Abstract
An endogenous meiotic driver in the dengue and yellow fever vector mosquito Aedes aegypti can cause highly male-biased sex ratio distortion in crosses from suitable genetic backgrounds. We previously selected a strain that carries a strong meiotic drive gene (D) linked with the male-determining allele (M) on chromosome 1 in A. aegypti. Here, we performed segregation analysis of the MD locus among backcross (BC1) progeny from a driver male and drive-sensitive females. Assessment of sex ratios among BC2 progeny showed ∼5.2% recombination between the MD locus and the sex determination locus. Multipoint linkage mapping across this region revealed consistent marker orders and recombination frequencies with the existing reference linkage map and placed the MD locus within a 6.5-cm interval defined by the LF159 locus and microsatellite marker 446GAA, which should facilitate future positional cloning efforts.
Keywords: gene drive, linkage map, microsatellite, segregation distortion, sex linkage, SSCP
Meiotic drive is one of the best known examples in which a selfish genetic element can increase in frequency and eventually become fixed in the population regardless of its phenotypic effect because its own fitness is partly uncoupled from that of the “host”’ individual (Hurst and Werren 2001). Meiotic drivers violate Mendel's first law of independent transmission of alleles to gametes, and instead, particular alleles are preferentially transmitted following meiosis. Such drive systems have the potential to also impact population dynamics of closely linked neutral polymorphisms due to a selective sweep or hitchhiking effect (Maynard-Smith and Haigh 1974). This phenomena has been confirmed in investigations of natural populations including, for example, the segregation distorter (SD) system of Drosophila melanogaster (Palopoli and Wu 1996), the mouse t-complex (Hammer and Silver 1994), and more recently for a sex-linked meiotic driver in D. simulans, wherein sex ratio drive induced a strong selective sweep in populations where the distorter was in high frequency (Derome et al. 2004).
The existence of an endogenous sex-linked meiotic drive system has been reported in 2 mosquito species, Aedes aegypti and Culex pipiens (Hickey and Craig 1966a; Sweeny and Barr 1978). Aedes aegypti is the primary vector of dengue, yellow fever, and Chikungunya viruses to humans in many subtropical and tropical regions. Given the general failure in development of vaccines or drugs to effectively prevent most mosquito-borne diseases, considerable interest has emerged in exploring the potential for employing transgenic insect approaches to disease control. This would entail introducing antipathogen effector genes into mosquitoes, thereby making them incompetent to transmit disease to humans followed by their release into natural populations. Inherent to the success of such population replacement efforts would be the identification of suitable gene drive systems that would ensure rapid transgene introgression into natural populations as well as persistent linkage disequilibrium between the driver locus and the antipathogen effector gene (James 2005). We previously reported selection for an A. aegypti strain (T37) from a population in Trinidad, West Indies that carries a strong meiotic driver (Mori et al. 2004) and subsequently observed evidence for a hitchhiking effect in controlled cage studies (Cha, Mori, et al. 2006). Computer simulations also suggested that under some conditions, for example, uniform population sensitivity and low fitness costs of associated genes, the A. aegypti drive system could facilitate near-fixation of an antipathogen transgene in natural populations (Huang et al. 2007).
The meiotic drive system in A. aegypti is known to be tightly linked with the sex determination locus on chromosome 1 (Hickey and Craig 1966a). Of note, sex determination in A. aegypti and other mosquitoes in the subfamily Culicinae does not involve an XY chromosome system but instead is determined by an autosomal gene with the male-determining allele (M) being dominant to the female-determining allele (m) and males being the heterogametic sex (Gilchrist and Haldane 1947). Because the meiotic drive gene (D) only functions when in cis configuration with the male-determining allele, it is generally symbolized as a superscript (MD) after Hickey and Craig (1966a). Furthermore, the MD gene product acts in trans with a responder locus tightly linked with the m allele, which shows variable response to MD from sensitive (ms) to insensitive (mi) (Suguna et al. 1977; Wood and Newton 1991; Cha, Chadee, et al. 2006), to promote breakage of ms allele–carrying chromosomes during meiosis (Newton et al. 1976). This results in a highly male-biased sex ratio in the resulting offspring (Hickey and Craig 1966b), for example, with appropriate crosses with T37 strain males, we obtain ∼85% male progeny (Mori et al. 2004).
At present, little is known about the molecular basis for the MD system in A. aegypti. However, other meiotic drive systems have been well characterized. The SD system in D. melanogaster has been shown to be determined by a duplicated but truncated RanGAP gene (Merrill et al. 1999), a regulatory gene of the small GTPase Ran. The mouse t-complex contains multiple distorters (Tcd loci) that act in trans on a single responder locus (Smok1). The Tcd1 locus encodes a Rho GTPase-activating protein, whereas theTcd2 locus contains the Fgd2 gene that encodes a truncated guanine nucleotide exchange factor (GEF) for Rho small G proteins (Bauer et al. 2005, 2007). These likely regulate different pathways in the Rho signaling cascade. Therefore, meiotic drive in both D. melanogaster and mouse has been associated with components of the Ras superfamily of regulatory small GTPases (Wennerberg et al. 2005). We recently employed suppressive subtractive hybridization to isolate transcripts enriched in testes from the A. aegypti T37 strain and identified a number of genes associated with signal transduction and cell cycle that included several Ras-related genes (Shin et al. 2011). Here, we take advantage of the existing linkage map (Severson et al. 2002) and whole genome sequence (Nene et al. 2007) for A. aegypti to localize the genetic map position for the MD locus as a foundation for ultimately isolating the MD gene and responder locus using positional cloning techniques.
Materials and Methods
Genetic Crosses
Two A. aegypti strains were used to generate mapping populations segregating for the MD locus. The T37 strain carries a strong MD gene and insensitive mi allele (Mori et al. 2004). The RED strain is fixed for highly sensitive ms alleles (Hickey and Craig 1966a; Mori et al. 2004). Mosquitoes were reared and maintained in an environmental chamber following our standard procedures (Schneider et al. 2007). Adult females were blood fed on anesthetized rats ∼1 week after emergence. Our protocol for maintenance and care of experimental animals was reviewed and approved by the Institutional Animal Care and Use Committee at the University of Notre Dame.
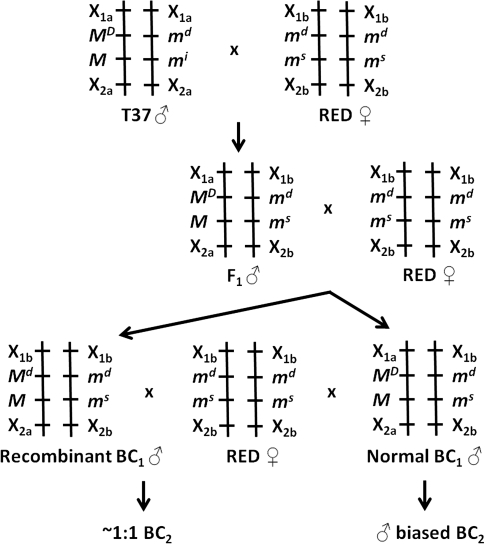
To prepare an F1 generation from pairwise matings, a single T37 strain male and 5 virgin RED strain females were placed in a 450 ml mesh-covered carton for 4 days. After blood feeding, individual females were transferred to 12.5 ml mesh-covered glass vials containing 1 ml water and a strip of paper towel as the oviposition substrate. F1 progeny was reared from eggs from a single pairwise mating, and backcross (BC1) populations were prepared by mating each F1 male with 5 virgin RED strain females as previously described. A single BC1 population was subsequently selected for genetic mapping. To assess the MD phenotype of individual BC1 males, the sex ratios observed in BC2 populations were determined. BC2 populations were prepared by mating each BC1 male with 5 virgin RED strain females as previously described. Sex ratios were determined for progeny from each of the females and tested for departure from the expected 1:1 using the X2 test. All parental F1 and BC1 adults were frozen and stored at −80 °C. The general crossing scheme is shown in Figure 1.
Figure 1.
Crossing scheme for segregation analysis of meiotic drive (MD) with the sex determination locus (M) and DNA-based genetic markers Xij. Male-determining (M) alleles are dominant to female-determining alleles (m). Female-determining chromosomes carry drive sensitive (ms) or drive insensitive (mi) responder loci. Recombination between MD and the M locus was revealed by sex ratios observed among BC2 progeny.
DNA Extraction and Genotyping
Genomic DNA was extracted from individual carcasses using our standard protocol (Severson et al. 1993). DNA from each mosquito was resuspended in tris–ethylenediaminetetraacetic acid (10 mM Tris–HCl, 1 mM EDTA, pH 8.0).
Single-strand conformational polymorphism (SSCP) marker analysis was performed following Bosio et al. (2000). Previously, reported individual loci across the region flanking the sex determination locus on chromosome 1 (Severson et al. 2002) were initially screened for polymorphism with BC1 parental individuals from the T37 and RED strains. Markers showing strain-specific polymorphism were then used to genotype BC1 male progeny.
Preliminary screenings and linkage analysis of microsatellite marker polymorphisms were performed using denaturing polyacrylamide gel electrophoresis following Chambers et al. (2007). In silico identification of putative single-copy microsatellite loci within A. aegypti genome supercontig assemblies (Nene et al. 2007) that were determined by BLASTn analysis at VectorBase (http://aaegypti.vectorbase.org) to carry genetic markers on chromosome 1 (Severson et al. 2002) were identified using the Tandem Repeats Finder program (Benson 1999) and evaluated as described in Lovin et al. (2009).
Linkage Analysis
SSCP and microsatellite marker genotypes and MD phenotypes for individual BC1 males were scored, and multipoint linkage analysis was performed using the MAPMAKER computer program (Lander et al. 1987) with an LOD of 3.0 as the threshold for significance. Pairwise recombination distances were converted to Kosambi centiMorgans (Kosambi 1944).
Results and Discussion
The observed segregation ratio for the F1 generation from the T37 male × RED female cross was 0.54:0.46 (male:female) among 102 total progeny. Because T37 males carry an insensitive responder allele (mi) and A. aegypti progeny from individual females tend to normally be slightly male based, these results were expected. For example, RED strain by RED strain crosses typically produce ∼54.5% male progeny (Mori et al. 2004). However, for the BC1 generation, we obtained 203 progeny that showed a highly male-biased sex ratio (0.81:0.19). Each of the BC1 males was allowed to mate with up to 5 RED strain females, and the BC2 generation eggs were then collected and hatched separately for each female and sex ratios determined.
The MD phenotype of 135 BC1 males was determined based on sex ratios observed among 1–5 BC2 families per male (Supplementary Table 1). An arbitrary threshold of at least 20 progeny per BC2 family was used for statistical comparisons of sex ratios. Most BC2 families for individual BC1 males showed significant male-biased sex ratio distortion, but for 7 of the 135 BC1 males, the BC2 families did not show sex ratio distortion, thus indicating 5.19% recombination between the MD gene and the male-determining locus.
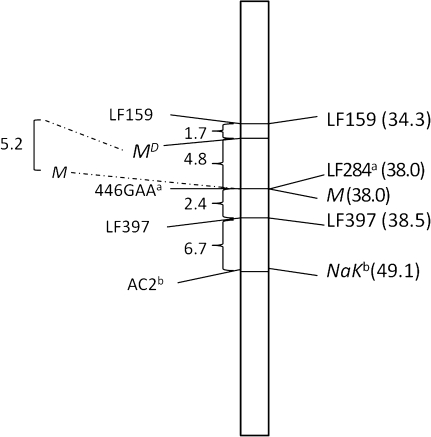
Multipoint linkage mapping of the MD locus was successfully performed relative to 2 SSCP and 2 microsatellite loci (Figure 2). The MD locus is located within a 6.5-cm interval defined by the SSCP marker LF159 (GenBank accession T58320) located within supercontig 1.388 and the microsatellite marker 446GAA (primers—F: TGACTTCCCTGGGCATAGAG, R: GCCCCATCATGCAACATAGT). The 446GAA locus is located within supercontig 1.446, which also contains the previously reported LF284 locus (BM005502) (Severson et al. 2002). Microsatellite locus AC2 was identified previously (Slotman et al. 2007), and BLASTn analysis placed it within supercontig 1.650, which also contains the NaK locus (AF393727) (Severson et al. 2002). As indicated in Figure 2, linear orders and map distances between markers were consistent with the composite linkage map (Severson et al. 2002). None of the other loci known to reside within the interval containing the MD locus were informative in the mapping population, nor were we able to identify informative microsatellite loci within supercontigs containing these loci.
Figure 2.
Linkage map of the meiotic drive (MD) locus on chromosome 1. Map distances are listed in Kosambi centiMorgans. The left side of the map shows the markers utilized and map distances from this study. Observed recombination between the sex determination locus (M) and MD is shown to the far left. The right side shows marker information from the existing reference map (Severson et al. 2002). aBoth loci are in supercontig 1.446; bboth loci are in supercontig 1.65.
In general, SDs are often found in genome regions with low recombination, such as heterochromatin associated with pericentromeric regions or with chromosomal inversions (Lyttle 1991). This results in reduced recombination between the distorter gene and the responder locus. This is particularly important as linkage disequilibrium across the drive system complex is critical to its function and long-term stability. For example, the SD system in D. melanogaster and t-haplotype in mouse each shows reduced recombination caused by inversions that promotes strong linkage disequilibrium across broad genome regions around the drive complex (Hammer and Silver 1994; Palopoli and Wu 1996). Although the presence and genome structure of potential inversions in the A. aegypti genome is largely unknown, there is evidence for differences in heterochromatin structure between the male- and female-determining chromosomes that could impact recombination frequency around the genome region containing the sex determination locus and the MD system (Wallace and Newton 1987). The sex determination locus is tightly coupled with the centromere (Newton et al. 1974), and the MD locus is associated with an intercalary band when present (Newton et al. 1974, 1976).
In conclusion, our mapping efforts have placed the MD system to a 6.5-cm interval in the A. aegypti genome, thus defining physical landmarks for future positional cloning efforts. A more fine-scale resolution was not possible due to low polymorphism observed among known genetic markers in this region and the incomplete status of the present A. aegypti genome assembly. Assignment to and orientation of supercontig assemblies to their respective chromosome positions is presently limited (Nene et al. 2007). Enhancement of the genome assembly is needed for identification of candidate genes for the MD locus within a minimum genome region for functional analysis. Ongoing efforts to identify physical positions and relative orientations of supercontigs on A. aegypti chromosomes (Sharakhova et al. 2011) coupled with availability of a well-characterized bacterial artificial chromosome library (Jiménez et al. 2004) should facilitate this process. Successful molecular characterization of the meiotic drive system would promote efforts to better understand the observed phenotypic outcomes and to evaluate mechanisms for effectively coupling effector genes with the drive system as a novel mechanism for disease control via population replacement.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Institute of Allergy and Infectious Diseases; National Institutes of Health grants PO1-AI45123 and R01-AI33127; and Grand Challenges in Global Health initiative.
Supplementary Material
References
- Bauer H, Veron N, Willert J, Herrmann BG. The t-complex-encoded guanine nucleotide exchange factor Fgd2 reveals that the two opposing signaling pathways promote transmission ratio distortion in the mouse. Genes Dev. 2007;21:143–147. doi: 10.1101/gad.414807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Willert J, Koschorz B, Herrmann BG. The t complex-encoded GTPase-activating protein Tapgap1 acts as a transmission ratio distorter in mice. Nat Genet. 2005;37:969–973. doi: 10.1038/ng1617. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WC., IV Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha SJ, Chadee DD, Severson DW. Population dynamics of an endogenous meiotic drive system in Aedes aegypti in Trinidad. Am J Trop Med Hyg. 2006b;75:70–77. [PubMed] [Google Scholar]
- Cha SJ, Mori A, Chadee DD, Severson DW. Cage trials using an endogenous meiotic drive gene in the mosquito, Aedes aegypti, to promote population replacement. Am J Trop Med Hyg. 2006a;74:62–68. [PubMed] [Google Scholar]
- Chambers EW, Meece JK, McGowan JA, Lovin DD, Hemme RR, McAbee K, Brown SE, Knudson DL, Severson DW. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J Hered. 2007;98:202–210. doi: 10.1093/jhered/esm015. [DOI] [PubMed] [Google Scholar]
- Derome N, Métayer K, Montchamp-Moreau C, Veuille M. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics. 2004;166:1357–1366. doi: 10.1534/genetics.166.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist BM, Haldane JBS. Sex linkage and sex determination in a mosquito Culex molestus. Hereditas. 1947;33:175–190. [Google Scholar]
- Hammer MF, Silver LM. Phylogenetic analysis of the alpha-globin pseudogene-4 (Hba-ps4) locus in the house mouse species complex reveals a stepwise evolution of t haplotypes. Mol Biol Evol. 1994;10:971–1001. doi: 10.1093/oxfordjournals.molbev.a040051. [DOI] [PubMed] [Google Scholar]
- Hickey WA, Craig GB. Genetic distortion of sex ratio in a mosquito Aedes aegypti. Genetics. 1966a;53:1177–1196. doi: 10.1093/genetics/53.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey WA, Craig GB. Distortion of sex ratio in populations of Aedes aegypti. Can J Genet Cytol. 1966b;8:260–278. doi: 10.1139/g66-033. [DOI] [PubMed] [Google Scholar]
- Huang Y, Magori K, Lloyd AL, Gould F. Introducing desirable transgenes into insect populations using Y-linked meiotic drive—a theoretical assessment. Evolution. 2007;61(4):717–726. doi: 10.1111/j.1558-5646.2007.00075.x. [DOI] [PubMed] [Google Scholar]
- Hurst GDD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- James AA. Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 2005;21:64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jiménez LV, Kang B-K, deBruyn B, Lovin DD, Severson DW. Characterization of an Aedes aegypti BAC library and chromosomal assignment of BAC clones for physical mapping. Insect Mol Biol. 2004;13:37–44. doi: 10.1046/j.0962-1075.2004.00456.x. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SF, Newburg L. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lovin DD, Washington KO, deBruyn B, Hemme RR, Mori A, Epstein SR, Harker BW, Streit TG, Severson DW. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 2009;10:590. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith SJ, Haigh J. The hitchhiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- Mori A, Chadee DD, Graham DH, Severson DW. Reinvestigation of an endogenous meiotic drive system in the mosquito Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2004;41:1027–1033. doi: 10.1603/0022-2585-41.6.1027. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, Loftus B, Xi Z, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1722. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton ME, Southern DI, Wood RJ. X and Y chromosomes of Aedes aegypti (L.) distinguished by Giemsa C-banding. Chromosoma. 1974;49:41–49. doi: 10.1007/BF00284986. [DOI] [PubMed] [Google Scholar]
- Newton ME, Wood RJ, Southern DI. A cytological analysis of meiotic drive in the mosquito Aedes aegypti (L.) Genetica. 1976;46:297–318. [Google Scholar]
- Palopoli MF, Wu CI. Rapid evolution of a coadapted gene complex: evidence from the Segregation Distorter (SD) system of meiotic drive in Drosophila melanogaster. Genetics. 1996;143:1675–1688. doi: 10.1093/genetics/143.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JR, Mori A, Romero-Severson J, Chadee DD, Severson DW. Investigations of dengue-2 susceptibility and body size among Aedes aegypti populations. Med Vet Entomol. 2007;21:370–376. doi: 10.1111/j.1365-2915.2007.00699.x. [DOI] [PubMed] [Google Scholar]
- Severson DW, Meece JK, Lovin DD, Saha G, Morlais I. Linkage map organization of expressed sequence tags and sequence tagged sites in the mosquito, Aedes aegypti. Insect Mol Biol. 2002;11:371–378. doi: 10.1046/j.1365-2583.2002.00347.x. [DOI] [PubMed] [Google Scholar]
- Severson DW, Mori A, Zhang Y, Christensen BW. Linkage map for Aedes aegypti using restriction fragment length polymorphism. J Hered. 1993;84:241–247. doi: 10.1093/oxfordjournals.jhered.a111333. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, Timoshevskiy VA, Yang F, Demin SI, Severson DW, Sharakhov IV. Imaginal discs—a new source of chromosomes for genome mapping of the yellow fever mosquito Aedes aegypti. PLoS Negl Trop Dis. 5:e1335. 2011 doi: 10.1371/journal.pntd.0001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Jin L, Lobo NF, Severson DW. Transcript profiling of the meiotic drive phenotype in testes of Aedes aegypti using suppressive subtractive hybridization. J Insect Physiol. 2011;57:1220–1226. doi: 10.1016/j.jinsphys.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, Kelly NB, Harrington LC, Kitthawee S, Jones JW, Scott TW, Caccone A, Powell JR. Polymorphic microsatellite markers for studies of Aedes aegypti (Diptera: Culicidae), the vector of dengue and yellow fever. Mol Ecol Notes. 2007;7:168–171. [Google Scholar]
- Suguna SG, Wood RJ, Curtis CF, Whitelaw A, Kazmi SJ. Resistance to meiotic drive at the MD locus in an Indian wild population of Aedes aegypti. Genet Res. 1977;29:123–132. doi: 10.1017/s0016672300017195. [DOI] [PubMed] [Google Scholar]
- Sweeny TL, Barr AR. Sex ratio distortion caused by meiotic drive in a mosquito, Culex pipiens L. Genetics. 1978;88:427–446. doi: 10.1093/genetics/88.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AJ, Newton ME. Heterochromatin diversity and cyclic response to selective silver staining in Aedes aegypti (L.) Chromosoma. 1987;95:89–93. doi: 10.1007/BF00293847. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- Wood RJ, Newton ME. Sex-ratio distortion caused by meiotic drive in mosquitoes. Am Nat. 1991;137:379–391. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.