Abstract
The molecular conformation of proteins is sensitive to the nature of the aqueous environment. In particular, the presence of ions can stabilize or destabilize (denature) protein secondary structure. The underlying mechanisms of these actions are still not fully understood. Here, we combine circular dichroism (CD), single-molecule Förster resonance energy transfer, and atomistic computer simulations to elucidate salt-specific effects on the structure of three peptides with large α-helical propensity. CD indicates a complex ion-specific destabilization of the α-helix that can be rationalized by using a single salt-free computer simulation in combination with the recently introduced scheme of ion-partitioning between nonpolar and polar peptide surfaces. Simulations including salt provide a molecular underpinning of this partitioning concept. Furthermore, our single-molecule Förster resonance energy transfer measurements reveal highly compressed peptide conformations in molar concentrations of NaClO4 in contrast to strong swelling in the presence of GdmCl. The compacted states observed in the presence of NaClO4 originate from a tight ion-backbone network that leads to a highly heterogeneous secondary structure distribution and an overall lower α-helical content that would be estimated from CD. Thus, NaClO4 denatures by inducing a molten globule-like structure that seems completely off-pathway between a fully folded helix and a coil state.
Introduction
Since Franz Hofmeister quantified the effectiveness of salts on the precipitation of proteins in 1888 (1), a coherent understanding of ion-specific (Hofmeister) effects in biochemical processes remains a challenging task (2–9). Salt effects are in particular important for the characterization of intermediate states during the process of protein folding. For instance, salts such as NaSO4 or KF typically stabilize native protein conformations, whereas GdmCl is a widely used unfolding agent whose molecular mechanism of action is still being debated (10). Other ions, such as ClO4−, I−, or SCN− are known to promote denatured states. The structural character of these denatured states, however, remains obscure and is often loosely interpreted as compact dry molten globule (11–14). Due to the complexity of competing nonpolar and polar solvation at biological surfaces, the detailed molecular understanding of the action of those salts on protein structures is still a subject of intense investigation (9).
Pegram and Record (8) recently proposed a framework for predicting the thermodynamics of Hofmeister effects based on partition coefficients, Ki, characterizing the distribution of specific ions between the surface of biomolecular compounds and bulk water. The Ki describe the binding beyond the (nonspecific) Coulomb interaction and have been coarsely separated into nonpolar “np” (hydrocarbon), polar “p” (amide), and ester oxygen “eo” components. The protein unfolding event changes the balance between polar versus nonpolar solvent accessible surface area (ASA) determining the accumulation or exclusion of ions at the surface. The measured Ki suggest that most cations are accumulated at the amide group while anions, apart from ClO4−, are excluded (8). Gdm+ is argued not to be excluded from nonpolar surfaces and its high accumulation at the amide group salts-in the backbone, i.e., drags it into solution. In contrast, NaSO4 or KF are preferentially excluded from protein surfaces leading to salting-out effects, i.e., precipitation and compaction (15). The partitioning concept, under the assumption that cation and anion effects are independent and additive (8), seems to successfully predict the Hofmeister effects on the protein stability of the lac repressor as well as DNA duplex formation/melting (16).
Unfortunately, local ion partitioning over the folded or unfolded protein surface cannot be measured directly. Typically, ion binding and structural characteristics must be indirectly inferred from spectral measurements such as NMR, circular dichroism (CD), or adsorption isotherms. In particular, CD is a standard technique to estimate the content of secondary structure within a polypeptide by analyzing specific spectral signatures (17). Another increasingly popular method to characterize the unfolded states of proteins is single-molecule Förster resonance energy transfer (smFRET) (18). Results for GdmCl, for instance, support the prevailing idea that unfolded proteins have the properties of random coil-like polymers (18–20). This does not necessarily imply that strong direct ion-protein interactions lead to swelled coil-like unfolding with vanishing secondary structure. Hence, it is unclear to what extent other strong denaturing salts such as NaClO4 unfold by swelling.
Here, we combine CD and atomistic computer simulations to elucidate the ion-specific effect of various salts (KF, KCl, NaCl, NaClO4, and GdmCl) on the structure of three model peptides: the neutral, salt bridge-supported [A(EAAAK)2A] peptide, and two net-charged peptides [(AE)6 and (AK)6]. In addition, smFRET experiments as a function of salt concentration were performed for various salts on a positively charged (AK)14 peptide. We demonstrate that the complex ion-specific action indicated by CD can be rationalized by a combination of salt-free simulations and the ion partitioning concept. Further molecular dynamics (MD) simulations including salt give molecular insight into the binding mechanism. At the same time, MD and Förster resonance energy transfer (FRET) experiments strongly indicate that these denatured states may exhibit a strong structural diversity (extended versus compact) specified by the nature of the salt.
Materials and Methods
Peptide synthesis and purification
All peptides (A(EAAAK)2A, (AE)6, (AK)6, and (AK)14C) were synthesized using standard Fmoc chemistry with N-terminal acetylation and C-terminal amidation. Peptide purification was done by C8 reverse phase HPLC and the sequence was confirmed by electrospray mass spectroscopy. Labeling was done in solution with atto488 succinimidyl esther and atto647N maleimide fluorophores. Labeled peptides were purified via anion exchange chromatography followed by liquid condensed/lung surfactant mass spectrometry. Details of CD spectroscopy are in the Supporting Material.
MD computer simulations
MD simulations were performed as described previously for brute-force (21) and replica-exchange MD (21). Full details of methods and analysis are in the Supporting Material.
Single-molecule fluorescence spectroscopy
Single-molecule FRET experiments with pulsed interleaved excitation (23,24) were carried out on a home-built confocal multiparameter fluorescence detection setup as described elsewhere (25) and the Supporting Material). Full details of analysis and calculation of RDA based on MD-derived structures are in the Supporting Material.
Results and Discussion
To investigate the effect of added salts (KF, KCl, NaCl, NaClO4, and GdmCl) on the secondary structure of model α-helices (a salt-bridge mediated A(EAAAK)2A and two net-charged [(AE)6 and (AK)6]), we first started by performing CD measurements.
Ion-specific destabilization of a salt-bridge supported α-helix
Fig. 1, A and B, show the CD-derived helicity of the salt-bridge supported EK-peptide. Without salt at T = 273 K, the peptide features 45% α-helicity compared to the longer version of the peptide (17 residues) with 70% helicity (26). The helicity follows a typical temperature denaturation curve with only 18% helicity remaining at 330 K. At a fixed temperature T = 273 K (Fig. 1 B and Fig. S2), all salts increasingly destabilized the secondary structure of the peptide for concentrations c > 0.5M, where screening of nonspecific electrostatic interactions, such as helix dipole-ion interactions (27,28), is basically completed. Destabilization is highly specific in the order NaClO4 ∼ KF < KCl < NaCl < GdmCl. Judging from the CD spectra, KF and NaClO4 are similarly weak destabilizers. NaCl has a considerably stronger effect than KCl while GdmCl is the strongest destabilizer. The observed trends are independent of T (Fig. S2), whereas the loss of helicity with NaCl is fairly independent of peptide length (Fig. S3).
Figure 1.
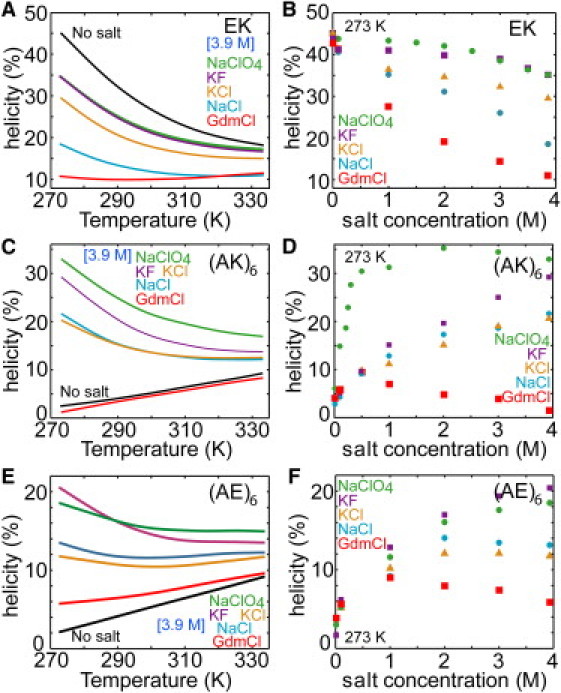
Salt-specific effects on α-helical secondary structure measured by CD. The thermal melting curves and effect of salt concentration at 273 K on the α-helicity of (A and B) EK, (C and D) (AK)6, and (E and F) (AE)6 peptides in 3.9 M concentration are plotted for various salts. Helicity was calculated from the CD signal at 222 nm (see Methods in the Supporting Material).
Ion-specific stabilization of net-charged α-helices
In contrast to the EK-peptide, the (AK)6 and (AE)6 peptides carry a net charge of +6 and −6 elementary charges, respectively. Thus, in the case of no salt, compact helical states are energetically forbidden due to electrostatic repulsion between the side chains. As anticipated the stabilization of the helical state due to electrostatic screening is observed in CD measurements for (AK)6 and (AE)6 (cf. Fig. 1, C–F), respectively. With the exception of (AK)6 in NaClO4, the stabilization of the peptides is nonspecific for salt concentrations c < 0.5 M. Hence, it is a purely screening effect and strong ion-peptide interactions can be ruled out for these concentrations. For higher concentrations, the stabilization is highly salt specific, and we observe the order NaClO4 ∼ KF > KCl ∼ NaCl > GdmCl. GdmCl destabilizes α-helical content for both peptides at concentrations above 0.5 M, giving rise to a remarkable nonmonotonic helicity versus concentration curve. In contrast to the EK-peptide, the difference between NaCl and KCl is small. Note the interesting asymmetry between the (AK)6 and (AE)6 peptides: although the charge pattern of the two peptides is the same, the particular chemistry of the side chains (K versus E) gives rise to a 10% better stabilization of (AK)6 versus (AE)6 at molar salt concentrations. These trends are independent of T (Figs. S4 and S5).
Submolar NaClO4 concentrations appear to be especially strong at stabilizing positively charged peptides ((AK)6 in Fig. 1 D), in line with previous work (29–31). This indicates that screening of the positively charged side chains by binding of the ClO4− anion is very effective. The absence of this strong stabilization for the negatively charged (AE)6 confirms this idea.
At molar concentrations, however, the behavior of NaClO4 for all three peptides, is in contrast to what is expected from the Hofmeister series ranking (2,5): a), The very weak destabilization of the EK peptide by NaClO4 is comparable to KF; and b), NaClO4 stabilization capacity is comparable with that of the strong stabilizer KF for (AE)6 and even much larger than for KF in the case of (AK)6. The expectation for NaClO4 comes from its high salting-in constant (2,3,5,8) and its strong interaction with the peptide (amide) group (5,8). This places the ClO4− ion in a similar location to Gdm+ and far from F− (5) in opposition with our observations.
Rationalization by all-atom MD computer simulations and ion partitioning
To gain additional insight into the molecular structure of the peptides measured experimentally, MD simulations were used.
Helicities and molecular structure
The helicity for the EK-peptide, as derived from CD in a molar salt concentration of NaCl, KCl, and KF, agrees qualitatively with previous MD computer simulations (22,32). These simulations revealed that Na+ has a stronger affinity to side-chain carboxylates and backbone carbonyls than K+, thereby weakening salt bridges and secondary structure hydrogen bonds (32), which leads to the observed stronger destabilization of the α-helix.
We expanded on MD simulations of the EK-peptide to include GdmCl and NaClO4 at ∼3.0 M of salt. The calculated average helicities h for the EK-peptide at 300 K in the various salts are summarized in Table 1. The destabilization follows the expected Hofmeister ranking: NaClO4 > GdmCl > NaCl > KCl > KF. This series is clearly related to direct ionic binding to the side chains and peptide backbone as shown in Fig. 2, A and B, where we plot the normalized ionic density distribution g(r) of the cations and anions around the amide carbonyl (O) and nitrogen (N), respectively. Based on ion coordination numbers within the first solvation shell, ions can be arranged into the following order ClO4− (0.60) > Gdm+ (0.34) > Na+ (0.14) > K+ (0.07) > Cl− (0.04) > F− (0.01) at the polar amide groups. At the charged headgroups, the order is given by ClO4− (0.48) > Gdm+ (0.27) > Na+ (0.22) > F− (0.12) ∼ Cl− (0.10) ∼ K+ (0.08). The larger ions ClO4− and Gdm+ also have an affinity to favorably interact with the hydrophobic methyl group of the ALA side chain (Fig. 2, A and B insets).
Table 1.
Secondary structure elements and end-to-end distance Ree of the investigated peptides calculated from MD computer simulations at T = 300 K
| EK |
(AK)6 |
(AE)6 |
|
|---|---|---|---|
| h 310t Ree(nm) | h 310t Ree(nm) | h 310t | |
| No salt | 0.62 0.17 1.74 | − | |
| KF (4 M) | 0.59 0.19 1.46 | 0.58 0.19 1.46 | 0.48 0.20 |
| KCl (3.5 M) | 0.55 0.18 1.60 | 0.63 0.15 1.68 | 0.54 0.18 |
| NaCl (3.7 M) | 0.35 0.23 1.48 | 0.62 0.15 1.54 | 0.44 0.19 |
| GdmCl(3.0 M) | 0.29 0.16 1.62 | 0.23 0.25 1.70 | – |
| NaClO4 (3.0 M) | 0.19 0.38 1.57 | 0.29 0.38 1.61 | – |
h is the average α-helicity, whereas 310t represents the sum of 310 helices and turn structures. Absolute errors on 310t structure estimation are about ±0.1 calculated from block averages (33).
Figure 2.
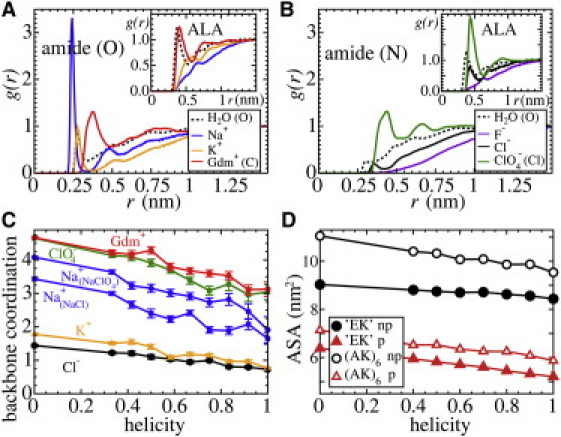
Salt-specific structures studied by MD simulations for T = 300 K. (A) Normalized density distribution g(r) of water (O) and cations (all from chloride salts) around the amide oxygen of the EK-peptide. Inset: g(r) for water and cations around the ALA methyl group. (B) Normalized density distribution g(r) of water (O) and anions around the amide nitrogen of the EK-peptide. Inset left: g(r) for water and anions around the ALA methyl group. (C) Ion coordination in a 0.5 nm shell around the backbone plotted versus average helicity h. Coordination increases upon unfolding due to direct binding to the backbone. (D) Nonpolar and polar ASA of the EK and (AK)6 peptides plotted versus helicity h. Both nonpolar and polar ASA linearly increase upon unfolding.
This direct binding to the peptide is strongly correlated with the α-helicity that is shown in Fig. 2 C. Here, we plot an estimate of the overall backbone coordination number of ions versus the peptide helicity by averaging the number of ions in a 0.5 nm thick shell around the backbone amide atoms for a fixed helicity (see the Supporting Material). Using this definition, the coordination reflects the integration of the g(r) (weighted by r2) in Fig. 2, A or B, up to r = 0.5 nm. For the strong binding ions (Gdm+, ClO4−, Na+), the backbone coordination increases by two molecules when going from the folded to completely denatured state. Thus, protein destabilization is clearly induced by preferential accumulation of ions to the backbone thereby perturbing secondary structure H-bonds. Cation coordination (Na+) quantitatively depends on the nature of the anion (Cl− versus ClO4−) (cf. Fig. 2 C), exemplifying a strong synergy in ionic binding.
The trends displayed by the calculated ion density distributions and coordination numbers are fully consistent with salting-in/out (1–3) and partition coefficients Kp and Knp for salts at the polar amide and the nonpolar hydrocarbon surface of peptides (8). The coefficients Kp and Knp for the investigated salts (given in Table S1) suggest that denaturants such as GdmCl and NaClO4 strongly accumulate at the backbone amide, whereas only weakly excluded from the nonpolar surface. In contrast, KF weakly interacts with the amide and is repelled from the aliphatic groups. Indeed, the ionic density distributions (Fig. 2, A and B), provide a consistent molecular picture: both Gdm+ and ClO4− strongly interact with the polar (amide) group and have a distinct affinity to hydrophobic groups. Representative MD snapshots for Gdm+ and ClO4− ions bound to the peptide with long lifetimes (∼0.1 ns) are shown in Fig. 3: Gdm+ or ClO4− hydrogen-bond to the amide while at the same time embedded between nonpolar groups, such as ALA methyl groups or the greasy lysine side chain. For NaClO4, both ions have a strong affinity to the backbone resulting in a concerted binding to multiple peptide groups (Fig. 3 D). A consequence of this network of Na+ and ClO4− ions is a remarkable anomalous binding time distribution Pb(t) (Fig. S6 and (22)), which is absent for all other investigated ions.
Figure 3.
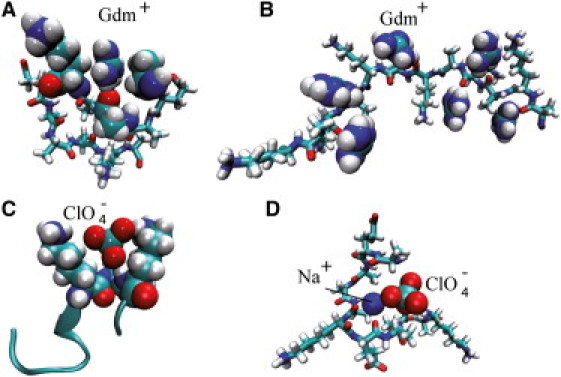
Representative MD simulations snapshots of Gdm+ and ClO4− binding to the EK and (AK)6 peptides. (A) A Gdm+-ion (spheres) hydrogen bonded to the backbone (stick structure) amide and embedded by a nonpolar lysine and alanine methyl groups of the EK peptide. (B) Gdm+ binding to (AK)6 in the coil state. (C) A ClO4− -ion hydrogen bonded to the backbone amide and embedded by lysine side chains in the (AK)6 peptide. (D) ClO4− and Na+ displaying concerted binding to the backbone in a compact state of the EK-peptide. Snapshots in A and B were chosen using the kinetic criterion such that ion binding was longer than 0.1 ns. In D the ionic complex was bound for >1 ns.
Ion partitioning concept and m-value analysis
To compare m-values predicted from the experimental partitioning concept for the EK peptide, we calculated the ASA for the polar and nonpolar peptide parts. The ASAs is roughly linear in helicity (Fig. 2 D). Upon unfolding, the calculation yields ΔASA of polar and nonpolar surfaces of 1.2 and 0.6 nm2, respectively. The resulting m-values, corrected for the salt-bridge contribution (see Methods in the Supporting Material), are given in Table 2. These m-values, based on experimental partitioning coefficients, follow the Hofmeister series for destabilization NaClO4 > GdmCl > NaCl > KCl ∼ KF.
Table 2.
m-values
| m (kBT M−1) | mcorr (kBT M−1) | mMD (kBT M−1) | |
|---|---|---|---|
| KF | −0.35(0.02) | −0.32(0.02) | −0.30(0.06) |
| KCl | −0.38(0.02) | −0.38(0.02) | −0.14(0.07) |
| NaCl | −0.38(0.02) | −0.42(0.02) | −0.78(0.07) |
| GdmCl | −0.44(0.04) | −0.51(0.04) | −0.97(0.09) |
| NaClO4 | −0.79(0.08) | −0.97(0.08) | −1.27(0.09) |
The m-value is determined using Eq. 1 in the Supporting Material, the MD-derived Δ(ASA)i for the EK-peptide and reported values for partial mi-values per unit ASA for polar “p”, nonpolar “np”, and ester oxygen “eo” compounds (see Table S1 and (8,16)). mcorr is the corrected value including the contribution of the salt bridge (see Supporting Material). mMD is the m-value derived from MD simulations including salt. Errors are shown in parenthesis (see Methods in the Supporting Material).
Next, we calculated m-values from the MD simulations in the presence of salts (see Supporting Methods for details and Table 2). Comparison of the simulation calculated m-values to those derived from ΔASA leads to a few important conclusions: first, the overall order in helical destabilization is consistent with what is predicted by the combination of ΔASA and the experimental partitioning (see above). All derived m-values have a similar order of magnitude indicating that the concept of m-values is reasonable, despite the assumptions made (e.g., force fields in simulations, additivity of partition coefficients, etc.) and methods used. Comparing values quantitatively, it appears that the MD calculated interaction of KCl with carbonyl/carboxyl groups is too weak, while NaCl and GdmCl interact too strong, pointing to the need for forcefield optimization. The agreement for KF may be fortuitous as it is a problematic salt both in experiments (8) and simulations. A striking result of this analysis, however, is the strong qualitative discrepancy of the large m-values for NaClO4 in both approaches compared to the small helicity change measured by CD.
For the charged peptides, previous MD simulations on (AK)6 and (AE)6 in the presence of NaCl and KCl revealed a high helical stability at molar salt concentration ((33) and Table 1). This was accompanied by a higher helicity for (AK)6 compared to (AE)6 in the same salt environment and a smaller difference between the influence of Na+ versus K+ on (AK)6. These findings are in qualitative agreement with the current CD data (Fig. 1). New simulations were performed for (AK)6 in molar GdmCl and NaClO4. As in the case of the EK-peptide, GdmCl and NaClO4 destabilize the α-helical structure compared to NaCl or KCl (Table 1). The individual (ΔASA)i upon unfolding are very similar in magnitude to the EK-peptide (Fig. 2 D) and our qualitative analysis of m-values above should also be valid here. The only difference is that no salt bridges are involved. The absence of salt bridges in (AK)6 and (AE)6 explains the vanishing difference between NaCl and KCl in their (de)stabilizing action on these peptides. Finally, ion partitioning and MD simulations are again in stark contrast to the strong stabilization of the (AK)6 peptide by molar NaClO4 indicated by CD (Fig. 2 D).
Single-molecule FRET shows a highly collapsed state of (AK)14 in molar NaClO4
To further elucidate structural characteristics of the peptides and the curious behavior of NaClO4 in its impact of the α-helical structure, we monitored the global conformation of a (AK)14 peptide via smFRET. We performed burst analysis (23) measurements with ∼10 pM amounts of atto488-(AK)14C-atto647N [(AK)14] in solution with increasing amounts of NaClO4 and compared them with measurements in the presence of KCl or GdmCl.
Using the photon distribution analysis (35,36) on the experimentally measured FRET efficiency histograms, we estimated the donor-acceptor separation RDA (Fig. 4 A and Fig. S7) and the residual distribution width beyond shot noise (Fig. S8). In the absence of any salt, the RDA is 4.7 nm corresponding to a slightly swollen coil due to the electrostatic repulsion. For comparison, FRET measurements have also been performed in a reference state at pH = 11 (where deprotonation of lysine residues should allow full α-helix formation) yielding a RDA = 4.9 nm. Thus, α-helical states without salt seem to have extensions similar to a coil state. Adding GdmCl first increases RDA to 5.0 nm and then decreases and settles at an average distance of 4.9 nm. At molar GdmCl, the RDA is 5.1 nm implying slight swelling of the coil. Submolar amounts of KCl have a similar behavior as GdmCl, initially increasing RDA with salt and then reducing it at higher salt concentrations. Molar amounts of KCl compact the peptide to 4.1 nm (Fig. 4 A) while increasing helix formation (Fig. 4, B and C). NaClO4 starts to collapse RDA even at submolar amounts. This compression is considerably stronger for molar NaClO4 where RDA further goes down to 3.6 nm. Although CD indicates a similar helicity for the peptide in NaClO4 at ∼1.5M (or KCl at 4 M) and pH = 11, FRET points to much more compact states in the NaClO4 solution.
Figure 4.
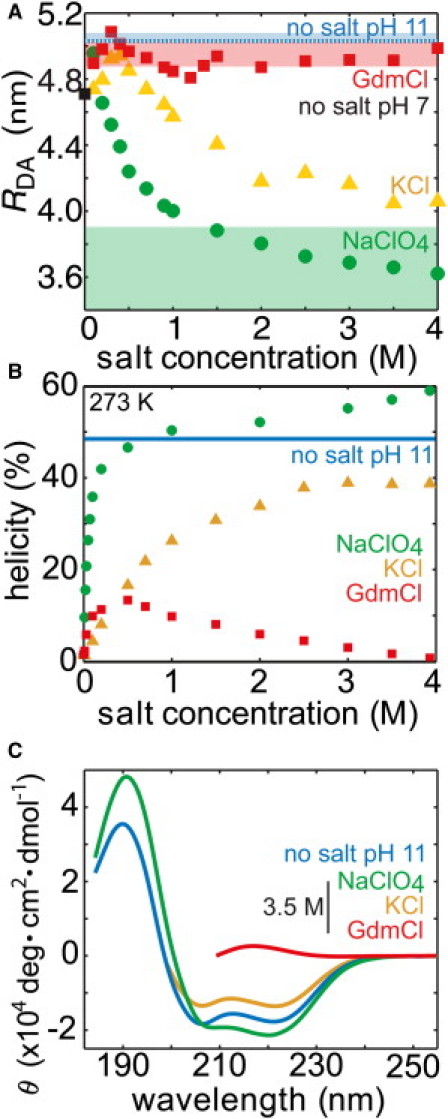
Ion-specific effect on the CD spectra and donor-acceptor fluorophore separation RDA of (AK)14. (A) Measured RDA (symbols) in the absence of salt at pH 7 (black), with KCl (yellow upper triangles), GdmCl (red squares), NaClO4 (green circles), and no salt pH 11 (light blue line). Standard deviations from several measurements are smaller than the size of the symbol (0.05 nm). Estimated range of RDA from MD-derived structures at 3.5M salt is represented as shaded areas depicting the upper and lower limits. (B) The salt-dependent helicity (derived from the signal at 222 nm) of (AK)14 in the absence of salt at pH 7 (black), with KCl (yellow), GdmCl (red), NaClO4 (green), and no salt pH 11 (light blue). The average value is a dotted line while the thickness of the shaded area represents the upper and lower boundaries. (C) CD spectra of (AK)14 in the absence of salt at pH 7 (black), with KCl (yellow), GdmCl (red), NaClO4 (green), and no salt pH 11 (light blue).
To compare the FRET measurements to the results from CD we measured the CD spectrum of the (AK)14 peptide. For this double labeled and longer peptide, the helical signature from CD remains qualitatively the same as for (AK)6 (Fig. 4 B at T = 273 K). Although GdmCl decreases helicity beyond a concentration of ∼1 M to <10%, KCl stabilizes the helix up to 40%, and NaClO4 again shows an extremely large stabilization action with 50–60% helicity at molar concentrations. As a reference measurement, we performed CD for (AK)14 in the absence of salt at a pH = 11 where all side chains are deprotonated and the peptide is net neutral, i.e., perfectly screened. In this case, the peptide displays ∼50% helicity, less than in molar NaClO4 at pH = 7. In Fig. 4 C, the raw CD spectrum in 3.5 M salt solution is shown. In comparison to the salt-free data at pH = 11, NaClO4 displays unusual features: the maximum around 190 nm is red-shifted and the helix-typical double-wells between 200 and 230 nm are less resolved. This red-shift has been interpreted as originating from helix distortion (37).
To corroborate the interpretation of the FRET data with the simulations, we took the following approach (see Supporting Methods for details and Fig. S9 for snapshots): i), the following three, highly complementary structural states were taken: 1), a fully α-helical structure, 2), fully coiled structures, and 3), compact structure with a minimum radius of gyration; ii), the accessible volume of linker and dye at each end were calculated; iii), the mean distance between dyes was then estimated and corrected for dye linker length and dynamics (38). The range of distances for each structural state 1)–3) is depicted in Fig. 4 A. The fully helical state has a calculated RcalcDA of 5.12 nm, the coiled states feature very similar extensions with a mean of RcalcDA of 4.95 nm. The two compact state configurations (see Fig. S9) result in a mean RcalcDA of 3.63 nm. Therefore, the distances measured with FRET correspond extremely well with the MD data. The combined FRET and MD analysis thus provide significant evidence for an abundance of tightly packed peptide states at molar NaClO4 concentrations in contrast to swollen coils in molar GdmCl or extended helices at pH = 11. Comparison to pH 11 shows that the high compaction at molar NaClO4 cannot be explained by electrostatic screening of the side chains (33,39).
The previously described trends can be compared to the average end-to-end distances, Ree, for the well-converged simulations of the shorter EK and (AK)6 peptides (Table 1). The strongest compaction is observed for KF. This is in line with compaction of polymers by preferential exclusion of ions (15). In contrast to KF, the peptides in molar GdmCl showed the highest extension in molar salt. The swelling of peptides and proteins upon denaturation in molar GdmCl is in fact a common phenomenon (18,20,40–42) even observed for purely hydrophobic model polymers (43). Interestingly, the presence of NaCl and NaClO4 result in considerable compacted structures similar to that of KF, with the difference that NaCl and NaClO4 interact preferentially with the backbone. NaClO4-induced compact states are not new as they have been observed in what was interpreted as a molten globule state (13–16). Of importance, compression with NaClO4 is accompanied by a significant increase in 310 and turn secondary structure elements (Table 1).
Sodium perchlorate, CD spectra, and the diversity of denatured states
The CD analysis indicates high α-helicity in molar NaClO4, whereas ion partitioning and the MD simulations do not. Despite its popularity, the interpretation of CD spectra is not always unique and has led to controversies in the past (44,45). For instance, it is not possible to unambiguously distinguish between partial contributions of an α-helix and a 310-helix to a CD spectrum with multiple components (see e.g., Fig. S4 A and (44,45)). In particular, Scholtz and Baldwin (46) showed that for the ClO4−-induced denatured state of ribonuclease A, CD suggested secondary structure formation although it did not provide measurable protection against amide proton exchange, and it does not belong to the class of structured molten globules. Their interpretation was that compact states were induced possibly leading to a change in the far-ultraviolet CD spectra, and caution was advised in the interpretation of CD data in molar concentrations of ClO4−. Along this line, pioneering work from the 70s demonstrated that direct binding of ions may change the spectral and vibrational properties of the amide chromophore, i.e., the double bond character of the peptide group (47–51). The relative effectiveness of the salts in altering CD (or NMR) spectra was presumed to correspond to the strength of relative amide binding, as reflected in the salting in constants or the partitioning coefficients shown in Table 2 and Table S1. In fact, NaClO4 has one of the largest binding constants of typical Hofmeister salts even exceeding that of the divalent CaCl2 (2,3,5,8). Mattice showed that very short alanine peptides (1–3 residues), which are unable to form α-helical i,i+4 amide bonds, exhibit a decreasing ellipticity at λ = 222 nm with addition of molar amounts of NaClO4 (50). Similarly, Rao et al. (49) demonstrated a shift in the α-helical CD signature for benzamide in molar NaCl. Small effects are also visible for a (GS)-peptide (see Supplementary Results and Fig. S10). It may not be only CD that suffers from problems at molar concentrations of salt, because strong binding to the amide chromophore may also alter the physical properties of the molecule (47–52).
Judging from literature and our results, the following picture of peptide structure in molar concentration of NaClO4 emerges. Both ions, Na+ and ClO4− favorably bind to the backbone in a concerted fashion as reflected by the partitioning coefficient Kp (Table S1 and Fig. 3 D). The binding affects both the optical and structural properties of the peptide: first, spectral properties of the amide chromophore are altered, at least for some sequences, such that ellipticity curves are shifted toward more helical distributions (50). Second, the observed binding to multiple and adjacent amide groups induces bends and kinks in the chain, thereby leading to compaction and distortion of ordered fragments as demonstrated by FRET and MD. The red-shift in the CD spectrum (Fig. 4 C) supports the interpretation as α-helical distortion (29). The binding of ClO4− ions facilitates sampling of 310 and turn configurations (Table 1), which cannot easily be distinguished from the α-helix in CD measurements. The 310-helix is actually the most hydrophilic secondary structure for short peptide fragments (53), much more open to the solvent than an α-helix. The higher exposure of amide groups can better satisfy both internal H-bonds and an increase coordination number (i.e., the accumulation of the ions). Thus, the denatured state in molar NaClO4 must be regarded as a compact, highly heterogeneous mixture of α, 310, and turn elements with much less α-helical content as indicated by CD. On the basis of these results, the ClO4−-induced compact denatured state cannot be viewed as a canonical molten globule state (54,55).
Why is there such a big difference in the structure of the denatured states in molar GdmCl (swollen coil) versus NaClO4 (compact)? Partitioning and MD simulations provide two plausible explanations: first, GdmCl is hardly excluded from the nonpolar groups of the peptide. A coil state gives maximum exposure of favorable amide groups while hydrophobic parts are covered by Gdm+ (consistent with swelling of a purely hydrophobic polymer (43)). NaClO4 is more excluded from nonpolar regions while binding strongly to the peptide backbone (Table S1 and Table 2). Second, due to the large affinity of both Na+ and ClO4− to amide groups, binding to multiple backbone groups in a concerted fashion is possible (Fig. 3 D). This, in turn, leads to kinks and turns and overall compression of the chain. Thus, due to their specific and direct binding properties, GdmCl and NaClO4 are both destabilizers of ordered secondary structures but lead to extremely different denatured states, where one is close to a random coil, the other one is a compact, structurally heterogeneous state. Future work will show how prevalent either mechanism is by comparing it with other strong denaturing ions (e.g., I−, SCN−).
Conclusion
The action of specific ions on protein structure is a highly complex issue. However, our direct comparison between experimental CD data and MD simulations of model peptides shows that computer simulations are qualitative enough to provide the right trends and give molecular insights. Destabilization of secondary structure is therefore induced by individual ion binding to specific peptide sites thus perturbing hydrogen bonds and salt bridges and drawing peptide groups into solution. Strong ion-specific destabilization typically occurs at salt concentrations larger than 0.5M, where nonspecific charge effects, such as helix charge-ion or helix dipole-ion interactions (27,28), are screened. The m-value analysis based on detailed ASA estimates provides strong support of the ion-partitioning concept (8) as well as ion binding to specific sites.
Interestingly, the CD data contrast the MD and partitioning results for NaClO4. Subsequent use of smFRET allowed us to validate results from MD simulations versus those obtained from CD, which was unable to distinguish heterogeneous collapsed states from homogeneous secondary structure content. Our analysis, in combination with previously reported results, points to a highly disordered class of compacted states in strongly accumulating salts such as NaClO4. These collapsed states have been in the past erroneously taken as molten globule intermediaries during protein unfolding and not as a denatured end-species, as previously surmised by Baldwin and co-workers (46). Thus, although both NaClO4 and GdmCl interact strongly with peptides, this does not necessarily imply a swollen, random coil-like denatured state. As we have shown, the detailed, specific and local binding structure makes the difference.
Acknowledgments
J.D. is grateful to the Deutsche Forschungsgemeinschaft (DFG) for support within the Emmy-Noether-Program and the Leibniz-Rechenzentrum (LRZ) Garching for computing time. A.H.C. and J.D. thank the Micro-Chemistry Core Facility of the MPI-B for Peptide Synthesis. A.H.C. thanks Adam Muschielok for discussions on AV and FRET calculations.
This work was supported by grants from the DFG (SPP 1464), the Ludwig-Maximilians-Universität München (LMUInnovativ BioImaging Network), and the Nanosystems Initiative Munich (NIM).
Contributor Information
Alvaro H. Crevenna, Email: crevenna@biochem.mpg.de.
Joachim Dzubiella, Email: joachim.dzubiella@helmholtz-berlin.de.
Supporting Material
References
- 1.Hofmeister F. Zur Lehre von der wirkung der salze. Zweite Mittheilung. Arch. Exp. Pathol. Pharmakol. 1888;24:247–260. [Google Scholar]
- 2.Von Hippel P.H., Schleich T. Ion effects on solution structure of biological macromolecules. Acc. Chem. Res. 1969;2:257–265. [Google Scholar]
- 3.Nandi P.K., Robinson D.R. The effects of salts on the free energies of nonpolar groups in model peptides. J. Am. Chem. Soc. 1972;94:1308–1315. doi: 10.1021/ja00759a043. [DOI] [PubMed] [Google Scholar]
- 4.Nandi P.K., Robinson D.R. The effects of salts on the free energy of the peptide group. J. Am. Chem. Soc. 1972;94:1299–1308. doi: 10.1021/ja00759a042. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins K.D. Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Collins K.D. Ion hydration: implications for cellular function, polyelectrolytes, and protein crystallization. Biophys. Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Pegram L.M., Record M.T., Jr. Thermodynamic origin of hofmeister ion effects. J. Phys. Chem. B. 2008;112:9428–9436. doi: 10.1021/jp800816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Cremer P.S. Chemistry of Hofmeister anions and osmolytes. Annu. Rev. Phys. Chem. 2010;61:63–83. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien E.P., Dima R.I., Thirumalai D. Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: lessons for protein denaturation mechanism. J. Am. Chem. Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- 11.Hamada D., Kidokoro S., Goto Y. Salt-induced formation of the molten globule state of cytochrome c studied by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA. 1994;91:10325–10329. doi: 10.1073/pnas.91.22.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huyghues-Despointes B.M.P., Scholtz J.M., Baldwin R.L. Effect of a single aspartate on helix stability at different positions in a neutral alanine-based peptide. Protein Sci. 1993;2:1604–1611. doi: 10.1002/pro.5560021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maity H., Eftink M.R. Perchlorate-induced conformational transition of Staphylococcal nuclease: evidence for an equilibrium unfolding intermediate. Arch. Biochem. Biophys. 2004;431:119–123. doi: 10.1016/j.abb.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair J.F., Shortle D. Analysis of long-range interactions in a model denatured state of staphylococcal nuclease based on correlated changes in backbone dynamics. Protein Sci. 1999;8:991–1000. doi: 10.1110/ps.8.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh T., Kalra A., Garde S. On the salt-induced stabilization of pair and many-body hydrophobic interactions. J. Phys. Chem. B. 2005;109:642–651. doi: 10.1021/jp0475638. [DOI] [PubMed] [Google Scholar]
- 16.Pegram L.M., Wendorff T., Record M.T. Why Hofmeister effects of many salts favor protein folding but not DNA helix formation. Proc. Natl. Acad. Sci. USA. 2010;107:7716–7721. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly S.M., Jess T.J., Price N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Merchant K.A., Best R.B., Eaton W.A. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Möglich A., Joder K., Kiefhaber T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. USA. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiffany M.L., Krimm S. Extended conformations of polypeptides and proteins in urea and guanidine hydrochloride. Biopolymers. 1973;12:575–587. [Google Scholar]
- 21.Hinczewski M., von Hansen Y., Netz R.R. How the diffusivity profile reduces the arbitrariness of protein folding free energies. J. Chem. Phys. 2010;132:245103. doi: 10.1063/1.3442716. [DOI] [PubMed] [Google Scholar]
- 22.von Hansen Y., Kalcher I., Dzubiella J. Ion specificity in α-helical folding kinetics. J. Phys. Chem. B. 2010;114:13815–13822. doi: 10.1021/jp107495f. [DOI] [PubMed] [Google Scholar]
- 23.Kapanidis A.N., Lee N.K., Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller B.K., Zaychikov E., Lamb D.C. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mapa K., Sikor M., Mokranjac D. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell. 2010;38:89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Marqusee S., Baldwin R.L. Helix stabilization by Glu-…Lys+ salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ihara S., Ooi T., Takahashi S. Effects of salts on the nonequivalent stability of the α-helices of isomeric block copolypeptides. Biopolymers. 1982;21:131–145. [Google Scholar]
- 28.Takahashi S., Kim E.-H., Ooi T. Comparison of α-helix stability in peptides having a negatively or positively charged residue block attached either to the N- or C-terminus of an α-helix: the electrostatic contribution and anisotropic stability of the α-helix. Biopolymers. 1989;28:995–1009. doi: 10.1002/bip.360280507. [DOI] [PubMed] [Google Scholar]
- 29.Peggion E., Cosani A., Borin G. Conformational studies on polypeptides. The effect of sodium perchlorate on the conformation of poly-L-lysine and of random copolymers of L-lysine and L-phenylalanine in aqueous solution. Biopolymers. 1972;11:633–643. doi: 10.1002/bip.1972.360110308. [DOI] [PubMed] [Google Scholar]
- 30.Maison W., Kennedy R.J., Kemp D.S. Chaotropic anions strongly stabilize short, N-capped uncharged peptide helicies: a new look at the perchlorate effect. Angew. Chem. Int. Ed. Engl. 2001;40:3819–3821. doi: 10.1002/1521-3773(20011015)40:20<3819::AID-ANIE3819>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 31.Asciutto E.K., General I.J., Madura J.D. Sodium perchlorate effects on the helical stability of a mainly alanine peptide. Biophys. J. 2010;98:186–196. doi: 10.1016/j.bpj.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dzubiella J. Salt-specific stability and denaturation of a short salt-bridge-forming alpha-helix. J. Am. Chem. Soc. 2008;130:14000–14007. doi: 10.1021/ja805562g. [DOI] [PubMed] [Google Scholar]
- 33.Dzubiella J. Salt-specific stability of short and charged alanine-based alpha-helices. J. Phys. Chem. B. 2009;113:16689–16694. doi: 10.1021/jp9077932. [DOI] [PubMed] [Google Scholar]
- 34.Reference deleted in proof.
- 35.Antonik M., Felekyan S., Seidel C.A. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J. Phys. Chem. B. 2006;110:6970–6978. doi: 10.1021/jp057257+. [DOI] [PubMed] [Google Scholar]
- 36.Kalinin S., Felekyan S., Seidel C.A. Probability distribution analysis of single-molecule fluorescence anisotropy and resonance energy transfer. J. Phys. Chem. B. 2007;111:10253–10262. doi: 10.1021/jp072293p. [DOI] [PubMed] [Google Scholar]
- 37.Manning M.C., Illangasekare M., Woody R.W. Circular dichroism studies of distorted alpha-helices, twisted beta-sheets, and beta turns. Biophys. Chem. 1988;31:77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 38.Sindbert S., Kalinin S., Seidel C.A. Accurate distance determination of nucleic acids via Förster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 39.Scholtz J.M., Qian H., Baldwin R.L. The energetics of ion-pair and hydrogen-bonding interactions in a helical peptide. Biochemistry. 1993;32:9668–9676. doi: 10.1021/bi00088a019. [DOI] [PubMed] [Google Scholar]
- 40.Müller-Späth S., Soranno A., Schuler B. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman E., Haran G. Coil-globule transition in the denatured state of a small protein. Proc. Natl. Acad. Sci. USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fierz B., Reiner A., Kiefhaber T. Local conformational dynamics in alpha-helices measured by fast triplet transfer. Proc. Natl. Acad. Sci. USA. 2009;106:1057–1062. doi: 10.1073/pnas.0808581106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godawat R., Jamadagni S.N., Garde S. Unfolding of hydrophobic polymers in guanidinium chloride solutions. J. Phys. Chem. B. 2010;114:2246–2254. doi: 10.1021/jp906976q. [DOI] [PubMed] [Google Scholar]
- 44.Andersen N.H., Liu Z., Prickett K.S. Efforts toward deriving the CD spectrum of a 3(10) helix in aqueous medium. FEBS Lett. 1996;399:47–52. doi: 10.1016/s0014-5793(96)01279-3. [DOI] [PubMed] [Google Scholar]
- 45.Silva R.A., Yasui S.C., Keiderling T.A. Discriminating 3(10)- from alpha-helices: vibrational and electronic CD and IR absorption study of related Aib-containing oligopeptides. Biopolymers. 2002;65:229–243. doi: 10.1002/bip.10241. [DOI] [PubMed] [Google Scholar]
- 46.Scholtz J.M., Baldwin R.L. Perchlorate-induced denaturation of ribonuclease A: investigation of possible folding intermediates. Biochemistry. 1993;32:4604–4608. doi: 10.1021/bi00068a017. [DOI] [PubMed] [Google Scholar]
- 47.Schleich T., Von Hippel P. Specific ion effects on the solution conformation of poly-L-proline. Biopolymers. 1969;7:861–877. [Google Scholar]
- 48.Balasubramanian D., Misra B.C. Effects of metal ions on the structure and spectra of the peptide group. Biopolymers. 1975;14:1019–1026. doi: 10.1002/bip.1975.360140510. [DOI] [PubMed] [Google Scholar]
- 49.Rao C.N., Rao K.G., Balasubramanian D. Binding of alkali and alkaline earth cations and of protons to the peptide group. FEBS Lett. 1974;46:192–194. doi: 10.1016/0014-5793(74)80366-2. [DOI] [PubMed] [Google Scholar]
- 50.Mattice W.L. The effect of temperature and salt concentration on the circular dichroism exhibited by unionized derivatives of L-alanine in aqueous solution. Biopolymers. 1974;13:169–183. doi: 10.1002/bip.1974.360130111. [DOI] [PubMed] [Google Scholar]
- 51.Fussenegger R., Rode B. Effect of metal-ion bonding to amides on character of C-N bond of ligand molecule. Chem. Phys. Lett. 1976;44:95–99. [Google Scholar]
- 52.Balasubramian D., Shaikh R. Interaction of lithium salts with model amides. Biopolymers. 1973;12:1639–1650. [Google Scholar]
- 53.Sorin E.J., Rhee Y.M., Pande V.S. The solvation interface is a determining factor in peptide conformational preferences. J. Mol. Biol. 2006;356:248–256. doi: 10.1016/j.jmb.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 54.Finkelstein A.V., Shakhnovich E.I. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers. 1989;28:1681–1694. doi: 10.1002/bip.360281004. [DOI] [PubMed] [Google Scholar]
- 55.Shakhnovich E.I., Finkelstein A.V. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989;28:1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- 56.Muschielok A., Andrecka J., Michaelis J. A nano-positioning system for macromolecular structural analysis. Nat. Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 57.Schrodinger L.L.C. Schrodinger, LLC; Portland, OR: 2010. The PyMOL Molecular Graphics System, Version 1.3r1. [Google Scholar]
- 58.Scholtz J.M., Qian H., Baldwin R.L. Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 59.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 60.Zander C., Sauer M., Seidel C.A.M. Detection and characterization of single molecules in aqueous solution. Appl. Phys. B. 1996;63:517–523. [Google Scholar]
- 61.Eggeling C., Berger S., Seidel C.A. Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. J. Biotechnol. 2001;86:163–180. doi: 10.1016/s0168-1656(00)00412-0. [DOI] [PubMed] [Google Scholar]
- 62.Isaksson M., Norlin N., Johansson L.B. On the quantitative molecular analysis of electronic energy transfer within donor-acceptor pairs. Phys. Chem. Chem. Phys. 2007;9:1941–1951. doi: 10.1039/b614817d. [DOI] [PubMed] [Google Scholar]
- 63.Ivanov V., Li M., Mizuuchi K. Impact of emission anisotropy on fluorescence spectroscopy and FRET distance measurements. Biophys. J. 2009;97:922–929. doi: 10.1016/j.bpj.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dang L.X. Mechanism and thermodynamics of ion selectivity in aqueous solutions of 18-crown-6 ether: a molecular dynamics study. J. Am. Chem. Soc. 1995;117:6954–6960. [Google Scholar]
- 65.Best R.B., Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalcher I., Dzubiella J. Structure-thermodynamics relation of electrolyte solutions. J. Chem. Phys. 2009;130:134507. doi: 10.1063/1.3097530. [DOI] [PubMed] [Google Scholar]
- 67.Mason P.E., Dempsey C.E., Jungwirth P. Specificity of ion-protein interactions: complementary and competitive effects of tetrapropylammonium, guanidinium, sulfate, and chloride ions. J. Phys. Chem. B. 2009;113:3227–3234. doi: 10.1021/jp8112232. [DOI] [PubMed] [Google Scholar]
- 68.Kudryavtsev V., Sikor M., Lamb D.C. Combining MFD and PIE for accurate single-pair Förster resonance energy transfer measurements. ChemPhysChem. 2012 doi: 10.1002/cphc.201100822. In press. [DOI] [PubMed] [Google Scholar]
- 69.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


