Abstract
The zinc fingers of retroviral gag nucleocapsid proteins (NC) are required for the specific packaging of the dimeric RNA genome into virions. In vitro, NC proteins activate both dimerization of viral RNA and annealing of the replication primer tRNA onto viral RNA, two reactions necessary for the production of infectious virions. In this study the role of the zinc finger of Moloney murine leukemia virus (MoMuLV) NCp10 in RNA binding and annealing activities was investigated through modification or replacement of residues involved in zinc coordination. These alterations did not affect the ability of NCp10 to bind RNA and promote RNA annealing in vitro, despite a complete loss of zinc affinity. However mutation of two conserved lysine residues adjacent to the finger motif reduced both RNA binding and annealing activities of NCp10. These findings suggest that the complexed NC zinc finger is not directly involved in RNA-protein interactions but more probably in a zinc dependent conformation of NC protein modulating viral protein-protein interactions, essential to the process of viral RNA selection and virion assembly. Then the NC zinc finger may cooperate to select the viral RNA genome to be packaged into virions.
Full text
PDF

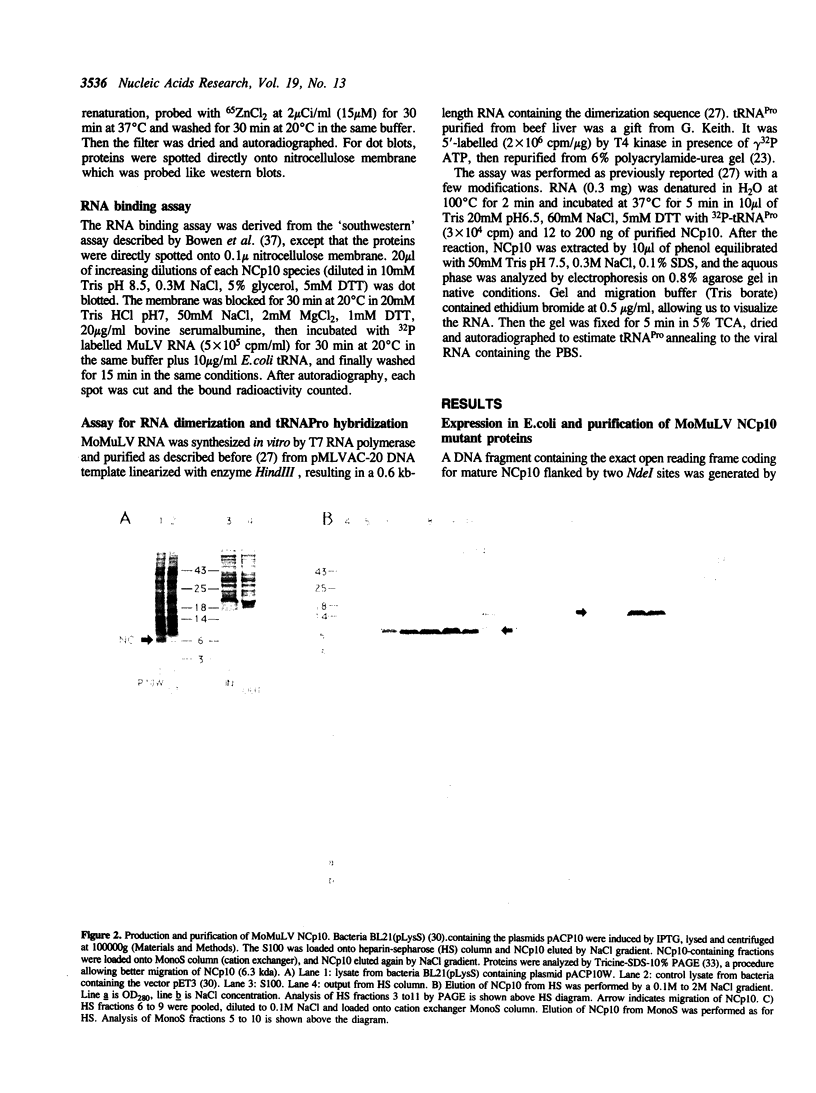
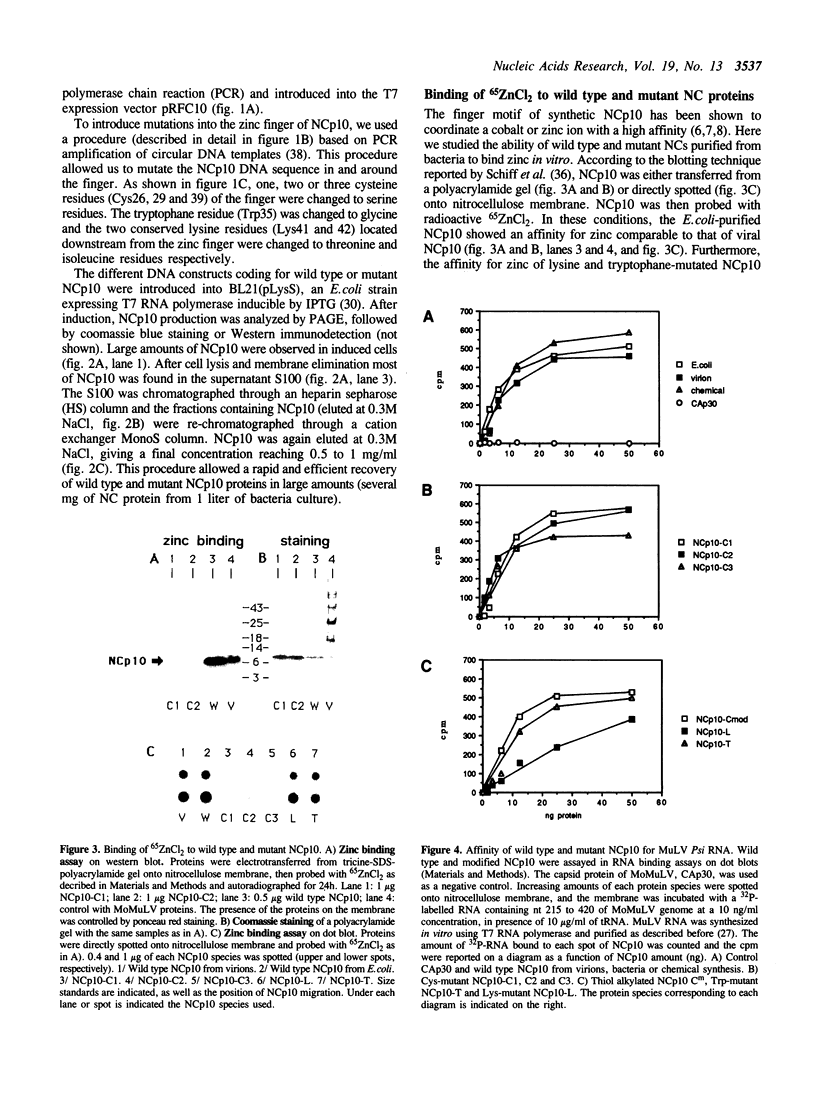

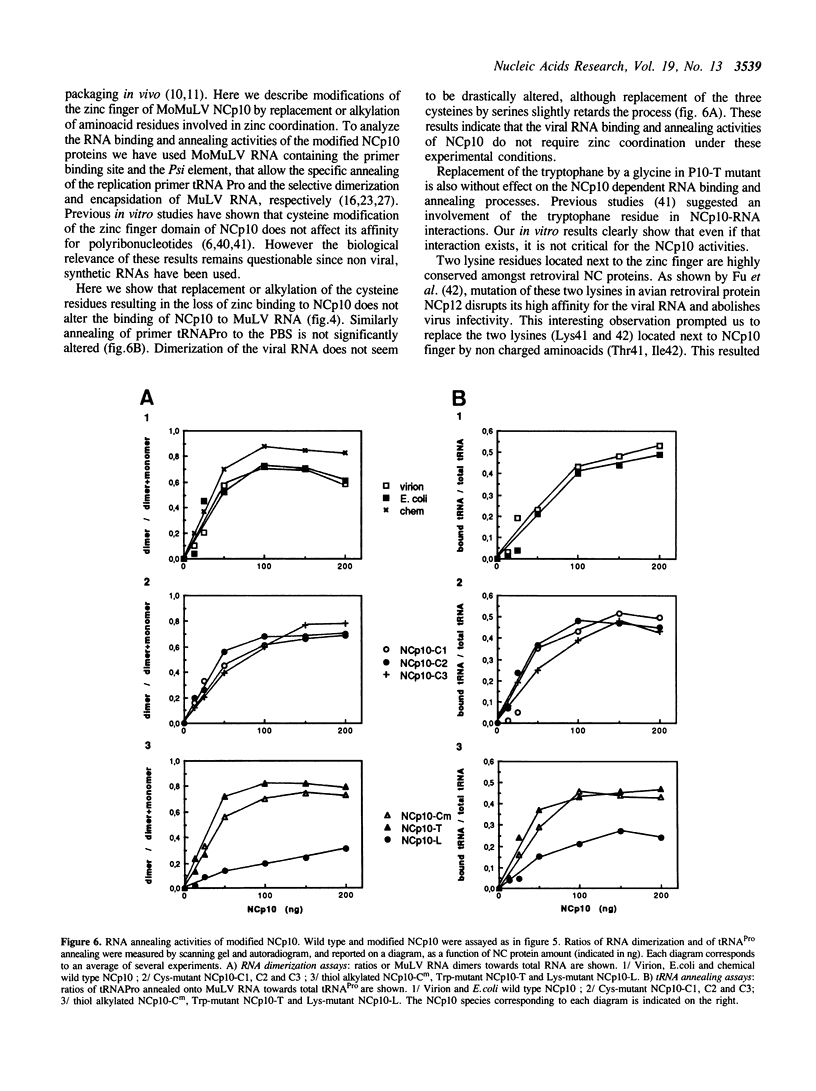


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldovini A., Young R. A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990 May;64(5):1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976 Apr;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Bieth E., Gabus C., Darlix J. L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990 Jan 11;18(1):119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen B., Steinberg J., Laemmli U. K., Weintraub H. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 1980 Jan 11;8(1):1–20. doi: 10.1093/nar/8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRESTFIELD A. M., MOORE S., STEIN W. H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J Biol Chem. 1963 Feb;238:622–627. [PubMed] [Google Scholar]
- Casas-Finet J. R., Jhon N. I., Maki A. H. p10, a low molecular weight single-stranded nucleic acid binding protein of murine leukemia retroviruses, shows stacking interactions of its single tryptophan residue with nucleotide bases. Biochemistry. 1988 Feb 23;27(4):1172–1178. doi: 10.1021/bi00404a016. [DOI] [PubMed] [Google Scholar]
- Cornille F., Mely Y., Ficheux D., Savignol I., Gerard D., Darlix J. L., Fournie-Zaluski M. C., Roques B. P. Solid phase synthesis of the retroviral nucleocapsid protein NCp10 of Moloney murine leukaemia virus and related "zinc-fingers" in free SH forms. Influence of zinc chelation on structural and biochemical properties. Int J Pept Protein Res. 1990 Dec;36(6):551–558. [PubMed] [Google Scholar]
- Crawford S., Goff S. P. A deletion mutation in the 5' part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985 Mar;53(3):899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J. L., Gabus C., Nugeyre M. T., Clavel F., Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990 Dec 5;216(3):689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Spahr P. F. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J Mol Biol. 1982 Sep 15;160(2):147–161. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- Davis J., Scherer M., Tsai W. P., Long C. Low-molecular- weight Rauscher leukemia virus protein with preferential binding for single-stranded RNA and DNA. J Virol. 1976 May;18(2):709–718. doi: 10.1128/jvi.18.2.709-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupraz P., Oertle S., Meric C., Damay P., Spahr P. F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990 Oct;64(10):4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Baltimore D. RNA metabolism of murine leukemia virus: detection of virus-specific RNA sequences in infected and uninfected cells and identification of virus-specific messenger RNA. J Mol Biol. 1973 Oct 15;80(1):93–117. doi: 10.1016/0022-2836(73)90235-0. [DOI] [PubMed] [Google Scholar]
- Fleissner E., Tress E. Isolation of a ribonucleoprotein structure from oncornaviruses. J Virol. 1973 Dec;12(6):1612–1615. doi: 10.1128/jvi.12.6.1612-1615.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Leis J. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp 12. Mutation which affects RNA binding in vitro blocks viral replication. J Biol Chem. 1988 Feb 15;263(5):2140–2145. [PubMed] [Google Scholar]
- Gorelick R. J., Henderson L. E., Hanser J. P., Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a "zinc finger-like" protein sequence. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L. M., Berg J. M. Retroviral nucleocapsid protein-metal ion interactions: folding and sequence variants. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6403–6407. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S., Kumar V., Theulaz I., Wahli W., Chambon P. The N-terminal DNA-binding 'zinc finger' of the oestrogen and glucocorticoid receptors determines target gene specificity. EMBO J. 1988 Oct;7(10):3037–3044. doi: 10.1002/j.1460-2075.1988.tb03168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989 Aug 25;17(16):6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel R. L., Henderson L. E., Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987 Apr 15;262(11):4961–4967. [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989 Sep;63(9):4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985 May;54(2):401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Méric C., Darlix J. L., Spahr P. F. It is Rous sarcoma virus protein P12 and not P19 that binds tightly to Rous sarcoma virus RNA. J Mol Biol. 1984 Mar 15;173(4):531–538. doi: 10.1016/0022-2836(84)90396-6. [DOI] [PubMed] [Google Scholar]
- Méric C., Goff S. P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989 Apr;63(4):1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C., Spahr P. F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986 Nov;60(2):450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen-Meyer J., Abraham A. K. Specificity of RNA binding by the structural protein (p10) of Friend murine leukemia virus. J Mol Biol. 1980 Sep 5;142(1):19–28. doi: 10.1016/0022-2836(80)90203-x. [DOI] [PubMed] [Google Scholar]
- Pan T., Giedroc D. P., Coleman J. E. 1H NMR studies of T4 gene 32 protein: effects of zinc removal and reconstitution. Biochemistry. 1989 Oct 31;28(22):8828–8832. doi: 10.1021/bi00448a022. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988 Aug 26;241(4869):1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Peters G., Harada F., Dahlberg J. E., Panet A., Haseltine W. A., Baltimore D. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J Virol. 1977 Mar;21(3):1031–1041. doi: 10.1128/jvi.21.3.1031-1041.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Roy C., Wang P. A., Erard M., Housset V., Gabus C., Paoletti C., Darlix J. L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990 Feb;64(2):774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugatsch T., Stacey D. W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983 Jul 30;128(2):505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- Roberts W. J., Pan T., Elliott J. I., Coleman J. E., Williams K. R. p10 single-stranded nucleic acid binding protein from murine leukemia virus binds metal ions via the peptide sequence Cys26-X2-Cys29-X4-His34-X4-Cys39. Biochemistry. 1989 Dec 26;28(26):10043–10047. doi: 10.1021/bi00452a024. [DOI] [PubMed] [Google Scholar]
- Roy C., Tounekti N., Mougel M., Darlix J. L., Paoletti C., Ehresmann C., Ehresmann B., Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990 Dec 25;18(24):7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff L. A., Nibert M. L., Fields B. N. Characterization of a zinc blotting technique: evidence that a retroviral gag protein binds zinc. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4195–4199. doi: 10.1073/pnas.85.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulein M., Burnette W. N., August J. T. Stoichiometry and specificity of binding of Rauscher oncovirus 10,000-dalton (p10) structural protein to nucleic acids. J Virol. 1978 Apr;26(1):54–60. doi: 10.1128/jvi.26.1.54-60.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J Virol. 1983 May;46(2):538–546. doi: 10.1128/jvi.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Strand M., August J. T. Structural proteins of ribonucleic acid tumor viruses. Purification of envelope, core, and internal components. J Biol Chem. 1976 Jan 25;251(2):559–564. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5' long terminal repeat and the start of the gag gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]