Abstract
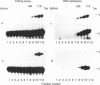
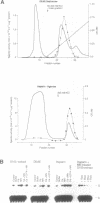
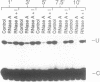
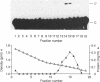
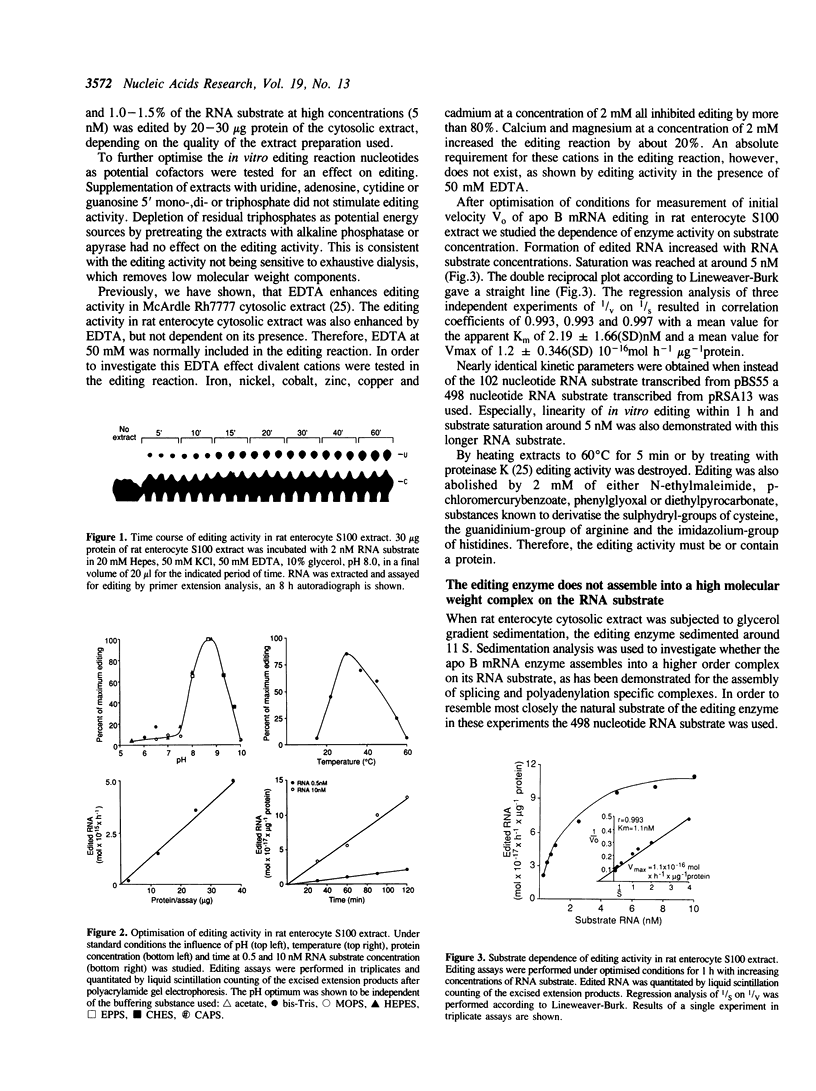
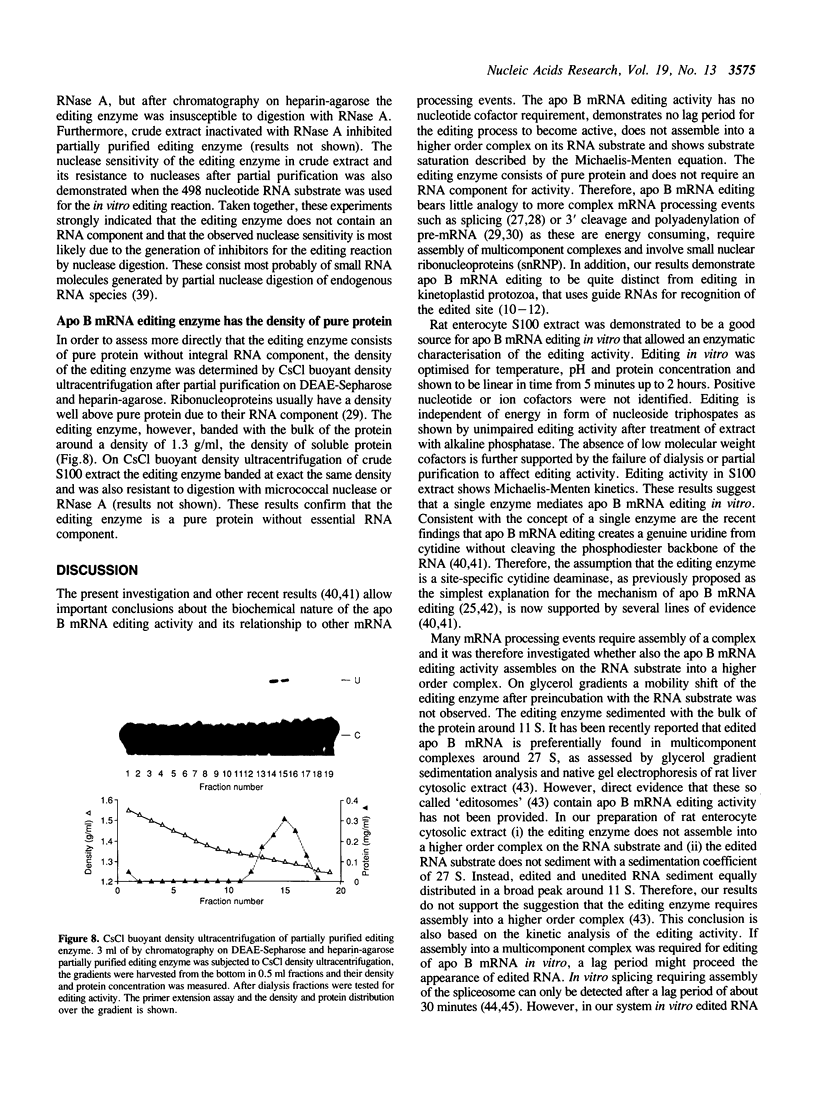
Intestinal apolipoprotein B mRNA is edited at nucleotide 6666 by a C to U transition resulting in a translational stop codon. The enzymatic properties of the editing activity were characterised in vitro using rat enterocyte cytosolic extract. The editing activity has no nucleotide or ion cofactor requirement. It shows substrate saturation with an apparent Km for the RNA substrate of 2.2 nM. The editing enzyme requires no lag period prior to catalysis, and does not assemble into a higher order complex on the RNA substrate. In crude cytosolic extract editing activity is completely abolished by treatment with micrococcal nuclease or RNAse A. Partially purified editing enzyme is no longer sensitive to nucleases, but is inhibited in a dose dependent manner by nuclease inactivated crude extract. The buoyant density of partially purified editing enzyme is 1.3 g/ml, that of pure protein. Therefore, the apolipoprotein B mRNA editing activity consists of a well defined enzyme with no RNA component. The nuclease sensitivity in crude cytosolic extract is explained by the generation of inhibitors for the editing enzyme. The editing of apo B mRNA has little similarity to complex mRNA processing events such as splicing and unlike editing in kinetoplastid protozoa does not utilise guide RNAs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baum C. L., Teng B. B., Davidson N. O. Apolipoprotein B messenger RNA editing in the rat liver. Modulation by fasting and refeeding a high carbohydrate diet. J Biol Chem. 1990 Nov 5;265(31):19263–19270. [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bhat G. J., Koslowsky D. J., Feagin J. E., Smiley B. L., Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990 Jun 1;61(5):885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990 Jan 26;60(2):189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Blum B., Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3' oligo(U) tail involved in recognition of the preedited region. Cell. 1990 Jul 27;62(2):391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Boström K., Garcia Z., Poksay K. S., Johnson D. F., Lusis A. J., Innerarity T. L. Apolipoprotein B mRNA editing. Direct determination of the edited base and occurrence in non-apolipoprotein B-producing cell lines. J Biol Chem. 1990 Dec 25;265(36):22446–22452. [PubMed] [Google Scholar]
- Boström K., Lauer S. J., Poksay K. S., Garcia Z., Taylor J. M., Innerarity T. L. Apolipoprotein B48 RNA editing in chimeric apolipoprotein EB mRNA. J Biol Chem. 1989 Sep 15;264(26):15701–15708. [PubMed] [Google Scholar]
- Burns D., Lewin A. Inhibition of the import of mitochondrial proteins by RNase. J Biol Chem. 1986 May 15;261(14):6153–6155. [PubMed] [Google Scholar]
- Cech T. R. RNA editing: world's smallest introns? Cell. 1991 Feb 22;64(4):667–669. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- Chabot B., Black D. L., LeMaster D. M., Steitz J. A. The 3' splice site of pre-messenger RNA is recognized by a small nuclear ribonucleoprotein. Science. 1985 Dec 20;230(4732):1344–1349. doi: 10.1126/science.2933810. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., Weng S. A., Silberman S. R., Cai S. J., Deslypere J. P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987 Oct 16;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Li X. X., Liao W. S., Wu J. H., Chan L. RNA editing of apolipoprotein B mRNA. Sequence specificity determined by in vitro coupled transcription editing. J Biol Chem. 1990 Apr 25;265(12):6811–6816. [PubMed] [Google Scholar]
- Christofori G., Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988 Sep 9;54(6):875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Davidson N. O., Powell L. M., Wallis S. C., Scott J. Thyroid hormone modulates the introduction of a stop codon in rat liver apolipoprotein B messenger RNA. J Biol Chem. 1988 Sep 25;263(27):13482–13485. [PubMed] [Google Scholar]
- Decker C. J., Sollner-Webb B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990 Jun 15;61(6):1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D. M., Casanova E. Characterization of the apolipoprotein B mRNA editing activity in enterocyte extracts. J Biol Chem. 1990 Dec 15;265(35):21401–21403. [PubMed] [Google Scholar]
- Driscoll D. M., Wynne J. K., Wallis S. C., Scott J. An in vitro system for the editing of apolipoprotein B mRNA. Cell. 1989 Aug 11;58(3):519–525. doi: 10.1016/0092-8674(89)90432-7. [DOI] [PubMed] [Google Scholar]
- Firgaira F. A., Hendrick J. P., Kalousek F., Kraus J. P., Rosenberg L. E. RNA required for import of precursor proteins into mitochondria. Science. 1984 Dec 14;226(4680):1319–1322. doi: 10.1126/science.6209799. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Weil J. H., Grienenberger J. M. Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res. 1990 Jul 11;18(13):3771–3776. doi: 10.1093/nar/18.13.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983 Nov;35(1):89–99. doi: 10.1016/0092-8674(83)90211-8. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Hodges P. E., Navaratnam N., Greeve J. C., Scott J. Site-specific creation of uridine from cytidine in apolipoprotein B mRNA editing. Nucleic Acids Res. 1991 Mar 25;19(6):1197–1201. doi: 10.1093/nar/19.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G. F. RNA evolution and the origins of life. Nature. 1989 Mar 16;338(6212):217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- Kane J. P. Apolipoprotein B: structural and metabolic heterogeneity. Annu Rev Physiol. 1983;45:637–650. doi: 10.1146/annurev.ph.45.030183.003225. [DOI] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985 Oct;42(3):725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krämer A. Site-specific degradation of RNA of small nuclear ribonucleoprotein particles with complementary oligodeoxynucleotides and RNase H. Methods Enzymol. 1990;181:284–292. doi: 10.1016/0076-6879(90)81129-i. [DOI] [PubMed] [Google Scholar]
- Mahendran R., Spottswood M. R., Miller D. L. RNA editing by cytidine insertion in mitochondria of Physarum polycephalum. Nature. 1991 Jan 31;349(6308):434–438. doi: 10.1038/349434a0. [DOI] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Identification of the human U7 snRNP as one of several factors involved in the 3' end maturation of histone premessenger RNA's. Science. 1987 Dec 18;238(4834):1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Ryner L. C., Manley J. L. Requirements for accurate and efficient mRNA 3' end cleavage and polyadenylation of a simian virus 40 early pre-RNA in vitro. Mol Cell Biol. 1987 Jan;7(1):495–503. doi: 10.1128/mcb.7.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Hiesel R., Wissinger B., Brennicke A. RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U-to-C transition. Mol Cell Biol. 1990 May;10(5):2428–2431. doi: 10.1128/mcb.10.5.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L. RNA editing--a novel genetic phenomenon? Science. 1990 Oct 26;250(4980):512–513. doi: 10.1126/science.1700474. [DOI] [PubMed] [Google Scholar]
- Simpson L., Shaw J. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell. 1989 May 5;57(3):355–366. doi: 10.1016/0092-8674(89)90911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. C., Kuo S. R., Backus J. W., Harris S. G., Sparks C. E., Sparks J. D. In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J. E., Adams D. E. Assembly of a polyadenylation-specific 25S ribonucleoprotein complex in vitro. Mol Cell Biol. 1988 May;8(5):2052–2062. doi: 10.1128/mcb.8.5.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm N. R., Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990 Jun 1;61(5):879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- Walbot V. RNA editing fixes problems in plant mitochondrial transcripts. Trends Genet. 1991 Feb;7(2):37–39. doi: 10.1016/0168-9525(91)90225-F. [DOI] [PubMed] [Google Scholar]
- Wang M. J., Davis N. W., Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988 Jun;7(6):1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. J., Gegenheimer P. Substrate masking: binding of RNA by EGTA-inactivated micrococcal nuclease results in artifactual inhibition of RNA processing reactions. Nucleic Acids Res. 1990 Nov 25;18(22):6625–6631. doi: 10.1093/nar/18.22.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. RNA editing: guided but not templated? Cell. 1990 Jun 15;61(6):917–920. doi: 10.1016/0092-8674(90)90053-h. [DOI] [PubMed] [Google Scholar]
- Weiser M. M. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem. 1973 Apr 10;248(7):2536–2541. [PubMed] [Google Scholar]
- Weissmann C., Cattaneo R., Billeter M. A. RNA editing. Sometimes an editor makes sense. Nature. 1990 Feb 22;343(6260):697–699. doi: 10.1038/343697a0. [DOI] [PubMed] [Google Scholar]
- Wu J. H., Semenkovich C. F., Chen S. H., Li W. H., Chan L. Apolipoprotein B mRNA editing. Validation of a sensitive assay and developmental biology of RNA editing in the rat. J Biol Chem. 1990 Jul 25;265(21):12312–12316. [PubMed] [Google Scholar]