Abstract
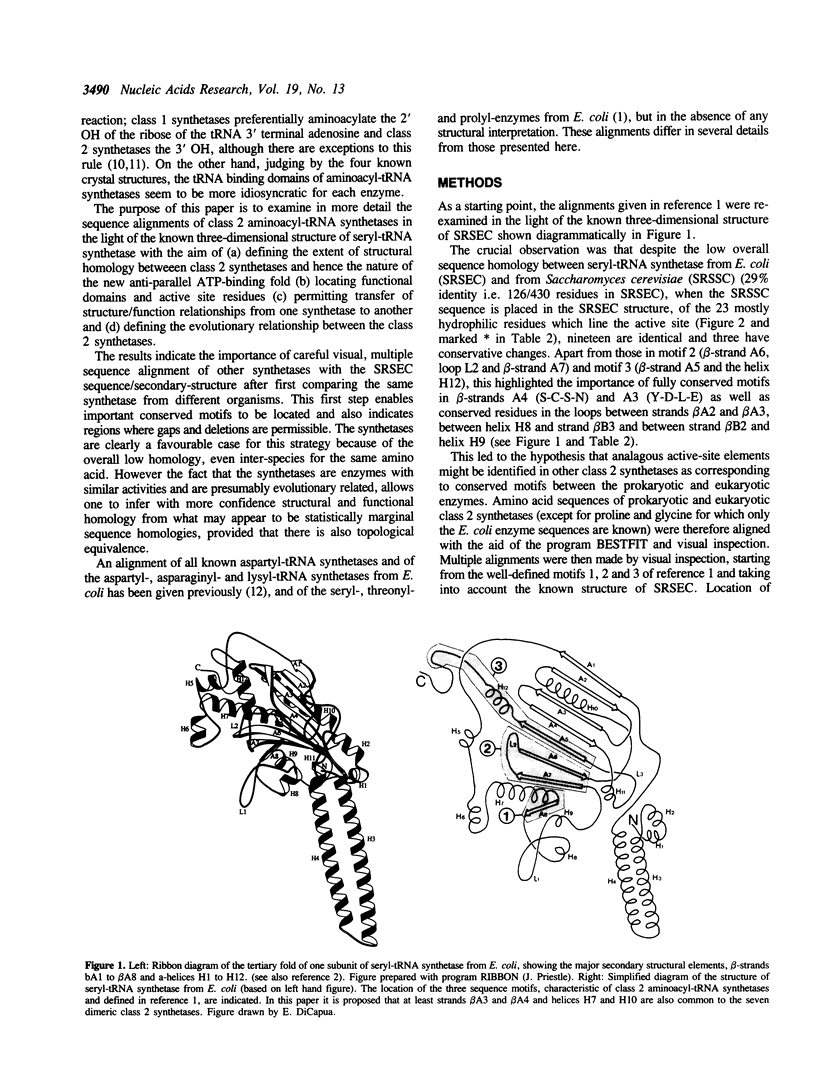
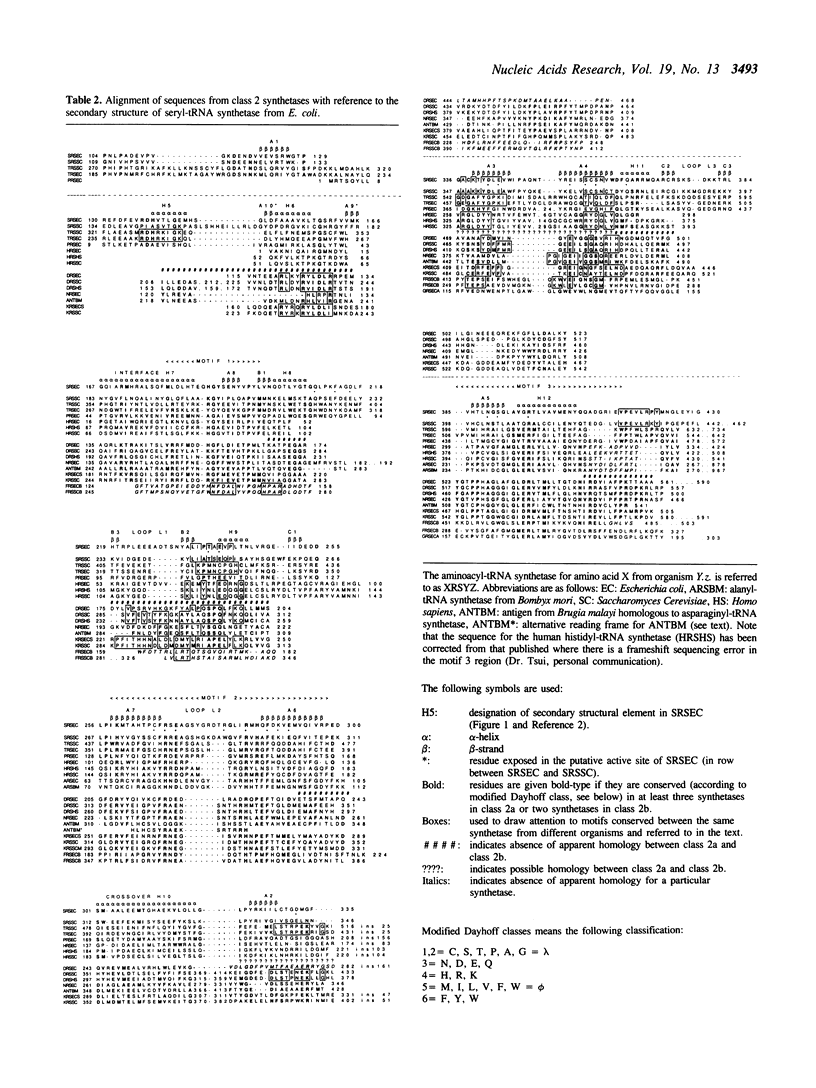
Class 2 aminoacyl-tRNA synthetases, which include the enzymes for alanine, aspartic acid, asparagine, glycine, histidine, lysine, phenylalanine, proline, serine and threonine, are characterised by three distinct sequence motifs 1,2 and 3 (reference 1). The structural and evolutionary relatedness of these ten enzymes are examined using alignments of primary sequences from prokaryotic and eukaryotic sources and the known three dimensional structure of seryl-tRNA synthetase from E. coli. It is shown that motif 1 forms part of the dimer interface of seryl-tRNA synthetase and motifs 2 and 3 part of the putative active site. It is further shown that the seven alpha 2 dimeric synthetases can be subdivided into class 2a (proline, threonine, histidine and serine) and class 2b (aspartic acid, asparagine and lysine), each subclass sharing several important characteristic sequence motifs in addition to those characteristic of class 2 enzymes in general. The alpha 2 beta 2 tetrameric enzymes (for glycine and phenylalanine) show certain special features in common as well as some of the class 2b motifs. In the alanyl-tRNA synthetase only motif 3 and possibly motif 2 can be identified. The sequence alignments suggest that the catalytic domain of other class 2 synthetases should resemble the antiparallel domain found in seryl-tRNA synthetase. Predictions are made about the sequence location of certain important helices and beta-strands in this domain as well as suggestions concerning which residues are important in ATP and amino acid binding. Strong homologies are found in the N-terminal extensions of class 2b synthetases and in the C-terminal extensions of class 2a synthetases suggesting that these putative tRNA binding domains have been added at a later stage in evolution to the catalytic domain.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anselme J., Härtlein M. Asparaginyl-tRNA synthetase from Escherichia coli has significant sequence homologies with yeast aspartyl-tRNA synthetase. Gene. 1989 Dec 14;84(2):481–485. doi: 10.1016/0378-1119(89)90524-6. [DOI] [PubMed] [Google Scholar]
- Anselme J., Härtlein M. Tyr-426 of the Escherichia coli asparaginyl-tRNA synthetase, an amino acid in a C-terminal conserved motif, is involved in ATP binding. FEBS Lett. 1991 Mar 11;280(1):163–166. doi: 10.1016/0014-5793(91)80228-u. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Starzyk R. M., Schimmel P. Understanding structural relationships in proteins of unsolved three-dimensional structure. Proteins. 1990;7(2):99–111. doi: 10.1002/prot.340070202. [DOI] [PubMed] [Google Scholar]
- Chang P. K., Dignam J. D. Primary structure of alanyl-tRNA synthetase and the regulation of its mRNA levels in Bombyx mori. J Biol Chem. 1990 Dec 5;265(34):20898–20906. [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Cysteinyl-tRNA synthetase: determination of the last E. coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 1991 Jan 25;19(2):265–269. doi: 10.1093/nar/19.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987 Dec 15;26(25):8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Leinfelder W., Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989 Nov 23;342(6248):453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- Fraser T. H., Rich A. Amino acids are not all initially attached to the same position on transfer RNA molecules. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3044–3048. doi: 10.1073/pnas.72.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman R., Gibson B., Donovan D., Biemann K., Eisenbeis S., Parker J., Schimmel P. Primary structure of histidine-tRNA synthetase and characterization of hisS transcripts. J Biol Chem. 1985 Aug 25;260(18):10063–10068. [PubMed] [Google Scholar]
- Gampel A., Tzagoloff A. Homology of aspartyl- and lysyl-tRNA synthetases. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6023–6027. doi: 10.1073/pnas.86.16.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. L., Tzagoloff A. Structure and evolution of a group of related aminoacyl-tRNA synthetases. J Mol Biol. 1991 Apr 5;218(3):557–568. doi: 10.1016/0022-2836(91)90701-7. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Härtlein M., Madern D., Leberman R. Cloning and characterization of the gene for Escherichia coli seryl-tRNA synthetase. Nucleic Acids Res. 1987 Feb 11;15(3):1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Peterson R., Yang D. C. cDNA sequence, predicted primary structure, and evolving amphiphilic helix of human aspartyl-tRNA synthetase. J Biol Chem. 1989 Oct 5;264(28):16608–16612. [PubMed] [Google Scholar]
- Koerner T. J., Myers A. M., Lee S., Tzagoloff A. Isolation and characterization of the yeast gene coding for the alpha subunit of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 1987 Mar 15;262(8):3690–3696. [PubMed] [Google Scholar]
- Leinfelder W., Zehelein E., Mandrand-Berthelot M. A., Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988 Feb 25;331(6158):723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- Lévêque F., Plateau P., Dessen P., Blanquet S. Homology of lysS and lysU, the two Escherichia coli genes encoding distinct lysyl-tRNA synthetase species. Nucleic Acids Res. 1990 Jan 25;18(2):305–312. doi: 10.1093/nar/18.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayaux J. F., Fayat G., Fromant M., Springer M., Grunberg-Manago M., Blanquet S. Structural and transcriptional evidence for related thrS and infC expression. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6152–6156. doi: 10.1073/pnas.80.20.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechulam Y., Fayat G., Blanquet S. Sequence of the Escherichia coli pheST operon and identification of the himA gene. J Bacteriol. 1985 Aug;163(2):787–791. doi: 10.1128/jb.163.2.787-791.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirande M., Waller J. P. The yeast lysyl-tRNA synthetase gene. Evidence for general amino acid control of its expression and domain structure of the encoded protein. J Biol Chem. 1988 Dec 5;263(34):18443–18451. [PubMed] [Google Scholar]
- Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J. P., Ehresmann B., Ehresmann C. Escherichia coli threonyl-tRNA synthetase and tRNA(Thr) modulate the binding of the ribosome to the translational initiation site of the thrS mRNA. J Mol Biol. 1990 Nov 20;216(2):299–310. doi: 10.1016/S0022-2836(05)80321-3. [DOI] [PubMed] [Google Scholar]
- Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J. P., Ehresmann C., Ehresmann B. Messenger RNA structure and gene regulation at the translational level in Escherichia coli: the case of threonine:tRNAThr ligase. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7892–7896. doi: 10.1073/pnas.85.21.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G., Hilger F., Fink G. R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986 Jul 18;46(2):235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Nilsen T. W., Maroney P. A., Goodwin R. G., Perrine K. G., Denker J. A., Nanduri J., Kazura J. W. Cloning and characterization of a potentially protective antigen in lymphatic filariasis. Proc Natl Acad Sci U S A. 1988 May;85(10):3604–3607. doi: 10.1073/pnas.85.10.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape L. K., Koerner T. J., Tzagoloff A. Characterization of a yeast nuclear gene (MST1) coding for the mitochondrial threonyl-tRNA1 synthetase. J Biol Chem. 1985 Dec 5;260(28):15362–15370. [PubMed] [Google Scholar]
- Pape L. K., Tzagoloff A. Cloning and characterization of the gene for the yeast cytoplasmic threonyl-tRNA synthetase. Nucleic Acids Res. 1985 Sep 11;13(17):6171–6183. doi: 10.1093/nar/13.17.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney S. D., Sauer R. T., Schimmel P. R. Purification and properties of alanine tRNA synthetase from Escherichia coli A tetramer of identical subunits. J Biol Chem. 1981 Jan 10;256(1):198–204. [PubMed] [Google Scholar]
- Putzer H., Brakhage A. A., Grunberg-Manago M. Independent genes for two threonyl-tRNA synthetases in Bacillus subtilis. J Bacteriol. 1990 Aug;172(8):4593–4602. doi: 10.1128/jb.172.8.4593-4602.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Sanni A., Mirande M., Ebel J. P., Boulanger Y., Waller J. P., Fasiolo F. Structure and expression of the genes encoding the alpha and beta subunits of yeast phenylalanyl-tRNA synthetase. J Biol Chem. 1988 Oct 25;263(30):15407–15415. [PubMed] [Google Scholar]
- Sellami M., Fasiolo F., Dirheimer G., Ebel J. P., Gangloff J. Nucleotide sequence of the gene coding for yeast cytoplasmic aspartyl-tRNA synthetase (APS); mapping of the 5' and 3' termini of AspRS mRNA. Nucleic Acids Res. 1986 Feb 25;14(4):1657–1666. doi: 10.1093/nar/14.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Graffe M., Dondon J., Grunberg-Manago M. tRNA-like structures and gene regulation at the translational level: a case of molecular mimicry in Escherichia coli. EMBO J. 1989 Aug;8(8):2417–2424. doi: 10.1002/j.1460-2075.1989.tb08372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Cramer F. Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2'- or 3'-hydroxyl group of the terminal adenosine. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3049–3053. doi: 10.1073/pnas.72.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui F. W., Siminovitch L. Isolation, structure and expression of mammalian genes for histidyl-tRNA synthetase. Nucleic Acids Res. 1987 Apr 24;15(8):3349–3367. doi: 10.1093/nar/15.8.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T. A., Gibson B. W., Keng T., Biemann K., Schimmel P. Primary structures of both subunits of Escherichia coli glycyl-tRNA synthetase. J Biol Chem. 1983 Sep 10;258(17):10637–10641. [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. tRNA-like structures tag the 3' ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weygand-Durasevic I., Johnson-Burke D., Söll D. Cloning and characterization of the gene coding for cytoplasmic seryl-tRNA synthetase from Saccharomyces cerevisiae. Nucleic Acids Res. 1987 Mar 11;15(5):1887–1904. doi: 10.1093/nar/15.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. T. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]



