Abstract
Saccharomyces cerevisiae SO4= transport is regulated over a wide dynamic range. Sulfur starvation causes ∼10,000-fold increase in the 35SO4= influx mediated by transporters Sul1p and Sul2p; >80% of the influx is via Sul2p. Adding methionine to S-starved cells causes a 50-fold decline (t1/2 ∼5 min) in SUL1 and SUL2 mRNA but a slower decline (t1/2 ∼1 h) in transport. In contrast, SO4= addition does not affect mRNA but causes a rapid (t1/2 = 2–4 min) decrease in transport. In met3Δ cells (unable to metabolize SO4=), addition of SO4= to S-starved cells causes inactivation of 35SO4= influx over times in which cellular SO4= contents are nearly constant. The relationship between cellular SO4= and transport inactivation shows that cellular SO4= is not the signal for Sul2p inactivation. Instead, the transport inactivation rate has the same dependence on extracellular SO4= as 35SO4= influx, indicating that Sul2p exhibits use-dependent inactivation; the transport process itself increases the probability of Sul2p inactivation and degradation. In addition, there is a transient efflux of SO4= shortly after adding >0.02 mM SO4= to S-starved met3Δ cells. This transient efflux provides further protection against excessive SO4= influx and may represent an alternate transport mode of Sul2p.
Introduction
A central function of biological membranes is regulation of the solute compositions of cytosol and organelles. The influx and efflux of a given solute are mediated by some combination of ATP-driven pumps, cotransporters, exchangers, carriers, and channels. Transporters are regulated at numerous levels, including transcription, phosphorylation, other posttranslational modifications, trafficking/targeting, degradation, protein-protein interactions, and small molecule regulators. In addition to these regulatory mechanisms involving separate molecular partners, some transport proteins have intrinsic kinetic properties that result in autoregulation, e.g., inactivation of voltage-gated cation channels.
An autoregulatory property of many coupled cotransporters is transinhibition, i.e., inhibition of influx by cytosolic substrate binding directly to the transporter. Transinhibition is detected as a time-dependent decrease in tracer influx following the addition of nonradioactive substrate (1). The physiological consequence of transinhibition is a reduced influx as cytosolic substrate increases. Therefore, transinhibition is a potentially powerful mechanism to limit net solute influx and thereby regulate cellular solute levels. There are many examples of transinhibition in mammalian (1–3), fungal (4–6), and bacterial (7–10) cotransporters.
Another way that cells limit solute influx is by reducing the number of functioning transporters, either by endocytosis from the plasma membrane or trafficking the transporter from the trans Golgi directly to a degradation pathway. Examples of substrate-regulated trafficking are the Saccharomyces cerevisiae transporters for manganese (11–13), copper (14), zinc (15,16), uracil (17,18), and amino acids (19,20). Substrate-induced downregulation of yeast transporters generally depends on ubiquitylation, and the cellular machinery for ubiquitin-mediated degradation of yeast transporters is becoming increasingly well understood (21).
The primary signal by which substrate causes altered trafficking and ultimate degradation of a transporter could be the size of the cytosolic pool of substrate (12,15,17,22). Recently, however, there is evidence that it is the transport process itself rather than the cellular pool of substrate that initiates transporter degradation. This mechanism, termed use-dependent or activity-dependent degradation, was proposed for the yeast iron transporter Fet3p/Ftr1p to account for the fact that intracellular iron does not trigger transporter degradation unless the iron has been transported inward by the transporter itself rather than some other pathway (23). The same kind of mechanism regulates the general amino acid transporter Gap1p; nontransportable substrates do not cause inactivation of Gap1p, indicating that an intermediate in the transport catalytic cycle is part of the signal for transporter inactivation (24,25). Other recent evidence for use-dependent transporter inactivation has been obtained for Aspergillus nidulans purine transport (26) and Arabidopsis thaliana NH4+ transport (27).
This work concerns regulation of SO4= transport in S. cerevisiae, which has two closely related SO4= transporters, Sul1p and Sul2p (28–31). After entering the cytosol, SO4= is metabolized to adenosine-5′-phosphosulfate (APS), 3′-phosphoadenosine-5′-phosphosulfate (PAPS), sulfite (SO3=), sulfide (HS−), and homocysteine, which is used to synthesize cysteine, methionine, and other S-containing compounds (32).
It has been known for many years that addition of SO4= causes downregulation of subsequent 35SO4= transport in S. cerevisiae (28), Penicillium notatum (33), and Neurospora crassa (4). It is not known, however, to what extent this inhibition is caused by 1), direct transinhibition by cytosolic SO4= binding to the transporter; 2), sensing of cytosolic SO4= or downstream metabolites; or 3), use-dependent inactivation of the kind recently described for Gap1p (24) and Fet3p/Ftr1p (23).
The data presented here show that addition of SO4= causes a more rapid and extensive downregulation of 35SO4= influx (mediated mainly by Sul2p) than has been observed previously in yeast and other fungi (4,28,33). Simultaneous measurement of 35SO4= influx and cellular SO4= contents under a variety of conditions indicate that downregulation of influx is not caused by mass action transinhibition or by sensing cellular SO4=. Instead, Sul2p exhibits what appears to be use-dependent inactivation, in which either binding or influx of extracellular SO4= increases the probability that the transporter will be inactivated and degraded. The data also show that use-dependent inactivation of Sul2p results in autoregulation of cellular SO4= contents.
In addition to use-dependent inactivation, a new mode of SO4= transport was observed: transient net efflux of SO4= following a sudden increase in extracellular [SO4=]. This transient efflux is not triggered by the size of the cellular SO4= load but instead appears to depend on transporter activity and may be an additional aspect of use-dependent inactivation. The transient efflux takes place even in the presence of relatively high extracellular [SO4=] and low extracellular pH, which would normally promote net SO4= influx. The transient efflux mode of Sul2p may be an additional mechanism to protect the cell from a SO4= overload.
Materials and Methods
Yeast
Haploid strains of S. cerevisiae from the Euroscarf deletion project were obtained from Invitrogen (Carlsbad, CA) or Open Biosystems (Thermo Fisher Scientific, Pittsburgh, PA). All strains are in the background of BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), which can metabolize SO4= only to HS− and not to homocysteine, methionine, etc (32).
Media
SO4=-free synthetic APG (arginine phosphate glucose) medium (34,35) consisted of 10 mM arginine free base, 8 mM H3PO4, 2 mM KH2PO4, 2 mM MgCl2, 0.2 mM CaCl2, NaOH to pH 6. In addition to arginine, the medium contained the amino acids, vitamins, and trace minerals (as Cl− rather than SO4= salts) in YNB (36), with double the normal amounts of leucine and histidine. The only sulfur added to the medium was 0.4 mM methionine. For transport experiments, media were buffered with 5 mM K+-citrate, pH 4.5.
Growth and sulfur starvation
Yeast were grown aerobically at 30°C with shaking at 225 RPM for 16 h in APG/methionine. For S-starvation, the medium was replaced by S-free APG for the times indicated in the figure legends.
RNA isolation and reverse transcription-polymerase chain reaction
RNA was isolated from 108 cells and treated with DNase 1 (Ambion; Austin, TX). RNA (1 μg) was reverse transcribed using BioRad reverse transcriptase (RNase H+) with oligo dT and random primers. Real time reverse transcription-polymerase chain reaction was performed with a BioRad iCycler using the following primers (IDT, Coralville, IA). SUL1 forward: TCGGGCTTAAATGAGGTGGGAGTT. SUL1 reverse: ATCCATGATCGAACGGAACCCACT. SUL2 forward: AAGGGAGAACGACCCTGAAT. SUL2 reverse: TGGCCTTTCTCAAATCAACC. Actin forward: GCCTTCTACGTTTCCATCCA. Actin reverse: GGCCAAATCGATTCTCAAAA.
Western blots
Rabbit polyclonal antibody was prepared and affinity purified by Proteintech Group (Chicago, IL) from a peptide near the N-terminal sequence of Sul2p (E4-V24; EGYPNFEEVEIPDFQETNNTV). Whole cell extracts were prepared by incubating cells for 5 min at room temperature in 0.1 N NaOH, centrifuging, and immediately suspending the pellet in hot 4% sodium dodecyl sulfate, 2% β-mercaptoethanol, and incubating 3 min at 100°C (37). Extracts were run on 5–12% gradient polyacrylamide gels using Laemmli (38) buffers. Protein was transferred (39) to PVDF, blocked overnight with 3% bovine serum albumin in phosphate buffered saline, 0.1% Tween 20, and incubated 6 h with 1:10,000 dilution of antibody, followed by 2 h in 1:100 dilution of peroxidase-conjugated secondary antibody (Sigma, St. Louis, MO). Blots were developed using Pierce ECL Plus reagents (Thermo-Fisher) and a FluorChem HD2 imager with AlphaView SA software (Protein Simple, Santa Clara, CA).
35SO4= influx
Transport activity was measured as initial influx of [35S] SO4= at 30°C in APG containing 0.1 mM SO4= unless otherwise specified. To measure initial tracer influx, 0.1–1 μCi [35S] Na2SO4 (Dupont-NEN, Boston, MA) was added per ml of suspension, and, after a specified time (usually 0.5 min), the suspension was filtered through ATTP filters (Millipore, Bedford, MA), washed with cold water, and counted. If no SO4= had been added to the suspension before tracer, 0.1 mM K2SO4 was added simultaneously with 35S. If K2SO4 had been added before the 35S, the tracer was supplemented with sufficient K2SO4 to bring the final extracellular concentration to 0.1 mM.
To measure cellular SO4= contents, K2SO4 + [35S] Na2SO4 was added at the concentration specified in the figure legends at t = 0. At the SO4= concentrations and cell densities that were used (see figure legends), sufficient SO4= was taken up by the cells that the cellular SO4= contents could be determined accurately by centrifuging aliquots and counting the supernatants. Cellular SO4= contents (μmol/ml cells) and initial 35SO4= influx (μmol/ml cells-min) were calculated from the cellular radioactivity (cellular CPM/ml suspension), cell density (ml cells/ml suspension), and specific activity (CPM/μmol SO4=).
The time course of the decline in 35SO4= transport activity following addition of K2SO4 (see Figs. 1–3) deviates slightly from a single exponential for the late time points. The deviation from a single exponential could have many causes, and there is no reason to expect such a complex process to have a single exponential time course. Nonetheless, to provide an estimate of the rate of transport inactivation as a single parameter, the rate constant (min−1) for inactivation was estimated by fitting the time course of decrease in 35SO4= influx to a single exponential (no additive term) over the first 15–25 min after addition of K2SO4. Curve fitting was with Sigmaplot Version 8, default settings.
Figure 1.
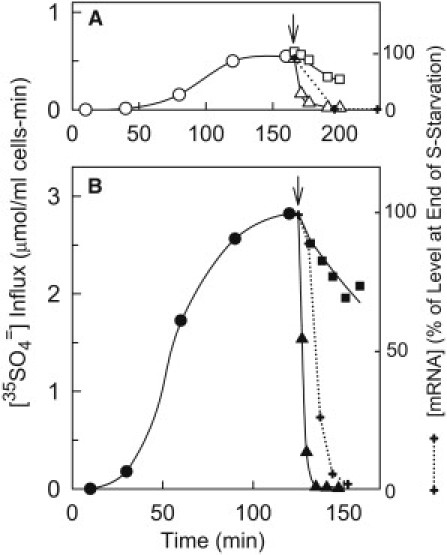
Effect of S-starvation and addition of methionine and SO4= on Sul1p (A) and Sul2p (B) function, measured as influx of 35SO4=. Cells of the sul2Δ (A) or sul1Δ (B) strains (BY4741 background) were grown overnight in APG medium containing 0.4 mM methionine and no other sulfur. The cells were centrifuged, resuspended in S-free APG at t = 0, and incubated aerobically at 30°C. Aliquots were withdrawn at the indicated times for measurement of initial influx of 35SO4= in APG buffered with citrate at pH 4.5 and containing 0.1 mM K2SO4 (○,●). At the arrows, 0.4 mM methionine was added to the suspensions, either alone (□,■) or with 0.1 mM K2SO4 (▵,▴), and aliquots were withdrawn for 35SO4= influx (0.1 mM SO4=) measurement at the indicated times. Additional experiments (one with sul2Δ and three with sul1Δ strain) confirm the time course of derepression during S-starvation and inactivation of transport after addition of SO4=. The dotted lines show the relative amounts of SUL1 (A) and SUL2 (B) mRNA following addition of methionine to S-starved cells.
Figure 2.
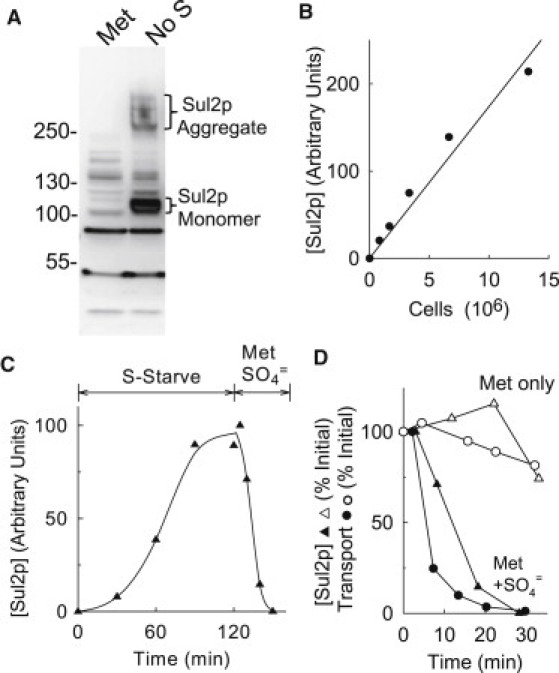
(A) Western blot of whole cell extracts of met3Δ cells grown in APG methionine followed by 2 h S-starvation (No S). Positions of Sul2p monomer and aggregate are indicated. (B) Total intensity of Sul2p monomer and aggregate bands as a function of loading for serial dilutions of an extract prepared as in the No S lane in A. (C) Time course of increase in total Sul2p in met3Δ cells during S-starvation in APG starting at t = 0 and after addition of 0.4 mM methionine and 0.1 mM K2SO4 at t = 121 min. (D) Time course of total cellular Sul2p (▴,▵) and transport activity (●,○) in met3Δ cells following addition of 0.4 mM methionine, either alone (▵,○) or with 0.1 mM K2SO4 (▴,●). Cells were grown in APG/methionine as in Fig. 1 and then resuspended in S-free medium for S-starvation. After 2 h, cells were resuspended at 30°C in fresh medium, and methionine with or without K2SO4 was added. Solid triangles represent the same data as in Fig. 2C.
Figure 3.
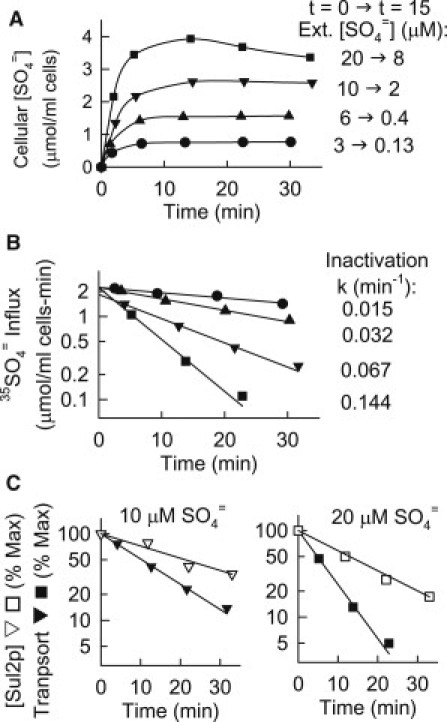
(A) Time course of accumulation of 35SO4= in met3Δ cells. At t = 0, after 2 h of S-starvation in S-free APG, 0.4 mM methionine plus the indicated concentration of [35S] K2SO4 were added, and the cellular 35S contents were determined by centrifugation and counting supernatants; cell density 0.30–0.35%. The initial and minimum extracellular [SO4=] (at t = 15 min) are indicated at the right. (B) Transport activity in the same cell preparations as in A, except that K2SO4 without 35SO4= was added at t = 0. At the indicated time, an aliquot was withdrawn for measurement of 0.5 min influx of 35SO4= at a total extracellular SO4= concentration of 0.1 mM. The medium for both accumulation and initial influx was APG buffered with 5 mM K-citrate at pH 4.5, 30°C. The rate constants for inactivation derived from each of the single exponential fits are listed at the right. (C) Time course of decrease in total Sul2p (▿,□) measured on Western blots of met3Δ cells that had been S-starved 2 h followed by addition of 0.4 mM methionine and 10μM or 20 μM K2SO4 as indicated at t = 0. For comparison, transport data for cells treated in the same way are replotted from B (▾,▪).
Results
Rapid downregulation of transport following addition of SO4= but not methionine to S-starved cells
Both SUL1 and SUL2 are repressed in growth medium containing >0.1 mM methionine (28,29). Following methionine removal (S-starvation), SUL2 is derepressed more rapidly than SUL1, as measured by 35SO4= influx (Fig. 1). After 2 h of S-starvation, the Sul2p-mediated influx (Fig. 1 B) is over 5 times larger than the Sul1p influx (Fig. 1 A), and 10,000-fold larger than the influx in cells that were not S-starved. Adding methionine to S-starved cells causes a relatively slow drop (t1/2 ∼50 min) in the influx mediated by both transporters. Under the same conditions, both SUL1 and SUL2 mRNA levels drop 50-fold in 30 min (Fig. 1, dotted lines). The half-time for the decrease in mRNA is ∼5 min, which is toward the short end of the range of yeast mRNA half-lives (40). Therefore, in the presence of methionine alone, both Sul1p and Sul2p continue to function in the plasma membrane long after the mRNA has been degraded; this finding is in agreement with early work by Breton and Surdin-Kerjan (28).
Addition of 0.1 mM K2SO4 with methionine to S-starved cells causes a rapid decrease in 35SO4= influx mediated by both Sul1p and Sul2p. Activity of Sul2p declines 100-fold in the 20 min following addition of K2SO4 (Fig. 1 B). Addition of K2SO4 alone, without methionine, causes rapid inactivation of Sul2p-mediated 35SO4= influx and no change in mRNA levels (data not shown). The decline in the flux is not quite as complete because continued transcription in the absence of methionine results in a steady state with low but detectable transport. The decline in Sul1p activity is slightly slower than Sul2p but still has a half-time of <5 min (Fig. 1 A).
SO4=-induced inactivation of transport and degradation of Sul2p in the absence of SO4= metabolism
To determine whether the rapid inactivation of SO4= transport following SO4= addition requires APS, PAPS, or other SO4= metabolites, further experiments were performed with the met3Δ strain, which lacks ATP sulfurylase and cannot convert SO4= to APS. To confirm the met3Δ phenotype, liquid chromatography (41) was used to demonstrate that APS and PAPS are not formed from SO4= in this strain (data not shown).
Western blots of whole cell extracts of met3Δ cells with Sul2p antibody show ∼100 kDa and ∼300 kDa bands that are induced by S-starvation (Fig. 2 A), along with nonspecific bands that are unaffected by sulfur status. The ∼100 kDa and ∼300 kDa bands are absent in the sul2Δ strain (data not shown), and we believe that the two bands represent respectively the Sul2p monomer (predicted Mr = 99,500) and a sodium dodecyl sulfate-stable aggregate of unknown composition. The sum of the intensities of the two bands is a reasonably linear function of loading for extracts of up to107 cells (Fig. 2 B); the sum of the intensities of the two bands was used as a comparative measure of total cellular Sul2p. Sul1p is more difficult to detect with the antibodies we have prepared, and we do not have quantitative information on the levels of Sul1p; transport under these conditions is mediated mainly (>80%) by Sul2p in any case (Fig. 1).
The time course of increase of total cellular Sul2p in met3Δ cells during S-starvation (Fig. 2 C) is indistinguishable from the time course of increase in Sul2p-mediated transport shown in Fig. 1 B. Addition of methionine causes a slow decrease in the levels of Sul2p and of transport (Fig. 2 D), indicating that in met3Δ cells as in sul1Δ cells, the Sul2p polypeptide is reasonably stable after transcription is repressed by methionine. Addition of methionine plus 0.1 mM K2SO4 causes rapid decay of both transport and Sul2p (Fig. 2, C and D). The time course of decrease in Sul2p appears to be slower than the decay of transport. The inactivation and degradation of Sul2p shown in Fig. 2 do not require a signal derived from formation of APS, PAPS, or other metabolite, because met3Δ cells do not make APS from SO4=. The remaining experiments described here use the met3Δ strain to examine SO4= itself as a potential signal for transport inactivation.
Rate of transport inactivation as a function of SO4= concentration
Fig. 3 shows that the rate of transport inactivation can be measured reasonably accurately under conditions in which total cellular SO4= contents are nearly constant. These data were obtained by measuring 35SO4= influx and cellular SO4= contents in parallel suspensions after adding methionine and K2SO4 to S-starved met3Δ cells. In one suspension, the K2SO4 was labeled with 35S, and the 35SO4= contents of the cells were measured at subsequent times (Fig. 3 A). In the other suspension, no 35S was added until transport activity (0.5 min influx; 0.1 mM [SO4=]) was measured on aliquots of suspension at the indicated times (Fig. 3 B).
The extent of SO4= uptake at low extracellular [SO4=] is remarkable. At an initial concentration of 3 μM, the S-starved cells take up over 95% of the [SO4=] (Fig. 3 A, solid circles), and the steady-state cellular SO4= content per ml cell water is over 5000 times that in the extracellular medium. As the SO4= concentration is raised, the proportion of SO4= taken up by the cells decreases (Fig. 3 A). At an initial extracellular [SO4=] of 20 μM, the cells take up about half the total, and the maximum cells/medium ratio is <500. A major reason for the lower cells/medium ratio is that transport activity progressively decreases with time, and the rate of decrease is higher at higher SO4= concentrations (Fig. 3 B). The rate of degradation of Sul2p also is an increasing function of the initial SO4= concentration in the medium. As was observed at 100 μM SO4=, the rate of degradation of Sul2p at initial [SO4=] of 10 μM or 20 μM is somewhat slower than the rate of transport inactivation under the same conditions (Fig. 3 C).
Lack of major sequestration of SO4=
Vesicles prepared from yeast vacuoles can concentrate SO4= (42), and it is quite possible that some of the cellular SO4= in the experiments presented here is sequestered in the vacuole. To estimate the extent of sequestration of SO4= into a slowly exchanging compartment, 10 μM 35SO4= was added to S-starved met3Δ cells, and pairs of aliquots were centrifuged at 10 min intervals to determine extracellular 35SO4= (Fig. 4 A). In one aliquot of each pair, 0.1 mM nonradioactive K2SO4 was added 1 min before centrifugation. In these aliquots there is a sizable efflux of 35SO4= as a consequence of exchange mediated by Sul2p. This 1 min exchange efflux decreases by ∼25% over a period of 45 min. In the same cell preparation, transport activity (measured as 0.5 min 35SO4= influx at an extracellular concentration of 0.1 mM; Fig. 4 B) decreases ∼10% over the time of the experiment.
Figure 4.
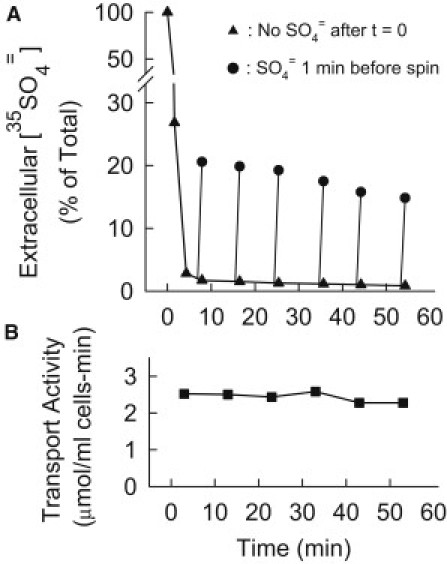
(A) Efflux of 35SO4= induced by addition of extracellular K2SO4 to 35SO4=-loaded met3Δ cells. Cells were grown and S-starved 2 h as in Fig. 3. At t = 0, 10 μM [35S] K2SO4 was added to a 1% suspension in APG, and aliquots were centrifuged at the indicated times and the 35S in the supernatant measured (▴). For aliquots represented by ●, 0.1 mM unlabeled K2SO4 was added 1 min before centrifugation. Addition of K2SO4 results in the 1 min loss of 20% of the cellular 35S for the first addition (t = 7 min) and 15% of the 35S for the last addition (t = 53 min). (B) Transport activity in the same cell preparations as in A, except that 10 μM unlabeled K2SO4 was added at t = 0. At the indicated times, aliquots were withdrawn for measurement of 0.5 min influx of 35SO4= (0.1 mM extracellular [SO4=]).
It should be noted that transport activity decreases more slowly in Fig. 4 than in Fig. 3, for two reasons. In Fig. 4, cell density is higher (less SO4= per cell) and no methionine was added; therefore new transporters were still being synthesized during the incubation. In a separate experiment, methionine was added with 10 μM K2SO4, and both the exchange efflux and transporter activity (measured as influx) declined by ∼60–70% in 50 min because of the lack of synthesis of new transporters. In this experiment as well as the one in Fig. 4 with no methionine, the similar decline in exchange efflux and transporter activity is consistent with a lack of major sequestration of cellular SO4= on this timescale.
Evidence against a feedback mechanism based on a cytosolic SO4= sensor
The time-dependent decrease in transport activity shown in Fig. 3 is not a result of transinhibition by cytosolic SO4=, because, over times in which cellular [SO4=] is very nearly constant (5–30 min), tracer influx continues to decline. The inactivation of transport also does not appear to be a consequence of negative feedback mechanism based on a sensor for cytosolic SO4=. Fig. 5 shows the rate constant (min−1) for transport inactivation plotted as a function of either the cellular or extracellular [SO4=] for experiments similar to (and including those) in Fig. 3. The solid circles are data from cells that were S-starved 2 h; the rate of inactivation for this subset of the data is a supralinear function of cellular SO4= (Fig. 5 A). However, for cells that had been S-starved only 40 min (with much lower poststarvation transport activity), a higher extracellular [SO4=] is needed to produce a given cellular [SO4=]. In these cells, transport inactivates rapidly even though total cellular [SO4=] is relatively low (Fig. 5 A, open triangles).
Figure 5.
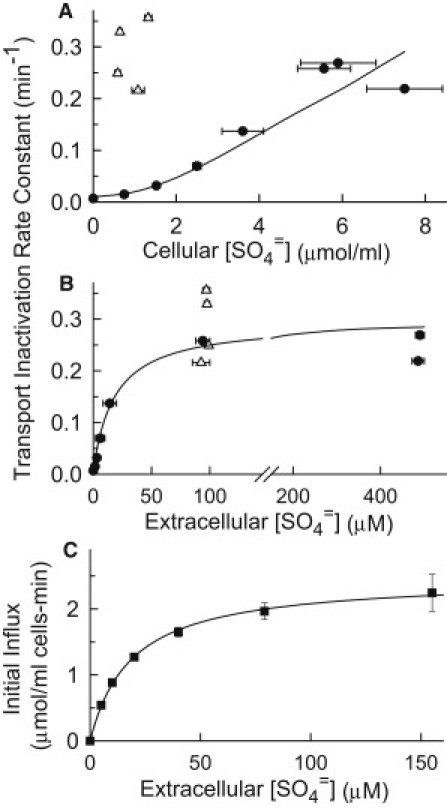
(A) Rate constant (min−1) for inactivation of 35SO4= influx, plotted as a function of the cellular SO4= contents (μmol/ml cells). Cells (met3Δ) were grown in APG/methionine and S-starved 40 min (▵) or 2 h (●). 0.4 mM methionine was then added with 0–500 μM K2SO4, and cellular SO4= contents and initial influx of 35SO4= were measured as in Fig. 3. The tracer influx was measured at the SO4= concentration in the incubation medium for concentrations of 100 or 500 μM. If the concentration in the incubation medium was lower, K2SO4 was added to the 35SO4= influx medium to make the total extracellular SO4= concentration 100 μM. (B) The transport inactivation data in A are plotted as a function of extracellular [SO4=]. (C) Initial 35SO4= influx as a function of extracellular [SO4=] concentration for met3Δ cells grown exactly as above and S-starved 2.5 h before influx measurement. The curves through the data in both B and C are rectangular hyperbolic (Michaelis-Menten) functions, with a Km of 19 μM.
The inactivation rate constants, plotted against extracellular SO4= (Fig. 5 B), can be fit reasonably well by a hyperbolic function with half-maximal [SO4=] of 19 μM, which is the Km for initial 35SO4= influx measured immediately after S-starvation under identical conditions (Fig. 5 C). These data show that the rate of inactivation of SO4= transport has the same dependence on extracellular [SO4=] as transport. This finding, and the fact that cellular SO4= is not the driver of transport inactivation, suggests that it is the transport process itself that causes the transporters to be inactivated.
Transient net SO4= efflux following acute loading
In SO4= accumulation experiments such as those in Fig. 3, we consistently observed that, at higher extracellular [SO4=], the cellular SO4= contents at the latest time points decline slightly. To investigate this efflux further, accumulation experiments were carried out, again with met3Δ cells, but for longer periods of time (Fig. 5). At 10 μM initial extracellular [SO4=] (Fig. 6 A), the cells take up 98% the SO4=, and a steady state lasts ∼2 h as Sul2p and Sul1p slowly inactivate. (The proportion of SO4= taken up in this experiment is higher than that in Fig. 3 A at the same initial concentration because the cell density is 1% rather than 0.3%.) After nearly all transporters have been inactivated, there is slow SO4= loss; later time points (t > 180 min; not shown) can be fit by a single exponential with rate constant 0.24/h. We believe that this rate represents passive efflux of SO4= across the yeast plasma membrane, driven mainly by the negative membrane potential.
Figure 6.
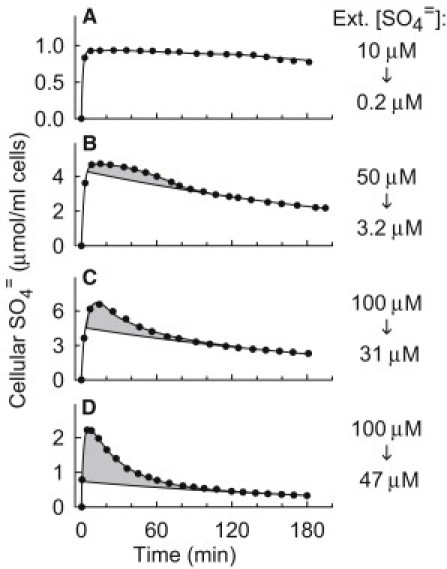
Transient efflux of SO4= after SO4= addition to S-starved met3Δ cells. Cells were grown in APG/methionine and S-starved 2 h (A–C) or 40 min (D) in S-free APG as in the earlier figures. Cells were then resuspended in fresh APG medium containing methionine to repress transcription, and 10 μM (A), 50 μM (B), or 100 μM (C and D) [35S]K2SO4 was added at t = 0. Cellular 35SO4= was measured by centrifuging aliquots and determining the radioactivity in the supernatant. Cell density was 1%. The curves below the shaded portions of the graphs represent single exponential functions with rate constant 0.22–0.26/h. The exponential fits of the transient effluxes are as follows. (B) k = 2.63/h (fit starting at t = 50 min); (C) k = 2.07/h (fit starting at t = 14 min); (D) k = 2.4/h (fit starting at t = 10 min).
At an initial extracellular [SO4=] of 50 μM (Fig. 6 B), the cells still take up over 90% of the SO4=. Transport inactivates more rapidly than in Fig. 6 A, because the extracellular SO4= concentration is higher. After transporter inactivation, there is an exponential loss of SO4=, again with a rate constant of ∼0.24/h. In the first hour, however, there is a slight transient loss of SO4= (shaded area of Fig. 6 B) that precedes the exponential loss. The transient loss is larger at higher SO4= concentration (Fig. 6 C), and the net loss takes place over times in which there is a strong driving force (with the inward H+ gradient) for influx. After the transient loss, the slow further loss of SO4= has the same time course as in Fig. 6 B. An even more pronounced transient efflux is observed in cells that had been S-starved only 40 min and have lower initial influx (Fig. 6 D). The transient efflux is not a result of cellular SO4= overload, because the maximum cellular [SO4=] in these cells is much lower than in Fig. 6, B or C, even though the transient efflux is considerably larger. After the transient efflux, the cells slowly lose remaining SO4= at the same rate as in the other experiments. The nature of the transient net efflux is discussed below.
Discussion
Use-dependent transporter inactivation versus sensors for cytosolic substrate
The original goal of this work was to determine whether yeast cells regulate SO4= influx by sensing cytosolic SO4=. Our data show that, contrary to what would be expected for a cytosolic SO4= sensor, the rate of inactivation of Sul2p depends on extracellular rather than cellular [SO4=] (Fig. 5). These data add to recent evidence for autoregulatory inactivation of transporters triggered by the transport process rather than the products of transport. The emphasis of the published work on use-dependent inactivation has been on the effects of mutations on trafficking and degradation of transporters (23–26). This work used a kinetic approach and is the first, to our knowledge, to show that the rate of transport inactivation has an extracellular substrate concentration dependence that is indistinguishable from that of influx, which is consistent with the idea that the rate of transporter inactivation is directly related to transport.
Use-dependent transport inactivation as autoregulatory mechanism
As pointed out by Kaplan and co-workers (23) in reference to Fet3p-Ftr1p, “Linking transport activity to degradation rate provides a simple feedback mechanism that ensures tight control of cytosolic metal levels…”. The data in Fig. 6 provide an experimental test of this idea. In Fig. 6 B, the maximum SO4= accumulation is ∼4.7 μmol/ml cells when extracellular [SO4=] is 3.2 μM. In Fig. 6 C, at a 10-fold higher extracellular [SO4=], the maximum accumulation is only 1.4-fold higher, indicating significant autoregulation of cellular SO4= contents. One of the reasons that net SO4= accumulation ceases in Fig. 6 C even though there is still a strong driving force for influx is that transport inactivates rapidly when the extracellular SO4= concentration is high (Fig. 5). By the time cellular SO4= has reached a maximum in Fig. 6 C, transport activity (measured as tracer influx) has decreased to ∼3% of what it was at t = 0, thereby limiting further influx. The inactivation of transport therefore autoregulates the accumulation of cellular SO4=, without the need for a sensing system for cytosolic SO4=.
A use-dependent mechanism for Sul2p downregulation is very well suited for rapid regulation. All that is required for downregulation is that an intermediate in the catalytic cycle is in a conformation that exposes the protein to an event (e.g., phosphorylation, ubiquitylation) that results in inactivation and degradation (24). In the case of Sul2p, the degradation signal is unknown, but Sul2p has a strong PEST sequence that could become exposed in the presence of extracellular SO4=, resulting in phosphorylation-induced degradation (43,44).
Timing of transport inactivation and Sul2p degradation
From comparison of blots and transport data (Figs. 2 and 3), it appears that inactivation of transport is more rapid than degradation of Sul2p, although both processes take place at all SO4= concentrations studied thus far (10–100 μM). Sul2p therefore may be regulated differently from Gap1p, in which inactivation at the plasma membrane and trafficking to the vacuole have different sensitivities to amino acid concentrations (25). Much more needs to be learned about trafficking of Sul2p before a more detailed comparison of its regulation with that of Gap1p can be made.
Why should a transporter have use-dependent inactivation?
It may seem inefficient for a cell to synthesize a transporter only to degrade it shortly after it starts performing its function. Why not produce fewer transporters and let them function longer? The answer to this question may have to do with the fact that toxins such as selenate and chromate are substrates for SO4= transporters (29,45). Placing an inherent limit on the number of turnovers of the catalytic cycle over the life of a SO4= transporter may be a mechanism for protecting the cell against excessive influx of toxic anions. Protection against iron toxicity may be the reason for a similar mechanism for Fet3p-Ftr1p (23).
Use it and lose it versus Use it or lose it
The mammalian serotonin transporter is phosphorylated and degraded when it is not transporting substrate; this regulation has been described as Use it or lose it (46,47). With yeast Sul2p, the regulation is in the opposite direction, i.e., Use it and lose it. However, both processes could be very similar mechanistically in that, in both cases, only certain conformational states of the transporter are targeted for degradation, and the proportion of transporters in the targeted state depends on functional status. For Use it or lose, the down-regulated conformation could be a form of the transporter that has no substrate bound, whereas for Sul2p, the degradable form requires the presence of extracellular SO4=.
Transient efflux mode of Sul2p
In addition to use-dependent transport inactivation, there is another mechanism to limit net SO4= accumulation: the transient SO4= efflux following SO4= addition to S-starved cells (Fig. 6). The overshoot in cellular SO4= (Fig. 6, C and D), cannot be explained by the fact that influx is progressively inactivating. Passive efflux under these conditions is very reproducible and has a rate constant of ∼0.24/h; the rate constant for transient efflux in Fig. 6 D is over 2/h. There is pronounced transient efflux in 40 min S-starved cells even though the total cellular SO4= is relatively low. Therefore, the transient efflux is not a cellular response to a high cellular SO4= load.
One possible mechanism for the transient efflux is that it represents an alternate transport mode. Yeast Sul2p cotransports H+ with SO4=, although the stoichiometry is not known with certainty. The transient SO4= efflux cannot be simply the reversal of normal (n)H+/SO4= cotransport, because there is still a strong driving force for influx during the transient net efflux. There is probably a transient cytosolic pH drop associated with H+/SO4= influx that would be expected to lower the driving force for further influx. However, in the 40 min S-starved cells (Fig. 5 D) the pH drop must be smaller because the influx is lower, and the transient efflux is nonetheless large, indicating that is not caused by a cytosolic pH drop. Therefore, the transient efflux, if it is mediated by Sul2p, represents a mode of transport with either altered H+/SO4= stoichiometry, (1:1 vs. 2:1 or 3:1) or an uncoupled channel-like mode. This possible alternate mode may represent an additional protection against excessive influx of SO4= (or chromate, selenate) beyond that provided by inactivation of the transporter.
It is also possible that, during the process of rapid internalization of transporters, the membrane is leakier than it had been before SO4= was added. The data in Fig. 6 indicate that the passive permeability of the yeast plasma membrane to SO4= is quite low. With such a low basal permeability, it is possible that increased endocytosis during Sul2p internalization could increase the outward leak. Therefore, even though the leak may be caused by Sul2p inactivation, it is not necessarily an alternate mode of the transporter.
Is there any true transinhibition of 35SO4= influx by cytosolic SO4=?
Early work on SO4= transport in S. cerevisiae (28), N. crassa (4), and P. notatum (33) provided evidence for transinhibition of SO4= influx by cytosolic SO4= binding either to the inward-facing transport site itself or to a separate inhibitory site. Because of the rapid time-dependent inactivation of influx under the conditions of our experiments, it is difficult to say whether there are acute effects of cytosolic SO4= on 35SO4= influx through functioning copies of Sul2p. There does not appear to be much transinhibition of 35SO4= influx for cellular [SO4=] <∼2 mM (2 μmol/ml). In Fig. 3, for example, with an initial extracellular SO4= concentration of 6 μM, the tracer influx is relatively high at t = 5–10 min even though cellular SO4= is ∼1.5 mM. That is, the tracer influx is not strongly affected by a sudden increase in cellular SO4= from near zero to 1.5 mM.
Even though there does not appear to be much transinhibition of tracer influx by cytosolic SO4=, there still could be significant inhibition of net SO4= influx by cytosolic SO4=. The extent of transinhibition of tracer versus net influx depends on the quantitative parameters in the catalytic cycle, i.e., binding affinities and translocation rates of loaded and empty transporters, none of which are known. It is therefore possible for Sul2p to exhibit no transinhibition of 35SO4= influx even though there is significant transinhibition of net flux. Accordingly, there may be three mechanisms for protecting cells against excessive SO4= influx: use-dependent transport inactivation; transient SO4= efflux triggered by sudden influx; and, possibly, direct transinhibition of net influx by cytosolic SO4=.
Conclusion
The yeast SO4= transporter Sul2p is very rapidly regulated for the apparent purpose of minimizing the chances of excessive influx of SO4= or a toxic homolog. Sul2p transport activity is downregulated as a consequence not of the accumulation of cellular SO4= but rather because of the transport process itself. The rate of inactivation has the same extracellular concentration dependence as the influx. In addition to use-dependent inactivation of Sul2p, a sudden influx of SO4= triggers a transient efflux of SO4= that may represent an alternative mode of Sul2p. If so, this system is an example of a transporter changing stoichiometries as an autoregulatory mechanism to prevent excessive substrate influx.
Acknowledgments
Vladimir Lupashin, Department of Physiology and Biophysics, University of Arkansas for Medical Sciences, provided valuable advice regarding yeast protocols. Discussions with Harel Weinstein (Weill Cornell College of Medicine) about mammalian cotransporter regulation mechanisms and with Clifford Slayman (Yale University School of Medicine) about yeast transport were also helpful.
This work was supported in part by National Institutes of Health grant R01 GM026861-26 to M.L.J.
Footnotes
Jian Cui is deceased.
References
- 1.Kelley D.S., Potter V.R. Regulation of amino acid transport systems by amino acid depletion and supplementation in monolayer cultures of rat hepatocytes. J. Biol. Chem. 1978;253:9009–9017. [PubMed] [Google Scholar]
- 2.Debernardi R., Magistretti P.J., Pellerin L. Trans-inhibition of glutamate transport prevents excitatory amino acid-induced glycolysis in astrocytes. Brain Res. 1999;850:39–46. doi: 10.1016/s0006-8993(99)02022-3. [DOI] [PubMed] [Google Scholar]
- 3.Devés R., Boyd C.A.R. Transporters for cationic amino acids in animal cells: discovery, structure, and function. Physiol. Rev. 1998;78:487–545. doi: 10.1152/physrev.1998.78.2.487. [DOI] [PubMed] [Google Scholar]
- 4.Marzluf G.A. Regulation of sulfate transport in neurospora by transinhibition and by inositol depletion. Arch. Biochem. Biophys. 1973;156:244–254. doi: 10.1016/0003-9861(73)90362-7. [DOI] [PubMed] [Google Scholar]
- 5.Hunter D.R., Segel I.H. Control of the general amino acid permease of Penicillium chrysogenum by transinhibition and turnover. Arch. Biochem. Biophys. 1973;154:387–399. doi: 10.1016/0003-9861(73)90071-4. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Sinani D., Lee J. Copper transport activity of yeast Ctr1 is down-regulated via its C-terminus in response to excess copper. J. Biol. Chem. 2009;284:4112–4122. doi: 10.1074/jbc.M807909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadner R.J. Regulation of methionine transport activity in Escherichia coli. J. Bacteriol. 1975;122:110–119. doi: 10.1128/jb.122.1.110-119.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verheul A., Glaasker E., Abee T. Betaine and L-carnitine transport by Listeria monocytogenes Scott A in response to osmotic signals. J. Bacteriol. 1997;179:6979–6985. doi: 10.1128/jb.179.22.6979-6985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber S., Comellas-Bigler M., Locher K.P. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 2008;321:246–250. doi: 10.1126/science.1156213. [DOI] [PubMed] [Google Scholar]
- 10.Parcej D., Tampé R. Solute-binding sites in ABC transporters for recognition, occlusion and trans-inhibition. Chem. Med. Chem. 2009;4:25–28. doi: 10.1002/cmdc.200800328. [DOI] [PubMed] [Google Scholar]
- 11.Liu X.F., Culotta V.C. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J. Biol. Chem. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- 12.Jensen L.T., Carroll M.C., Culotta V.C. Down-regulation of a manganese transporter in the face of metal toxicity. Mol. Biol. Cell. 2009;20:2810–2819. doi: 10.1091/mbc.E08-10-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portnoy M.E., Jensen L.T., Culotta V.C. The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem. J. 2002;362:119–124. doi: 10.1042/0264-6021:3620119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Sitaram A., Burd C.G. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic. 2007;8:1375–1384. doi: 10.1111/j.1600-0854.2007.00616.x. [DOI] [PubMed] [Google Scholar]
- 15.Gitan R.S., Luo H., Eide D. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 1998;273:28617–28624. doi: 10.1074/jbc.273.44.28617. [DOI] [PubMed] [Google Scholar]
- 16.Gitan R.S., Eide D.J. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 2000;346:329–336. [PMC free article] [PubMed] [Google Scholar]
- 17.Blondel M.-O., Morvan J., Volland C. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell. 2004;15:883–895. doi: 10.1091/mbc.E03-04-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Séron K., Blondel M.-O., Volland C. Uracil-induced down-regulation of the yeast uracil permease. J. Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanbrough M., Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen E.J., Kaiser C.A. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:14837–14842. doi: 10.1073/pnas.232591899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauwers E., Erpapazoglou Z., André B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Kaouass M., Gamache I., Poulin R. The spermidine transport system is regulated by ligand inactivation, endocytosis, and by the Npr1p Ser/Thr protein kinase in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:2109–2117. doi: 10.1074/jbc.273.4.2109. [DOI] [PubMed] [Google Scholar]
- 23.Felice M.R., De Domenico I., Kaplan J. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 2005;280:22181–22190. doi: 10.1074/jbc.M414663200. [DOI] [PubMed] [Google Scholar]
- 24.Cain N.E., Kaiser C.A. Transport activity-dependent intracellular sorting of the yeast general amino acid permease. Mol. Biol. Cell. 2011;22:1919–1929. doi: 10.1091/mbc.E10-10-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risinger A.L., Cain N.E., Kaiser C.A. Activity-dependent reversible inactivation of the general amino acid permease. Mol. Biol. Cell. 2006;17:4411–4419. doi: 10.1091/mbc.E06-06-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gournas C., Amillis S., Diallinas G. Transport-dependent endocytosis and turnover of a uric acid-xanthine permease. Mol. Microbiol. 2010;75:246–260. doi: 10.1111/j.1365-2958.2009.06997.x. [DOI] [PubMed] [Google Scholar]
- 27.Lanquar V., Loqué D., Frommer W.B. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell. 2009;21:3610–3622. doi: 10.1105/tpc.109.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breton A., Surdin-Kerjan Y. Sulfate uptake in Saccharomyces cerevisiae: biochemical and genetic study. J. Bacteriol. 1977;132:224–232. doi: 10.1128/jb.132.1.224-232.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherest H., Davidian J.C., Surdin-Kerjan Y. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics. 1997;145:627–635. doi: 10.1093/genetics/145.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith F.W., Hawkesford M.J., Clarkson D.T. Isolation of a cDNA from Saccharomyces cerevisiae that encodes a high affinity sulphate transporter at the plasma membrane. Mol. Gen. Genet. 1995;247:709–715. doi: 10.1007/BF00290402. [DOI] [PubMed] [Google Scholar]
- 31.Khurana O.K., Coupland L.A., Howitt S.M. Homologous mutations in two diverse sulphate transporters have similar effects. FEBS Lett. 2000;477:118–122. doi: 10.1016/s0014-5793(00)01783-x. [DOI] [PubMed] [Google Scholar]
- 32.Thomas D., Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuppoletti J., Segel I.H. Transinhibition kinetics of the sulfate transport system of Penicillium notatum: analysis based on an iso uni uni velocity equation. J. Membr. Biol. 1974;17:239–252. doi: 10.1007/BF01870185. [DOI] [PubMed] [Google Scholar]
- 34.Nass R., Cunningham K.W., Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J. Biol. Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Navarro A., Ramos J. Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 37.Kushnirov V.V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caponigro G., Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong K.O., Wong K.P. Direct measurement and regulation of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) generation in vitro. Biochem. Pharmacol. 1996;52:1187–1194. doi: 10.1016/0006-2952(96)00461-3. [DOI] [PubMed] [Google Scholar]
- 42.Hirata T., Wada Y., Futai M. Sodium and sulfate ion transport in yeast vacuoles. J. Biochem. 2002;131:261–265. doi: 10.1093/oxfordjournals.jbchem.a003097. [DOI] [PubMed] [Google Scholar]
- 43.Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- 44.Rechsteiner M., Rogers S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 45.Marzluf G.A. Genetic and metabolic controls for sulfate metabolism in Neurospora crassa: isolation and study of chromate-resistant and sulfate transport-negative mutants. J. Bacteriol. 1970;102:716–721. doi: 10.1128/jb.102.3.716-721.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramamoorthy S., Blakely R.D. Phosphorylation and sequestration of serotonin transporters differentially modulated by psychostimulants. Science. 1999;285:763–766. doi: 10.1126/science.285.5428.763. [DOI] [PubMed] [Google Scholar]
- 47.Steiner J.A., Carneiro A.M.D., Blakely R.D. Going with the flow: trafficking-dependent and -independent regulation of serotonin transport. Traffic. 2008;9:1393–1402. doi: 10.1111/j.1600-0854.2008.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


