Abstract
We demonstrate an in situ transmission electron microscopy technique for imaging proteins in liquid water at room temperature. Liquid samples are loaded into a microfabricated environmental cell that isolates the sample from the vacuum with thin silicon nitride windows. We show that electron micrographs of acrosomal bundles in water are similar to bundles imaged in ice, and we determined the resolution to be at least 2.7 nm at doses of ∼35 e/Å2. The resolution was limited by the thickness of the window and radiation damage. Surprisingly, we observed a smaller fall-off in the intensity of reflections in room-temperature water than in 98 K ice. Thus, our technique extends imaging of unstained and unlabeled macromolecular assemblies in water from the resolution of the light microscope to the nanometer resolution of the electron microscope. Our results suggest that real-time imaging of protein dynamics is conceptually feasible.
“The colder the better” is the common wisdom in structural biology, and thus data are conventionally collected at cryogenic temperatures for x-ray crystallography and transmission electron microscopy (TEM). Several decades of research have shown that radiation damage is reduced significantly at liquid nitrogen (1) or helium (2) temperatures. Typically, a high-resolution structure is degraded through bond breakage and rearrangements, as well as interactions with highly reactive free radicals from the surrounding solvent (3). It is believed these effects are minimized in cryoelectron microscopy by preserving structure in vitreous ice of either cells by high-pressure freezing or molecules by plunge freezing (4). However, one consequence is that structures are immobile and the mechanisms underlying function are inferred from static states. Although dynamic processes such as assembly and disassembly of proteins, and in vitro motility of motor proteins are easily detected at the single-molecule level by high-resolution fluorescence light microscopy, the molecular structure of proteins can only be achieved at nanometer resolution by EM, NMR, or x-ray methods.
Forty years after Matricardi et al.'s (5) report on the electron diffraction patterns of hydrated flat catalase crystals in the presence of water vapor, little progress has been made in the direct imaging of proteins in liquids. Recently developed techniques allow biological imaging of micron-scale objects or labeled specimens by scanning EM (SEM) and TEM in liquid (6–8). However, the direct imaging of nanometer-sized proteins has not been achieved yet. Here, we show that protein structures can be directly imaged in liquid water at ambient temperature by TEM, where radiation damage to protein structure in water is unexpectedly less significant than that observed at 98 K. To study proteins in their native environment, we adapt methods from material sciences for studying nanoparticle growth in solution at room temperature using a liquid environmental cell operating in a 120 keV transmission electron microscope (Tecnai T12; FEI, Hillsboro, OR) equipped with a 4096×4096 pixel camera (Ultrascan; Gatan, Warrendale, PA) (9) (Supporting Material). Other groups used a similar approach to image whole cells in liquid water by scanning transmission electron microscopy (STEM), which visualizes gold nanoparticle labels to provide structural information about cells with thicknesses of a few micrometers (7). In this work, we focus on the direct imaging of protein structures in liquid water by TEM without the use of labels. The technique involves a liquid cell that is microfabricated from silicon and has an electron translucent silicon nitride (Si3N4) membrane window with lateral dimensions of ∼3 × 50 μm and a thickness of only 10 nm (Fig. 1 A). The use of ultrathin (∼10 nm) Si3N4 membranes for developing liquid environmental cells has proven to be critical for direct TEM imaging of protein structures in liquid water. The protein solution is loaded into the liquid cell and forms a thin liquid film (80–300 nm depending on the thickness of the indium spacer) between the silicon nitride membranes (Supporting Material). In the electron microscope, the electron beam penetrates through the top and bottom Si3N4 membranes and the enclosed thin layer of liquid sample.
Figure 1.
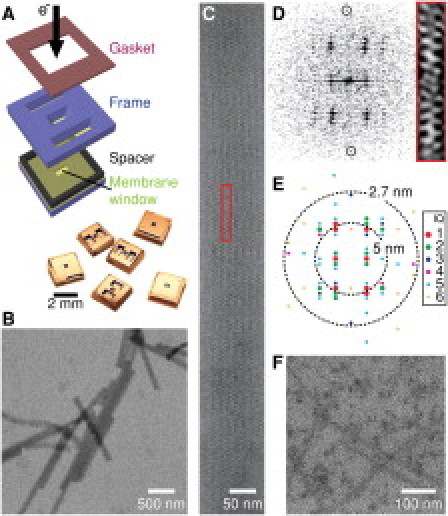
Imaging proteins in a liquid cell. (A) The liquid cell is assembled and a protein solution is loaded through two large reservoirs that are then sealed by a gasket. The protein solution is drawn by capillary force into the liquid cell and forms a thin film between two 10-nm-thick Si3N4 membranes. (B) Low-magnification image of 80-nm-diameter acrosomal bundles in liquid. (C) An acrosomal bundle in h0l orientation. The unit cell is boxed. (D) Fourier transform pattern from a portion of the image of the acrosomal bundle in C. The meridional reflection at 2.7 nm is circled. Red box: Unit cell reconstructed from reflections with S/N ratio > 1.2. (E) IQ plot of the Fourier transform and the resolution shells at 5 and 2.7 nm. (F) MAP-assembled microtubules show rod-shaped structures (enhanced contrast), whereas the protofilament substructure is not resolved.
We imaged an acrosomal process (a crystalline bundle of actin filaments) in liquid water by TEM, because its structure can be preserved to >7 Å in vitreous ice under a transmission electron microscope (10), and we previously characterized its radiation hardiness extensively at liquid nitrogen temperatures (11). Initially, we were unable to detect the acrosomes in liquid cells with a 0.5- to 1.0-μm gap and 50- to 100-nm-thick windows due to scatter and absorption from water and Si3N4. However, when the thicknesses of water and membranes were reduced, we were able to clearly detect the acrosomes and identify the characteristic view along the h0l projection with a = b = 147 Å, c = 762 Å (10,11). Fig. 1, B and C, show the familiar patterns of bands and stripes and well-defined zigzag features from actin subunits in the filaments (10). Compared with images in vitreous ice, the image contrast of the acrosomes in liquid water is lower because of the additional thickness of the Si3N4 windows.
To assess the quality and resolution of the images, we measured the intensity of reflections from their Fourier transforms (Fig. 1 D), displayed by an IQ plot (11,12) (Fig. 1 E). The IQ values shown in Fig. 1 range from 1 (signal/noise (S/N) ratio ≥ 7) to 6 (S/N ratio > 1.1) (12). The highest-resolution reflection (S/N ratio > 2) detected is the 2.7-nm meridional arising from the helical rise of the actin subunits in filaments. Thus, at this resolution, when imaging at 120 keV, actin structure is qualitatively preserved to a similar extent in room-temperature water as in ice. Similarly, MAP-stabilized microtubules (diameter: 25 nm) are also imaged in 80-nm-thick water (Fig. 1 F). However, we are unable to see the protofilament substructures, which are visible at higher magnifications in ice. These images demonstrate the feasibility of imaging proteins in water, but immediately show that resolution is limited.
One major limit to resolution is the significant scattering from the window that reduces the S/N ratio (Si3N4: Z = 10.6; water: Z = 4.8). As the membrane thickness increases, we find that the contrast and resolution of the acrosome images degrade. Here, we define the image resolution to be the largest extent of the Fourier transform of the image with amplitude with S/N ratio > 2. Thus, we obtained resolutions of 2.7 nm, 5.1 nm, and 12.4 nm corresponding to membrane thicknesses of 10 nm, 14 nm, and 25 nm, respectively (with the same water thickness of ∼300 nm; Supporting Material).
A second major limit to resolution is radiation damage caused by high-energy electrons. Radiation damage to a structure is first revealed by degradation or alteration of the highest-resolution features, which is more pronounced for organic polymers and biological specimens than for inorganic materials (13). At very high electron doses, the acrosomes in ice develop bubbles, whereas in water the structure dries and darkens (Supporting Material). For periodic structures and at lower electron doses, a more quantitative measure of radiation damage is defined by the decay of the Fourier peak amplitude. Our previous studies on imaging of acrosomal bundles indicate that the tolerable dose (D1/e), at which the amplitude of Fourier peaks (A/A0) drops by a factor of e, is ∼25 e/Å2 under 400 keV electron beam and at cryogenic temperature (11). It is noted that the tolerable electron doses for imaging powder samples and gelatin-encapsulated samples are several times lower at room temperature than in cryogenic conditions (1). To assess the extent of structural damage of the bundle in liquid water at room temperature, we measure the fall-off in intensity of reflection as a function of electron dose as the same acrosome is sequentially imaged. The amplitudes are normalized and arranged into two groups, (i.e., 27–50 Å and >50 Å) according to their reciprocal lattice sizes, and are plotted as a function of the electron dose (Fig. 2; also see Supporting Material). The plots are fitted to a simple exponential decay function for each case: A/A0 = exp[−(D − D0)/D1/e], where D0 is the electron dose delivered for the first image. For acrosomes imaged in ice at 120 keV, D1/e(98 K) = 31 ± 3 e/Å2 for 27–50 Å and D1/e(98 K) = 68 ± 4 e/Å2 for >50 Å. However, when imaging in liquid water, we find that contrary to expectations, the radiation damage is less in water than in ice: D1/e(293 K) = 35 ± 2 e/Å2 for 27–50 Å and D1/e(293 K) = 110 ± 5 e/Å2 for >50 Å. Because of the limit in resolution from the Si3N4 window, we are currently unable to measure radiation damage on the high-resolution features of the acrosome structure. Thus, it is not feasible to directly compare the radiation damage with our previous measurements for acrosomes imaged at 400 keV.
Figure 2.
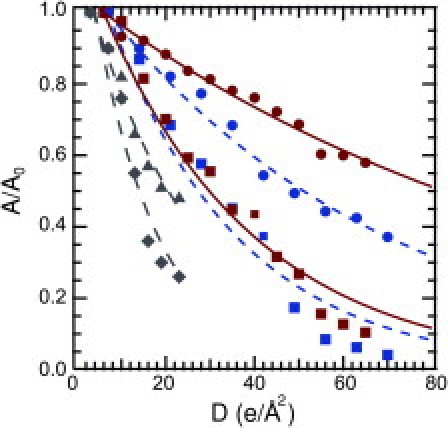
Radiation damage. A fall-off in resolution is shown at 293 K (aqueous) for resolution shells of 27–50 Å (red square) and >50 Å (red circle), 98 K (cryo) for resolution shells of 27–50 Å (blue square), and >50 Å (blue circle) at 120 keV for resolution shells of 27–50 Å and >50 Å (Fig. 1E), compared with damage at 400 keV and at 98 K for resolution shells of 22–37 Å (gray diamond) and >37 Å (gray triangle; from Schmid et al. (11). Experimental data are fitted (lines) to exponential decay.
Previous studies reported increased radiation damage to dried or glucose-embedded biological specimens at room versus cryogenic temperatures (1,4,14). The similarity between water and ice in tolerable electron dose for imaging proteins is contrary to the previous expectation. We suggest the difference lies in the mechanisms of radiation damage in water versus in ice caused by radicals generated during electron irradiation. Free radicals have slower reaction rates (15) and diffuse faster in 293 K water than in 98 K ice (16,17) (see Supporting Material). Additionally, specimen movements seen in ice are caused by two mechanisms: charging and gaseous hydrogen buildup induced during radiolysis of water inside the ice (18). These movements should be nonexistent in liquid water. The combination of the slower reaction rate, mobility of free radicals, and absence of distortion in water may account for the comparable tolerable dose of electrons. Another mechanism of damage is covalent bond breakage from inelastic scattering of electrons. However, this damage should be equivalent in aqueous and frozen water because the scattering cross section is determined by the atomic composition of protein and beam parameters. Thus, to our knowledge, our study provides the first direct comparison of radiation damage in water at room temperature versus in ice at liquid nitrogen temperature for biological TEM.
Our results illustrate that unlabeled protein structures can be imaged directly in water with a resolution of at least 2.7 nm, and the radiation tolerance of an acrosomal bundle is higher in liquid water than in frozen ice. Our ability to directly image proteins in water suggests that it may be possible to study protein dynamics (e.g., the assembly/disassembly or translocation of proteins) under physiologically relevant aqueous conditions with nanometer resolution. For decades, investigators have used biological labels to study these events with fluorescence light microscopy methods. This study provides the groundwork for future nanometer-scale dynamic imaging without labels, and opens what to our knowledge is a new avenue for biological TEM.
Acknowledgments
We thank Robert Glaeser and Wah Chiu for helpful discussions on radiation damage and Les Wilson for the gift of microtubules. H.Z. thanks the support from Laboratory Directed Research and Development of Lawrence Berkeley National Laboratory.
Supporting Material
References and Footnotes
- 1.Hayward S.B., Glaeser R.M. Radiation damage of purple membrane at low temperature. Ultramicroscopy. 1979;4:201–210. doi: 10.1016/s0304-3991(79)90211-0. [DOI] [PubMed] [Google Scholar]
- 2.Chiu W., Knapek E., Dietrick I. Electron radiation damage of a thin protein crystal at 4K. Ultramicroscopy. 1981;6:291–295. [Google Scholar]
- 3.Glaeser R.M. Retrospective: radiation damage and its associated “information limitations”. J. Struct. Biol. 2008;163:271–276. doi: 10.1016/j.jsb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Dubochet J., Adrian M., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 5.Matricardi V.R., Moretz R.C., Parson D.F. Electron diffraction of wet proteins: catalase. Science. 1972;21:268–270. doi: 10.1126/science.177.4045.268. [DOI] [PubMed] [Google Scholar]
- 6.de Jonge N., Ross F.M. Electron microscopy of specimens in liquid. Nat. Nanotechnol. 2011;6:695–704. doi: 10.1038/nnano.2011.161. [DOI] [PubMed] [Google Scholar]
- 7.de Jonge N., Peckys D.B., Piston D.W. Electron microscopy of whole cells in liquid with nanometer resolution. Proc. Natl. Acad. Sci. USA. 2009;106:2159–2164. doi: 10.1073/pnas.0809567106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiberge S., Nechushtan A., Moses E. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc. Natl. Acad. Sci. USA. 2004;101:3346–3351. doi: 10.1073/pnas.0400088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng H.R., Smith R.K., Alivisatos A.P. Observation of single colloidal platinum nanocrystal growth trajectories. Science. 2009;324:1309–1312. doi: 10.1126/science.1172104. [DOI] [PubMed] [Google Scholar]
- 10.Schmid M.F., Jakana J., Chiu W. Imaging frozen, hydrated acrosomal bundle from Limulus sperm at 7 A resolution with a 400 kV electron cryomicroscope. J. Mol. Biol. 1993;230:384–386. doi: 10.1006/jmbi.1993.1155. [DOI] [PubMed] [Google Scholar]
- 11.Schmid M.F., Jakana J., Chiu W. Effects of radiation damage with 400-kV electrons on frozen, hydrated actin bundles. J. Struct. Biol. 1992;108:62–68. doi: 10.1016/1047-8477(92)90007-w. [DOI] [PubMed] [Google Scholar]
- 12.Henderson R., Baldwin J.M., Zemlin F. Structure of purple membrane from Halobacterium halobium: recording, measurement, and evaluation of electron micrographs at 3.5Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- 13.Spence J.C.H. 3rd ed. Oxford University Press; New York: 2003. High-Resolution Electron Microscopy (Monographs on the Physics and Chemistry of Materials) [Google Scholar]
- 14.Stark H., Zemlin F., Boettcher C. Electron radiation damage to protein crystals of bacteriorhodopsin at different temperatures. Ultramicroscopy. 1996;63:75–79. [Google Scholar]
- 15.Vöhringer-Martinez E., Hansmann B., Abel B. Water catalysis of a radical-molecule gas-phase reaction. Science. 2007;315:497–501. doi: 10.1126/science.1134494. Erratum in Science 2007;316:691. [DOI] [PubMed] [Google Scholar]
- 16.Patten F., Gordy W. Temperature effects on free radical formation and electron migration in irradiated proteins. Proc. Natl. Acad. Sci. USA. 1960;46:1137–1144. doi: 10.1073/pnas.46.8.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Symons M.C.R. Radiation processes in frozen aqueous systems. Ultramicroscopy. 1982;10:97–103. [Google Scholar]
- 18.Meents A., Gutmann S., Schulze-Briese C. Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc. Natl. Acad. Sci. USA. 2010;107:1094–1099. doi: 10.1073/pnas.0905481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


