis
Once the oxygen present in the medium has been depleted by the bd oxidase (see section III.E; Fig. 12a), Ba. subtilis has the ability to exploit the use of an alternative final electron acceptor such as nitrate (158). Two distinct operons specifying two nitrate reductases have been characterized: (i) narGHJI encoding a respiratory nitrate reductase, which is required for nitrate respiration and is dependent on Fnr, and (ii) nasBC encoding a assimilatory nitrate reductase involved in the assimilation of nitrogen from nitrate under oxic conditions (159) and references therein) (Fig. 16). Ba. subtilis does not contain nirK, nor, or nos denitrification genes. Under anoxic conditions with nitrate or nitrite, this bacterium reduces nitrite to ammonium by the Nir NasDE encoded by nasD and nasE (Fig. 16) (i.e., reactions 5 and 6). The NasDE Nir was previously identified as an assimilatory enzyme [(157) and references therein]. Under aerobic conditions of nitrogen limitation, this enzyme catalyzes the reduction of nitrite to ammonia for the anabolic incorporation of nitrogen into biomolecules. It also functions catabolically in anaerobic respiration, which involves the use of nitrite as a terminal electron acceptor (157). Nitrite reduction catalyzed by Ba. subtilis Nir does not result in a proton gradient and coupled ATP generation. Instead, nitrite enhances anaerobic growth by serving as an electron sink to recycle the cellular pyridine nucleotide pool (157).
FIG. 16.
Schematic representation of denitrification control in Ba. subtilis. ResDE regulates gene transcription in response to both redox/oxygen changes and presence of NO, Fnr provides control in response to low oxygen concentration, NsrR in response to NO, and TnrA in response to high oxygen concentration and limited nitrogen availability. narGHJI encodes a respiratory nitrate reductase. nasBC encodes an assimilatory nitrate reductase; nasDEF, a nitrite reductase; and hmp, a flavohemoglobin. Positive regulation is denoted by arrows, and negative regulation is indicated by perpendicular lines.
The nasDE genes, together with nasBC (encoding assimilatory nitrate reductase) and nasF (required for Nir siroheme cofactor formation), constitute the nas operon (157). Transcription of nasDEF is driven not only by the nasBCDEF operon promoter, but also from an internal promoter residing between the nasC and nasD genes. Transcription from both promoters is activated by nitrogen limitation during aerobic growth by the nitrogen regulator, TnrA. However, under conditions of oxygen limitation, nasDEF expression and Nir activity were significantly induced (157, 158).
The expression of genes involved in nitrate respiration in Ba. subtilis is also regulated by the ResDE two-component signal transduction system (Fig. 16). To successfully switch from aerobic growth to nitrate respiration, the ResDE two-component signal transduction system should be activated, which allows induction of genes that function in anaerobic metabolism including fnr, nasDEF, and hmp (encoding flavohemoglobin) [(90) and references therein]. Footprinting analysis revealed that ResD tandemly binds to the −41 to −83 region of hmp and the −46 to −92 region of nasD (90). In vitro run-off transcription experiments showed that ResD is necessary and sufficient to activate transcription of the ResDE regulon. Although phosphorylation of ResD by ResE kinase greatly stimulated transcription, unphosphorylated ResD, and ResD with a phosphorylation site (Asp57) mutation, it was able to activate transcription at a low level (90). These results suggest that ResD itself, in addition to its activation through phosphorylation-mediated conformation change, senses oxygen limitation via an unknown mechanism leading to anoxic gene activation. Although ResDE was initially believed to sense anoxic conditions, subsequent studies have revealed that NO is also required (151, 155). It has been proposed that Ba. subtilis senses NO and responds through activation of ResDE-dependent transcription (156). In this context, it has been recently discovered that the NO-sensitive transcription factor NsrR is involved in ResDE-dependent gene regulation (156). In Ba. subtilis, NsrR represses transcription of the Nir (nasDEF) genes that are under positive control of ResDE (Fig. 16). Derepression is achieved by reaction of NO with NsrR. Anaerobically purified Ba. subtilis NsrR (BsNsrR) bears a [4Fe–4S] cluster that, on exposure to NO, forms dinitrosyl iron complexes (263). [4Fe–4S]-NsrR binds around the −35 element of the nasD promoter with much higher affinity than apo-NsrR, and this binding is sensitive to NO. RNAP and phosphorylated ResD make a ternary complex at the nasD promoter, and NsrR dissociates the preformed ternary complex (123).
VI. Concluding Remarks
Genetic, biochemical, and transcriptomic analyses have revealed that in many bacterial species, the adaptation of respiratory metabolism to changing environments with different oxygen conditions is controlled by Fnr-type transcriptional regulators that directly sense O2 or by the Fnr paralog, FixK; which senses O2 through the FixLJ two-component system. In addition to external oxygen concentration, other signals such as redox changes can regulate the expression of genes involved in respiration. Two-component regulatory systems (e.g., ArcBA, RegBA/PrrBA, RoxSR, RegSR, ResDE, and ActSR) from different bacterial species integrate the response to multiple redox signals. The redox state of the membrane-localized UQ/ubiquinol pool as well as the redox state of the conserved cysteine265 were considered as major indicators of redox variations detected by these regulatory systems. Recently, it has been proposed that both signals are able of functioning independently (259). In addition, a third model has been proposed in Rho. sphaeroides and Pseudomonas, where redox changes are sensed by the terminal oxidase cbb3, which modulates PrrB and RoxS kinase/phosphatase activity (47, 122, 152). Those redox-dependent responses are complex, as recent reports have proposed that the activity of the sensor kinase ArcA from E. coli can also be modulated by the redox state of the MK pool (18) or the presence of fermentation products (208). Particulary interesting is the case for the Gram-positive bacteria such as Ba. subtilis, where, in addition to the Fnr-type and the ResDE two-component system, the respiratory shift from oxic to microoxic conditions is also controlled by the repressor Rex, whose activity is influenced by the balance of the NADH/NAD+ pool in the cell (31, 92).
The respiratory shift in response to changes in oxygen concentration becomes even more sophisticated in bacterial species that use nitrate as respiratory substrate through denitrification or ammonification pathways. This shift is controlled not only by oxygen limitation (FixL and Fnr-type), but also by the presence of nitrate/nitrite (NarXL, NarQP, and NarR) or/and NO (NnrR, NorR, and NsrR). Furthermore, recent findings have shown that redox-responsive two-component regulatory systems (e.g., PrrBA, ActSR, RegSR, and ResDE) are also involved in the regulation of genes needed for anaerobic nitrate respiration. Intriguingly, Ba. subtilis ResDE requires anaerobiosis and the presence of NO to activate transcription of nitrate respiration genes (151, 155, 156). In this context, it has been recently discovered that the NO-sensitive transcription factor NsrR plays a role in ResDE-dependent gene regulation (123, 156). However, in Gram negative bacteria, the involvement of such NO-sensitive repressor in regulation of denitrification genes by others redox-responsive regulators (PrrBA, ActSR, RegSR, etc) is at the moment unknown.
In summary, it is clear that bacteria adopt a range of strategies to monitor the redox cellular state to coordinate the respiratory shift from oxic to anoxic environments. This control is based on a complex regulatory network where different regulators responding to diverse redox signals interact and control expression of target genes. The study and comparison of regulatory pathways in different bacterial groups is very useful to better understand how bacteria integrate different signals and how the regulatory systems respond to these signals. In spite of their diversity, regulatory networks from different bacteria share common proteins as well as their complexity. With the advent of the post-genomic era, and the move toward more predictive biology, a better understanding of how these complex regulatory circuits interact to integrate transcriptional responses to multiple environmental cues will be achieved.
Abbreviations Used
- Arc
aerobic respiration control
- cd1Nir
cd1-type nitrite reductase
- CIO
cyanide insensitive oxidase
- Crp
cAMP receptor protein
- CuNir
Cu-type nitrite reductase
- Cys
cysteine
- Cyt
cytochrome
- DMSO
dimethyl sulfoxide
- ETC
electron transport chain
- Fnr
fumarate and nitrate reductase regulatory protein
- HCO
heme-copper oxidase
- His
histidine
- H-T-H
helix-turn-helix motif
- ITC
isothermal titration calorimetry
- MK
menaquinone
- MKH2
menahydroquinone
- NDH
NADH dehydrogenase
- Nap
periplasmic nitrate reductase
- Nar
membrane-bound nitrate reductase
- Nir
nitrite reductase
- NnrR
nitrite and nitric oxide reductase regulator
- Nor
nitric oxide reductase
- Nos
nitrous oxide reductase
- NrfA
cytochrome c nitrite reductase
- PMF
proton motive force
- RNAP
RNA polymerase
- SDH
succinate dehydrogenase
- TM
transmembrane
- UQ
ubiquinone
- UQH2
ubihydroquinone
Acknowledgments
The authors would like to thank all of their colleagues who have contributed to the research arising from their laboratories that is discussed in this article and also Junta de Andalucía (grant RMN-4746 partially funded by FEDER), CYTED (grant 107PICO312), and Ministerio de Ciencia e Innovación (grants AGL2010-18607 and 200940I116 partially funded by FEDER) for financial support. Support from the Junta de Andalucía (BIO-275) is also acknowledged. D.J.R. thanks the Royal Society and Wolfson Foundation for a Merit Award Fellowship and the Biotechnology and Biological Research Council for financial support for N-cycle research.
References
- 1.Alvarez-Ortega C. Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez AF. Georgellis D. In vitro and in vivo analysis of the ArcB/A redox signaling pathway. Methods Enzymol. 2010;471:205–228. doi: 10.1016/S0076-6879(10)71012-0. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H. Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai H. Kodama T. Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 5.Arnoux P. Sabaty M. Alric J. Frangioni B. Guigliarelli B. Adriano JM. Pignol D. Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat Struct Biol. 2003;10:928–934. doi: 10.1038/nsb994. [DOI] [PubMed] [Google Scholar]
- 6.Azarkina N. Siletsky S. Borisov V. von Wachenfeldt C. Hederstedt L. Konstantinov AA. A cytochrome bb’-type quinol oxidase in Bacillus subtilis strain 168. J Biol Chem. 1999;274:32810–32817. doi: 10.1074/jbc.274.46.32810. [DOI] [PubMed] [Google Scholar]
- 7.Badrick AC. Hamilton AJ. Bernhardt PV. Jones CE. Kappler U. Jennings MP. McEwan AG. PrrC, a Sco homologue from Rhodobacter sphaeroides, possesses thiol-disulfide oxidoreductase activity. FEBS Lett. 2007;581:4663–4667. doi: 10.1016/j.febslet.2007.08.058. [DOI] [PubMed] [Google Scholar]
- 8.Baek SH. Hartsock A. Shapleigh JP. Agrobacterium tumefaciens C58 uses ActR and FnrN to control nirK and nor expression. J Bacteriol. 2008;190:78–86. doi: 10.1128/JB.00792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek SH. Shapleigh JP. Expression of nitrite and nitric oxide reductases in free-living and plant-associated Agrobacterium tumefaciens C58 cells. Appl Environ Microbiol. 2005;71:4427–4436. doi: 10.1128/AEM.71.8.4427-4436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baggs EM. Philippot L. Nitrous oxide production in the terrestrial environment. In: Moir JWB, editor. Nitrogen Cycling in Bacteria. Norkfolk, UK: Caister Academic Press; 2011. [Google Scholar]
- 11.Baker SC. Ferguson SJ. Ludwig B. Page MD. Richter OM. van Spanning RJ. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol Mol Biol Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balatri E. Banci L. Bertini I. Cantini F. Ciofi-Baffoni S. Solution structure of Sco1: a thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure. 2003;11:1431–1443. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Bates BC. Climate Change and Water, edited by IPCC Secretariat 2008. http://www.ipcc.ch/meetings/session28/doc13.pdf http://www.ipcc.ch/meetings/session28/doc13.pdf
- 14.Bauer CE. Elsen S. Bird TH. Mechanisms for redox control of gene expression. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 15.Bauer E. Kaspar T. Fischer HM. Hennecke H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J Bacteriol. 1998;180:3853–3863. doi: 10.1128/jb.180.15.3853-3863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedmar EJ. Mesa S. Velasco L. Robles EF. Bonnard N. Delgado MJ. Vigara AJ. Vílchez C. Garbayo I. Desnitrificación en Bradyrhizobium japonicum: Un misterio resuelto. In: Vega JM, editor; Márquez AJ, editor; Avances en el Metabolismo del Nitrógeno: De los Microorganismos a las Plantas. Huelva, Spain: Servicio de Publicaciones de la Universidad de Huelva; 2005. pp. 35–44. [Google Scholar]
- 17.Bedzyk L. Wang T. Ye RW. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J Bacteriol. 1999;181:2802–2806. doi: 10.1128/jb.181.9.2802-2806.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bekker M. Alexeeva S. Laan W. Sawers G. Teixeira de Mattos J. Hellingwerf K. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J Bacteriol. 2010;192:746–754. doi: 10.1128/JB.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belevich I. Borisov VB. Konstantinov AA. Verkhovsky MI. Oxygenated complex of cytochrome bd from Escherichia coli: stability and photolability. FEBS Lett. 2005;579:4567–4570. doi: 10.1016/j.febslet.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Bell LC. Richardson DJ. Ferguson SJ. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- 21.Bergaust L. Shapleigh J. Frostegard A. Bakken L. Transcription and activities of NOx reductases in Agrobacterium tumefaciens: the influence of nitrate, nitrite and oxygen availability. Environ Microbiol. 2008;10:3070–3081. doi: 10.1111/j.1462-2920.2007.01557.x. [DOI] [PubMed] [Google Scholar]
- 22.Berks BC. Baratta D. Richardson J. Ferguson SJ. Purification and characterization of a nitrous oxide reductase from Thiosphaera pantotropha. Implications for the mechanism of aerobic nitrous oxide reduction. Eur J Biochem. 1993;212:467–476. doi: 10.1111/j.1432-1033.1993.tb17683.x. [DOI] [PubMed] [Google Scholar]
- 23.Berks BC. Page MD. Richardson DJ. Reilly A. Cavill A. Outen F. Ferguson SJ. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol Microbiol. 1995;15:319–331. doi: 10.1111/j.1365-2958.1995.tb02246.x. [DOI] [PubMed] [Google Scholar]
- 24.Bertero MG. Rothery RA. Palak M. Hou C. Lim D. Blasco F. Weiner JH. Strynadka NC. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 25.Binnerup SJ. Sorensen J. Nitrate and nitrite microgradients in barley rhizosphere as detected by a highly sensitive denitrification bioassay. Appl Environ Microbiol. 1992;58:2375–2380. doi: 10.1128/aem.58.8.2375-2380.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blasco F. Iobbi C. Ratouchniak J. Bonnefoy V. Chippaux M. Nitrate reductases of Escherichia coli: sequence of the second nitrate reductase and comparison with that encoded by the narGHJI operon. Mol Gen Genet. 1990;222:104–111. doi: 10.1007/BF00283030. [DOI] [PubMed] [Google Scholar]
- 27.Blasco F. Pommier J. Augier V. Chippaux M. Giordano G. Involvement of the narJ or narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol Microbiol. 1992;6:221–230. doi: 10.1111/j.1365-2958.1992.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 28.Bobik C. Meilhoc E. Batut J. FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J Bacteriol. 2006;188:4890–4902. doi: 10.1128/JB.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borisov VB. Gennis RB. Hemp J. Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouchal P. Struharova I. Budinska E. Sedo O. Vyhlidalova T. Zdrahal Z. van Spanning R. Kucera I. Unraveling an FNR based regulatory circuit in Paracoccus denitrificans using a proteomics-based approach. Biochim Biophys Acta. 2010;1804:1350–1358. doi: 10.1016/j.bbapap.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Brekasis D. Paget MS. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2) EMBO J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brondijk TH. Fiegen D. Richardson DJ. Cole JA. Roles of NapF, NapG and NapH, subunits of the Escherichia coli periplasmic nitrate reductase, in ubiquinol oxidation. Mol Microbiol. 2002;44:245–255. doi: 10.1046/j.1365-2958.2002.02875.x. [DOI] [PubMed] [Google Scholar]
- 33.Brondijk TH. Nilavongse A. Filenko N. Richardson DJ. Cole JA. NapGH components of the periplasmic nitrate reductase of Escherichia coli K-12: location, topology and physiological roles in quinol oxidation and redox balancing. Biochem J. 2004;379:47–55. doi: 10.1042/BJ20031115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruscella P. Eraso JM. Roh JH. Kaplan S. The use of chromatin immunoprecipitation to define PpsR binding activity in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 2008;190:6817–6828. doi: 10.1128/JB.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bueno E. Bedmar EJ. Richardson DJ. Delgado MJ. Role of Bradyrhizobium japonicum cytochrome c550 in nitrite and nitrate respiration. FEMS Microbiol Lett. 2008;279:188–194. doi: 10.1111/j.1574-6968.2007.01034.x. [DOI] [PubMed] [Google Scholar]
- 36.Bueno E. Mesa S. Sanchez C. Bedmar EJ. Delgado MJ. NifA is required for maximal expression of denitrification genes in Bradyrhizobium japonicum. Environ Microbiol. 2010;12:393–400. doi: 10.1111/j.1462-2920.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- 37.Bueno E. Richardson DJ. Bedmar EJ. Delgado MJ. Expression of Bradyrhizobium japonicum cbb3 terminal oxidase under denitrifying conditions is subjected to redox control. FEMS Microbiol Lett. 2009;298:20–28. doi: 10.1111/j.1574-6968.2009.01711.x. [DOI] [PubMed] [Google Scholar]
- 38.Bühler D. Rossmann R. Landolt S. Balsiger S. Fischer HM. Hennecke H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J Biol Chem. 2010;285:15704–15713. doi: 10.1074/jbc.M109.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Büsch A. Strube K. Friedrich B. Cramm R. Transcriptional regulation of nitric oxide reduction in Ralstonia eutropha H16. Biochem Soc Trans. 2005;33:193–194. doi: 10.1042/BST0330193. [DOI] [PubMed] [Google Scholar]
- 40.Buschmann S. Warkentin E. Xie H. Langer JD. Ermler U. Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 41.Butland G. Spiro S. Watmough NJ. Richardson DJ. Two conserved glutamates in the bacterial nitric oxide reductase are essential for activity but not assembly of the enzyme. J Bacteriol. 2001;183:189–199. doi: 10.1128/JB.183.1.189-199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter JP. Hsaio YH. Spiro S. Richardson DJ. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl Environ Microbiol. 1995;61:2852–2858. doi: 10.1128/aem.61.8.2852-2858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cartron ML. Roldan MD. Ferguson SJ. Berks BC. Richardson DJ. Identification of two domains and distal histidine ligands to the four haems in the bacterial c-type cytochrome NapC; the prototype connector between quinol/quinone and periplasmic oxido-reductases. Biochem J. 2002;368:425–432. doi: 10.1042/BJ20020865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cebron A. Garnier J. Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower Seine River (France) Water Res. 2005;39:4979–4992. doi: 10.1016/j.watres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Cole JA. Richardson DJ. Respiration of nitrate and nitrite. In: Böck A, editor; Curtiss R III, editor; Kaper JB, editor; Karp PD, editor; Neidhardt FC, editor; Nyström T, editor; Slauch JM, editor; Squires CL, editor. EcoSal-Escherichia coli and Salmonella. Washington DC: ASM Press; 2008. [Google Scholar]
- 46.Comolli JC. Donohue TJ. Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol Microbiol. 2002;45:755–768. doi: 10.1046/j.1365-2958.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 47.Comolli JC. Donohue TJ. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol Microbiol. 2004;51:1193–1203. doi: 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 48.Constantinidou C. Hobman JL. Griffiths L. Patel MD. Penn CW. Cole JA. Overton TW. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 49.Cooper M. Tavankar GR. Williams HD. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology. 2003;149:1275–1284. doi: 10.1099/mic.0.26017-0. [DOI] [PubMed] [Google Scholar]
- 50.Cosseau C. Batut J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch Microbiol. 2004;181:89–96. doi: 10.1007/s00203-003-0641-5. [DOI] [PubMed] [Google Scholar]
- 51.Cotter PA. Melville SB. Albrecht JA. Gunsalus RP. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 52.Crack J. Green J. Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 53.Crack JC. Gaskell AA. Green J. Cheesman MR. Le Brun NE. Thomson AJ. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J Am Chem Soc. 2008;130:1749–1758. doi: 10.1021/ja077455+. [DOI] [PubMed] [Google Scholar]
- 54.Crack JC. Green J. Cheesman MR. Le Brun NE. Thomson AJ. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci U S A. 2007;104:2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cramm R. Busch A. Strube K. NO-dependent transcriptional activation of gene expression in Ralstonia eutropha H16. Biochem Soc Trans. 2006;34:182–184. doi: 10.1042/BST0340182. [DOI] [PubMed] [Google Scholar]
- 56.Crosson S. McGrath PT. Stephens C. McAdams HH. Shapiro L. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc Natl Acad Sci U S A. 2005;102:8018–8123. doi: 10.1073/pnas.0503022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruz-Ramos H. Crack J. Wu G. Hughes MN. Scott C. Thomson AJ. Green J. Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darwin AJ. Ziegelhoffer EC. Kiley PJ. Stewart V. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J Bacteriol. 1998;180:4192–4198. doi: 10.1128/jb.180.16.4192-4198.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Vries R. Suharti S. Pouvreau LAM. Nitric oxide reductase: structural variations and catalytic mechanism. In: Bothe H, editor; Ferguson SJ, editor; Newton WE, editor. Biology of the Nitrogen Cycle. The Netherlands: Elsevier; 2007. [Google Scholar]
- 60.de Vries S. Strampraad MJ. Lu S. Moenne-Loccoz P. Schroder I. Purification and characterization of the MQH2:NO oxidoreductase from the hyperthermophilic archaeon Pyrobaculum aerophilum. J Biol Chem. 2003;278:35861–35868. doi: 10.1074/jbc.M300857200. [DOI] [PubMed] [Google Scholar]
- 61.Delgado MJ. Bedmar EJ. Downie JA. Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv Microb Physiol. 1998;40:191–231. doi: 10.1016/s0065-2911(08)60132-0. [DOI] [PubMed] [Google Scholar]
- 62.Delgado MJ. Bonnard N. Tresierra-Ayala A. Bedmar EJ. Muller P. The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology. 2003;149:3395–3403. doi: 10.1099/mic.0.26620-0. [DOI] [PubMed] [Google Scholar]
- 63.Delledonne M. Zeier J. Marocco A. Lamb C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci U S A. 2001;98:13454–13459. doi: 10.1073/pnas.231178298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dias JM. Than ME. Humm A. Huber R. Bourenkov GP. Bartunik HD. Bursakov S. Calvete J. Caldeira J. Carneiro C. Moura JJ. Moura I. Romao MJ. Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 65.Donohue TJ. Kiley PJ. Bacterial responses to O2 limitation. In: Storz G, editor; Hengge D, editor. Bacterial Stress Responses. Washington, DC: American Society for Microbiology; 2010. pp. 175–189. [Google Scholar]
- 66.Downie JA. Legume haemoglobins: symbiotic nitrogen fixation needs bloody nodules. Curr Biol. 2005;15:R196–R198. doi: 10.1016/j.cub.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 67.Ducluzeau AL. Ouchane S. Nitschke W. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 68.Dufour YS. Kiley PJ. Donohue TJ. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet. 2010;6:e1001027. doi: 10.1371/journal.pgen.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellington MJ. Bhakoo KK. Sawers G. Richardson DJ. Ferguson SJ. Hierarchy of carbon source selection in Paracoccus pantotrophus: strict correlation between reduction state of the carbon substrate and aerobic expression of the nap operon. J Bacteriol. 2002;184:4767–4774. doi: 10.1128/JB.184.17.4767-4774.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellington MJ. Fosdike WL. Sawers RG. Richardson DJ. Ferguson SJ. Regulation of the nap operon encoding the periplasmic nitrate reductase of Paracoccus pantotrophus: delineation of DNA sequences required for redox control. Arch Microbiol. 2006;184:298–304. doi: 10.1007/s00203-005-0044-x. [DOI] [PubMed] [Google Scholar]
- 71.Ellington MJ. Sawers G. Sears HJ. Spiro S. Richardson DJ. Ferguson SJ. Characterization of the expression and activity of the periplasmic nitrate reductase of Paracoccus pantotrophus in chemostat cultures. Microbiology. 2003;149:1533–1540. doi: 10.1099/mic.0.26277-0. [DOI] [PubMed] [Google Scholar]
- 72.Elsen S. Swem LR. Swem DL. Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emmerich R. Hennecke H. Fischer HM. Evidence for a functional similarity between the two-component regulatory systems RegSR, ActSR, and RegBA (PrrBA) in a-Proteobacteria. Arch Microbiol. 2000;174:307–313. doi: 10.1007/s002030000207. [DOI] [PubMed] [Google Scholar]
- 74.Emmerich R. Panglungtshang K. Strehler P. Hennecke H. Fischer HM. Phosphorylation, dephosphorylation and DNA-binding of the Bradyrhizobium japonicum RegSR two-component regulatory proteins. Eur J Biochem. 1999;263:455–463. doi: 10.1046/j.1432-1327.1999.00517.x. [DOI] [PubMed] [Google Scholar]
- 75.Eraso JM. Kaplan S. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry. 2000;39:2052–2062. doi: 10.1021/bi9923858. [DOI] [PubMed] [Google Scholar]
- 76.Eraso JM. Kaplan S. Half-Site DNA sequence and spacing length contributions to PrrA binding to PrrA site 2 of RSP3361 in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 2009;191:4353–4364. doi: 10.1128/JB.00244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eraso JM. Roh JH. Zeng X. Callister SJ. Lipton MS. Kaplan S. Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J Bacteriol. 2008;190:4831–4848. doi: 10.1128/JB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandez-Piñar R. Ramos JL. Rodríguez-Hervá JJ. Espinosa-Urgel M. A two-component regulatory system integrates redox state and population density sensing in Pseudomonas putida. J Bacteriol. 2008;190:7666–7674. doi: 10.1128/JB.00868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferrarini A. De Stefano M. Baudouin E. Pucciariello C. Polverari A. Puppo A. Delledonne M. Expression of Medicago truncatula genes responsive to nitric oxide in pathogenic and symbiotic conditions. Mol Plant-Microbe Interact. 2008;21:781–790. doi: 10.1094/MPMI-21-6-0781. [DOI] [PubMed] [Google Scholar]
- 80.Field SJ. Roldan MD. Marritt SJ. Butt JN. Richardson DJ. Watmough NJ. Electron transfer to the active site of the bacterial nitric oxide reductase is controlled by ligand binding to heme b3. Biochim Biophys Acta. 2011;1807:451–457. doi: 10.1016/j.bbabio.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Field SJ. Thorndycroft FH. Matorin AD. Richardson DJ. Watmough NJ. The respiratory nitric oxide reductase (NorBC) from Paracoccus denitrificans. Methods Enzymol. 2008;437:79–101. doi: 10.1016/S0076-6879(07)37005-5. [DOI] [PubMed] [Google Scholar]
- 82.Filenko N. Spiro S. Browning DF. Squire D. Overton TW. Cole J. Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol. 2007;189:4410–4417. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fischer HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fleischhacker AS. Kiley PJ. Iron-containing transcription factors and their roles as sensors. Curr Opin Chem Biol. 2011;15:335–341. doi: 10.1016/j.cbpa.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flock U. Lachmann P. Reimann J. Watmough NJ. Adelroth P. Exploring the terminal region of the proton pathway in the bacterial nitric oxide reductase. J Inorg Biochem. 2009;103:845–850. doi: 10.1016/j.jinorgbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 86.Flock U. Thorndycroft FH. Matorin AD. Richardson DJ. Watmough NJ. Adelroth P. Defining the proton entry point in the bacterial respiratory nitric-oxide reductase. J Biol Chem. 2008;283:3839–3845. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 87.Garcia-Horsman JA. Barquera B. Rumbley J. Ma J. Gennis RB. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gates AJ. Richardson DJ. Butt JN. Voltammetric characterization of the aerobic energy-dissipating nitrate reductase of Paracoccus pantotrophus: exploring the activity of a redox-balancing enzyme as a function of electrochemical potential. Biochem J. 2008;409:159–168. doi: 10.1042/BJ20071088. [DOI] [PubMed] [Google Scholar]
- 89.Gavira M. Roldan MD. Castillo F. Moreno-Vivian C. Regulation of nap gene expression and periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J Bacteriol. 2002;184:1693–1702. doi: 10.1128/JB.184.6.1693-1702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geng H. Nakano S. Nakano MM. Transcriptional activation by Bacillus subtilis ResD: tandem binding to target elements and phosphorylation-dependent and -independent transcriptional activation. J Bacteriol. 2004;186:2028–2037. doi: 10.1128/JB.186.7.2028-2037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Geng H. Zhu Y. Mullen K. Zuber CS. Nakano MM. Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. J Bacteriol. 2007;189:1745–1755. doi: 10.1128/JB.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Geng H. Zuber P. Nakano MM. Regulation of respiratory genes by ResD-ResE signal transduction system in Bacillus subtilis. Methods Enzymol. 2007;422:448–464. doi: 10.1016/S0076-6879(06)22023-8. [DOI] [PubMed] [Google Scholar]
- 93.Giardina G. Castiglione N. Caruso M. Cutruzzola F. Rinaldo S. The Pseudomonas aeruginosa DNR transcription factor: light and shade of nitric oxide-sensing mechanisms. Biochem Soc Trans. 2011;39:294–298. doi: 10.1042/BST0390294. [DOI] [PubMed] [Google Scholar]
- 94.Giardina G. Rinaldo S. Johnson KA. Di Matteo A. Brunori M. Cutruzzola F. NO sensing in Pseudomonas aeruginosa: structure of the transcriptional regulator DNR. J Mol Biol. 2008;378:1002–1015. doi: 10.1016/j.jmb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 95.Gilberthorpe NJ. Poole RK. Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J Biol Chem. 2008;283:11146–11154. doi: 10.1074/jbc.M708019200. [DOI] [PubMed] [Google Scholar]
- 96.Gilles-Gonzalez MA. Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 97.Goddard AD. Moir JW. Richardson DJ. Ferguson SJ. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol Microbiol. 2008;70:667–681. doi: 10.1111/j.1365-2958.2008.06436.x. [DOI] [PubMed] [Google Scholar]
- 98.Gonzalez PJ. Correia C. Moura I. Brondino CD. Moura JJ. Bacterial nitrate reductases: Molecular and biological aspects of nitrate reduction. J Inorg Biochem. 2006;100:1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 99.Govantes F. Orjalo AV. Gunsalus RP. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon. Mol Microbiol. 2000;38:1061–1073. doi: 10.1046/j.1365-2958.2000.02215.x. [DOI] [PubMed] [Google Scholar]
- 100.Granados-Baeza MJ. Gomez-Hernandez N. Mora Y. Delgado MJ. Romero D. Girard L. Novel reiterated Fnr-type proteins control the production of the symbiotic terminal oxidase cbb3 in Rhizobium etli CFN42. Mol Plant-Microbe Interact. 2007;20:1241–1249. doi: 10.1094/MPMI-20-10-1241. [DOI] [PubMed] [Google Scholar]
- 101.Green J. Crack JC. Thomson AJ. LeBrun NE. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12:145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 102.Green J. Paget MS. Bacterial redox sensors. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 103.Green J. Scott C. Guest JR. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv Microb Physiol. 2001;44:1–34. doi: 10.1016/s0065-2911(01)44010-0. [DOI] [PubMed] [Google Scholar]
- 104.Gyan S. Shiohira Y. Sato I. Takeuchi M. Sato T. Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J Bacteriol. 2006;188:7062–7071. doi: 10.1128/JB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanke A. Strous M. Climate, fertilization, and the nitrogen cycle. J Cosmol. 2010;8:1838–1845. [Google Scholar]
- 106.Happ HN. Braatsch S. Broschek V. Osterloh L. Klug G. Light-dependent regulation of photosynthesis genes in Rhodobacter sphaeroides 2.4.1 is coordinately controlled by photosynthetic electron transport via the PrrBA two-component system and the photoreceptor AppA. Mol Microbiol. 2005;58:903–914. doi: 10.1111/j.1365-2958.2005.04882.x. [DOI] [PubMed] [Google Scholar]
- 107.Hartsock A. Shapleigh JP. Identification, functional studies, and genomic comparisons of new members of the NnrR regulon in Rhodobacter sphaeroides. J Bacteriol. 2010;192:903–911. doi: 10.1128/JB.01026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hauser F. Pessi G. Friberg M. Weber C. Rusca N. Lindemann A. Fischer HM. Hennecke H. Dissection of the Bradyrhizobium japonicum NifA+s54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol Genet Genomics. 2007;278:255–271. doi: 10.1007/s00438-007-0246-9. [DOI] [PubMed] [Google Scholar]
- 109.Hayatsu M. Tago K. Saito M. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr. 2008;54:33–45. [Google Scholar]
- 110.Hendriks J. Oubrie A. Castresana J. Urbani A. Gemeinhardt S. Saraste M. Nitric oxide reductases in bacteria. Biochim Biophys Acta. 2000;1459:266–273. doi: 10.1016/s0005-2728(00)00161-4. [DOI] [PubMed] [Google Scholar]
- 111.Heurlier K. Thomson MJ. Aziz N. Moir JW. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J Bacteriol. 2008;190:2488–2495. doi: 10.1128/JB.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hino T. Matsumoto Y. Nagano S. Sugimoto H. Fukumori Y. Murata T. Iwata S. Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 113.Isabella VM. Lapek JD., Jr. Kennedy EM. Clark VL. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jepson BJ. Marietou A. Mohan S. Cole JA. Butler CS. Richardson DJ. Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem Soc Trans. 2006;34:122–126. doi: 10.1042/BST0340122. [DOI] [PubMed] [Google Scholar]
- 115.Jepson BJ. Mohan S. Clarke TA. Gates AJ. Cole JA. Butler CS. Butt JN. Hemmings AM. Richardson DJ. Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli. J Biol Chem. 2007;282:6425–6437. doi: 10.1074/jbc.M607353200. [DOI] [PubMed] [Google Scholar]
- 116.Jones KM. Kobayashi H. Davies BW. Taga ME. Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jones MR. Richardson DJ. McEwan AG. Jackson JB. Ferguson SJ. In vivo redox poising of the cyclic electron transport system of Rhodobacter capsulatus and the effects of the auxiliary oxidants, nitrate, nitrous oxide and trimethylamine N-oxide, as revealed by multiple short flash excitation. Biochem Biophys Acta. 1990;1017:209–216. [Google Scholar]
- 118.Jormakka M. Richardson D. Byrne B. Iwata S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure. 2004;12:95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 119.Justino MC. Vicente JB. Teixeira M. Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- 120.Kaplan S. Eraso J. Roh JH. Interacting regulatory networks in the facultative photosynthetic bacterium, Rhodobacter sphaeroides 2.4.1. Biochem Soc Trans. 2005;33:51–55. doi: 10.1042/BST0330051. [DOI] [PubMed] [Google Scholar]
- 121.Kawakami T. Kuroki M. Ishii M. Igarashi Y. Arai H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 122.Kim YJ. Ko IJ. Lee JM. Kang HY. Kim YM. Kaplan S. Oh JI. Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 2007;189:5617–5625. doi: 10.1128/JB.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kommineni S. Yukl E. Hayashi T. Delepine J. Geng H. Moenne-Loccoz P. Nakano MM. Nitric oxide-sensitive and -insensitive interaction of Bacillus subtilis NsrR with a ResDE-controlled promoter. Mol Microbiol. 2010;78:1280–1293. doi: 10.1111/j.1365-2958.2010.07407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Körner H. Sofia HJ. Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 125.Kraft B. Strous M. Tegetmeyer HE. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–117. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 126.Kulajta C. Thumfart JO. Haid S. Daldal F. Koch HG. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J Mol Biol. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 127.Kullik I. Fritsche S. Knobel H. Sanjuan J. Hennecke H. Fischer HM. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the s54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Laguri C. Phillips-Jones MK. Williamson MP. Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity. Nucleic Acids Res. 2003;31:6778–6787. doi: 10.1093/nar/gkg891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Laguri C. Stenzel RA. Donohue TJ. Phillips-Jones MK. Williamson MP. Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides. Biochemistry. 2006;45:7872–7881. doi: 10.1021/bi060683g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laratta WP. Choi PS. Tosques IE. Shapleigh JP. Involvement of the PrrB/PrrA two-component system in nitrite respiration in Rhodobacter sphaeroides 2.4.3: evidence for transcriptional regulation. J Bacteriol. 2002;184:3521–3529. doi: 10.1128/JB.184.13.3521-3529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laratta WP. Shapleigh JP. Site-directed mutagenesis of NnrR: a transcriptional regulator of nitrite and nitric oxide reductase in Rhodobacter sphaeroides. FEMS Microbiol Lett. 2003;229:173–178. doi: 10.1016/S0378-1097(03)00821-8. [DOI] [PubMed] [Google Scholar]
- 132.Lazazzera BA. Beinert H. Khoroshilova N. Kennedy MC. Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 133.Lindemann A. Koch M. Pessi G. Muller AJ. Balsiger S. Hennecke H. Fischer HM. Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum. FEMS Microbiol Lett. 2010;312:184–191. doi: 10.1111/j.1574-6968.2010.02115.x. [DOI] [PubMed] [Google Scholar]
- 134.Lindemann A. Moser A. Pessi G. Hauser F. Friberg M. Hennecke H. Fischer HM. New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J Bacteriol. 2007;189:8928–8943. doi: 10.1128/JB.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu HP. Takio S. Satoh T. Yamamoto I. Involvement in denitrification of the napKEFDABC genes encoding the periplasmic nitrate reductase system in the denitrifying phototrophic bacterium Rhodobacter sphaeroides f. sp. denitrificans. Biosci Biotechnol Biochem. 1999;63:530–536. doi: 10.1271/bbb.63.530. [DOI] [PubMed] [Google Scholar]
- 136.Liu X. De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 137.Malpica R. Franco B. Rodriguez C. Kwon O. Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci U S A. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Malpica R. Sandoval GR. Rodriguez C. Franco B. Georgellis D. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal. 2006;8:781–795. doi: 10.1089/ars.2006.8.781. [DOI] [PubMed] [Google Scholar]
- 139.Mao L. Mackenzie C. Roh JH. Eraso JM. Kaplan S. Resat H. Combining microarray and genomic data to predict DNA binding motifs. Microbiology. 2005;151:3197–3213. doi: 10.1099/mic.0.28167-0. [DOI] [PubMed] [Google Scholar]
- 140.Marietou A. Richardson D. Cole J. Mohan S. Nitrate reduction by Desulfovibrio desulfuricans: a periplasmic nitrate reductase system that lacks NapB, but includes a unique tetraheme c-type cytochrome, NapM. FEMS Microbiol Lett. 2005;248:217–225. doi: 10.1016/j.femsle.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 141.Martinez-Espinosa RM. Cole JA. Richardson DJ. Watmough NJ. Enzymology and ecology of the nitrogen cycle. Biochem Soc Trans. 2011;39:175–178. doi: 10.1042/BST0390175. [DOI] [PubMed] [Google Scholar]
- 142.Martínez-Espinosa RM. Dridge EJ. Bonete MJ. Butt JN. Butler CS. Sargent F. Richardson DJ. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol Lett. 2007;276:129–139. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 143.Matsuda Y. Inamori K. Osaki T. Eguchi A. Watanabe A. Kawabata S. Iba K. Arata H. Nitric oxide-reductase homologue that contains a copper atom and has cytochrome c-oxidase activity from an aerobic phototrophic bacterium Roseobacter denitrificans. J Biochem. 2002;131:791–800. doi: 10.1093/oxfordjournals.jbchem.a003167. [DOI] [PubMed] [Google Scholar]
- 144.Mesa S. Bedmar EJ. Chanfon A. Hennecke H. Fischer HM. Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J Bacteriol. 2003;185:3978–3982. doi: 10.1128/JB.185.13.3978-3982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mesa S. Hauser F. Friberg M. Malaguti E. Fischer HM. Hennecke H. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol. 2008;190:6568–6579. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mesa S. Hennecke H. Fischer HM. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem Soc Trans. 2006;34:156–159. doi: 10.1042/BST0340156. [DOI] [PubMed] [Google Scholar]
- 147.Mesa S. Reutimann L. Fischer HM. Hennecke H. Posttranslational control of transcription factor FixK2, a key regulator for the Bradyrhizobium japonicum-soybean symbiosis. Proc Natl Acad Sci U S A. 2009;106:21860–21865. doi: 10.1073/pnas.0908097106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mesa S. Ucurum Z. Hennecke H. Fischer HM. Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2. J Bacteriol. 2005;187:3329–3338. doi: 10.1128/JB.187.10.3329-3338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mills PC. Rowley G. Spiro S. Hinton JC. Richardson DJ. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology. 2008;154:1218–1228. doi: 10.1099/mic.0.2007/014290-0. [DOI] [PubMed] [Google Scholar]
- 150.Minchin FR. James EK. Becana M. Oxygen diffusion, production of reactive oxygen and nitrogen species, and antioxidants in legume nodules. In: Dilworth MJ, editor; James EK, editor; Sprent JI, editor; Newton WE, editor. Nitrogen-Fixing Leguminous Symbioses. New York: Springer Science; 2008. pp. 321–362. [Google Scholar]
- 151.Moore CM. Nakano MM. Wang T. Ye RW. Helmann JD. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J Bacteriol. 2004;186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Morales G. Ugidos A. Rojo F. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ Microbiol. 2006;8:1764–1774. doi: 10.1111/j.1462-2920.2006.01061.x. [DOI] [PubMed] [Google Scholar]
- 153.Moreno-Vivián C. Cabello P. Martinez-Luque M. Blasco R. Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moreno-Vivián C. Ferguson SJ. Definition and distinction between assimilatory, dissimilatory and respiratory pathways. Mol Microbiol. 1998;29:664–666. doi: 10.1046/j.1365-2958.1998.00946.x. [DOI] [PubMed] [Google Scholar]
- 155.Nakano MM. Induction of ResDE-dependent gene expression in Bacillus subtilis in response to nitric oxide and nitrosative stress. J Bacteriol. 2002;184:1783–1787. doi: 10.1128/JB.184.6.1783-1787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nakano MM. Geng H. Nakano S. Kobayashi K. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J Bacteriol. 2006;188:5878–5887. doi: 10.1128/JB.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakano MM. Hoffmann T. Zhu Y. Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nakano MM. Zuber P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 159.Nakano MM. Zuber P. Glaser P. Danchin A. Hulett FM. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nellen-Anthamatten D. Rossi P. Preisig O. Kullik I. Babst M. Fischer HM. Hennecke H. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol. 1998;180:5251–5255. doi: 10.1128/jb.180.19.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nelson KE. Weinel C. Paulsen IT. Dodson RJ. Hilbert H. Martins dos Santos VA. Fouts DE. Gill SR. Pop M. Holmes M. Brinkac L. Beanan M. DeBoy RT. Daugherty S. Kolonay J. Madupu R. Nelson W. White O. Peterson J. Khouri H. Hance I. Chris Lee P. Holtzapple E. Scanlan D. Tran K. Moazzez A. Utterback T. Rizzo M. Lee K. Kosack D. Moestl D. Wedler H. Lauber J. Stjepandic D. Hoheisel J. Straetz M. Heim S. Kiewitz C. Eisen JA. Timmis KN. Dusterhoft A. Tummler B. Fraser CM. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 162.Nienaber A. Huber A. Gottfert M. Hennecke H. Fischer HM. Three new NifA-regulated genes in the Bradyrhizobium japonicum symbiotic gene region discovered by competitive DNA-RNA hybridization. J Bacteriol. 2000;182:1472–1480. doi: 10.1128/jb.182.6.1472-1480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nikel PI. Zhu J. San KY. Mendez BS. Bennett GN. Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. J Bacteriol. 2009;191:5538–5548. doi: 10.1128/JB.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nilavongse A. Brondijk TH. Overton TW. Richardson DJ. Leach ER. Cole JA. The NapF protein of the Escherichia coli periplasmic nitrate reductase system: demonstration of a cytoplasmic location and interaction with the catalytic subunit, NapA. Microbiology. 2006;152:3227–3237. doi: 10.1099/mic.0.29157-0. [DOI] [PubMed] [Google Scholar]
- 165.Oh JI. Kaplan S. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry. 1999;38:2688–2696. doi: 10.1021/bi9825100. [DOI] [PubMed] [Google Scholar]
- 166.Oh JI. Kaplan S. Redox signaling: globalization of gene expression. EMBO J. 2000;19:4237–4247. doi: 10.1093/emboj/19.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oh JI. Ko IJ. Kaplan S. The default state of the membrane-localized histidine kinase PrrB of Rhodobacter sphaeroides 2.4.1 is in the kinase-positive mode. J Bacteriol. 2001;183:6807–6814. doi: 10.1128/JB.183.23.6807-6814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oh JI. Ko IJ. Kaplan S. Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry. 2004;43:7915–7923. doi: 10.1021/bi0496440. [DOI] [PubMed] [Google Scholar]
- 169.Oldroyd GE. Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 170.Olmo-Mira MF. Gavira M. Richardson DJ. Castillo F. Moreno-Vivian C. Roldan MD. NapF is a cytoplasmic iron-sulfur protein required for Fe-S cluster assembly in the periplasmic nitrate reductase. J Biol Chem. 2004;279:49727–49735. doi: 10.1074/jbc.M406502200. [DOI] [PubMed] [Google Scholar]
- 171.Overton TW. Whitehead R. Li Y. Snyder LA. Saunders NJ. Smith H. Cole JA. Coordinated regulation of the Neisseria gonorrhoeae-truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J Biol Chem. 2006;281:33115–33126. doi: 10.1074/jbc.M607056200. [DOI] [PubMed] [Google Scholar]
- 172.Pagels M. Fuchs S. Pane-Farre J. Kohler C. Menschner L. Hecker M. McNamarra PJ. Bauer MC. von Wachenfeldt C. Liebeke M. Lalk M. Sander G. von Eiff C. Proctor RA. Engelmann S. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol. 2010;76:1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pawlik G. Kulajta C. Sachelaru I. Schroder S. Waidner B. Hellwig P. Daldal F. Koch HG. The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. J Bacteriol. 2010;192:6378–6389. doi: 10.1128/JB.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pearson IV. Page MD. van Spanning RJ. Ferguson SJ. A mutant of Paracoccus denitrificans with disrupted genes coding for cytochrome c550 and pseudoazurin establishes these two proteins as the in vivo electron donors to cytochrome cd1 nitrite reductase. J. Bacteriol. 2003;185:6308–6315. doi: 10.1128/JB.185.21.6308-6315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Peña-Sandoval GR. Georgellis D. The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction. J Bacteriol. 2010;192:1735–1739. doi: 10.1128/JB.01401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pereira MM. Santana M. Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 177.Pereira MM. Sousa FL. Verissimo AF. Teixeira M. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- 178.Peters A. Kulajta C. Pawlik G. Daldal F. Koch HG. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J Bacteriol. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pitcher RS. Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 180.Pohlmann A. Cramm R. Schmelz K. Friedrich B. A novel NO-responding regulator controls the reduction of nitric oxide in Ralstonia eutropha. Mol Microbiol. 2000;38:626–638. doi: 10.1046/j.1365-2958.2000.02157.x. [DOI] [PubMed] [Google Scholar]
- 181.Pomowski A. Zumft WG. Kroneck PM. Einsle O. N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase. Nature. 2011;477:234–237. doi: 10.1038/nature10332. [DOI] [PubMed] [Google Scholar]
- 182.Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 183.Poole RK. Cook GM. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 184.Potter L. Angove H. Richardson D. Cole J. Nitrate reduction in the periplasm of gram-negative bacteria. Adv Microb Physiol. 2001;45:51–112. doi: 10.1016/s0065-2911(01)45002-8. [DOI] [PubMed] [Google Scholar]
- 185.Preisig O. Zufferey R. Thöny-Meyer L. Appleby CA. Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Price CE. Driessen AJ. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim Biophys Acta. 2010;1803:748–766. doi: 10.1016/j.bbamcr.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 187.Pullan ST. Gidley MD. Jones RA. Barrett J. Stevanin TM. Read RC. Green J. Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Puri-Taneja A. Schau M. Chen Y. Hulett FM. Regulators of the Bacillus subtilis cydABCD operon: identification of a negative regulator, CcpA, and a positive regulator, ResD. J Bacteriol. 2007;189:3348–3358. doi: 10.1128/JB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ranson-Olson B. Jones DF. Donohue TJ. Zeilstra-Ryalls JH. In vitro and in vivo analysis of the role of PrrA in Rhodobacter sphaeroides 2.4.1 hemA gene expression. J Bacteriol. 2006;188:3208–3218. doi: 10.1128/JB.188.9.3208-3218.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ranson-Olson B. Zeilstra-Ryalls JH. Regulation of the Rhodobacter sphaeroides 2.4.1 hemA gene by PrrA and FnrL. J Bacteriol. 2008;190:6769–6778. doi: 10.1128/JB.00828-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rauhamäki V. Bloch DA. Verkhovsky MI. Wikstrom M. Active site of cytochrome cbb3. J Biol Chem. 2009;284:11301–11308. doi: 10.1074/jbc.M808839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ravishankara AR. Daniel JS. Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 193.Reents H. Munch R. Dammeyer T. Jahn D. Hartig E. The Fnr regulon of Bacillus subtilis. J Bacteriol. 2006;188:1103–1112. doi: 10.1128/JB.188.3.1103-1112.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rey FE. Harwood CS. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol Microbiol. 2010;75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 195.Richardson D. Felgate H. Watmough N. Thomson A. Baggs E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle - could enzymic regulation hold the key? Trends Biotechnol. 2009;27:388–397. doi: 10.1016/j.tibtech.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 196.Richardson D. Ferguson SJ. The influence of carbonsubstrate on the activity of the periplasmic nitrate reductase in aerobically grown Thiosphaera pantotropha. Arch Microbiol. 1992;157:535–537. [Google Scholar]
- 197.Richardson DJ. Bacterial respiration: a flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 198.Richardson DJ. Redox complexes of the nitrogen cycle. In: Moir JWB, editor. Nitrogen Cycling in Bacteria. Norkfolk, UK: Caister Academic Press; 2011. pp. 23–39. [Google Scholar]
- 199.Richardson DJ. Berks BC. Russell DA. Spiro S. Taylor CJ. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci. 2001;58:165–178. doi: 10.1007/PL00000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Richardson DJ. King GF. Kelly DJ. MacEwan AG. Ferguson SJ. Jackson JB. The role of auxiliary oxidants in maintaining redox balance during phototrophic growth of Rhodobacter capsulatus on propionate or butyrate. Arch Microbiol. 1988;150:131–137. [Google Scholar]
- 201.Richardson DJ. van Spannig RJ. Ferguson SJ. The prokaryotic nitrate reductases. In: Bothe H, editor; Ferguson SJ, editor; Newton WE, editor. Biology of the Nitrogen Cycle. The Netherlands: Elsevier; 2007. pp. 21–35. [Google Scholar]
- 202.Rinaldo S. Arcovito A. Giardina G. Castiglione N. Brunori M. Cutruzzola F. New insights into the activity of Pseudomonas aeruginosa cd1 nitrite reductase. Biochem Soc Trans. 2008;36:1155–1159. doi: 10.1042/BST0361155. [DOI] [PubMed] [Google Scholar]
- 203.Rinaldo S. Cutruzzola F. Nitrite reductases in denitrification. In: Bothe H, editor; Ferguson SJ, editor; Newton WE, editor. Biology of the Nitrogen Cycle. The Netherlands: Elsevier; 2007. pp. 37–56. [Google Scholar]
- 204.Robles EF. Sanchez C. Bonnard N. Delgado MJ. Bedmar EJ. The Bradyrhizobium japonicum napEDABC genes are controlled by the FixLJ-FixK2-NnrR regulatory cascade. Biochem Soc Trans. 2006;34:108–110. doi: 10.1042/BST0340108. [DOI] [PubMed] [Google Scholar]
- 205.Rodgers KR. Lukat-Rodgers GS. Insights into heme-based O2 sensing from structure-function relationships in the FixL proteins. J Inorg Biochem. 2005;99:963–977. doi: 10.1016/j.jinorgbio.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 206.Rodionov DA. Dubchak IL. Arkin AP. Alm EJ. Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Roldán MD. Sears HJ. Cheesman MR. Ferguson SJ. Thomson AJ. Berks BC. Richardson DJ. Spectroscopic characterization of a novel multiheme c-type cytochrome widely implicated in bacterial electron transport. J Biol Chem. 1998;273:28785–28790. doi: 10.1074/jbc.273.44.28785. [DOI] [PubMed] [Google Scholar]
- 208.Rolfe MD. Ter Beek A. Graham AI. Trotter EW. Asif HM. Sumo S. Sanguinetti G. Teixeira de Mattos J. Poole RK. Green J. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem. 2011;286:10147–10154. doi: 10.1074/jbc.M110.211144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Salmon K. Hung SP. Mekjian K. Baldi P. Hatfield GW. Gunsalus RP. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- 210.Salmon KA. Hung SP. Steffen NR. Krupp R. Baldi P. Hatfield GW. Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 211.Satterlee JD. Suquet C. Bidwai AK. Erman JE. Schwall L. Jimenez R. Mass instability in isolated recombinant FixL heme domains of Bradyrhizobium japonicum. Biochemistry. 2008;47:1540–1553. doi: 10.1021/bi701672m. [DOI] [PubMed] [Google Scholar]
- 212.Schau M. Chen Y. Hulett FM. Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J Bacteriol. 2004;186:4585–4595. doi: 10.1128/JB.186.14.4585-4595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schreiber K. Krieger R. Benkert B. Eschbach M. Arai H. Schobert M. Jahn D. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol. 2007;189:4310–4314. doi: 10.1128/JB.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sciotti MA. Chanfon A. Hennecke H. Fischer HM. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic Genes in Bradyrhizobium japonicum. J Bacteriol. 2003;185:5639–5642. doi: 10.1128/JB.185.18.5639-5642.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sears HJ. Sawers G. Berks BC. Ferguson SJ. Richardson DJ. Control of periplasmic nitrate reductase gene expression napEDABC from Paracoccus pantotrophus in response to oxygen and carbon substrates. Microbiology. 2000;146(Pt 11):2977–2985. doi: 10.1099/00221287-146-11-2977. [DOI] [PubMed] [Google Scholar]
- 216.Sears HJ. Spiro S. Richardson DJ. Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology. 1997;143:3767–3774. doi: 10.1099/00221287-143-12-3767. [DOI] [PubMed] [Google Scholar]
- 217.Shalel-Levanon S. San KY. Bennett GN. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab Eng. 2005;7:364–374. doi: 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 218.Shapleigh JP. Oxygen control of nitrogen oxide respiration, focusing on a-proteobacteria. Biochem Soc Trans. 2011;39:179–183. doi: 10.1042/BST0390179. [DOI] [PubMed] [Google Scholar]
- 219.Sharma V. Wikstrom M. Kaila VR. Redox-coupled proton transfer in the active site of cytochrome cbb3. Biochim Biophys Acta. 2010;1797:1512–1520. doi: 10.1016/j.bbabio.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 220.Sickmier EA. Brekasis D. Paranawithana S. Bonanno JB. Paget MS. Burley SK. Kielkopf CL. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 221.Simon J. van Spanning RJ. Richardson DJ. The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim Biophys Acta. 2008;1777:1480–1490. doi: 10.1016/j.bbabio.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 222.Smith MS. Nitrous oxide production by Escherichia coli is correlated with nitrate reductase activity. Appl Environ Microbiol. 1983;45:1545–1547. doi: 10.1128/aem.45.5.1545-1547.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sousa EH. Tuckerman JR. Gonzalez G. Gilles-Gonzalez MA. A memory of oxygen binding explains the dose response of the heme-based sensor FixL. Biochemistry. 2007;46:6249–6257. doi: 10.1021/bi7003334. [DOI] [PubMed] [Google Scholar]
- 224.Spector MP. Garcia del Portillo F. Bearson SM. Mahmud A. Magut M. Finlay BB. Dougan G. Foster JW. Pallen MJ. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology. 1999;145:3035–3045. doi: 10.1099/00221287-145-11-3035. [DOI] [PubMed] [Google Scholar]
- 225.Spiro S. Nitric oxide-sensing mechanisms in Escherichia coli. Biochem Soc Trans. 2006;34:200–202. doi: 10.1042/BST0340200. [DOI] [PubMed] [Google Scholar]
- 226.Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 227.Stewart V. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem Soc Trans. 2003;31:1–10. doi: 10.1042/bst0310001. [DOI] [PubMed] [Google Scholar]
- 228.Stewart V. Bledsoe PJ. Fnr-, NarP- and NarL-dependent regulation of transcription initiation from the Haemophilus influenzae Rd napF (periplasmic nitrate reductase) promoter in Escherichia coli K-12. J Bacteriol. 2005;187:6928–6935. doi: 10.1128/JB.187.20.6928-6935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stock AM. Robinson VL. Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 230.Suharti Heering HA. de Vries S. NO reductase from Bacillus azotoformans is a bifunctional enzyme accepting electrons from menaquinol and a specific endogenous membrane-bound cytochrome c551. Biochemistry. 2004;43:13487–13495. doi: 10.1021/bi0488101. [DOI] [PubMed] [Google Scholar]
- 231.Swem DL. Bauer CE. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J Bacteriol. 2002;184:2815–2820. doi: 10.1128/JB.184.10.2815-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Swem DL. Swem LR. Setterdahl A. Bauer CE. Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. J Bacteriol. 2005;187:8081–8087. doi: 10.1128/JB.187.23.8081-8087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Swem LR. Elsen S. Bird TH. Swem DL. Koch HG. Myllykallio H. Daldal F. Bauer CE. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J Mol Biol. 2001;309:121–138. doi: 10.1006/jmbi.2001.4652. [DOI] [PubMed] [Google Scholar]
- 234.Swem LR. Gong X. Yu CA. Bauer CE. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem. 2006;281:6768–6775. doi: 10.1074/jbc.M509687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Swem LR. Kraft BJ. Swem DL. Setterdahl AT. Masuda S. Knaff DB. Zaleski JM. Bauer CE. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 2003;22:4699–4708. doi: 10.1093/emboj/cdg461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Takaya N. Dissimilatory nitrate reduction metabolisms and their control in fungi. J Biosci Bioeng. 2002;94:506–510. doi: 10.1016/s1389-1723(02)80187-6. [DOI] [PubMed] [Google Scholar]
- 237.Thorndycroft FH. Butland G. Richardson DJ. Watmough NJ. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem J. 2007;401:111–119. doi: 10.1042/BJ20060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tolla DA. Savageau MA. Phenotypic repertoire of the FNR regulatory network in Escherichia coli. Mol Microbiol. 2011;79:149–165. doi: 10.1111/j.1365-2958.2010.07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Torres MJ. Bueno E. Mesa S. Bedmar EJ. Delgado MJ. Emerging complexity in the denitrification regulatory network of Bradyrhizobium japonicum. Biochem Soc Trans. 2011;39:284–288. doi: 10.1042/BST0390284. [DOI] [PubMed] [Google Scholar]
- 240.Tosques IE. Shi J. Shapleigh JP. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1996;178:4958–4964. doi: 10.1128/jb.178.16.4958-4964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tseng CP. Albrecht J. Gunsalus RP. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tucker NP. D'Autreaux B. Yousafzai FK. Fairhurst SA. Spiro S. Dixon R. Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J Biol Chem. 2008;283:908–918. doi: 10.1074/jbc.M705850200. [DOI] [PubMed] [Google Scholar]
- 243.Tucker NP. Le Brun NE. Dixon R. Hutchings MI. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010;18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 244.Ugidos A. Morales G. Rial E. Williams HD. Rojo F. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ Microbiol. 2008;10:1690–1702. doi: 10.1111/j.1462-2920.2008.01586.x. [DOI] [PubMed] [Google Scholar]
- 245.Van Alst NE. Sherrill LA. Iglewski BH. Haidaris CG. Compensatory periplasmic nitrate reductase activity supports anaerobic growth of Pseudomonas aeruginosa PAO1 in the absence of membrane nitrate reductase. Can J Microbiol. 2009;55:1133–1144. doi: 10.1139/w09-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.van Spannig RJ. Structure, function, regulation and evolution of the nitrite and nitrous oxide reductases: denitrification enzymes with a b-propeller fold. In: Moir JWB, editor. Nitrogen Cycling in Bacteria. Norkfolk, UK: Caister Academic Press; 2011. pp. 135–161. [Google Scholar]
- 247.van Spannig RJ. Delgado MJ. Richardson DJ. The nitrogen cycle: denitrification and its relationship to N2 fixation. In: Werner D, editor; Newton WE, editor. Fixation in Agriculture, Forestry, Ecology and the Environment. The Netherlands: Elsevier; 2005. pp. 277–342. [Google Scholar]
- 248.van Spannig RJ. Richardson DJ. Ferguson SJ. Introduction to the biochemistry and molecular biology of denitrification. In: Bothe H, editor; Ferguson SJ, editor; Newton WE, editor. Biology of the Nitrogen Cycle. The Netherlands: Elsevier; 2007. pp. 83–93. [Google Scholar]
- 249.Veldman R. Reijnders WN. van Spanning RJ. Specificity of FNR-type regulators in Paracoccus denitrificans. Biochem Soc Trans. 2006;34:94–96. doi: 10.1042/BST0340094. [DOI] [PubMed] [Google Scholar]
- 250.von Wachenfeldt C. Hederstedt L. Respiratory cytochromes, other heme-proteins and heme biosynthesis. In: Hoch JA, editor; Losick R, editor; Sonenshein AL, editor. Bacillus subtilis and Its Close Relatives: from Genes to Cells. Washington DC: American Society for Microbiology Publications; 2001. pp. 163–179. [Google Scholar]
- 251.Wang E. Ikonen TP. Knaapila M. Svergun D. Logan DT. von Wachenfeldt C. Small-angle X-ray scattering study of a Rex family repressor: conformational response to NADH and NAD+ binding in solution. J Mol Biol. 2011;408:670–683. doi: 10.1016/j.jmb.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 252.Watmough NJ. Field SJ. Hughes RJ. Richardson DJ. The bacterial respiratory nitric oxide reductase. Biochem Soc Trans. 2009;37:392–399. doi: 10.1042/BST0370392. [DOI] [PubMed] [Google Scholar]
- 253.Willett J. Smart JL. Bauer CE. RegA control of bacteriochlorophyll and carotenoid synthesis in Rhodobacter capsulatus. J Bacteriol. 2007;189:7765–7773. doi: 10.1128/JB.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Williams HD. Zlosnik JE. Ryall B. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv Microb Physiol. 2007;52:1–71. doi: 10.1016/S0065-2911(06)52001-6. [DOI] [PubMed] [Google Scholar]
- 255.Winstedt L. Yoshida K. Fujita Y. von Wachenfeldt C. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J Bacteriol. 1998;180:6571–6580. doi: 10.1128/jb.180.24.6571-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wood NJ. Alizadeh T. Bennett S. Pearce J. Ferguson SJ. Richardson DJ. Moir JW. Maximal expression of membrane-bound nitrate reductase in Paracoccus is induced by nitrate via a third FNR-like regulator named NarR. J Bacteriol. 2001;183:3606–3613. doi: 10.1128/JB.183.12.3606-3613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wood NJ. Alizadeh T. Richardson DJ. Ferguson SJ. Moir JW. Two domains of a dual-function NarK protein are required for nitrate uptake, the first step of denitrification in Paracoccus pantotrophus. Mol Microbiol. 2002;44:157–170. doi: 10.1046/j.1365-2958.2002.02859.x. [DOI] [PubMed] [Google Scholar]
- 258.Wu J. Bauer CE. RegB/RegA, a global redox-responding two-component system. Adv Exp Med Biol. 2008;631:131–148. doi: 10.1007/978-0-387-78885-2_9. [DOI] [PubMed] [Google Scholar]
- 259.Wu J. Bauer CE. RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. MBio 1. 2010;pii:e00272–10. doi: 10.1128/mBio.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wunsch P. Zumft WG. Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J Bacteriol. 2005;187:1992–2001. doi: 10.1128/JB.187.6.1992-2001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yan A. Kiley PJ. Techniques to isolate O2-sensitive proteins: [4Fe-4S]-FNR as an example. Methods Enzymol. 2009;463:787–805. doi: 10.1016/S0076-6879(09)63042-1. [DOI] [PubMed] [Google Scholar]
- 262.Yoon SS. Karabulut AC. Lipscomb JD. Hennigan RF. Lymar SV. Groce SL. Herr AB. Howell ML. Kiley PJ. Schurr MJ. Gaston B. Choi KH. Schweizer HP. Hassett DJ. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J. 2007;26:3662–3672. doi: 10.1038/sj.emboj.7601787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yukl ET. Elbaz MA. Nakano MM. Moenne-Loccoz P. Transcription factor NsrR from Bacillus subtilis senses nitric oxide with a 4Fe-4S cluster. Biochemistry. 2008;47:13084–13092. doi: 10.1021/bi801342x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhang X. Hulett FM. ResD signal transduction regulator of aerobic respiration in Bacillus subtilis: ctaA promoter regulation. Mol Microbiol. 2000;37:1208–1219. doi: 10.1046/j.1365-2958.2000.02076.x. [DOI] [PubMed] [Google Scholar]
- 265.Zufferey R. Preisig O. Hennecke H. Thony-Meyer L. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J Biol Chem. 1996;271:9114–9119. doi: 10.1074/jbc.271.15.9114. [DOI] [PubMed] [Google Scholar]
- 266.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zumft WG. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J Inorg Biochem. 2005;99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 268.Zumft WG. Kroneck PM. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv Microb Physiol. 2007;52:107–227. doi: 10.1016/S0065-2911(06)52003-X. [DOI] [PubMed] [Google Scholar]