Abstract
Background
Cluster thinning is an agronomic practice in which a proportion of berry clusters are removed from the vine to increase the source/sink ratio and improve the quality of the remaining berries. Until now no transcriptomic data have been reported describing the mechanisms that underlie the agronomic and biochemical effects of thinning.
Results
We profiled the transcriptome of Vitis vinifera cv. Sangiovese berries before and after thinning at veraison using a genome-wide microarray representing all grapevine genes listed in the latest V1 gene prediction. Thinning increased the source/sink ratio from 0.6 to 1.2 m2 leaf area per kg of berries and boosted the sugar and anthocyanin content at harvest. Extensive transcriptome remodeling was observed in thinned vines 2 weeks after thinning and at ripening. This included the enhanced modulation of genes that are normally regulated during berry development and the induction of a large set of genes that are not usually expressed.
Conclusion
Cluster thinning has a profound effect on several important cellular processes and metabolic pathways including carbohydrate metabolism and the synthesis and transport of secondary products. The integrated agronomic, biochemical and transcriptomic data revealed that the positive impact of cluster thinning on final berry composition reflects a much more complex outcome than simply enhancing the normal ripening process.
Background
Many agronomic practices are employed to maximize grape berry quality in the highly competitive wine industry, including the control of bud load during winter pruning and cluster thinning during berry development. Cluster thinning acts directly to increase the source/sink balance of grapevine plants and the technique is used to prevent overcropping in varieties characterized by excessive bud fertility or in areas where reduced yield is a prerequisite for high-quality wine production. Under such conditions, cluster thinning is performed to obtain a leaf area/yield ratio of 0.8-1.2 m2/kg. Below this threshold value several authors [1,2] reported a positive correlation between berry juice soluble sugars and leaf area/crop weight ratio. Similar results have been achieved following cluster thinning in different varieties and sites [3-7]. Thinning also increases the anthocyanin content of berries, which is an important quality determinant of red wines [8-10]. The anthocyanin composition is also affected, e.g. cluster thinning induced the accumulation of 3',4'-substituted anthocyanins in Sangiovese and Nebbiolo varieties [3,5].
Although it is well known that many different factors influence flavonoid and anthocyanin biosynthesis [11], sugar is likely to play a prominent role because of the concomitant accumulation of soluble solids in the berry flesh and anthocyanins in the skin of red grape varieties. This relationship was first proposed by Pirie and Mullins [12], who suggested that the sugar content of red berry flesh could regulate anthocyanin production, and this was supported by in vitro experiments showing an increase in phenylalanine ammonia-lyase (PAL) activity [13]. Increased anthocyanin accumulation after treatment with sucrose and other sugars has been already demonstrated in grapevine and in a variety of other plant species [14-17]. Several sugars have been shown to induce genes encoding enzymes in the anthocyanin biosynthesis pathway, such as chalcone synthase (CHS), dihydroflavonol reductase (DFR), leucoanthocyanidin dioxygenase (LDOX) [18,19] and flavanone 3-hydroxylase (F3H) [20]. Sucrose boxes have been identified in the promoters of some of these genes [18,19].
Little is currently known about the regulation of gene expression when the source/sink ratio is deliberately altered in the field. We report the first transcriptomic analysis (integrated with agronomic and biochemical data) aiming to determine the mechanisms that control Sangiovese berry composition by comparing gene expression profiles of thinned and control vines. Berry transcriptional profiles were analyzed during ripening using the most comprehensive grapevine microarray available to date, representing 29,549 genes from the most recent 12X grapevine V1 gene prediction http://srs.ebi.ac.uk/. We observed substantial transcriptomic remodeling in berries from thinned vines which became evident by 2 weeks post-treatment and persisted until ripening, with particular impact on genes involved in carbohydrate metabolism, flavonoid biosynthesis and transport. The results from these studies provide insight into the molecular basis of berry ripening induced by vineyard management techniques.
Results
The effect of cluster thinning on yield and berry ripening
Cluster thinning (CT) was carried out to remove approximately 50% of the bunches on each vine, leaving approximately eight clusters per vine in comparison with 16 on control (C) plants, thus reducing the yield by ~54% (Table 1). The average bunch and berry weight remained the same in CT and C plants (Table 1).
Table 1.
Influence of cluster thinning (CT) on yield component and berry composition at harvest.
| Yield/ vine (kg) |
Cluster/ vine (n) |
Cluster weight (g) |
Berry weight (g) |
Leaf area/ vine (m2) |
Leaf area/ yield (m2/kg) |
°Brix | TA (g/L) |
pH | |
|---|---|---|---|---|---|---|---|---|---|
| C | 6.3 a | 16 a | 386 | 2.37 | 3.84 | 0.6 b | 20.8 b | 7.6 a | 3.4 b |
| CT | 2.9 b | 8 b | 353 | 2.24 | 3.43 | 1.2 a | 22.7 a | 6.8 b | 3.5 a |
| Significancezy | * | * | ns | ns | ns | * | * | * | * |
z Means separated within columns by the Student-Newman-Keuls test (n = 12 except for Brix, TA and pH were n = 3).
y *, significant at p ≤ 0.05; ns, not significant.
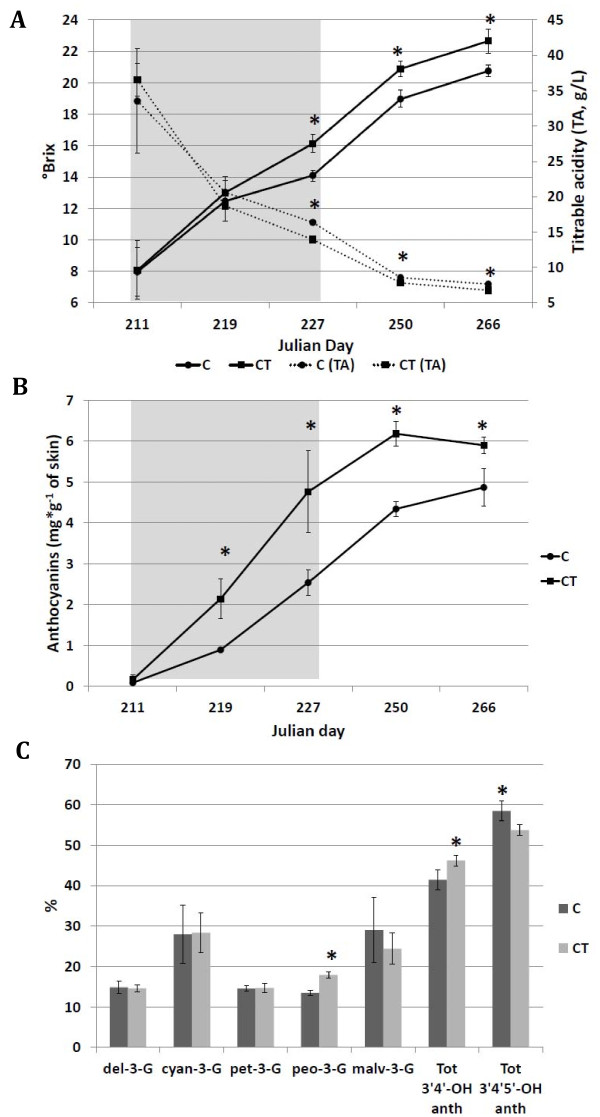
The leaf area per vine was similar in CT and C plants at harvest, indicating that cluster thinning increased the leaf area/yield ratio from 0.6 m2/kg in C plants to 1.2 m2/kg in CT plants (Table 1). CT berries also accumulated more total soluble solids than C berries from full veraison (8 d after cluster thinning) until harvest, and were less acidic and had higher °Brix values than controls at harvest (Figure 1A and Table 1).
Figure 1.
Agronomic effects of cluster thinning. (A) Accumulation of soluble sugars (°Brix) and titratable acidity (TA) in berries from C and CT vines (n = 3). (B) Anthocyanin concentration during ripening and (C) anthocyanin composition at harvest in berry skins from C and CT vines (n = 3). Bars represent ± SE. Asterisks indicate significant differences between the treatments at the same date using the Student-Newman-Keuls test (*P < 0.05). Gray background represents the veraison phase.
The anthocyanin content of berry skin was analyzed by HPLC over the same period, showing that anthocyanins accumulated more rapidly in CT berries compared to controls (Figure 1B), and the difference in anthocyanin content between the two samples was already significantly higher by the second sampling date (JD 219) corresponding to full veraison, and gradually declined but was still evident at harvest (Figure 1B). HPLC analysis also revealed that the increase in total anthocyanins was not evenly distributed among the five main glucosylated species that characterize the Sangiovese cultivar (Figure 1C). We observed a significant increase in the levels of 3'4'-OH anthocyanins associated to cluster thinning, which modified the anthocyanin profile of CT berries, particularly increasing levels of the glucosylated form of peonidin compared to C berries.
Transcriptional modulation induced by cluster thinning
To investigate the molecular changes that take place in response to cluster thinning, we carried out a comparative microarray analysis of CT and C berries at time points JD 211, 227 and 266, corresponding to the beginning of veraison (BV), the end of veraison (EV) and harvest (H).
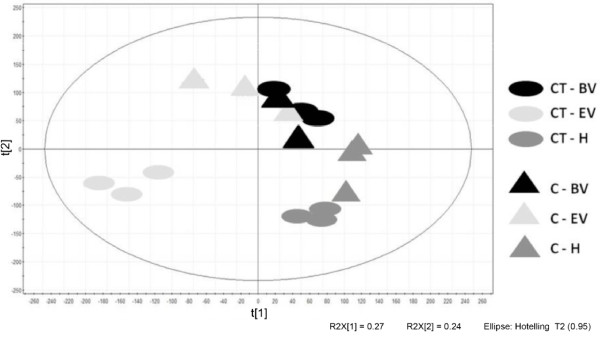
Principal Component Analysis (PCA) of the global transcriptomic data revealed enough uniformity among the three biological replicates to defined associations between treatments (Figure 2). The two principal components, explaining about the 50% of the overall variance, allowed us to clearly separate C and CT at the EV stage, whereas the separation was less clear-cut at the BV and H stages suggesting that the main transcriptomic changes induced by cluster thinning occurred at the EV stage.
Figure 2.
Principal component analysis (PCA) shows that the most severe changes are at the end of veraison. Biological replicates relative to time point beginning of veraison (BV), end of veraison (EV) and harvest (H) are represented as circles for CT and as triangles for C.
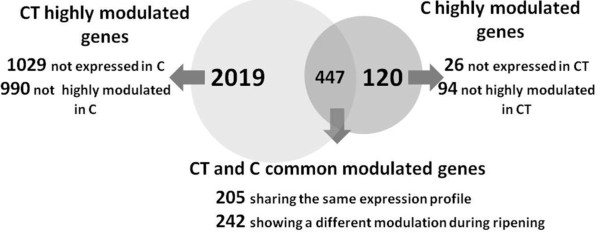
To identify the gene expression profiles with the greatest contribution to the differences between the C and CT transcriptomes, a multiclass comparison analysis was carried out using Significance Analysis of Microarray (SAM) with a false discovery rate (FDR) of 2% (TMev 4.3). We identified 1626 genes modulated during C berry development and 6033 modulated during CT berry development, with a fold change ≥ 2 in at least one comparison (Additional File 1). To evaluate the principal modifications triggered by cluster thinning, we focused on genes with a fold change ≥ 5, narrowing the analysis to 567 genes modulated during C berry development and 2466 genes modulated during CT berry development. A comparison of these datasets indicated three different sets of modulated transcripts. The first grouped 447 genes modulated in both treatments, the second grouped 2019 genes modulated only in CT berries and the third grouped 120 genes modulated only in C berries (Figure 3). For convenience, genes that showed less than a five-fold change in expression were described as 'not highly modulated'.
Figure 3.
Differentially expressed genes during ripening in CT and C berries. Transcripts are divided into three different gene datasets according to their high modulation in CT, in C or in both. The number of CT highly modulated genes either not expressed in control berries or expressed but not highly modulated is shown. Similarly the number of C highly modulated genes either not highly expressed in CT berries or expressed but not highly modulated is shown. The number of common modulated transcripts with equal or different modulation is specified.
Clustering analysis using Pearson's correlation distance divided the common, CT highly modulated and C highly modulated transcripts (Tables 2, 3 and 4) into eight groups representing the minimum number of profiles required to describe the three sampling time points. Clusters 1-4 represent genes that are downregulated during at least one analyzed time point compared to the BV stage, whereas clusters 5-8 represent genes that are upregulated during at least one analyzed time point compared to the BV stage.
Table 2.
Cluster distribution of genes highly modulated both in C and CT berries.
| Cluster number |
Expression profile | Common modulated genes | |
|---|---|---|---|
| C | CT | ||
| 1 |

|
5 | 8 |
| 2 |

|
80 | 179 |
| 3 |

|
216 | 90 |
| 4 |
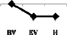
|
14 | 40 |
| 5 |

|
1 | 2 |
| 6 |

|
40 | 44 |
| 7 |
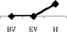
|
78 | 70 |
| 8 |
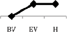
|
13 | 14 |
Analysis was performed separately for C and CT berries at the beginning of veraison (BV), end of veraison (EV) and harvest (H). The expression profiles of the 447 common modulated genes during berry ripening were clustered in eight groups obtained by the k-means method using Pearson's correlation distance. The representative profile and the number of genes for each treatment in every cluster are indicated.
Table 3.
Distribution of 192 annotated transcripts (common to C and CT berries but differentially modulated during ripening) into eight clusters according to the gene expression trends.
| CLUSTER 1 | CLUSTER 2 | CLUSTER 3 | CLUSTER 4 | CLUSTER 5 | CLUSTER 6 | CLUSTER 7 | CLUSTER 8 | ||
|---|---|---|---|---|---|---|---|---|---|
|

|
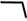
|
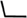
|
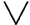
|
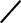
|
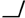
|

|
||
| Carbohydrate metabolic process | C | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 0 |
| CT | 0 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | |
| Cell wall organization or biogenesis | C | 0 | 4 | 6 | 0 | 0 | 1 | 0 | 0 |
| CT | 0 | 6 | 3 | 1 | 0 | 0 | 1 | 0 | |
| Cellular amino acid and derivative metabolic process | C | 0 | 2 | 5 | 0 | 0 | 1 | 1 | 0 |
| CT | 1 | 5 | 1 | 1 | 0 | 0 | 0 | 1 | |
| Cellular homeostasis | C | 0 | 2 | 3 | 0 | 0 | 1 | 0 | 0 |
| CT | 0 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | |
| Cellular process | C | 1 | 2 | 6 | 0 | 0 | 2 | 2 | 1 |
| CT | 0 | 6 | 1 | 2 | 0 | 2 | 2 | 1 | |
| Developmental process | C | 0 | 1 | 4 | 1 | 0 | 3 | 0 | 1 |
| CT | 0 | 5 | 0 | 1 | 0 | 1 | 2 | 1 | |
| Generation of precursor metabolites and energy | C | 0 | 0 | 27 | 1 | 0 | 0 | 0 | 0 |
| CT | 1 | 25 | 0 | 2 | 0 | 0 | 0 | 0 | |
| Hormone metabolic process | C | 0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 |
| CT | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | |
| Lipid metabolic process | C | 0 | 2 | 4 | 0 | 0 | 1 | 0 | 1 |
| CT | 0 | 2 | 1 | 3 | 0 | 1 | 1 | 0 | |
| Nitrogen compound metabolic process | C | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| CT | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Nucleic acid metabolic process | C | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| CT | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Protein metabolic process | C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Response to hormone stimulus | C | 0 | 0 | 9 | 0 | 0 | 0 | 1 | 0 |
| CT | 0 | 7 | 0 | 2 | 0 | 1 | 0 | 0 | |
| Response to stress | C | 0 | 1 | 7 | 0 | 0 | 2 | 4 | 2 |
| CT | 1 | 7 | 1 | 0 | 0 | 2 | 2 | 3 | |
| Secondary metabolic process | C | 0 | 3 | 3 | 2 | 0 | 0 | 10 | 0 |
| CT | 0 | 4 | 2 | 2 | 0 | 9 | 0 | 1 | |
| Signal transduction | C | 0 | 2 | 9 | 0 | 0 | 2 | 0 | 0 |
| CT | 0 | 8 | 1 | 2 | 0 | 0 | 2 | 0 | |
| Transcription | C | 0 | 1 | 11 | 0 | 0 | 1 | 2 | 0 |
| CT | 0 | 9 | 0 | 3 | 0 | 2 | 1 | 0 | |
| Transport | C | 0 | 3 | 8 | 3 | 0 | 3 | 3 | 1 |
| CT | 0 | 11 | 0 | 3 | 0 | 3 | 3 | 1 | |
| TOTAL DIFFERENTIAL MODULATED GENES per CLUSTER | C | 1 | 24 | 112 | 7 | 0 | 19 | 23 | 6 |
| CT | 3 | 108 | 11 | 24 | 0 | 21 | 17 | 8 | |
In each cluster, transcripts are divided according to their functional categories.
Table 4.
Cluster distribution of highly modulated genes in CT and C berries.
| CT | C | ||||||
| Cluster number | Expression profile | Total genes | Genes not expressed in C | Genes not highly modulated in C | Total genes | Genes not expressed in CT | Genes not highly modulated in CT |
| 1 |

|
404 | 23 | 381 | 3 | 0 | 3 |
| 2 |

|
251 | 30 | 221 | 21 | 5 | 16 |
| 3 |

|
58 | 4 | 54 | 16 | 1 | 15 |
| 4 |
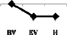
|
172 | 25 | 147 | 1 | 0 | 1 |
| 5 |

|
357 | 303 | 54 | 5 | 2 | 3 |
| 6 |

|
196 | 164 | 32 | 38 | 7 | 31 |
| 7 |
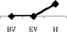
|
89 | 54 | 35 | 34 | 10 | 24 |
| 8 |
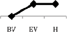
|
492 | 426 | 66 | 2 | 1 | 1 |
Expression profiles of the 2019 CT and 120 C highly modulated genes during berry ripening were clustered in eight groups obtained by the k-means method using Pearson's correlation distance. The representative profile and the number of genes in each cluster are indicated. For each cluster, the CT highly modulated genes that are expressed without high modulation or not expressed at all in control berries are indicated, and similarly the C highly modulated genes that are expressed without high modulation in CT berries or not expressed at all are indicated.
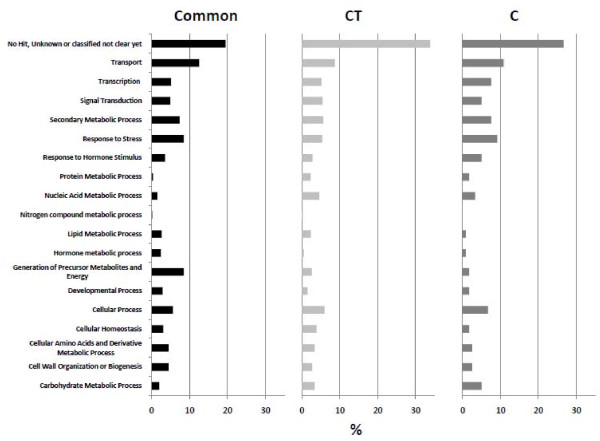
All the transcripts were annotated against the V1 version of the 12X draft annotation of the grapevine genome http://genomes.cribi.unipd.it/DATA/ allowing 70% of the modulated genes to be identified (Additional File 2). To investigate the functional distribution of commonly and specifically modulated transcripts, we distributed them into 18 Gene Ontology (GO) functional categories and determined the percentage of genes in each category for each of the three data sets (Figure 4). The most represented functional categories, shared among the three datasets, were "Transport", "Transcription", "Secondary Metabolic Process", "Response to Stress" and "Cellular Process", which included the main genes involved in the physiology of berry ripening. The functional distribution of modulated genes was similar in the CT and C specific gene sets, suggesting that the CT treatment has a widespread effect on transcription rather than impacting on a specific functional category. The results for the common, CT highly modulated and C highly modulated genes are discussed in more detail below.
Figure 4.
Distribution of common, CT and C highly modulated transcripts into 18 GO functional categories. Transport: GO:0006810; Transcription: GO:0006350; Signal Transduction: GO:0007165; Secondary Metabolic Process: GO:0019748; Response to Stress: GO:0006950; Response to Hormone Stimulus: GO:0009725; Protein Metabolic Process: GO:0019538; Nucleic Acid Metabolic Process: GO:0090304; Nitrogen Compound Metabolic Process: GO:0006807; Lipid Metabolic Process: GO:0006629; Hormone Metabolic Process: GO:0042445; Generation of precursor metabolites and energy: GO:0006091; Developmental Process: GO:0032502; Cellular Process: GO:0009987; Cellular Homeostasis: GO:0019725; Cellular amino acid and derivative metabolic process: GO:0006519; Cell wall organization or biogenesis: GO:0071554; Carbohydrate metabolic process: GO: 0005975. Percentages were calculated on the total number of modulated genes for each dataset.
Common modulated genes
The first gene set contains 447 genes that are modulated in both C and CT berries, representing 79% of the genes modulated in C berries and 22% of those modulated in CT berries. Approximately equal numbers of genes for each treatment were upregulated in comparison to the BV stage and were therefore distributed into clusters 5-8 (132 for C berries and 130 for CT berries, Table 2). Similarly, there were approximately equal numbers of downregulated genes distributed into clusters 1-4 (315 for C berries and 317 for CT berries) although the nature of the distribution was distinct (Table 2). In particular, there were more genes downregulated throughout ripening (cluster 2) and specifically between BV to EV (cluster 4) in CT berries, whereas there were more genes downregulated specifically between EV and H (cluster 3) in C berries. Interestingly, very few common genes were detected in clusters 1 and 5, which represent EV-specific downregulation (1) and upregulation (5) in both treatments.
We identified 205 genes with similar expression profiles in both treatments and 242 with expression profiles that differed between C and CT berries during ripening (Figure 3, Additional File 2). The cluster distribution for the 192 genes with successful functional annotations belonging to the latter group is shown in Table 3.
Many genes belonging to "Generation of Precursor Metabolites and Energy", the most representative functional category, and to several others such as "Carbohydrate Metabolic Process", "Cell Wall Organization or Biogenesis", "Hormone Metabolic Process", "Response to Hormone Stimulus", "Response to Stress" and "Transport", were allocated to cluster 3 in C berries (downregulated specifically between EV and H) but shifted to cluster 2 in CT berries (downregulated throughout ripening). This pattern was particularly evident among the 24 genes with a role in photosynthesis, the six involved in the cell wall changes associated with berry softening (e.g. pectate lyase, pectinesterase and xyloglucan endotransglucosylase), the three involved in sugar metabolism (fructose-bisphosphate aldolase, the vacuolar invertase GIN1 and galactinol synthase) and the two involved in ethylene biosynthesis, i.e. 1-aminocyclopropane-1-carboxylic acid synthase (ACS) and 1-aminocyclopropane-1-carboxylic acid oxidase (ACO). These data are presented in Additional File 2. For the vacuolar invertase GIN1 (VIT_160022g00670), the expression profiles in C and CT berries were confirmed by real-time RT-PCR (Additional File 3).
We also noted several genes that were allocated to cluster 7 in C berries (upregulated specifically between EV to H) but to cluster 6 in CT berries (upregulated throughout ripening) (Table 3). These genes were predominantly involved in secondary metabolic processes, e.g. five PAL isogenes (VIT_00s2508g00010, VIT_16s0039g01120, VIT_16s0039g01320, VIT_16s0039g01300, VIT_00s2849g00010) and two stilbene synthases (STSs, VIT_16s0100g01070, VIT_16s0100g00850). We confirmed the expression profile of one PAL (VIT_16s0039g01120) in C and CT berries by real-time RT-PCR (Additional File 3).
In addition, several of the 205 genes whose expression profile did not change qualitatively after cluster thinning showed marked quantitative differences in expression (Additional File 4). In particular, we observed stronger downregulation of several genes involved in photosynthesis, cell wall metabolism, stress responses and hormone metabolism in CT berries. Notable examples included genes involved in the dynamic remodeling of cell wall polysaccharides, such as a xyloglucan endotransglucosylase/hydrolase (VIT_00s0386g00050), which was downregulated 10-fold more strongly in CT berries compared to controls, and genes related to hormone metabolism, such as a cytokinin dehydrogenase (VIT_18s0001g13200).
Whereas many common genes were downregulated more strongly in CT berries compared to controls, very few were induced more strongly after cluster thinning. However, examples included galactinol synthase (VIT_07s0005g0970), which is an important regulator of carbon partitioning, as well as a STS (VIT_09s0018g01490) and a PAL involved in the phenylpropanoid/flavonoid pathway (VIT_16s0039g01170). Remarkably, the glutathione-S-transferase VvGST4 (VIT_04s0079g00690), whose involvement in berry ripening has been reported previously [21], was upregulated throughout ripening in CT berries compared to controls. We validated the expression profile of one gene in this group by real-time RT-PCR, the flavonol synthase VIT_18s0001g03430 (Additional File 3).
Genes highly modulated in response to cluster thinning
The second group of modulated genes comprised 2019 CT highly modulated transcripts that could be divided into eight clusters according to their expression profiles (Table 4). Approximately equal numbers of genes were downregulated (885, clusters 1-4) and upregulated (1134, clusters 5-8). Unlike the cluster distribution of the common genes discussed above, many CT highly modulated genes were assigned to clusters 1 and 5, and most were assigned to clusters 1, 2, 4, 5, 6 and 8, characterized by prompt modulation in response to cluster thinning (i.e. changes already visible at EV). This suggests that physiological and biochemical changes in the berry almost occur immediately after the treatment. We investigated the expression of the CT highly modulated genes in C berries (Table 4), which allowed us to distinguish between those not expressed in C berries and those expressed but not highly modulated in C berries. Among the 885 genes grouped in clusters 1-4, 82 were not expressed in the controls and 803 were expressed but not highly modulated during ripening. The expression of 82 genes in CT berries but not in C berries at the earliest sampling stage is unlikely to reflect a response to treatment and is probably the consequence of inter-vine variability at the BV stage. Certain agronomic parameters such as titratable acidity and soluble sugar content showed higher standard deviations at BV than the subsequent stages (Figure 1A), confirming that samples collected at BV could be more heterogeneous than those collected later. This could likewise explain the presence of C highly downregulated transcripts that are not expressed in CT berries (Table 4). Surprisingly, a significantly majority of the 1134 genes in clusters 5-8 were not expressed in C berries, while only a minority was expressed but not highly modulated in C berries.
The 2019 CT highly modulated genes were assigned to 18 functional categories as shown in Figure 4. Given the major influence of carbohydrate metabolism, secondary metabolism and the transport of carbohydrate and secondary metabolites on berry quality traits, we will focus on genes in the categories "Carbohydrate Metabolic Process", "Secondary Metabolism" and "Transport", which represent 18% of all the genes modulated in CT berries following the treatment.
Genes belonging to the "Carbohydrate Metabolic Process" category
We identified 68 CT highly modulated genes involved in carbohydrate metabolism. These are listed in Table 5, which provides the Gene_ID, annotation, metabolic process, cluster assignment, expression in C berries, and the EV/BV and H/BV fold change. We found that 28 of these genes grouped in clusters 1-4 and 40 (including 33 not expressed in control berries) grouped in clusters 5-8. The modulated genes are involved in various primary metabolic pathways, including sucrose and starch metabolism, glycolysis, the pentose phosphate pathway and the Krebs cycle, indicating a large-scale reprograming of carbohydrate metabolism in response to cluster thinning. This includes the downregulation of two invertases (VIT_18s0072g01040 and VIT_00s2527g00010) and one sucrose-phosphate synthase (VIT_05s0029g01140) probably representing the decline in photosynthesis during berry ripening. The downregulation of VIT_00s2527g00010 was confirmed by real time RT-PCR (Additional File 3).
Table 5.
CT highly modulated genes in the "Carbohydrate metabolic process" functional category.
| Gene_ID | Description | Metabolic Process | Profile | Expression in C |
FC EV/BV | FC H/BV |
|---|---|---|---|---|---|---|
| VIT_18s0072g00770 | fructose-1,6-bisphosphatase, cytosolic | Gluconeogenesis | 1 | E | -12.1 | -4.3 |
| VIT_07s0205g00090 | glycogen synthase 2 | Glycogenesis | 1 | E | -6.6 | -1.2 |
| VIT_18s0001g15580 | glycogenin glucosyltransferase (glycogenin) | Glycogenesis | 1 | E | -6.5 | -2.1 |
| VIT_06s0004g06920 | fructose-6-phosphate-2-kinase | Glycolysis/Gluconeogenesis | 1 | E | -6.6 | -1.6 |
| VIT_06s0004g05900 | Phosphopyruvate hydratase. | Glycolysis/Gluconeogenesis | 1 | E | -5.6 | -1.5 |
| VIT_01s0011g00250 | 6-phosphogluconolactonase | Pentose phosphate pathway | 1 | E | -5.9 | -1.5 |
| VIT_14s0030g01900 | ribose-5-phosphate isomerase | Pentose phosphate pathway/Calvin cycle | 1 | E | -7.8 | -2.1 |
| VIT_02s0087g00440 | Beta-amylase 8 | Starch metabolism | 1 | E | -5.4 | -1.7 |
| VIT_17s0000g07680 | alpha-N-aEtylglucosaminidase | Sugar metabolism | 1 | E | -5.09 | -1.37 |
| VIT_18s0072g01040 | Invertase, neutral/alkaline | Sugar metabolism | 1 | E | -5.3 | -1.9 |
| VIT_00s1530g00010 | stachyose synthase precursor | Sugar metabolism | 1 | E | -5.7 | -3.7 |
| VIT_05s0029g01140 | sucrose-phosphate synthase | Sugar metabolism | 1 | E | -6.5 | -0.5 |
| VIT_11s0065g00150 | glycogen synthase | Glycogenesis | 1 | NE | -5.1 | -1.1 |
| VIT_03s0038g04570 | ADP-glucose pyrophosphorylase large subunit 1 | Starch metabolism | 1 | NE | -8.3 | -2.2 |
| VIT_04s0023g03010 | fructose-bisphosphate aldolase, chloroplast precursor | Glycolysis | 2 | E | -4.0 | -5.6 |
| VIT_14s0068g00680 | glyEraldehyde-3-phosphate dehydrogenase A, chloroplast precursor | Glycolysis | 2 | E | -6.7 | -9.8 |
| VIT_14s0108g00270 | aldose 1-epimerase | Glycolysis/Gluconeogenesis | 2 | E | -1.9 | -5.2 |
| VIT_03s0088g01190 | malate dehydrogenase, glyoxysomal precursor | Malic acid metabolism | 2 | E | -1.6 | -5.5 |
| VIT_08s0007g01570 | fructose 1,6-bisphosphatase | Pentose phosphate pathway | 2 | E | -6.9 | -13.1 |
| VIT_11s0078g00310 | isoamylase-type starch-debranching enzyme 1 | Starch metabolism | 2 | E | -3.3 | -7.6 |
| VIT_05s0049g01130 | aldo/keto reductase | Sugar metabolism | 2 | E | -6.2 | -7.1 |
| VIT_00s2527g00010 | beta-fructosidase/invertase | Sugar metabolism | 2 | E | -3.7 | -5.2 |
| VIT_14s0060g00740 | galactinol synthase [Vitis riparia] | Sugar metabolism | 2 | NE | -5.3 | -8.1 |
| VIT_05s0094g00930 | Phosphoglucomutase/phosphomannomutase C terminal | Carbohydrate metabolic proEss | 4 | E | -10.6 | -10.3 |
| VIT_08s0007g07600 | pyruvate kinase, cytosolic isozyme | Glycolysis | 4 | E | -6.9 | -5.5 |
| VIT_15s0048g00370 | transketolase, chloroplast precursor | Glycolysis | 4 | E | -12.5 | -9.9 |
| VIT_05s0020g02310 | pyruvate, orthophosphate dikinase | Pyruvate metabolism | 4 | E | -15.0 | -9.7 |
| VIT_13s0019g02330 | GDP-mannose pyrophosphorylase (GMP1) | Sugar metabolism | 4 | E | -4.7 | -5.2 |
| VIT_18s0089g00590 | GlyEraldehyde-3-phosphate dehydrogenase, cytosolic | Glycolysis | 5 | NE | 5.1 | 0.6 |
| VIT_10s0003g05550 | carbohydrate oxidase | Pentose phosphate pathway | 5 | NE | 16.3 | 0.8 |
| VIT_14s0060g00730 | galactinol synthase | Sugar metabolism | 5 | NE | 10.2 | 1.2 |
| VIT_05s0062g00990 | aldo/keto reductase AKR | Aldehyde detoxification pathways (oxidative stress) | 6 | E | 4.3 | 6.0 |
| VIT_02s0025g01560 | UDP-glucose 4-epimerase GEPI48 | Sugar metabolism | 6 | E | 6.2 | 7.6 |
| VIT_06s0004g02060 | aldehyde dehydrogenase 3B1 | Aldehyde detoxification pathways (oxidative stress) | 6 | NE | 2.3 | 7.2 |
| VIT_08s0007g01010 | aldo/keto reductase | Aldehyde detoxification pathways (oxidative stress) | 6 | NE | 10.6 | 16.8 |
| VIT_13s0064g01420 | succinate dehydrogenase [ubiquinone] flavoprotein subunit | Citric acid cycle | 6 | NE | 4.5 | 7.0 |
| VIT_07s0005g00440 | pyruvate kinase | Glycolysis | 6 | NE | 5.7 | 15.3 |
| VIT_00s0233g00030 | trehalose-6-phosphate phosphatase | Stress toleranE | 6 | NE | 2.2 | 7.9 |
| VIT_02s0241g00180 | UDP-D-GLUCURONATE 4-EPIMERASE 5 GAE5 | Carbohydrate metabolic proEss | 7 | E | 1.4 | 13.8 |
| VIT_14s0036g01210 | trehalose 6-phosphate synthase | Stress toleranE | 7 | E | 0.8 | 5.9 |
| VIT_13s0019g04370 | phosphoglucomutase/phosphomannomutase | Carbohydrate metabolic proEss | 7 | NE | 1.6 | 9.6 |
| VIT_00s0173g00110 | Trehalose-phosphatase | Starch and sucrose metabolism | 7 | NE | 1.4 | 8.2 |
| VIT_11s0037g00710 | trehalose-phosphate phosphatase | Starch and sucrose metabolism | 7 | NE | 1.4 | 13.9 |
| VIT_03s0063g00410 | Alpha-amylase | Starch metabolism | 7 | NE | 1.2 | 7.5 |
| VIT_04s0044g01130 | alcohol dehydrogenase [Vitis vinifera] | Fermentative metabolism | 8 | E | 7.1 | 2.4 |
| VIT_04s0044g01120 | alcohol dehydrogenase [Vitis vinifera] | Fermentative metabolism | 8 | E | 12.9 | 5.1 |
| VIT_07s0005g03360 | malate dehydrogenase, cytosolic | Malic acid metabolism | 8 | E | 5.8 | 4.3 |
| VIT_19s0085g01240 | gamma hydroxybutyrate dehydrogenase-like protein | Butanoate metabolism | 8 | NE | 7.9 | 5.6 |
| VIT_15s0046g00910 | serine/threonine protein phosphatase 1; PP1 | Carbohydrate metabolic proEss | 8 | NE | 7.6 | 3.0 |
| VIT_04s0008g02300 | pyruvate dehydrogenase E1 beta subunit | Fermentative metabolism | 8 | NE | 11.3 | 11.2 |
| VIT_07s0205g00070 | phosphoenolpyruvate carboxykinase | Gluconeogenesis | 8 | NE | 41.7 | 17.4 |
| VIT_14s0171g00440 | GlyEraldehyde-3-phosphate dehydrogenase GAPC3, cytosolic | Glycolysis | 8 | NE | 27.4 | 3.8 |
| VIT_01s0137g00090 | aldehyde dehydrogenase (NAD+) | Glycolysis/Gluconeogenesis | 8 | NE | 5.9 | 7.2 |
| VIT_01s0137g00080 | aldehyde dehydrogenase (NAD+) | Glycolysis/Gluconeogenesis | 8 | NE | 14.0 | 12.7 |
| VIT_07s0005g00430 | pyruvate kinase | Glycolysis/Gluconeogenesis | 8 | NE | 6.9 | 5.7 |
| VIT_07s0005g03350 | malate dehydrogenase, cytosolic | Malic acid metabolism | 8 | NE | 7.7 | 2.7 |
| VIT_03s0038g00040 | NADP dependent malic enzyme | Malic acid metabolism | 8 | NE | 21.5 | 3.2 |
| VIT_16s0039g01050 | NADP dependent malic enzyme | Malic acid metabolism | 8 | NE | 34.2 | 9.6 |
| VIT_16s0039g00580 | NADP dependent malic enzyme | Malic acid metabolism | 8 | NE | 8.1 | 6.2 |
| VIT_15s0045g00190 | NADP dependent malic enzyme | Malic acid metabolism | 8 | NE | 40.8 | 6.5 |
| VIT_04s0008g00180 | NADP-dependent malic enzyme | Malic acid metabolism | 8 | NE | 14.0 | 3.0 |
| VIT_16s0013g01670 | 6-phosphogluconate dehydrogenase, cytosolic | Pentose phosphate pathway | 8 | NE | 34.3 | 27.4 |
| VIT_02s0012g03060 | 6-phosphogluconate dehydrogenase, decarboxylating | Pentose phosphate pathway | 8 | NE | 13.2 | 12.8 |
| VIT_05s0051g00010 | beta-amylase 1 | Starch metabolism | 8 | NE | 8.1 | 5.4 |
| VIT_16s0022g00740 | granule-bound starch synthase Ib precursor | Starch metabolism | 8 | NE | 34.1 | 36.4 |
| VIT_00s0131g00420 | Isoamylase isoform 3 | Starch metabolism | 8 | NE | 6.4 | 3.2 |
| VIT_00s1562g00010 | Sucrose synthase 2 | Sugar metabolism | 8 | NE | 19.9 | 3.2 |
| VIT_18s0075g00330 | sucrose-phosphate synthase | Sugar metabolism | 8 | NE | 11.4 | 8.2 |
For each gene (Gene_ID) the annotation (Description), the function (Metabolic Process), the cluster number (Profile), the expression in C (E = expressed but not highly modulated; NE = not expressed) and the Fold Change (FC) referred to BV are reported.
We identified several genes involved in the synthesis and/or degradation of sugars and starch that were specifically induced by cluster thinning, including two genes involved in sucrose re-synthesis (the sucrose synthase VIT_00s1562g00010, and the sucrose-phosphate synthase VIT_18s0075g00330), four genes involved in glycolysis (the glyceraldehyde-3-phosphate dehydrogenases VIT_14s0171g00440 and VIT_18s0089g00590, and the pyruvate kinases VIT_07s0005g00440 and VIT_07s0005g00430), four involved in starch metabolism (the α-amylase VIT_03s0063g00410, the β-amylase, VIT_05s0051g00010, the isoamylase VIT_00s0131g00420 and the granule-bound starch synthase VIT_16s0022g00740), and four involved in the production of osmolytes such as trehalose (VIT_00s0233g00030, VIT_14s0036g01210, VIT_00s0173g00110 and VIT_11s0037g00710).
Microarray analysis also revealed that two CT highly modulated malate dehydrogenase (MDH) isogenes in two different cellular compartments showed opposite expression profiles during ripening, i.e. downregulation of the glyoxysomal isoform compared to control berries (VIT_03s0088g01190) but upregulation of the cytosolic isoform (VIT_07s0005g03360). Interestingly, another cytosolic MDH (VIT_07s0005g03350) was upregulated at EV and thereafter in CT berries but not expressed in controls, along with five NADP-dependent malic enzyme (ME) isogenes (VIT_03s0038g00040, VIT_16s0039g01050, VIT_16s0039g00580, VIT_15s0045g00190, VIT_04s0008g00180) and one phosphoenolpyruvate carboxykinase (VIT_07s0205g00070). Altogether these data suggest that the malate degradation is specifically influenced by cluster thinning, a hypothesis supported by the lower titratable acidity of CT berries compared to controls during ripening (Figure 1A and Table 1).
The oxidative burst that occurs in berries at the onset of ripening [22] seems to be enhanced by the thinning treatment, as supported by the upregulation of two alcohol dehydrogenases (ADHs, VIT_04s0044g01130 and VIT_04s0044g01120), three aldehyde dehydrogenases (ALDHs, VIT_01s0137g00090, VIT_01s0137g00080 and VIT_06s0004g02060) and two aldo/keto reductases (VIT_05s0062g00990 and VIT_08s0070g01010).
Genes belonging to the "Secondary Metabolism Process" category
We identified 108 CT highly modulated genes putatively involved in secondary metabolism (Table 6), 25 of which were downregulated (clusters 1-4), and 83 (including 70 not expressed in control berries) of which were upregulated (clusters 5-8). The downregulated genes included those related to the general phenylpropanoid pathway and phenolic acid metabolism. In particular, we detected two isoforms of 4-coumarate-CoA ligase (4CLs, VIT_17s0000g01790 and VIT_14s0171g00300), one caffeic acid 3-O-methyltransferase (COMT, VIT_08s0007g04520), one cinnamoyl-CoA reductase (VIT_16s0039g01670) and one cinnamyl alchohol dehydrogenase (VIT_19s0014g04980). A small number of genes involved in the flavonoid pathway were also downregulated in CT berries, including a flavonoid 3'-hydroxylase (F3'H, VIT_09s0002g01090), a F3H (VIT_16s0098g00860), a LDOX (VIT_08s0105g00380) and the leucoanthocyanidin reductase LAR1 (VIT_01s0011g02960). The downregulation of such genes may reflect the more robust suppression of proanthocyanidin biosynthesis in CT berries compared to controls, which may also account for the downregulation of a putative UDP-glucose:flavonoid glucosyltrasferase (VIT_16s0115g00340). This gene could be related to the reduced glucosylation of flavonoid compounds such as proanthocyanidin monomers produced during the herbaceous berry growth phase. Glucosylated proanthocyanidin does not accumulate in grapevine tissues but transient glucosylation might be necessary for the vacuolar import of monomers as reported in Medicago truncatula [23]. The downregulation of a geranylgeranyl diphosphatase synthase (VIT_15s0024g00850) and a geranylgeranyl reductase (VIT_17s0000g06280) in CT berries suggests that terpenoid metabolism is also affected by the treatment.
Table 6.
CT highly modulated genes in the "Secondary metabolic process" functional category.
| Gene_ID | Description | Metabolic Process | Profile | Expression in C |
FC EV/BV | FC H/BV |
|---|---|---|---|---|---|---|
| VIT_02s0025g04020 | S-N-methylcoclaurine 3'-hydroxylase | Alkaloid biosynthesis | 1 | E | -5.2 | -2.4 |
| VIT_12s0035g01080 | carotenoid isomerase 1, chloroplast precursor | Carotenoid biosynthesis | 1 | E | -5.9 | -1.7 |
| VIT_06s0080g00810 | lycopene beta cyclase (LYC) | Carotenoid biosynthesis | 1 | E | -6.1 | -1.1 |
| VIT_17s0000g09610 | CYP71D10 | Electron transport | 1 | E | -16.9 | -5.2 |
| VIT_09s0002g01090 | flavonoid 3-monooxygenase | Flavonoid metabolism | 1 | E | -6.4 | -2.9 |
| VIT_16s0115g00340 | UDP-glucose:flavonoid glucosyltransferase | Flavonoid metabolism | 1 | E | -9.8 | -5.2 |
| VIT_08s0007g04520 | caffeic acid 3-O-methyltransferase | Phenolic acid metabolism | 1 | E | -6.2 | -1.7 |
| VIT_17s0000g01790 | 4-coumarate-CoA ligase 2 | Phenylpropanoid metabolism | 1 | E | -5.4 | -1.2 |
| VIT_15s0024g00850 | Geranylgeranyl diphosphate synthase | Terpenoid metabolism | 1 | E | -5.9 | -1.2 |
| VIT_17s0000g06280 | geranylgeranyl reductase | Terpenoid metabolism | 1 | E | -5.0 | -2.2 |
| VIT_15s0046g03570 | Salutaridine reductase | Alkaloid biosynthesis | 2 | E | -2.5 | -6.6 |
| VIT_15s0021g01060 | CYP72A1 | Electron transport | 2 | E | -3.4 | -7.5 |
| VIT_18s0001g09650 | CYP81E1 | Electron transport | 2 | E | -1.8 | -5.1 |
| VIT_16s0098g00860 | Flavanone 3-hydroxylase | Flavonoid metabolism | 2 | E | -3.0 | -7.7 |
| VIT_08s0105g00380 | Leucoanthocyanidin dioxygenase | Flavonoid metabolism | 2 | E | -3.7 | -8.7 |
| VIT_16s0039g01670 | cinnamoyl-CoA reductase | Phenolic acid metabolism | 2 | E | -2.9 | -5.4 |
| VIT_14s0171g00300 | 4-coumarate-CoA ligase | Phenylpropanoid metabolism | 2 | E | -1.8 | -6.0 |
| VIT_16s0039g00990 | Glutathione S-transferase 8 GSTU8 | Secondary metabolism/Oxidative stress | 2 | E | -1.5 | -5.1 |
| VIT_19s0015g02500 | CYP72A1 | Electron transport | 3 | E | -1.2 | -7.0 |
| VIT_19s0014g04980 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 3 | E | -1.6 | -6.9 |
| VIT_07s0031g00620 | zeaxanthin epoxidase (ZEP) (ABA1) | Carotenoids biosynthesis | 4 | E | -13.8 | -9.0 |
| VIT_03s0063g01590 | CYP82C2 | Electron transport | 4 | E | -6.8 | -3.6 |
| VIT_01s0011g02960 | Leucoanthocyanidin reductase 1 | Flavonoid metabolism | 4 | E | -4.6 | -6.5 |
| VIT_05s0049g01120 | Glutathione S-transferase 25 GSTU7 | Secondary metabolism/Oxidative stress | 4 | E | -5.2 | -6.4 |
| VIT_04s0023g01640 | steroid 22-alpha-hydroxylase (CYP90B1) (DWF4) | Electron transport | 4 | NE | -10.0 | -13.1 |
| VIT_12s0134g00620 | UDP-glucose:flavonoid glucosyltransferase | Flavonoid metabolism | 5 | E | 10.3 | 0.5 |
| VIT_06s0004g06400 | UDP-glucose:flavonoid glucosyltransferase | Flavonoid metabolism | 5 | E | 9.3 | 1.2 |
| VIT_19s0015g02950 | Secologanin synthase CYP72A1 | Alkaloid biosynthesis | 5 | NE | 7.4 | 1.4 |
| VIT_04s0210g00030 | strictosidine synthase | Alkaloid biosynthesis | 5 | NE | 7.0 | 1.4 |
| VIT_15s0048g01970 | CYP708A1 | Electron transport | 5 | NE | 9.3 | 0.8 |
| VIT_19s0027g00040 | CYP72A59 | Electron transport | 5 | NE | 5.7 | 1.8 |
| VIT_02s0012g02340 | CYP76C4 | Electron transport | 5 | NE | 5.9 | 1.1 |
| VIT_07s0129g00810 | CYP81E8 | Electron transport | 5 | NE | 5.7 | 1.5 |
| VIT_19s0090g01620 | CYP89H3 | Electron transport | 5 | NE | 5.8 | 0.5 |
| VIT_11s0016g01040 | CYP92A2v4 | Electron transport | 5 | NE | 18.3 | 2.0 |
| VIT_06s0009g02910 | Flavonoid 3',5'-hydroxylase | Flavonoid metabolism | 5 | NE | 18.7 | 1.7 |
| VIT_06s0009g03010 | Flavonoid 3',5'-hydroxylase [Vitis vinifera] | Flavonoid metabolism | 5 | NE | 6.3 | 1.2 |
| VIT_03s0110g00340 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 5 | NE | 15.2 | 1.3 |
| VIT_11s0016g01050 | ferulate 5-hydroxylase | Phenolic acid metabolism | 5 | NE | 8.2 | 0.7 |
| VIT_08s0007g05050 | 4-coumarate--CoA ligase | Phenylpropanoid metabolism | 5 | NE | 5.4 | 0.5 |
| VIT_00s0240g00020 | Glutathione S-transferase 23 GSTU23 | Secondary metabolism/Oxidative stress | 5 | NE | 21.1 | 2.7 |
| VIT_01s0026g01370 | glutathione S-transferase 29 GSTU18 | Secondary metabolism/Oxidative stress | 5 | NE | 5.8 | 0.7 |
| VIT_19s0093g00290 | GLUTATHIONE S-TRANSFERASE TAU 25 | Secondary metabolism/Oxidative stress | 5 | NE | 39.3 | 2.2 |
| VIT_19s0014g02550 | (-)-germacrene D synthase | Sesquiterpene biosynthesis | 5 | NE | 5.5 | 0.6 |
| VIT_16s0050g01590 | UDP-glucose:flavonoid glucosyltransferase | Flavonoid metabolism | 6 | E | 3.5 | 6.0 |
| VIT_16s0039g01100 | phenylalanine ammonia-lyase [Vitis vinifera] | Phenylpropanoid metabolism | 6 | E | 2.6 | 7.0 |
| VIT_16s0039g01240 | phenylalanine ammonia-lyase [Vitis vinifera] | Phenylpropanoid metabolism | 6 | E | 2.7 | 6.5 |
| VIT_17s0000g09550 | CYP71A26 | Electron transport | 6 | NE | 3.1 | 9.3 |
| VIT_06s0009g03130 | CYP79A2 | Electron transport | 6 | NE | 3.9 | 10.9 |
| VIT_13s0106g00280 | CYP79A2 | Electron transport | 6 | NE | 2.6 | 23.9 |
| VIT_15s0048g01480 | geraniol 10-hydroxylase | Monoterpenoids biosynthesis | 6 | NE | 3.0 | 12.7 |
| VIT_08s0040g03040 | Glutathione S-transferase GSTL1 | Secondary metabolism/Oxidative stress | 6 | NE | 4.9 | 7.5 |
| VIT_12s0034g01650 | glutathione S-transferase Z2 GSTZ2 | Secondary metabolism/Oxidative stress | 6 | NE | 4.2 | 7.6 |
| VIT_16s0100g01120 | Stilbene synthase | Stilbene metabolism | 6 | NE | 6.2 | 9.7 |
| VIT_16s0100g01010 | Stilbene synthase | Stilbene metabolism | 6 | NE | 20.7 | 19.9 |
| VIT_16s0100g00830 | Stilbene synthase | Stilbene metabolism | 6 | NE | 3.3 | 5.7 |
| VIT_16s0100g00750 | Stilbene synthase | Stilbene metabolism | 6 | NE | 11.8 | 12.6 |
| VIT_10s0042g00920 | Stilbene synthase | Stilbene metabolism | 6 | NE | 15.3 | 19.3 |
| VIT_16s0100g00780 | Stilbene synthase | Stilbene metabolism | 6 | NE | 13.7 | 25.1 |
| VIT_16s0100g01040 | stilbene synthase - grape | Stilbene metabolism | 6 | NE | 6.2 | 8.3 |
| VIT_16s0100g00910 | stilbene synthase - grape | Stilbene metabolism | 6 | NE | 8.2 | 15.3 |
| VIT_16s0100g01020 | stilbene synthase [Vitis pseudoreticulata] | Stilbene metabolism | 6 | NE | 9.8 | 15.4 |
| VIT_16s0100g00960 | stilbene synthase [Vitis pseudoreticulata] | Stilbene metabolism | 6 | NE | 9.7 | 15.0 |
| VIT_10s0042g00840 | stilbene synthase [Vitis pseudoreticulata] | Stilbene metabolism | 6 | NE | 25.6 | 38.3 |
| VIT_16s0100g01030 | stilbene synthase [Vitis quinquangularis] | Stilbene metabolism | 6 | NE | 7.5 | 16.4 |
| VIT_16s0100g01160 | stilbene synthase [Vitis vinifera] | Stilbene metabolism | 6 | NE | 9.2 | 13.4 |
| VIT_16s0100g00810 | stilbene synthase [Vitis vinifera] | Stilbene metabolism | 6 | NE | 12.5 | 12.7 |
| VIT_16s0100g01170 | stilbene synthase 1 [Vitis vinifera] | Stilbene metabolism | 6 | NE | 6.4 | 6.8 |
| VIT_16s0100g00930 | Stilbene synthase 2 | Stilbene metabolism | 6 | NE | 10.3 | 11.5 |
| VIT_16s0100g00760 | stilbene synthase 2 [Vitis sp. cv. 'Norton'] | Stilbene metabolism | 6 | NE | 20.4 | 25.5 |
| VIT_01s0137g00560 | CYP71B34 | Electron transport | 7 | E | 1.0 | 5.1 |
| VIT_18s0001g12800 | Dihydroflavonol 4-reductase | Flavonoid metabolism | 7 | E | 1.0 | 5.9 |
| VIT_17s0000g07210 | flavonoid 3'-hydroxylase [Vitis vinifera] | Flavonoid metabolism | 7 | E | 1.7 | 7.4 |
| VIT_16s0039g01280 | phenylalanine ammonia-lyase [Vitis vinifera] | Phenylpropanoid metabolism | 7 | E | 2.3 | 8.7 |
| VIT_12s0028g00920 | Glutathione S-transferase 9 GSTF9 | Secondary metabolism/Oxidative stress | 7 | E | 1.3 | 5.6 |
| VIT_19s0090g00140 | 5-alpha-taxadienol-10-beta-hydroxylase | Diterpenoid biosynthesis | 7 | NE | 1.8 | 10.1 |
| VIT_03s0091g00040 | limonoid UDP-glucosyltransferase | Limonoids metablolism | 7 | NE | 1.3 | 5.6 |
| VIT_16s0039g01130 | phenylalanine ammonia-lyase [Vitis vinifera] | Phenylpropanoid metabolism | 7 | NE | 4.6 | 24.4 |
| VIT_16s0100g01200 | stilbene synthase | Stilbene metabolism | 7 | NE | 3.6 | 12.9 |
| VIT_19s0015g02700 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | E | 9.9 | 4.1 |
| VIT_19s0015g02890 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | E | 10.8 | 7.0 |
| VIT_19s0093g00150 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | E | 8.7 | 5.8 |
| VIT_13s0064g00810 | 9,10[9',10']carotenoid cleavage dioxygenase [Vitis vinifera] | Carotenoid metabolism | 8 | NE | 10.0 | 7.2 |
| VIT_19s0015g02100 | CYP72A59 | Electron transport | 8 | NE | 27.5 | 6.2 |
| VIT_06s0009g02850 | CYP79A2 | Electron transport | 8 | NE | 6.2 | 2.0 |
| VIT_07s0129g00770 | CYP81D2 | Electron transport | 8 | NE | 61.2 | 8.7 |
| VIT_07s0129g00830 | CYP81D2 | Electron transport | 8 | NE | 15.4 | 6.0 |
| VIT_03s0038g04620 | isoflavone reductase | Isoflavone metabolism | 8 | NE | 14.6 | 4.7 |
| VIT_03s0038g04680 | isoflavone reductase Bet v 6.0101 | Isoflavone metabolism | 8 | NE | 10.0 | 6.9 |
| VIT_03s0038g04690 | Isoflavone reductase protein 6 | Isoflavone metabolism | 8 | NE | 11.5 | 8.6 |
| VIT_03s0038g04630 | isoflavone reductase related protein | Isoflavone metabolism | 8 | NE | 134.7 | 14.6 |
| VIT_02s0025g02940 | Caffeic acid O-3-methyltransferase | Phenolic acid metabolism | 8 | NE | 6.2 | 3.4 |
| VIT_15s0107g00210 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 8 | NE | 15.1 | 4.2 |
| VIT_13s0067g00560 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 8 | NE | 26.8 | 12.4 |
| VIT_02s0025g03110 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 8 | NE | 10.2 | 7.6 |
| VIT_13s0064g00290 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 8 | NE | 11.1 | 4.9 |
| VIT_19s0093g00110 | Glutathione S-transferase 22 GSTU22 | Secondary metabolism/Oxidative stress | 8 | NE | 101.3 | 29.5 |
| VIT_19s0015g02610 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | NE | 36.3 | 8.6 |
| VIT_19s0015g02730 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | NE | 13.7 | 3.5 |
| VIT_19s0027g00460 | Glutathione S-transferase 25 GSTU25 | Secondary metabolism/Oxidative stress | 8 | NE | 25.0 | 8.1 |
| VIT_06s0004g05670 | Glutathione S-transferase 25 GSTU7 | Secondary metabolism/Oxidative stress | 8 | NE | 10.4 | 2.5 |
| VIT_00s0240g00050 | Glutathione S-transferase 8 GSTU19 | Secondary metabolism/Oxidative stress | 8 | NE | 116.7 | 16.8 |
| VIT_19s0093g00160 | Glutathione S-transferase 8 GSTU19 | Secondary metabolism/Oxidative stress | 8 | NE | 11.9 | 3.5 |
| VIT_19s0093g00220 | Glutathione S-transferase 8 GSTU19 | Secondary metabolism/Oxidative stress | 8 | NE | 51.5 | 8.2 |
| VIT_19s0093g00310 | Glutathione S-transferase 8 GSTU19 | Secondary metabolism/Oxidative stress | 8 | NE | 10.7 | 6.4 |
| VIT_19s0015g02560 | GLUTATHIONE S-TRANSFERASE TAU 25 | Secondary metabolism/Oxidative stress | 8 | NE | 28.1 | 3.6 |
| VIT_12s0035g02100 | Glutathione S-transferase Z1 GSTZ1 | Secondary metabolism/Oxidative stress | 8 | NE | 12.8 | 3.6 |
| VIT_16s0100g00920 | stilbene synthase - grape | Stilbene metabolism | 8 | NE | 12.1 | 7.2 |
For each gene (Gene_ID) the annotation (Description), the function (Metabolic Process), the cluster number (Profile), the expression in C (E = expressed but not highly modulated; NE = not expressed) and the Fold Change (FC) referred to BV are reported.
The upregulated genes included several transcripts related to the general phenylpropanoid pathway, e.g. four PAL isogenes (VIT_16s0039g01100, VIT_16s0039g01240, VIT_16s0039g01280 and VIT_16s0039g01130) and one 4CL (VIT_08s0007g05050). This suggests more phenolic precursors enter the multibranched phenylpropanoid pathway in CT berries. Indeed, many downstream pathways appeared to be specifically activated after cluster thinning. The induction of stilbene synthesis is suggested by the high upregulation of 19 STSs that are not expressed in control berries (many induced as early as the EV stage), the induction of isoflavone synthesis is indicated by the upregulation of four isoflavone reductase genes (VIT_03s0038g04620, VIT_03s0038g04680, VIT_03s0038g04690 and VIT_03s0038g04630), and the activation of phenolic acid metabolism is indicated by the upregulation of a COMT (VIT_02s0025g02940), a ferulate 5-hydroxylase (VIT_11s0016g01050) and five cinnamyl alcohol dehydrogenase genes (VIT_03s0110g0034, VIT_15s0107g00210, VIT_13s0067g00560, VIT_02s0025g03110 and VIT_13s0064g00290).
A small number of genes from the flavonoid pathway were also induced by cluster thinning, including the flavonoid 3',5'-hydroxylase genes F3'5'Hi (VIT_06s0009g02910), F3'5'Hk (VIT_06s0009g03010), the F3'Hb (VIT_17s0000g07210) [24] and the DFR (VIT_18s0001g12800) described by [25]. The expression profile of F3'5'Hi was further confirmed by real time RT-PCR (Additional File 3). Three putative UDP-glucose:flavonoid glucosyltrasferase transcripts (VIT_12s0134g00620, VIT_06s0004g06400 and VIT_16s0050g01590) were also upregulated, and these could also be involved in anthocyanin synthesis although this has yet to be confirmed.
In addition to the cinnamyl alcohol dehydrogenase genes, a few other genes responsible for the synthesis of flavor compounds were also induced by cluster thinning, including germacrene D-synthase (VIT_19s0014g02550), geraniol 10-hydroxylase (VIT_15s0048g01480), limonoid UDP-glucosyltrasferase (VIT_03s0091g00040) and a carotenoid cleavage dioxygenase (VIT_13s0064g00810), suggesting that the aromatic profile of berries is also affected by the treatment.
A very important consequence of cluster thinning was the high modulation of several members of the GST gene family (Table 6). Twenty different GST genes were activated in CT berries, including 16 tau-type (U), one phi-type (F), one lambda-type (L) and two zeta-type (Z) enzymes, according to the classification devised by Edwards et al. [26]. Five U-type and one F-type CT highly modulated GSTs were expressed but not highly modulated in control berries, whereas 13 U-type GSTs were not expressed in control berries at all. Only one of these transcripts (VIT_19s0015g02610) has already been functionally characterized in grapevine and it corresponds to VvGST5 (VIT_19s0015g02610 [21]). This was strongly upregulated between the EV and H stages in CT berries, which parallels the accumulation of anthocyanins. However, VvGST5 could not induce anthocyanin accumulation in transient assays carried out by Conn et al. [21], indicating that the enzyme is unlikely to have a direct role in anthocyanin biosynthesis. Only two GST transcripts (VIT_16s0039g00990 and VIT_05s0049g01120) were highly downregulated in CT berries.
Genes belonging to the "Transport" category
We identified 175 CT highly modulated genes in the "Transport" category, 36 of which are putatively involved in the transport of carbohydrates (10) or secondary metabolites (26) (Table 7). Of the ten putative carbohydrate transporters, seven were expressed but not highly modulated during ripening in control berries but were highly downregulated after cluster thinning. These included the sucrose transporter VvSUC11 (VIT_18s0001g08220 [27]), the polyol transporter VvPMT5 (VIT_03s0063g02250 [28]), an ERD6-like sugar transporter (VIT_07s0104g00830 [29]), a glucose-6-phosphate/phosphate-translocator required for import into plastids (VIT_11s0052g00430 [30]) and a succinate/fumarate mitochondrial transporter (VIT_08s0217g00010), all of which were downregulated at the EV stage. The remaining two transcripts, representing sucrose transporter VvSUC27 (VIT_18s0076g00250 [27]) and a 2-oxoglutarate/malate carrier protein (VIT_18s0001g07320) were downregulated at both the EV and H stages. In contrast, three dicarboxylate/tricarboxylate carriers (VIT_00s0188g00090, VIT_07s0031g02470 and VIT_08s0007g07270) were strongly upregulated by cluster thinning. Together with the strong induction of malate-degrading enzymes, the induction of dicarboxylate/tricarboxylate carriers reinforces the idea that thinning has a direct and specific impact on malate metabolism.
Table 7.
CT highly modulated genes in "Transport" functional category specifically involved in carbohydrate and secondary metabolite translocation.
| Gene_ID | Description | Metabolic Process | Profile | Expression in C |
FC EV/BV | FC H/BV |
|---|---|---|---|---|---|---|
| VIT_11s0052g00430 | Glucose-6-phosphate/phosphate-translocator | Carbohydrate transport | 1 | E | -6.0 | -1.9 |
| VIT_08s0217g00010 | Succinate/fumarate mitochondrial transporter | Carbohydrate transport | 1 | E | -5.30 | -2.86 |
| VIT_03s0063g02250 | POLYOL TRANSPORTER 5 (VIT_PMT5) | Carbohydrate transport | 1 | E | -7.1 | -1.2 |
| VIT_07s0104g00830 | Sugar transporter ERD6-like 7 | Carbohydrate transport | 1 | E | -9.5 | -0.6 |
| VIT_18s0001g08220 | SUCROSE TRANSPORTER 11 (VIT_SUC11) | Carbohydrate transport | 1 | E | -5.6 | -1.4 |
| VIT_14s0108g00430 | ABC transporter B member 16 | Secondary metabolite transport | 2 | E | -3.9 | -7.3 |
| VIT_11s0052g01560 | MATE efflux family protein | Secondary metabolite transport | 2 | E | -1.5 | -6.1 |
| VIT_07s0031g02550 | ABC transporter G member 14 | Secondary metabolite transport | 2 | NE | -9.2 | -19.2 |
| VIT_08s0056g00780 | MATE efflux family protein | Secondary metabolite transport | 2 | NE | -5.8 | -12.5 |
| VIT_12s0059g02180 | MATE efflux family protein | Secondary metabolite transport | 2 | NE | -7.7 | -12.0 |
| VIT_18s0001g07320 | 2-oxoglutarate/malate carrier protein, Mitochondrial | Carbohydrate transport | 4 | E | -12.11 | -7.35 |
| VIT_18s0076g00250 | SUCROSE TRANSPORTER 27 (VIT_SUC27) | Carbohydrate transport | 4 | E | -16.5 | -17.2 |
| VIT_07s0031g02470 | dicarboxylate/tricarboxylate carrier (DTC) | Carbohydrate transport | 5 | NE | 19.03 | 2.69 |
| VIT_06s0061g01460 | ABC transporter G member 22 | Secondary metabolite transport | 5 | NE | 15.8 | 2.0 |
| VIT_07s0031g00750 | MATE efflux family protein | Secondary metabolite transport | 5 | NE | 8.6 | 0.9 |
| VIT_11s0052g01540 | MATE efflux family protein | Secondary metabolite transport | 5 | NE | 14.5 | 1.8 |
| VIT_07s0005g04680 | ABC transporter C member 9 | Secondary metabolite transport | 6 | NE | 4.6 | 6.7 |
| VIT_09s0002g05360 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 6 | NE | 5.2 | 16.0 |
| VIT_09s0002g05370 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 6 | NE | 9.7 | 17.0 |
| VIT_09s0002g05400 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 6 | NE | 9.5 | 15.4 |
| VIT_09s0002g05410 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 6 | NE | 3.3 | 8.4 |
| VIT_09s0002g05440 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 6 | NE | 4.0 | 10.6 |
| VIT_18s0001g11760 | MATE efflux family protein | Secondary metabolite transport | 7 | E | 0.8 | 5.1 |
| VIT_09s0002g05490 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 7 | NE | 1.8 | 15.6 |
| VIT_09s0002g02440 | ABC transporter C member 12 | Secondary metabolite transport | 8 | E | 5.8 | 3.3 |
| VIT_00s0188g00090 | dicarboxylate/tricarboxylate carrier (DTC) | Carbohydrate transport | 8 | NE | 32.96 | 15.30 |
| VIT_08s0007g07270 | dicarboxylate/tricarboxylate carrier (DTC) | Carbohydrate transport | 8 | NE | 5.95 | 3.07 |
| VIT_19s0015g00010 | ABC transporter C member 9 | Secondary metabolite transport | 8 | NE | 9.9 | 8.5 |
| VIT_19s0015g00040 | ABC transporter C member 9 | Secondary metabolite transport | 8 | NE | 8.0 | 6.9 |
| VIT_13s0101g00010 | ABC transporter F member 2 | Secondary metabolite transport | 8 | NE | 56.9 | 23.4 |
| VIT_03s0180g00300 | ABC transporter F member 2 | Secondary metabolite transport | 8 | NE | 23.2 | 7.0 |
| VIT_15s0024g00840 | ABC transporter F member 2 | Secondary metabolite transport | 8 | NE | 55.7 | 17.2 |
| VIT_09s0002g05430 | ABC transporter g family pleiotropic drug resistance 12 PDR12 | Secondary metabolite transport | 8 | NE | 5.9 | 4.4 |
| VIT_14s0068g01740 | ABCNAP14 | Secondary metabolite transport | 8 | NE | 12.5 | 7.1 |
| VIT_13s0019g05200 | MATE efflux family protein | Secondary metabolite transport | 8 | NE | 6.8 | 6.2 |
| VIT_10s0042g00310 | MRP-like ABC transporter | Secondary metabolite transport | 8 | NE | 23.2 | 13.4 |
For each gene (Gene_ID) the annotation (Description), the function (Metabolic Process), the cluster number (Profile), the expression in C (E = expressed but not highly modulated; NE = not expressed) and the Fold Change (FC) referred to BV are reported.
Among the 26 putative secondary metabolite transporters highly modulated in CT berries, 19 belonged to the ATP-Binding Cassette (ABC) transporter family and seven to the Multidrug and Toxic Compound Extrusion (MATE) transporter family. Five of the transcripts were downregulated in CT berries, while 21 were upregulated (19 of which were not expressed in control berries at all). The ABC and MATE transporters were analyzed by querying the protein sequence of the probe consensus against the NCBI databases using BLASTX (Additional File 5). This was necessary because ABC and MATE transporters play multiple roles at the cellular level. None of the sequences we identified matched the grapevine anthocyanin-acylglucoside MATE transporters that have been functionally characterized [31]. Using Arabidopsis thaliana as a reference organism, we found that most of the transporter sequences matched significantly (e-value < 10-50) with an Arabidopsis protein homolog (Additional File 5). Furthermore, two CT highly upregulated MATE transporters matched TRANSPARENT TESTA 12 (AtTT12), a MATE transporter associated with flavonoid sequestration in vacuoles [32]. These sequences were VIT_18s0001g11760 (e-value = 3 × 10-96), which was not highly modulated in control berries but was upregulated in CT berries in the EV and H stages, and VIT_11s0052g01540 (e-value = 5 × 10-74), which was not expressed in control berries but was upregulated at the EV stage in CT berries. Exploration of the latest release of the Vitis vinifera genome revealed that the two MATE transporters mentioned above are not the closest homologs of AtTT12 (data not shown). However, the potential role of these two TT12-like genes in flavonoid transport cannot be ruled out.
Genes highly modulated in control berries
The third group of 120 genes was highly modulated in control berries (Table 4). Although, like the common and CT highly modulated genes, these grouped into eight clusters, more than half of them were assigned to clusters 6 and 7, showing that most C highly modulated genes were upregulated during at least one ripening phase. There were 41 downregulated genes (clusters 1-4), six of which were not expressed at all in CT berries and 35 of which were expressed but not highly modulated during ripening after the cluster thinning treatment. There were 79 upregulated genes (clusters 5-8), 20 of which were not expressed in CT berries and 59 of which were expressed, but not highly modulated.
Using the annotation criteria described above, 15 C highly modulated genes were assigned to the "Transport" category, only one of which was involved in sugar transport (Table 8). This was the putative hexose transporter VvHT5 (VIT_05s0020g03140), which was induced strongly between the EV and H stages. Six genes involved in carbohydrate metabolism were highly modulated in control berries. We identified an ADH (VIT_06s0004g04320), an L-idonate dehydrogenase (VIT_16s0100g00290) which is involved in tartaric acid biosynthesis [33], a galactinol synthase (VIT_14s0060g00760) and a trehalose 6-phosphate phosphatase (VIT_18s0001g05300), all of which were downregulated, and an aldo-keto reductase (VIT_05s0077g01300) and a 2-phosphoglycerate kinase (VIT_06s0061g00280) that were upregulated. It is interesting to note that different isoforms of all these enzymes except L-idonate dehydrogenase were also found in the list of CT highly modulated genes. The ADH was highly downregulated in control berries but not highly modulated after cluster thinning, indicating that ADH activity is needed in CT berries as already suggested by the upregulation of two ADH genes in thinned vines.
Table 8.
C highly modulated genes in the "Carbohydrate metabolic process", "Secondary metabolic process" and "Transport" (carbohydrate and secondary metabolite) functional categories.
| Gene_ID | Description | Metabolic Process | Profile | Expression in C |
FC EV/BV | FC H/BV |
|---|---|---|---|---|---|---|
| VIT_16s0100g00290 | L-idonate dehydrogenase | Carbohydrate acid metabolism | 2 | E | -2.1 | -6.5 |
| VIT_18s0001g12180 | CYP721A1 | Electron transport | 2 | E | -3.9 | -11.2 |
| VIT_14s0060g00760 | galactinol synthase | Sugar metabolism | 2 | E | -2.3 | -5.2 |
| VIT_18s0001g11470 | CyP82A3 | Electron transport | 3 | E | -2.1 | -6.6 |
| VIT_06s0004g04320 | alcohol dehydrogenase | Fermentative metabolism | 3 | E | -1.8 | -8.7 |
| VIT_18s0001g05300 | trehalose-6-phosphate phosphatase | Starch and sucrose metabolism | 3 | NE | -1.1 | -5.7 |
| VIT_05s0077g01300 | Aldo-keto reductase | Aldehyde detoxification pathways (oxidative stress) | 6 | E | 5.1 | 5.0 |
| VIT_10s0003g05450 | reticuline oxidase precursor | Alkaloid biosynthesis | 6 | E | 2.8 | 10.1 |
| VIT_16s0039g00880 | CYP89H3 | Electron transport | 6 | E | 7.3 | 18.0 |
| VIT_18s0001g12690 | Isoflavone reductase protein 4 | Isoflavone metabolism | 6 | E | 2.0 | 6.2 |
| VIT_06s0061g00280 | 2-phosphoglycerate kinase | Glycolysis | 7 | E | 1.2 | 6.0 |
| VIT_11s0065g00350 | trans-cinnamate 4-monooxygenase (C4H) | Phenolic acid metabolism | 7 | E | 0.8 | 5.6 |
| VIT_05s0020g03140 | Hexose Transporter 5 (VvHT5) | Secondary metabolite transport | 7 | E | 1.1 | 9.0 |
| VIT_07s0151g01070 | copalyl diphosphate synthase | Diterpenoid biosynthesis | 7 | NE | 1.2 | 11.2 |
| VIT_04s0023g01290 | UDP-glucose:flavonoid glucosyltransferase | Flavonoid metabolism | 7 | NE | 1.8 | 18.9 |
For each gene (Gene_ID) the annotation (Description), the function (Metabolic Process), the cluster number (Profile), the expression in C (E = expressed but not highly modulated; NE = not expressed) and the Fold Change (FC) relative to the BV stage are reported.
Very few genes related to phenol metabolism were highly modulated in control berries, the exceptions being an isoflavone reductase (VIT_18s0001g12690), a cinnamate-4-hydroxylase (VIT_11s0065g00350) and one transcript putatively involved in anthocyanin modification (VIT_04s0023g01290) which were upregulated. The third of these transcripts was not expressed at all in CT berries as confirmed by real-time RT-PCR (Additional File 3). We did not identify any GST genes that were highly modulated in control berries, or any genes with a putative role in secondary metabolite transport or storage.
Cluster thinning mainly affects the berry transcriptome at the end of veraison
To complete the analysis of our microarray results, we compared the C and CT berry transcriptomes at each time point using a SAM unpaired comparison with a FDR of 2%. This revealed 4123 genes that were differentially expressed in C and CT berries at EV, and 178 at H, in each case with a fold change ≥ 2 (Additional File 6). No genes were found to be differentially expressed in C and CT berries at the BV stage. As anticipated by the previous PCA, these data indicate that C and CT berries at the BV stage are indistinguishable at the transcriptomic level, the main changes occurring at the EV stage, followed by minor further changes at the H stage. As described above, we then focused on genes with a fold change ≥ 5, narrowing the analysis to 1167 differentially-expressed genes at EV and 53 at H, 40 of which were common to both stages. We found that 833 genes were upregulated and 347 were downregulated in CT berries at EV and/or at H.
Approximately 80% (940) of these genes had already been identified as differentially expressed either in C or in CT berries by the cluster analysis based on SAM multiclass described above (Additional File 7). Because these genes were mined from the microarray data using two different approaches, we can have greater confidence that they represent a genuine molecular response to thinning and most (893) are indeed included in the list of genes highly modulated in response to CT. The remaining 20% of genes identified only by the direct comparison at each time point represent genes with a less than five-fold difference in expression between C and CT during ripening, and which were therefore excluded from the first cluster analysis.
As described for the first dataset, we determined the functional categories of the identified genes and focused on those related to carbohydrate metabolism, secondary metabolism and the transport of carbohydrates and secondary metabolites (Table 9). All genes belonging to these categories were differentially expressed in C and CT berries, but only at the EV stage. Only two genes were related to carbohydrate metabolism: a trehalose-phosphate synthase potentially involved in stress tolerance which was downregulated in CT berries (VIT_19s0014g00300), and a pyruvate decarboxylase involved in cellular respiration which was upregulated in CT berries (VIT_15s0024g00630). All the genes related to secondary metabolism were upregulated in CT berries, including two cinnamyl alcohol dehydrogenases (VIT_13s0047g00760 and VIT_00s0371g00010), two GSTs (VIT_07s0104g01810 and VIT_06s0004g05680) and two UDP-glucose:flavonoid 5,3-O-glucosyltransferases (VIT_18s0041g00840 and VIT_18s0041g00950). The induction of these genes reinforces our cluster analysis results, which already showed that the expression of several cinnamyl alcohol dehydrogenase, GST and putative UDP-glucose:flavonoid glucosyl transferase genes was triggered by thinning. The same holds true for the four secondary metabolite transporters, three of which were upregulated (VIT_15s0045g01030, VIT_14s0030g00900, VIT_10s0003g03470) and one of which was downregulated in CT berries (VIT_16s0100g00350).
Table 9.
Differentially expressed genes in C and CT berries at the EV stage belonging to "Carbohydrate metabolic process", "Secondary metabolic process" and "Transport" functional categories.
| Gene_ID | Description | Metabolic Process | FC CT/C at EV |
|---|---|---|---|
| VIT_19s0014g00300 | alpha, alpha-trehalose-phosphate synthase | Stress tolerance | -5.20 |
| VIT_15s0024g00630 | pyruvate decarboxylase isozyme 2 | Glycolysis | 5.52 |
| VIT_13s0047g00760 | Cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 6.58 |
| VIT_00s0371g00010 | cinnamyl alcohol dehydrogenase | Phenolic acid metabolism | 5.03 |
| VIT_19s0135g00230 | CYP72A1 | Electron transport | 7.69 |
| VIT_19s0015g02520 | CYP72A1 | Electron transport | 5.28 |
| VIT_19s0015g02540 | CYP72A59 | Electron transport | 7.30 |
| VIT_19s0015g02780 | CYP72A59 | Electron transport | 5.48 |
| VIT_07s0104g01810 | Glutathione S-transferase 13 GSTF13 | Secondary metabolism/Oxidative stress | 8.03 |
| VIT_06s0004g05680 | Glutathione S-transferase 25 GSTU7 | Secondary metabolism/Oxidative stress | 5.11 |
| VIT_18s0041g00840 | UDP-glucose: flavonoid 5,3-O-glucosyltransferase | Flavonoid metabolism | 10.10 |
| VIT_18s0041g00950 | UDP-glucose: flavonoid 5,3-O-glucosyltransferase | Flavonoid metabolism | 5.04 |
| VIT_16s0100g00350 | ABC transporter B member 8 | Secondary metabolite transport | -5.12 |
| VIT_15s0045g01030 | MRP-like ABC transporter MRP6 | Secondary metabolite transport | 9.44 |
| VIT_14s0030g00900 | MRP-like ABC transporter MRP6 | Secondary metabolite transport | 5.09 |
| VIT_10s0003g03470 | MRP5 | Secondary metabolite transport | 7.80 |
For each gene (Gene_ID) the annotation (Description), the function (Metabolic Process), and the Fold Change (FC) between CT and C at the EV stage are reported.
Discussion
Although many previous studies have considered the impact of increasing the source/sink ratio on grape berry composition, ours is the first investigation to look at the consequences of cluster thinning on global gene expression profiles, which is the basis of most of the observed physiological and biochemical changes.
Although there is only limited evidence for a strict relationship between yield and quality [34], in this study the effectiveness of thinning reflected the suboptimal leaf area to yield ratio of control vines (0.6 m2/kg). Increasing this ratio to 1.2 m2/kg by cluster thinning boosted the levels of sugars and anthocyanins and reduced acidity at harvest. Several authors have reported similar effects on sugars and anthocyanins [35,36], but there is disagreement on the impact of thinning on titratable acidity, with reports suggesting that acidity is unaffected [7,37], slightly increased [1] or decreased by yield reduction [2,38]. In all cases, the effect on acidity seems to be related to the impact of cluster thinning on ripening, particularly the soluble solids content, because thinning reduces acidity only when the soluble solids concentration is strongly and positively affected ([38,39] and this study).
The grouping of gene expression profiles by principal component analysis showed that C and CT berries could be distinguished as early as two weeks after treatment. Microarray data were then analyzed by two different approaches. Genes that were differentially expressed in C and CT berries were initially clustered on the basis of their expression profiles, and then we carried out a direct comparison of C and CT transcriptomes at each time point.
The first approach revealed such a large number of genes modulated during ripening that we applied a > 5 fold change cut-off threshold before assigning a gene to the modulated group. This resulted in 2466 transcripts that were considered to be modulated in CT berries and 567 that were modulated in C berries, including 447 that were common to both treatments. Approximately half of the common genes were more strongly modulated in CT berries, including several downregulated genes involved in photosynthesis, carbon utilization, carbohydrate metabolism, cell wall modifications and hormone metabolic processes that are already known to have a role in berry ripening [22,40], and several upregulated genes involved in the normal ripening process (e.g. genes related to secondary metabolite biosynthesis). These data strongly suggest that the entire course of berry ripening is enhanced by the cluster thinning.
An interesting and unexpected result of the microarray analysis was the relatively large number of genes (2019) highly modulated only in CT berries. More than a half of these genes were never expressed in control berries at any ripening stage, and were activated uniquely by the cluster thinning treatment. This shows that thinning is able to trigger the transcription of genes that otherwise would not be activated in untreated berries and therefore that the effect of thinning goes beyond the simple enhancement or acceleration of the normal ripening process. This appears to affect many different metabolic and cellular processes because the CT highly modulated genes are distributed throughout all 18 functional categories we considered.
The CT high modulation of genes involved in carbohydrate metabolism supports the impact of cluster thinning on sugar accumulation commencing at the EV stage. This reflects the achievement of an optimal balance between leaf area and yield in thinned vines (1.2 m2/kg) compared to controls (0.6 m2/kg) as previously reported [2-7]. Interestingly, all CT highly modulated genes involved in sugar transport (including VvSUC11 and VvSUC27) were downregulated in CT berries but expressed at a constant level throughout ripening in controls. The downregulation of sucrose transporters in CT berries contrasts with the increase in sucrose transporter mRNA during berry development reported by Davies et al. [27] and the general enhancing effect of thinning on the entire ripening process. One possible explanation is that the higher sugar concentration triggers negative feedback that affects the sucrose transporters. Indeed the presence of sugar-response elements in the promoters of various sucrose transporter genes, potentially acting as cis-regulatory elements involved in sugar signaling, has recently been reported [41].
Despite the significant decline in starch concentration following veraison, several genes involved in starch biosynthesis and modification are modulated during ripening [40]. We detected both highly up- and down-regulated genes in CT involved in starch degradation, and their role is unclear given that developing CT berries accumulate large amounts of sugar. Simultaneous starch synthesis and degradation may facilitate the unloading and storage of sugars in the ripening fruit [39]. Starch-degrading enzymes might also provide carbon backbones for the biosynthesis of secondary metabolites, which could also act as signaling molecules in the regulation of genes controlling phenylpropanoid synthesis [17,42].
The strong induction of genes encoding malate-degrading enzymes in CT berries, together with the induction of dicarboxylate-tricarboxylate carriers (which transport malate across the mitochondrial membrane thus supplying substrates for the Krebs cycle) supports the specific modulation of malate metabolism by cluster thinning. Malate, whose catabolism is considered responsible of total acidity reduction in grape berry after véraison [43] is liberated from the vacuole in post-véraison becoming available for catabolism through various avenues [44,45] and, with the advance of ripening, malic acid is likely a vital source of carbon.
Although advanced malate degradation is heavily dependent upon the extent to which berry temperature is elevated [46,47] in response to increased sunlight exposure, this condition was not tested in our experiment as the cluster microclimate was not modified by the removal of one cluster of each shoot, maintaining an unchanged canopy structure.
In this respect, our microarray data suggest that malic acid catabolism is accelerated by cluster thinning following the general enhanced ripening process as seen in other researches [48].
Genes involved in the metabolism of phenylpropanoids and aromatic compounds were strongly modulated in CT berries. Several transcripts involved in phenolic acid, stilbene, flavonoid and isoflavonoid metabolism were affected by the treatment, suggesting that these distinct branches of the phenylpropanoid pathway are directly affected by the increased source/sink balance. However it is possible that the synthesis of phenolic compounds such as stilbenes or isoflavonoids could be part of a systemic response to wounding resulting from the removal of berry clusters, since these compounds are normally produced by the plant in response to stress conditions such as wounding or interactions with pathogens [49,50].
The anthocyanin content of CT berries was higher than that of controls (Figure 1B and Table 1) but the only CT highly upregulated transcript related to the flavonoid/anthocyanin pathway that could explain this result was DFR. However, looking specifically for known anthocyanin-related transcripts, we found that VvGST4 (VIT_04s0079g00690) and VvMYBA1 (VIT_02s0033g00410) were also more strongly upregulated in CT berries compared to controls (Additional File 2). The high induction of three flavonoid glucosyltransferases and the large number of CT highly upregulated GSTs and transporters of the ABC and MATE families may also play an important role in triggering anthocyanin biosynthesis, although their precise functions in berry ripening remain to be determined. Several putative flavonoid-related transcripts such as F3H, LDOX, LAR1 and UDP-glucose:flavonoid glucosyltransferase were highly downregulated in CT berries, which may reflect the slowing down of the synthesis of non-anthocyanin flavonoid compounds such as proanthocyanidin. Several reports indicate that agronomic treatments such as water stress and cluster thinning can increase total anthocyanins and also produce a shift in their profile [3,4,51,52]. The anthocyanin composition of CT berries differed from controls in our study, with higher relative levels of peonidin 3-glucoside and total 3'4'-OH anthocyanins, in agreement with earlier reports [3,51]. This might reflect the specific upregulation of F3'Hb in CT berries during the EV and H phases. Although F3'5'Hi and F3'5'Hk were upregulated in CT berries and not expressed at all in controls, there was a significant decrease in the levels of 3'4'5'-OH anthocyanins at harvest, probably because this expression pattern was restricted just to the EV stage. This suggests that the biosynthesis of 3'4'-OH and 3'4'5'-OH anthocyanins is controlled independently [5].
Cluster thinning is known to change the aromatic profile of grape berries and increase the resveratrol content in wine [53,54]. In agreement, our microarray data reveal the induction of many putative aroma-modifying transcripts (e.g. geraniol 10-hydroxylase, germacrene D-synthase, as well as two isoforms each of ADH and ALDH) and the specific upregulation of 19 STSs.
The number of genes highly modulated in control berries (567) was small in comparison with previous studies [22,40] but this reflects our stringent application of the requirement for a fold change > 5. Only 120 of the control highly modulated genes were not highly modulated in CT berries, none of which were involved in carbohydrate or secondary product metabolism. Therefore we assume they play a relatively minor role in the control of berry quality traits at harvest.
The direct comparison of C and CT transcriptomes at each time point indicated that the greatest number of genes differentially expressed in C and CT berries were found at the EV stage, whereas only minor differences were detectable at harvest. This generally supported the cluster analysis results because almost all the differentially expressed genes had already been detected. The novel data obtained by direct comparison related mainly to genes whose expression was only slightly modulated during ripening.
Conclusions
We investigated the effect of cluster thinning on the Sangiovese berry transcriptome in association with agronomic parameters and the biochemical properties of berries during ripening. The increased source-sink ratio achieved by cluster thinning reflected the optimal leaf area/crop weight ratio in thinned vines (1.2 m2/kg) compared to 0.6 m2/kg in controls. This caused the sugar and anthocyanin content to increase from veraison to harvest at an accelerated rate, along with extensive transcriptomic reprogramming involving both the increased modulation of genes that are normally regulated during ripening and the induction of a large group of genes that are not expressed in ripening control berries. Cluster thinning therefore accelerates the normal ripening process but also superimposes a metabolic environment involving the induction of novel processes not found in ripening control berries and the repression of some pathways that are part of the normal ripening process. Although the possibility that the transcriptomic reprogramming was partially an effect and not only the cause of the observed metabolic changes cannot be excluded, our data provide a significant contribution to current understanding of the molecular consequences of cluster thinning, specifically the impact of increasing the source/sink ratio at veraison on carbohydrate and anthocyanin accumulation.
Methods
Plant material
The trial was carried out in 2008 on adult Sangiovese (Vitis vinifera L.) vines, clone 12T grafted to SO4, in a vineyard in Bologna, Italy (44°30'N, 11°24'E), with North-South oriented rows (vine spacing 1 m × 2.8 m, vertical shoot positioned spur pruned cordon training system (12 buds per vine), with cordon height at 1.0 m above ground and a canopy height of 1.3-1.4 m). Pest management was carried out according to Regione Emilia Romagna local practice. Shoots were manually trimmed when most started to outgrow the top wire, which occurred on Julian Day (JD) 192. Nine vines per treatment, with the same cluster number at flowering (16 per vine), were selected in a single uniform row and each vine was randomly assigned in three blocks to the two following groups: control (C, no treatment) and cluster thinning (CT, removal of 50% of total clusters per vine at veraison, JD 211).
Berry sampling
Samples of 40 berries, taken from three vines in each block, were collected at the following stages: a) beginning of veraison (BV, berries softening and °Brix ~8; JD 211); b) full veraison (one week later; JD 219); c) end of veraison (EV, soft and fully-colored berries; JD 227); d) ripening (two weeks after stage c, JD 250); e) full ripening (H, harvest, JD 266). The samples were divided in two and 20 berries were processed immediately in order to monitor berry ripening, while the rest were frozen at -20°C in preparation for HPLC analysis of anthocyanins. We also collect 30 additional samples at stages a, c and e (same method as above), which were immediately frozen in liquid nitrogen and stored at -80°C for subsequent microarray analysis.
Agronomic parameters and biochemical analysis
Final vine leaf area was estimated after the achievement of a linear relationship between shoot length (cm) and leaf area (cm2) for at least 15 shoots for both C and CT samples collected from vines at the end of shoot growth. Leaf area was measured with a leaf area meter (LI-3000A, Li-Cor Biosciences, Lincoln, Nebraska, USA), ensuring the contribution of the main and lateral leaf areas were kept separate. Two separate correlations for the main and lateral areas (y) and shoot lengths (x) were so established (C main: y = 16.212 x, R2 = 0.70; C laterals: y = 14.834x, R2 = 0.70; CT main: y = 13.441x, R2 = 0.71; CT laterals: y = 15.844x, R2 = 0.89). After leaf fall, each shoot per vine was then measured and the shoot lengths were used to estimate the corresponding pooled total leaf area per vine using the linear relationship calculated above.
The must obtained after crushing the first 20 berries per sample was immediately filtered through a strainer and a drop was used for °Brix analysis with a temperature-compensating CR50 refractometer (Maselli Misure Spa, PR, Italy). We then diluted 5 ml of the same must in seven volumes of double distilled water and titrated this against 1 N, 0.5 N or 0.25 N NaOH (Sigma-Aldrich, St. Louis, MO, USA) with a Crison Compact Tritator (Crison, Barcelona, Spain) according to the stage of berry ripening, to obtain pH and titratable acidity data (expressed as g/L of tartaric acid equivalents).
The second 20 berries were used to extract anthocyanins for HPLC analysis as described by Mattivi et al. [55], using a Waters 1525 instrument (Waters, Milford, MA) equipped with a diode array detector (DAD) and using a Phenomenex (Castel Maggiore, BO, Italy) reversed-phase column (RP18, 250 mm × 4 mm, 5 μM). Anthocyanins were quantified at 520 nm using an external calibration curve with malvidin-3-glucoside chloride as the standard (Sigma-Aldrich, St. Louis, MO).
We recorded cluster number and weight, berry weight and total yield per vine for each tested vine at harvest. Statistical analysis of agronomic parameters and biochemical data were carried out using the mixed General Linear Model (GLM) procedure of the SAS statistical package (SAS Institute, Cary, NC), and treatment comparisons were carried out using the Tukey test.
Microarray analysis
Total RNA for microarray analysis was isolated from ~200 mg of the ground berry tissue without seeds using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO). RNA quality and quantity were determined using a Nanodrop 2000 instrument (Thermo Scientific, Wilmington, DE) and a Bioanalyzer Chip RNA 7500 series II (Agilent, Santa Clara, CA). The cDNA synthesis, labeling, hybridization and washing reactions were performed according to the NimbleGen Arrays User's Guide (V 3.2). Each hybridization was carried out on a NimbleGen microarray 090818 Vitis exp HX12 (Roche, NimbleGen Inc., Madison, WI), representing 29,549 predicted genes on the basis of the 12X grapevine V1 gene prediction version http://srs.ebi.ac.uk/. The chip probe design is available at the following URL: http://ddlab.sci.univr.it/FunctionalGenomics/.
The microarray was scanned using a ScanArray 4000XL (Perkin-Elmer) at 532 nm (Cy-3 absorption peak) and GenePix Pro7 software (Molecular Devices, Sunnyvale, CA, USA) according to the manufacturers' instructions. Images were analyzed using NimbleScan v2.5 software (Roche), which produces Pair Files containing the raw signal intensity data for each probe and Calls Files with normalized expression data derived from the average of the intensities of the four probes for each gene. The normalized gene expression data were finally converted to log2 values. All microarray expression data are available at GEO under the series entry GPL13936.
Pearson Correlation analysis and Principal Component Analysis (PCA; SIMCA P+ Umetrics, Umea, Sweden) were carried out to evaluate the robustness of the three biological replicates in each sample. A gene was considered to be expressed if the normalized expression value was higher than the value obtained by averaging the fluorescence of the negative control present on the chip, for at least two of the three biological replicates. A Significance Analysis of Microarrays (SAM) was implemented using TMeV software http://www.tm4.org/mev, with a false discovery rate of 2%. Cluster analysis was performed by the k-means method with Pearson's correlation distance (TMeV) comparing EV and H gene expression to BV.
Real-time PCR
First strand cDNA was synthesized using 1 μg of total RNA as the template and the Improm-II™ Reverse Transcription system (Promega), according to the manufacturer's instructions. The total RNA was derived from the three biological replicates used for microarray analysis and all RNA samples were first treated with DNase I (Promega). Gene-specific primers were designed for six genes using the sequence information in the 3'-UTR, using actin and elongation factor 1 (EF1) genes as references [56] (Additional File 8). Primers and cDNA were mixed with the Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and the reaction was carried out on an ABI PRISM StepOne Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using the following cycling conditions: 50°C hold for 2 min and a 95°C hold for 10 min followed by 45 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 20 s. Nonspecific PCR products were identified by the dissociation curves. Amplification efficiency was calculated from raw data using LingRegPCR software [57]. The relative expression ratio was calculated for development time points relative to the first sampling time point (beginning of veraison, BV) according to the Pfaffl equation [58]. Standard error (SE) values were calculated according to Pfaffl et al. [59]. The final data was calculated as previously reported [60].
Authors' contributions
CP set up the RNA extraction, performed microarray and HPLC experiments, participated in data analysis, prepared figures and tables and wrote the initial manuscript draft. SZ set up the real-time RT-PCR protocol, performed data analysis and participated in the drafting of the manuscript. GBT participated in the interpretation of data and supervised and critically revised the manuscript. GA and GV sampled the material and performed the HPLC analysis. SDS set up the microarray experiment and participated in the real time RT-PCR experiments and gene annotation. CI conceived the study and participated in the project design. MP and IF conceived the study, participated in its design and coordination and contributed to and critically revised the manuscript. It is the authors' opinion that CP and SZ contributed equally as first authors to this manuscript. All authors have read and approved the final version of the manuscript.
Supplementary Material
Differentially expressed genes along berry development displaying a two-fold or greater change in transcript abundance between EV and BV or EV and H, in CT and C treatments. For each gene the annotation, the EV/BV and the H/BV Fold-change (FC) are indicated. Data obtained for CT and C are listed in two separate worksheets.
Differentially expressed genes along berry development displaying a five-fold or greater change in transcript abundance between EV and BV or EV and H, in CT and C treatments. For each gene the annotation, the gene ontology, the EV/BV and the H/BV Fold-change (FC), the expression profile and the expression behavior in the other treatment are reported. Data obtained for CT and C are listed in two separate worksheets.
Real time RT-PCR validation of six selected genes. Expression profiles measured by real time RT-PCR are determined by calculating the relative expression ratio value for each stage relative to the BV stage. Real time RT-PCR data are reported as means ± SE of three biological replicates, obtained by using two reference genes. An actin beta/gamma 1 (VIT_12s0178g00200) and an elongation factor 1 (VIT_06s0004g03220) were used as control genes. For each gene the expression profile obtained by microarray analysis is shown on the right side. A-B: Vacuolar invertase 1, GIN 1 (VIT_16s0022g00670); C-D: PAL [Vitis vinifera] (VIT_16s0039g01120); E-F: Flavonol synthase (VIT_18s0001g03430); G-H: β-Fructosidase -invertase (VIT_00s2527g00010); I-J: Flavonoid 3’5’-hydroxylase (VIT_06s0009g02910); K-L: UDP-glucose:flavonoid glucosyltransferase (VIT_04s0023g01290).
Differentially expressed genes along berry development displaying a similar expression profile between C and CT. For each gene, the annotation, the gene ontology, the profile and the CT/C Fold-change (FC) ratio at EV and at H are reported.
Differentially expressed genes along CT berry development encoding ABC and MATE transporters. Analysis was performed by querying the protein sequence of the probe consensus against the NCBI databases using BLASTX. For each gene the correspondent Arabidopsis thaliana homologue, the NCBI reference sequence and E-value are specified.
Differentially expressed genes displaying a two-fold or greater change in transcript abundance between C and CT at EV and H time points. For each gene the description and the CT/C Fold-change (FC) are indicated. Data obtained for EV and H are listed in two separate worksheets.
Differentially expressed genes displaying a five-fold or greater change in transcript abundance between C and CT at EV and /or H. For each gene the CT/C Fold-change (FC) at EV and /or H, the description, the gene ontology, the presence of the gene among genes identified by cluster analysis as CT specific, C specific, common to C and CT with a different expression profile and common to C and CT with the same expression profile, are reported..
List of the primers used for real time RT-PCR validation experiment.
Contributor Information
Chiara Pastore, Email: cpastore@agrsci.unibo.it.
Sara Zenoni, Email: sara.zenoni@univr.it.
Giovanni Battista Tornielli, Email: giovannibattista.tornielli@univr.it.
Gianluca Allegro, Email: gallegro@agrsci.unibo.it.
Silvia Dal Santo, Email: silvia.dalsanto@univr.it.
Gabriele Valentini, Email: gvalentini@agrsci.unibo.it.
Cesare Intrieri, Email: viticolt@agrsci.unibo.it.
Mario Pezzotti, Email: mario.pezzotti@univr.it.
Ilaria Filippetti, Email: ilaria.filippetti@unibo.it.
Acknowledgements
This work was supported by: Fondazione Cariverona (Completamento e attività del Centro di genomica Funzionale Vegetale), Verona, Italy. Special thanks to Alberto Ferrarini and Marianna Fasoli for their support in the hybridization analysis.
References
- Kliewer WM, Dokoozlian NK. Leaf Area/Crop Weight Ratios of Grapevines: Influence on Fruit Composition and Wine Quality. Am J Enol Vitic. 2005;56(2):170–181. [Google Scholar]
- Kliewer WM, Weaver RJ. Effect of Crop Level and Leaf Area on Growth, Composition, and Coloration of 'Tokay' Grapes. Am J Enol Vitic. 1971;22(3):172–177. [Google Scholar]
- Filippetti I, Ramazzotti S, Centinari M, Brucchetti B, Intrieri C. Esperienze triennali sugli effetti del diradamento dei grappoli sulla qualità delle uve della cultivar Sangiovese. Italus Hortus. 2007;14:412–416. [Google Scholar]
- Guidoni S, Allara P, Schubert A. Effect of Cluster Thinning on Berry Skin Anthocyanin Composition of Vitis vinifera cv. Nebbiolo. Am J Enol Vitic. 2002;53(3):224–226. [Google Scholar]
- Guidoni S, Ferrandino A, Novello V. Effects of Seasonal and Agronomical Practices on Skin Anthocyanin Profile of Nebbiolo Grapes. Am J Enol Vitic. 2008;59(1):22–29. [Google Scholar]
- Intrieri C Filippetti I Valentini G Seghetti L Il diradamento migliora la qualità del Montepulciano a pergola L'informatore Agrario 20054459–64.22201165 [Google Scholar]
- Keller M, Mills LJ, Wample RL, Spayd SE. Cluster Thinning Effects on Three Deficit-Irrigated Vitis vinifera Cultivars. Am J Enol Vitic. 2005;56(2):91–103. [Google Scholar]
- Dokoozlian NK, Hirschfelt DJ. The Influence of Cluster Thinning at Various Stages of Fruit Development on Flame Seedless Table Grapes. Am J Enol Vitic. 1995;46(4):429–436. [Google Scholar]
- Guidoni S, Argamante N. Influenza del diradamento dei grappoli sull'accumolo di antociani nelle uve. Quaderno di Viticoltura ed Enologia Università di Torino. 2003;26:27–42. [Google Scholar]
- Palliotti A, Cartechini A. Cluster thinning effetcts on yield and grape composition in different grapevine cultivars. Acta Hort. 2000;512:111–120. [Google Scholar]
- Downey MO, Dokoozlian NK, Krstic MP. Cultural Practice and Environmental Impacts on the Flavonoid Composition of Grapes and Wine: A Review of Recent Research. Am J Enol Vitic. 2006;57(3):257–268. [Google Scholar]
- Pirie A, Mullins MG. Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic Acid. Plant Physiol. 1976;58(4):468–472. doi: 10.1104/pp.58.4.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubelakis-Angelakis KA, Kliewer WM. Effects of Exogenous Factors on Phenylalanine Ammonia-Lyase Activity and Accumulation of Anthocyanins and Total Phenolics in Grape Berries. Am J Enol Vitic. 1986;37(4):275–280. [Google Scholar]
- Nagira Y, Ozeki Y. A system in which anthocyanin synthesis is induced in regenerated torenia shoots. J Plant Res. 2004;117(5):377–383. doi: 10.1007/s10265-004-0170-6. [DOI] [PubMed] [Google Scholar]
- Nagira Y, Shimamura K, Hirai S, Shimanuki M, Kodama H, Ozeki Y. Identification and characterization of genes induced for anthocyanin synthesis and chlorophyll degradation in regenerated torenia shoots using suppression subtractive hybridization, cDNA microarrays, and RNAi techniques. J Plant Res. 2006;119(3):217–230. doi: 10.1007/s10265-006-0264-4. [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140(2):637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Merillon JM. Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry. 2000;53(6):659–665. doi: 10.1016/S0031-9422(99)00620-2. [DOI] [PubMed] [Google Scholar]
- Gollop R, Even S, Colova-Tsolova V, Perl A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot. 2002;53(373):1397–1409. doi: 10.1093/jexbot/53.373.1397. [DOI] [PubMed] [Google Scholar]
- Gollop R, Farhi S, Perl A. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Science. 2001;161:579–588. doi: 10.1016/S0168-9452(01)00445-9. [DOI] [Google Scholar]
- Zheng Y, Tian L, Liu H, Pan Q, Zhan J, Huang W. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regulation. 2009;58(3):251–260. doi: 10.1007/s10725-009-9373-0. [DOI] [Google Scholar]
- Conn S, Curtin C, Bezier A, Franco C, Zhang W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Exp Bot. 2008;59(13):3621–3634. doi: 10.1093/jxb/ern217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S, Perazzolli M, Malossini A, Cestaro A, Dematte L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at veraison. BMC Genomics. 2007;8:428. doi: 10.1186/1471-2164-8-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3'-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21(8):2323–2340. doi: 10.1105/tpc.109.067819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falginella L, Castellarin SD, Testolin R, Gambetta GA, Morgante M, Di Gaspero G. Expansion and subfunctionalisation of flavonoid 3',5'-hydroxylases in the grapevine lineage. BMC Genomics. 2010;11:562. doi: 10.1186/1471-2164-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.) Plant Mol Biol. 1994;24(5):743–755. doi: 10.1007/BF00029856. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5(5):193–198. doi: 10.1016/S1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Davies C, Wolf T, Robinson SP. Three putative sucrose transporters are differentially expressed in grapevine tissues. Plant Science. 1999;147(2):93–100. doi: 10.1016/S0168-9452(99)00059-X. [DOI] [Google Scholar]
- Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-Symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell. 2005;17(1):204–218. doi: 10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 2007;581(12):2318–2324. doi: 10.1016/j.febslet.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flugge UI, Schneider A. The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell. 2005;17(3):760–775. doi: 10.1105/tpc.104.029124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Verries C, Souquet JM, Mazauric JP, Klein M, Cheynier V. et al. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 2009;150(1):402–415. doi: 10.1104/pp.109.135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJ, Leon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13(4):853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Cook DR, Ford CM. L-tartaric acid synthesis from vitamin C in higher plants. Proc Natl Acad Sci USA. 2006;103(14):5608–5613. doi: 10.1073/pnas.0510864103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DI, Lombard PB. Environmental and Management Practices Affecting Grape Composition and Wine Quality - A Review. American Journal of Enology and Viticulture. 1993;44(4):409–430. [Google Scholar]
- Mazza G, Fukumoto L, Delaquis P, Girard B, Ewert B. Anthocyanins, phenolics, and color of Cabernet Franc, Merlot, and Pinot Noir wines from British Columbia. J Agric Food Chem. 1999;47(10):4009–4017. doi: 10.1021/jf990449f. [DOI] [PubMed] [Google Scholar]
- Reynolds AG, Price SF, Wardle DA, Watson BT. Fruit Environment and Crop Level Effects on Pinot noir. I. Vine Performance and Fruit Composition in British Columbia. American Journal of Enology and Viticulture. 1994;45(4):452–459. [Google Scholar]
- Ough CS, Nagaoka R. Effect of Cluster Thinning and Vineyard Yields on Grape and Wine Composition and Wine Quality of Cabernet Sauvignon. American Journal of Enology and Viticulture. 1984;35(1):30–34. [Google Scholar]
- Di Profio F, Reynolds AG, Kasimos A. Canopy Management and Enzyme Impacts on Merlot, Cabernet franc, and Cabernet Sauvignon. I. Yield and Berry Composition. American Journal of Enology and Viticulture. 2011;62(2):139–151. doi: 10.5344/ajev.2010.10024. [DOI] [Google Scholar]
- Agasse A, Vignault G, Kappel C, Conde C, Gerò H, Delrot S. In: Grapevine Molecular Physiology Biotechnology. 2. Springer, editor. New York; 2009. Sugar transport & sugar sensing in grape; pp. 105–139. [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics. 2007;8:429. doi: 10.1186/1471-2164-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thevenot P, Lemoine R, Atanassova R, Laloi M. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biology. 2009;10(1):245. doi: 10.1186/1471-2229-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larronde F, Krisa S, Decendit A, Chèze C, Deffieux G, Mèrrilon JM. Regulation of polyphenols production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Reports. 1998;17:946–950. doi: 10.1007/s002990050515. [DOI] [PubMed] [Google Scholar]
- Ruffner HP, Possner D, Brem S, Rast DM. The physiological role of malic enzyme in grape ripening. Planta. 1984;160(5):444–448. doi: 10.1007/BF00429761. [DOI] [PubMed] [Google Scholar]
- Famiani F, Walker RP, Tecsi L, Chen ZH, Proietti P, Leegood RC. An immunohistochemical study of the compartmentation of metabolism during the development of grape (Vitis vinifera L.) berries. J Exp Bot. 2000;51(345):675–683. doi: 10.1093/jexbot/51.345.675. [DOI] [PubMed] [Google Scholar]
- Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70(11-12):1329–1344. doi: 10.1016/j.phytochem.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Lakso AN, Kliewer WM. The Influence of Temperature on Malic Acid Metabolism in Grape Berries. II. Temperature Responses of Net Dark CO2 Fixation and Malic Acid Pools. Am J Enol Vitic. 1978;29(3):145–149. [Google Scholar]
- Ruffner HP, Hawker JS, Hale CR. Temperature and enzymic control of malate metabolism in berries of Vitis vinifera. Phytochemistry. 1976;15(12):1877–1880. doi: 10.1016/S0031-9422(00)88835-4. [DOI] [Google Scholar]
- Dokoozlian NK, Kliewer WM. Influence of Light on Grape Berry Growth and Composition Varies during Fruit Development. Journal of the American Society for Horticultural Science. 1996;121(5):869–874. [Google Scholar]
- Gutierrez-Gonzalez JJ, Guttikonda SK, Tran LS, Aldrich DL, Zhong R, Yu O, Nguyen HT, Sleper DA. Differential expression of isoflavone biosynthetic genes in soybean during water deficits. Plant Cell Physiol. 2010;51(6):936–948. doi: 10.1093/pcp/pcq065. [DOI] [PubMed] [Google Scholar]
- Xu W, Yu Y, Zhou Q, Ding J, Dai L, Xie X, Xu Y, Zhang C, Wang Y. Expression pattern, genomic structure, and promoter analysis of the gene encoding stilbene synthase from Chinese wild Vitis pseudoreticulata. J Exp Bot. 2011;62(8):2745–2761. doi: 10.1093/jxb/erq447. [DOI] [PubMed] [Google Scholar]
- Bucelli P, Giannetti F. Incidenza del diradamento dei grappoli sulla composizione dell'uva e sulla qualità del vino. Rivista di Viticoltura ed Enologia. 1996;2:59–67. [Google Scholar]
- Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E. G DIG. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007;30(11):1381–1399. doi: 10.1111/j.1365-3040.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- Prajitna A, Dami IE, Steiner TE, Ferree DC, Scheerens JC, Schwartz SJ. Influence of Cluster Thinning on Phenolic Composition, Resveratrol, and Antioxidant Capacity in Chambourcin Wine. Am J Enol Vitic. 2007;58(3):346–350. [Google Scholar]
- Reynolds AG, Schlosser J, Sorokowsky D, Roberts R, Willwerth J, de Savigny C. Magnitude of Viticultural and Enological Effects. II. Relative Impacts of Cluster Thinning and Yeast Strain on Composition and Sensory Attributes of Chardonnay Musque. Am J Enol Vitic. 2007;58(1):25–41. [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R. Metabolite profiling of grape: Flavonols and anthocyanins. J Agric Food Chem. 2006;54(20):7692–7702. doi: 10.1021/jf061538c. [DOI] [PubMed] [Google Scholar]
- Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes along berry development displaying a two-fold or greater change in transcript abundance between EV and BV or EV and H, in CT and C treatments. For each gene the annotation, the EV/BV and the H/BV Fold-change (FC) are indicated. Data obtained for CT and C are listed in two separate worksheets.
Differentially expressed genes along berry development displaying a five-fold or greater change in transcript abundance between EV and BV or EV and H, in CT and C treatments. For each gene the annotation, the gene ontology, the EV/BV and the H/BV Fold-change (FC), the expression profile and the expression behavior in the other treatment are reported. Data obtained for CT and C are listed in two separate worksheets.
Real time RT-PCR validation of six selected genes. Expression profiles measured by real time RT-PCR are determined by calculating the relative expression ratio value for each stage relative to the BV stage. Real time RT-PCR data are reported as means ± SE of three biological replicates, obtained by using two reference genes. An actin beta/gamma 1 (VIT_12s0178g00200) and an elongation factor 1 (VIT_06s0004g03220) were used as control genes. For each gene the expression profile obtained by microarray analysis is shown on the right side. A-B: Vacuolar invertase 1, GIN 1 (VIT_16s0022g00670); C-D: PAL [Vitis vinifera] (VIT_16s0039g01120); E-F: Flavonol synthase (VIT_18s0001g03430); G-H: β-Fructosidase -invertase (VIT_00s2527g00010); I-J: Flavonoid 3’5’-hydroxylase (VIT_06s0009g02910); K-L: UDP-glucose:flavonoid glucosyltransferase (VIT_04s0023g01290).
Differentially expressed genes along berry development displaying a similar expression profile between C and CT. For each gene, the annotation, the gene ontology, the profile and the CT/C Fold-change (FC) ratio at EV and at H are reported.
Differentially expressed genes along CT berry development encoding ABC and MATE transporters. Analysis was performed by querying the protein sequence of the probe consensus against the NCBI databases using BLASTX. For each gene the correspondent Arabidopsis thaliana homologue, the NCBI reference sequence and E-value are specified.
Differentially expressed genes displaying a two-fold or greater change in transcript abundance between C and CT at EV and H time points. For each gene the description and the CT/C Fold-change (FC) are indicated. Data obtained for EV and H are listed in two separate worksheets.
Differentially expressed genes displaying a five-fold or greater change in transcript abundance between C and CT at EV and /or H. For each gene the CT/C Fold-change (FC) at EV and /or H, the description, the gene ontology, the presence of the gene among genes identified by cluster analysis as CT specific, C specific, common to C and CT with a different expression profile and common to C and CT with the same expression profile, are reported..
List of the primers used for real time RT-PCR validation experiment.