Abstract
In vivo analysis of Drosophila melanogaster has enhanced our understanding of many biological processes, notably the mechanisms of heredity and development. While in vivo analysis of mutants has been a strength of the field, analyzing fly cells in culture is valuable for cell biological, biochemical and whole genome approaches in which large numbers of homogeneous cells are required. An efficient genetic method to derive Drosophila cell lines using expression of an oncogenic form of Ras (RasV12) has been developed. Mutations in tumor suppressors, which are known to cause cell hyperproliferation in vivo, could provide another method for generating Drosophila cell lines. Here we screened Drosophila tumor suppressor mutations to test if they promoted cell proliferation in vitro. We generated primary cultures and determined when patches of proliferating cells first emerged. These cells emerged on average at 37 days in wild-type cultures. Using this assay we found that a Pten mutation had a strong effect. Patches of proliferating cells appeared on average at 11 days and the cultures became confluent in about 3 weeks, which is similar to the timeframe for cultures expressing RasV12. Three Pten mutant cell lines were generated and these have now been cultured for between 250 and 630 cell doublings suggesting the life of the mutant cells is likely to be indefinite. We conclude that the use of Pten mutants is a powerful means to derive new Drosophila cell lines.
Introduction
The establishment of cell lines from human tissues involves genetic manipulation of telomerase, tumor suppressors and oncogenes. Telomerase is required to circumvent the finite number of divisions most somatic cells experience due to telomere shortening [1]. In human cells, telomerase expression together with mutations in tumor suppressors leads to immortality [2]. Rodent cells, in contrast to human cells become immortal spontaneously at high frequency [3]. Expression of oncogenes such as Ras allows cells to be independent of growth factors [2]. Expression of oncogenic Ras in human primary cells that lack telomerase activity causes senescence, but we discovered expressing a Ras oncogene (RasV12) in Drosophila primary embryonic cells promotes cell proliferation to rapidly give rise to immortal cell lines [4], [5], [6]. This different response may be because Drosophila maintains telomere length without telomerase [7]. Expression of RasV12 has proved to be a useful genetic tool to create Drosophila mutant cell lines [4], [8], [9]. By analogy with mammalian cells, inactivation of tumor suppressors could provide another genetic means to immortalize Drosophila cells. To test this idea we surveyed a collection of Drosophila tumor suppressor mutants for their ability to promote proliferation of cells in culture.
Homologs of many mammalian tumor suppressor genes are conserved in Drosophila and new tumor suppressors have been discovered in genetic screens using flies. These include both whole organism screens for larval-pupal lethals with overgrowth phenotypes in the imaginal discs and screens for tumors that develop as clonal patches in adults (reviewed in [10]). Analysis of these genes in Drosophila has made important contributions to understanding the biology of tumor suppressors and in a number of cases has supported the involvement of these genes in human cancers (reviewed in [10]).
Drosophila tumor suppressors are broadly divided into two classes; neoplastic and hyperplastic that distinguish their different overgrowth phenotypes [10], [11]. The first Drosophila neoplastic tumor suppressor isolated, lethal (2) giant larvae (lgl) [12], [13], was later found to be part of complex called the Scribble complex (Scib/Dlg/Lgl), which is required for the establishment and maintenance of cell polarity in epithelia (reviewed in [11], [14], [15]). Loss of function mutations in these and other neoplastic tumor suppressor genes leads to an increase in cell number, and a failure to terminally differentiate. The second class, hyperplastic tumor suppressors, determine proper tissue size by regulating the number of cells in an organ or tissue (reviewed in [11]). Pten and some members of the Hippo pathway are well-characterized examples of hyperplastic tumor suppressors [11], [16], [17]. Loss of function mutations in hyperplastic tumor suppressors cause an increase in cell number, although the ability of the cells to differentiate is not compromised.
Here we tested mutations in both classes of tumor suppressor genes for their ability to promote proliferation of Drosophila cells in vitro. We found that a Pten mutation had a dramatic effect on primary cultures. The cultures rapidly became confluent and gave rise to continuous lines. This identifies Pten mutation as a second genetic approach for generating cell lines in Drosophila.
Results and Discussion
Primary-culture assay to determine when proliferating cells appear in tumor-suppressor mutant cultures relative to wild type and RasV12-expressing cultures
Wild-type embryonic primary cultures follow a pattern of development that initially involves the appearance of morphologically distinct types of terminally differentiated cell types such as muscle, nerve and fat body (reviewed in [18]). Patches of proliferating spindle-shaped, and more rarely epithelial-like, cells emerge much later than these differentiated cells types and are likely to be a major cell type that gives rise to continuous lines [18]. These proliferating cell patches appear in wild type cultures after a delay of several weeks, are often transient, and typically occur in waves over many months with only the later ones giving rise to persistent populations [18].
In this study, wild-type cultures followed the expected pattern with patches of proliferating cells emerging on average at day 37 with a range spanning a few days in individual cultures (Figure 1B and 1C). These serve as a control to identify genotypes in which these cells appear earlier, such as cultures expressing oncogenic RasV12 (Act5C-Gal4; UAS- RasV12). In RasV12 cultures, patches of proliferating cells appeared on average at about day 8 (Figure 1B). Changes in the timing of appearance and persistence of proliferating cells can be an indicator of genotypes that will readily give rise to continuous cell lines as we discovered for primary cultures expressing RasV12 that readily progressed to continuous cell lines [4]. Thus, cultures expressing transgenic RasV12 served as a positive control in these assays.
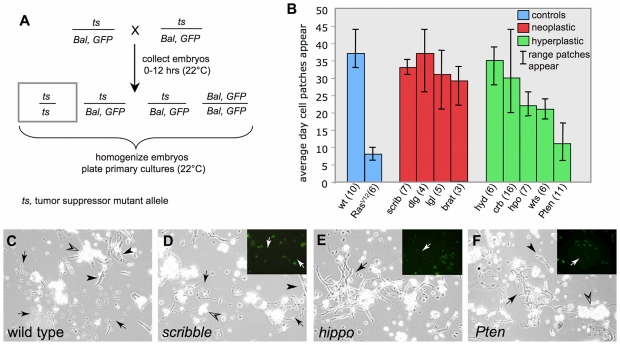
Figure 1. Time of appearance of proliferating cell patches in wild-type and tumor-suppressor mutant primary cultures.
(A) Cross to generate embryos for tumor suppressor primary cultures. Tumor suppressor (ts) mutants were maintained in stocks with a marked balancer chromosome that expresses a GFP transgene (Bal, GFP). One quarter of the progeny embryos are the desired class (boxed genotype). Primary cultures were established from mixed embryos and cells homozygous for the ts allele could be recognized because they are GFP negative. (B) The average day, and the range of days at which proliferating cells appear is shown. The number of cultures for each genotype is given (n). Wild-type control and Act-Gal4; UAS-RasV12 positive control (blue); neoplastic mutants (red); hyperplastic mutants (green). The appearance of proliferating cells in Pten and RasV12 cultures was significantly earlier than in wild-type control cultures (P<0.001). The appearance of proliferating cells in wts and hippo cultures was also significantly earlier than in wild-type control cultures (P<0.05). There were no significant differences between wild-type control cultures and any of the neoplastic mutants. See also Figure S1. (C–F) Examples of cultures showing proliferating patches that first appeared on average at about 33–37 days in wild type (C) and scribble (D), 21 days in hippo (E) and 11 days in Pten (F). Differentiated cell types such as muscle (arrowhead) and fat (open arrowhead) are present in all genotypes. Insets in D–F show a GFP image demonstrating that the indicated patches of proliferating cells (arrows) were negative for GFP and therefore of the mutant genotype.
We used the time of appearance of proliferating cell patches, in comparison with wild-type and RasV12-expressing cultures, as an assay for the effects of tumor suppressor mutations to discover which, if any, had positive effects and could provide a means for deriving cell lines. Cultures were established from the progeny of flies heterozygous for loss-of-function tumor suppressor mutations (Table 1; Figure 1A). The cultures were, therefore, derived from embryos of four genotypes but the homozygous mutant cells could be distinguished in the cultures, as all other genotypes expressed the marker gene Green Fluorescent Protein (GFP) under the control of the ubiquitous promoters Actin5C or Ubiquitin (Figure 1A). Several primary cultures were established for each genotype and examined every few days over a period of about 8 weeks. The range of days on which patches of proliferating cells appeared in a given culture was determined and the GFP marker allowed us to infer the genotype of the cells. The results are shown in Figure 1B and discussed below.
Table 1. Tumor suppressors tested in in vitro assays.
| Gene (allele used*) | Mammalian homolog | Tumor suppressor class and Complex or Pathway | GO classification** | |
| Molecular function | Biological Process | |||
| scribble (scrib1) | Scrib | neoplastic Scrib/Dlg/Lgl | kinase regulator | signal transduction cell adhesion |
| discs large 1 (dlgXI-2) | Dlg1 | neoplastic Scrib/Dlg/Lgl | unknown-protein interaction domains | signal transduction cell adhesion |
| lethal (2) giant larvae (lgl4) | Llgl1 | neoplastic Scrib/Dlg/Lgl | signal transduction cell adhesion | |
| brain tumor (brat11) | TRIM | neoplastic Myc regulation | E3 ubiquitin-protein ligase | cell cycle nervous system |
| hyperplastic discs (hyd35) | EDD | hyperplastic | ubiquitin-protein ligase activity | protein metabolic process |
| crumbs (crb2) | Crb1 | hyperplastic wts/hpo pathway | transmembrane EGF | apical/basal polarity |
| hippo (hpoBF33) | MST1/2 | hyperplastic wts/hpo pathway | kinase | signal transduction apoptosis cell cycle |
| warts (wtsMGH1) | LATS1/2 | hyperplastic wts/hpo pathway | kinase | signal transduction apoptosis cell cycle |
| Pten (Pten117) | Pten | hyperplastic insulin pathway | phosphatase | cell cycle, signal transduction |
*The allele used in this study is indicated. All alleles used were loss-of-function and when possible null alleles. The alleles and their sources are described in the methods section.
**GO classification adapted from Panther (http://www.pantherdb.org/).
Neoplastic tumor suppressor mutants have in vitro growth characteristics that are similar to wild type
We tested four tumor suppressor mutants, from the neoplastic class (Table 1). scribble (scrib), discs large 1 (dlg), and lgl, which are part of the Scribble complex, and brain tumor (brat). The mutants have overgrowth phenotypes in imaginal discs and neural tissues and can form tumors when implanted into host flies [10], [14], [19]. For each mutant strain, multiple primary cultures were established and the time at which patches of proliferating cells emerged was determined (Figure 1B). Cultures of each genotype developed patches of spindle-shaped cells on average between 29 and 37 days (scrib, 33; dlg, 37; lgl, 31; brat, 29; Figure 1B–D). The range of days over which these cells appeared overlapped that of wild-type cultures (Figure 1B). In the long term, the cultures grew slowly taking on average between 11 and 13 weeks to reach confluence. In contrast, RasV12-expressing cultures, and as we show below Pten mutant cultures, reach confluence in about three weeks [4]. We conclude use of the neoplastic mutants, which we tested, is not a practical approach for the development of cell lines.
Some hyperplastic tumor-suppressor mutants promote cell proliferation in vitro—Pten is the most potent
We tested 5 hyperplastic tumor suppressor mutants including the hyperplastic discs (hyd) mutant [20] and 4 genes from two major signaling pathways; the Hippo pathway (crumbs (crb), hippo (hpo) and warts (wts)) (reviewed in [17]) and the insulin pathway (Pten) [16], [21], [22].
hyd
hyd mutants show a variable disc overgrowth phenotype as homozygous mutants, which led to classification of the gene as a tumor suppressor [20]. In the in vitro assay, proliferating patches appeared on average at 35 days in hyd mutant cultures, which is similar to the time frame for wild type (Figure 1B). In keeping with the idea that hyd mutant cells do not have increased ability to proliferate, clonal mutant patches are small and out competed by surrounding wild-type cells and some discs in homozygous mutants are small [20] [23]. The gene has also been found to be overexpressed, rather than lost, in some human tumors [24].
Hippo pathway
crb mutants represent the group of hyperplastic tumor suppressors that function upstream in the Hippo pathway (crb, expanded, merlin and fat) [17], [25], [26], [27], [28]. Cultures derived from crb mutants develop proliferating patches on average at 30 days (Figure 1B). Primary cultures from core members of the Hippo pathways (hpo and wts) developed patches of proliferating cells earlier on average at 22 or 21 days, respectively (Figure 1B and 1E). This was significantly shorter than the time of appearance of these cells in wild-type cultures (P<0.005) (Figure 1B; Figure S1). The longer time for the appearance of proliferating patches in crb mutant cultures is in keeping with the evidence that genes acting upstream of the core Hippo pathway appear to play modulating roles which may be small and additive [17]. Finding that wts mutants lead to the earlier appearance of proliferating cells is in keeping with our finding that reducing wts function with RNAi also promoted the development of proliferating cell patches when compared to controls [4]. Thus mutants of core tumor suppressor genes in the Hippo pathway can be used to accelerate the derivation of new cell lines and indeed we have generated cell lines by expressing wtsRNAi [4].
Insulin pathway
The occurrence of proliferating cell patches was earliest and most pronounced in cultures derived from mutants of the insulin pathway gene Pten, where they appeared in large numbers on average by 11 days (Figure 1B and 1F). This was significantly shorter than the time of appearance of these cells in wild-type cultures (P<0.001) (Figure 1B; Figure S1). We tracked the development of Pten mutant primary cultures over longer periods and found they mirrored the development of cultures expressing RasV12: they reached confluence at about 3 weeks and the 10th passage by 5 months (Table 2; [4]). The time of the appearance of proliferating cells in Pten cultures compared with RasV12 cultures was not significantly different (Figure S1). Three cell lines were generated (Pten-1, -1A and -X; Table 2). These lines have been passaged multiple times representing between 250 and 630 cell doublings and thus can be considered immortal. The lines were established from the null allele, Pten117, which has a deletion in the coding sequence that causes a frame shift [29], and the cell line genotype was confirmed using PCR (see methods). Together these results support the use of a loss-of-function Pten mutation as a practical strategy for deriving new Drosophila cell lines.
Table 2. Generation and characteristics of Pten cell lines in reference to other cell lines.
| Cell line | Weeks to confluence | Months to passage 10 | Doubling time (h) (22°C) | Confluent density (×106)* |
| Pten 1 | 3 | 5 | 43 | 2.3 |
| Pten 1A | 3 | 5 | 40 | 3.2 |
| Pten X | N/D | N/D | 42 | 2.3 |
| Ras 3 | 3 | 4 | 31 | 7.0 |
| Ras 7 | 3 | 4 | 32 | 8.9 |
| Pten Ras 8 | 5 | 4 | 36 | 3.5 |
| Pten Ras 9 | 5 | 4 | 36 | 3.2 |
| S2 | N/A | N/A | 33 | 6.5 |
*surface area = 3.8 cm2.
ANOVA and a Tukey-Kramer multiple comparison test showed that Pten and Pten; Ras lines had a significantly lower confluent density than Ras lines (P<0.01) (F value = 31.052 = MStreatment/MSresidual).
Pten117 mutant and RasV12-expressing primary cells show comparable accelerated growth in culture, but the combination has no added effect
As loss of Pten accelerated the appearance of proliferating cells, when compared to wild type and all other tumor suppressor mutants tested (Figure 1B), the mutant primary cultures were examined more closely and compared with the development of RasV12 expressing cultures. We also tested whether simultaneous loss of Pten and expression of RasV12 had an additive effect. Cultures were established from controls, and stocks that gave rise to Pten mutant embryos, embryos expressing transgenic RasV12, and Pten mutant embryos expressing transgenic RasV12.
Initially, the cell types and their morphologies were very similar in all cultures (Figure 2A, 2D, 2G, 2J), however, after 10 days, there were marked differences between the mutant and control genotypes. Pten, RasV12, and Pten; RasV12 cultures had numerous small patches of spindle-shaped cells and more rarely patches of epithelial-like cells (Figure 2E, 2H, 2K), whereas control cultures had only differentiated cell types (Figure 2B). By 20 days these differences were more pronounced and the mutant cultures were densely populated with proliferating cells (Figure 2F, 2I, 2L). At this stage control cultures still remained sparse and comprised of differentiated cell types (Figure 2C). The Pten mutant cultures also expressing RasV12 developed in a similar fashion to those mutant for Pten alone or those just expressing RasV12. We generated two cell lines from Pten; RasV12 cultures and these also developed in a similar fashion to those derived from cells with the single genetic changes (Table 2).
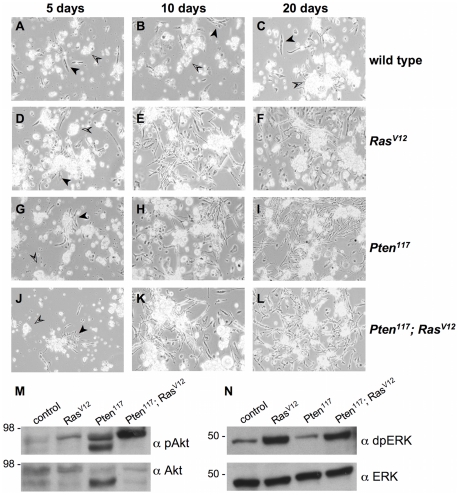
Figure 2. Pten mutant primary cultures mirror the development of primary cultures expressing oncogenic RasV12.
(A–C) Wild type. (D–F) RasV12 (Act5C-Gal4; UAS-RasV12). (G–I) Pten117. (J–L) Pten117; RasV12 (Pten117; Act5C-Gal4/Pten117; UAS-RasV12). After five days, primary cultures of all genotypes (A, D, G, J) had differentiated cell types including fat body (open arrowheads) and muscle (arrowheads). After ten days, wild-type cultures (B) had only the same differentiated cell types, whereas, cultures of the other genotypes (E, H, K) had patches of spindle-shaped cells. After 20 days, wild-type cultures (C) had only the same differentiated cell types, whereas, cultures of the other genotypes (F, I, L) were densely populated with spindle-shaped cells. (M and N) Western-blot analysis of primary culture extracts with cells of the indicated genotypes. (M) The Akt pathway is activated (pAkt) above control (wild type) levels in cultures with RasV12 expressing cells (RasV12), Pten mutant cells (Pten117) and Pten mutant cells expressing RasV12 (Pten117; RasV12). The Erk pathway is activated (dpErk) above control (wild type) levels in cultures with RasV12 expressing cells (RasV12) and Pten mutant cells expressing RasV12 (Pten117; RasV12). Total Akt and Erk, as detected by α-Akt and α-Erk were used as loading controls [40], [41]. Akt is detected as two bands (Cell Signaling Technology) [31]. For unknown reasons the lower band is more prominent in the Pten mutant cultures.
We examined pathway activation in the primary cultures and found as expected that Akt signaling was activated in Pten mutant cultures (Figure 2M). Pten is a key negative regulator of Akt signaling. We also examined MAPK/Erk signaling as determined by phosphorylation of Drosophila Erk a downstream effector of Ras signaling. Pten cultures showed wild-type levels of Erk activation (Figure 2N). As expected, primary cultures expressing RasV12 had high levels of Erk activation (Figure 2N). It is known that Drosophila RasV12, but probably not endogenous Ras, can also signal through the Akt pathway [4], [30], [31]. In keeping with our previous findings for RasV12 expression in vitro, we also observed enhanced Akt activation in RasV12-expressing primary cultures ([4]; Figure 3M). The Pten; RasV12 cultures showed strong activation of both pathways (Figure 2M, 2N). We conclude that the combination of molecular changes resulting from the different genetic backgrounds does not result in additive effects at the cellular level that can further enhance cell proliferation.
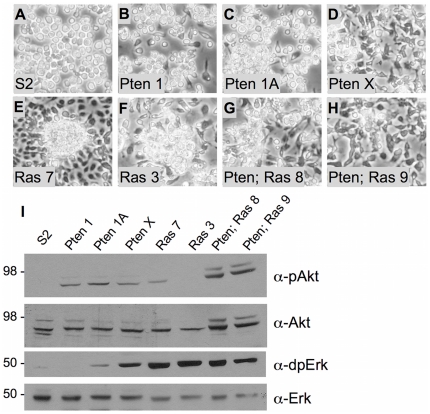
Figure 3. Morphological and molecular comparison of Pten117 cell lines with S2, RasV12 and Pten117; RasV12 cell lines.
(A–H) Cells are shown after 6 days of growth from the same starting cell number. (A) S2. Cells show the typical round morphology and are loosely attached to the surface. (B–D) Pten117. Pten 1 and 1A have round and spindle-shaped cells. Cells in the Pten 1 line are more loosely attached. Cells in line Pten X are primarily spindle shaped. (E–F) RasV12. Ras 7 and 3 have spindle shaped cells that form dense attached clusters. (G–H) Pten117; RasV12. Pten; Ras 8 and 9 have spindle shaped cells that form small clusters. (I) Western-blot analysis of cell extracts from the indicated lines. The Akt pathway is activated (pAkt) above control levels in all lines with Pten mutations. The Ras 7 line also shows elevated activation. The Erk pathway is activated (dpErk) in most lines with very low levels in S2 cells and undetectable levels in line Pten 1. Total Akt and Erk, as detected by α-Akt and α-Erk were used as loading controls [40], [41]. Akt is detected as two bands (Cell Signaling Technology) [31].
Characteristics of Pten, RasV12, and Pten; RasV12 cell lines
We compared characteristics of Pten117 and Pten117; RasV12 cell lines with RasV12 lines and cells of the commonly used Drosophila S2 line [32]. The cells have different morphologies likely reflecting different cell types (Figure 3A–H). All lines, including S2, were generated from whole embryos and, therefore, the cell type is not known a priori, although subsequent analysis of S2 cells suggest they are of macrophage origin [33]. All lines showed doubling times between 31 h and 43 h at 22°C during the steepest part of the growth curve and achieved a broad range of confluent densities (Table 2). Differences in doubling times and confluent densities could reflect in part the different cell types present in the lines, but it is also likely that there is a genetic component conferred by the Pten mutation. The Pten117 mutant lines had the longest doubling times (Table 2). The Pten117 and the Pten117; RasV12 expressing lines mutant lines also had significantly lower confluent densities than the RasV12 expressing lines (Table 2). Thus cells with a mutation in Pten fail to reach high confluent densities even in the presence of RasV12. Further analysis will be required to determine the molecular basis for the effect. By the measures of doubling time and confluent density alone, the Pten117 lines have less practical use because growing the cells takes longer and not as many cells can be generated in a culture vessel. However, as discussed below there are likely to be cases in which having an additional genetic method to generate cell lines will be critical. Pten117 mutant cell cultures will also be key reagents for studying the immortalization process itself.
Western analysis of the lines showed the expected pathways were strongly activated; Akt in Pten117 mutant lines, Erk in the RasV12 expressing lines and both in the Pten117; RasV12 lines (Figure 3I). S2 cells showed only basal activation of Erk (Figure 3I). The Ras 7 line showed activation of PI3K (Figure 3I), as has been noted for cells expressing the RasV12 allele in vivo [30], [31] and in vitro [4]. Cells of the line Pten X, showed higher levels of Erk activation than the other two Pten lines demonstrating that individual lines can differ presumably either due to the specific cell type present in the line or through additional genetic changes that occur during prolonged culture (Figure 3I). In S2 cells there was no activation of Akt, which is a target of insulin signaling, and only weak activity of Erk signaling (Figure 3I). Analysis of S2 cells has shown that the insulin receptor is expressed, but the pathway is unlikely to be active because the ligands are not expressed [34]. The PVR pathway is thought to be active and could be contributing to the activation of Erk we observed [34].
Concluding remarks
Our major finding is that Pten loss promotes cell proliferation in vitro and allows the rapid derivation of cell lines, providing another genetic method to generate cell lines in Drosophila in addition to the use of oncogenic RasV12 [4]. We have successfully used expression of transgenic RasV12 to derive cell lines from the rumi mutant [8]. The cells, which are null for rumi have been used to study Notch signaling in vitro [9]. In a similar fashion Pten mutations could be used to establish cell lines corresponding to mutants. This is important because it provides an alternate signaling context in which to study a mutant where having the Ras pathway active would interfere with the analysis.
Materials and Methods
Fly stocks
CyO, Act5C-GFP; TM6B, Ubi-GFP; FM7i, Act-GFP, crb2 and l(2)gl4 flies were obtained from the Bloomington Drosophila Stock Center. The GFP-marked balancer chromosomes were used to generate stocks of the tumor suppressor mutations. The following alleles were obtained from colleagues: hpoBF33, J. Jiang [35]; wtsMGH1, D.J. Pan [36]; brat11, D. Frank [37]; Pten117, H. Stocker [29]; hyd35, J. Treisman [20]; scrib 1, D. Bilder [38]; dlgXI-2, V. Budnik [39]. Pten117, UAS-RASV12 and Pten117, Act5C-Gal4 recombinant chromosomes were generated using standard crosses.
Cell culture
Primary cultures were established using a standard method [4]. Schneider's medium with 10% heat inactivated Fetal Bovine serum was used for all cell culture work. Multiple primary cultures were generated for each genotype and examined to document the development of various cells types. The time at which patches of dividing cells emerged was determined. Cultures of Pten 117 and Pten 117; RasV12 cells were maintained until they reached confluence and were sub-cultured to generate cell lines using standard methods [4].
Cell line characterization
For growth curves, cells were plated at 2.5×105 cells/well in a 12-well plate with 1 ml of medium. Duplicate cell counts were made over 7 days. Doubling time was determined during the period when cells numbers increased most steeply in a given line (typically days 3–5). Confluent densities were determined at the point when there was no further increase in cell number.
Genotyping of Pten mutant cell lines
The Pten117 allele has a deletion of 5 base pairs in the coding region of the gene that results in a frame-shift and insertion of a premature stop codon. DNA from Pten cultures was amplified using the primers shown below. The amplified DNA was purified and sequenced to confirm presence of the mutation.
5′-GCGAAAGTTCATAAATATCATGGC, 3′-CGGCGCTGAATGTGGCGC.
Western Analysis
Cells were lysed in TN1 buffer containing 125 mM NaCl, 50 mM Tris (pH = 8.0), 10 mM EDTA (pH = 8.0), 10 mM Na4P2O7 ·10H2O, 10 mM NaF, 1% Triton X-100, 3 mM Na3VO4 supplemented with protease inhibitor cocktail (Roche Diagnostics Corp. Indianapolis, IN), centrifuged, and supernatants were used for analysis. Total protein (10 µg) was separated on polyacrylamide gels and immunoblots were incubated with antibodies directed against Drosophila-specific Akt and pAkt (Ser 505) (Cell Signaling Technology; Danvers, MA), pan-Erk, (Santa Cruz Biotechnology; Santa Cruz, CA) and dpErk1/2 (E10) (Cell Signaling Technology; Danvers, MA).
Supporting Information
Statistical analysis of cell proliferation data. Box and whisker plot of the data from Figure 1B in the main text showing the time for proliferating cells to appear in primary cultures of different genotypes. Boxes are drawn between the mean and the median. The whiskers end at the maximum and minimum values in the sample population. Samples were analyzed using Dunn's multiple comparison test post the Kruskal-Wallis test. Pairs that were identified as significantly different are connected by solid lines. Both Pten mutant and RasV12 expressing cultures are significantly different than wild-type cultures (P<0.001). Both hpo and wts cultures are significantly different than wild-type cultures (P<0.05).
(PDF)
Acknowledgments
We thank the Bloomington Stock Center and colleagues listed in the text for stocks. We also thank our colleagues Harald Vaessin and Mark Seeger for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the National Institutes of Health (R01GM071856 and 1R21HD063119-01; http://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hayflick L. Mortality and immortality at the cellular level. A review. Biochemistry (Mosc) 1997;62:1180–1190. [PubMed] [Google Scholar]
- 2.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 3.Kraemer PM, Ray FA, Brothman AR, Bartholdi MF, Cram LS. Spontaneous immortalization rate of cultured Chinese hamster cells. J Natl Cancer Inst. 1986;76:703–709. doi: 10.1093/jnci/76.4.703. [DOI] [PubMed] [Google Scholar]
- 4.Simcox A, Mitra S, Truesdell S, Paul L, Chen T, et al. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PLoS Genet. 2008;4:e1000142. doi: 10.1371/journal.pgen.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 6.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 7.Pardue ML, Debaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simcox AA, Austin CL, Jacobsen TL, Jafar-Nejad H. Drosophila embryonic ‘fibroblasts’: Extending mutant analysis in vitro. Fly (Austin) 2008;2:306–309. doi: 10.4161/fly.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonardi J, Fernandez-Valdivia R, Li YD, Simcox AA, Jafar-Nejad H. Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling. Development. 2011;138:3569–3578. doi: 10.1242/dev.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 11.Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu Rev Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 12.Gateff E, Schneiderman HE. Developmental studies of a new mutant of Drosophila melanogaster: lethal malignant brain tumor [l(2)gl 4]. Am Zool. 1967;7:760. [Google Scholar]
- 13.Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- 14.Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, et al. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 15.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 16.Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echalier G. Drosophila cells in culture. New York: Academic Press; 1997. [Google Scholar]
- 19.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 20.Mansfield E, Hersperger E, Biggs J, Shearn A. Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogaster imaginal discs and germ cells. Dev Biol. 1994;165:507–526. doi: 10.1006/dbio.1994.1271. [DOI] [PubMed] [Google Scholar]
- 21.Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- 22.Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, et al. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 23.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 24.Clancy JL, Henderson MJ, Russell AJ, Anderson DW, Bova RJ, et al. EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene. 2003;22:5070–5081. doi: 10.1038/sj.onc.1206775. [DOI] [PubMed] [Google Scholar]
- 25.Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 2010;4:288–293. doi: 10.4161/fly.4.4.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling C, Zheng Y, Yin F, Yu J, Huang J, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 28.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldham S, Stocker H, Laffargue M, Wittwer F, Wymann M, et al. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development. 2002;129:4103–4109. doi: 10.1242/dev.129.17.4103. [DOI] [PubMed] [Google Scholar]
- 30.Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willecke M, Toggweiler J, Basler K. Loss of PI3K blocks cell-cycle progression in a Drosophila tumor model. Oncogene. 2011 doi: 10.1038/onc.2011.125. [DOI] [PubMed] [Google Scholar]
- 32.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 33.Ramet M, Pearson A, Manfruelli P, Li X, Koziel H, et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- 34.Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 37.Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- 38.Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 39.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 40.Gabay L, Seger R, Shilo B-Z. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 41.Kockel L, Kerr KS, Melnick M, Bruckner K, Hebrok M, et al. Dynamic switch of negative feedback regulation in Drosophila Akt-TOR signaling. PLoS Genet. 2010;6:e1000990. doi: 10.1371/journal.pgen.1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis of cell proliferation data. Box and whisker plot of the data from Figure 1B in the main text showing the time for proliferating cells to appear in primary cultures of different genotypes. Boxes are drawn between the mean and the median. The whiskers end at the maximum and minimum values in the sample population. Samples were analyzed using Dunn's multiple comparison test post the Kruskal-Wallis test. Pairs that were identified as significantly different are connected by solid lines. Both Pten mutant and RasV12 expressing cultures are significantly different than wild-type cultures (P<0.001). Both hpo and wts cultures are significantly different than wild-type cultures (P<0.05).
(PDF)