Abstract
Bundling of microtubules (MTs) is critical for the formation of complex MT arrays. In land plants, the interphase cortical MTs form bundles specifically following shallow-angle encounters between them. To investigate how cells select particular MT contact angles for bundling, we used an in vitro reconstitution approach consisting of dynamic MTs and the MT-cross-linking protein MAP65-1. We found that MAP65-1 binds to MTs as monomers and inherently targets antiparallel MTs for bundling. Dwell-time analysis showed that the affinity of MAP65-1 for antiparallel overlapping MTs is about three times higher than its affinity for single MTs and parallel overlapping MTs. We also found that purified MAP65-1 exclusively selects shallow-angle MT encounters for bundling, indicating that this activity is an intrinsic property of MAP65-1. Reconstitution experiments with mutant MAP65-1 proteins with different numbers of spectrin repeats within the N-terminal rod domain showed that the length of the rod domain is a major determinant of the range of MT bundling angles. The length of the rod domain also determined the distance between MTs within a bundle. Together, our data show that the rod domain of MAP65-1 acts both as a spacer and as a structural element that specifies the MT encounter angles that are conducive for bundling.
Introduction
Microtubule (MT) bundles play a crucial role in the formation and maintenance of organized MT arrays. In plant cells, the acentrosomal interphase MTs at the cell cortex are highly bundled, and their spatial organization dictates the direction of cell expansion (1). These so-called cortical MTs are nucleated from dispersed sites at the cell cortex (2–4) and are attached to the plasma membrane along their lengths (5,6). The cortical MTs are highly dynamic and treadmill along the plasma membrane surface, frequently interacting with other MTs (2). A subset of these interactions leads to the formation of cortical MT bundles. It is important to note that the encounter angle between interacting cortical MTs is a key determinant of the bundling probability (7). Specifically, bundling is observed to occur only after shallow-angle interactions (typically <40°) between cortical MTs (7). Steep-angle cortical MT interactions are followed by either MT crossover or depolymerization (7). The dependency of cortical MT bundling on the encounter angle appears to be important for the proper organization of the cortical MT array, because large shifts in the distribution of bundling angles were found to hinder cortical MT organization in computer simulations (8). However, the molecular basis for why only shallow-angle encounters lead to cortical MT bundling is unknown.
MT bundles are generated by the activity of MT-cross-linking proteins. The conserved MAP65/Ase1/PRC1 family of MT-cross-linking proteins plays a major role in the formation of both interphase and mitotic MT arrays. The Arabidopsis genome encodes nine MAP65 proteins, of which MAP65-1, MAP65-2, MAP65-5, and MAP65-8 localize to cortical MTs in vivo (9–11). Recently, genetic analyses have revealed that MAP65-1 and MAP65-2 together regulate cell growth during interphase (12) and play a role in cytokinesis (13). The Arabidopsis MAP65-1 is the most extensively studied isoform and is the focus of this study. Purified MAP65-1 bundles taxol-stabilized MTs in vitro and appears as filamentous cross-bridges that separate adjacent MTs by a distance of ∼25 nm (14,15). Electron microscopic observation of bundled cortical MTs in vivo shows that the spacing between adjacent MTs is also ∼25 nm (6,16,17), indicating that the MAP65 proteins are the major MT bundling proteins in this system. Similar to other MAP65/Ase1/PRC1 members, MAP65-1 is able to discriminate between parallel and antiparallel MTs in vitro and localizes to regions of antiparallel MT overlap with high specificity (15). Consistent with these results, MAP65-1 has been recently shown to preferentially label bundled cortical MTs in vivo, a significant subset of which contain antiparallel MTs (12).
Structural modeling of MAP65-1 based on fold recognition predicts the presence of four spectrin repeats that are thought to form an extended rodlike structure ∼25 nm in length (18). This N-terminal rod domain of MAP65-1 is thought to be flexible when bound to a single MT, based on the presence of several disordered domains in its sequence and on its hydrodynamic properties (15). Recent structural analysis of PRC1, the human MAP65 homolog, also suggests that the rod domain is likely to be flexible when PRC1 is bound to a single MT (19). Monomers of MAP65-1 are proposed to homodimerize through their rod domains, thus creating a cross-bridge between adjacent MTs (15). We hypothesize that a long and flexible rod domain might allow MAP65-1 to homodimerize within a particular range of angular orientations, thus specifying the range of bundling angles.
To investigate whether the rod domain of MAP65-1 is responsible for specifically selecting shallow-angle MT encounters for bundle formation, we developed a cell-free in vitro reconstitution assay consisting of dynamic MTs and purified MAP65-1. We found that MAP65-1 inherently selects shallow-angle encounters between antiparallel MTs for bundling. Time-lapse imaging of GFP-tagged MAP65-1 showed that MAP65-1 preferentially accumulates at and dynamically tracks with regions of antiparallel MT overlap. This property is associated with an increase in the dwell time of MAP65-1 within regions of antiparallel MT overlap. Reconstitution experiments with mutant versions of MAP65-1 that either lack a spectrin repeat or have additional spectrin repeats showed that the length of the rod domain determines both the spacing between cross-linked MTs and the range of encounter angles that lead to MT bundling. Together, these data provide a molecular mechanism for why only certain encounter angles lead to cortical MT bundling in plant cells.
Materials and Methods
Protein expression and purification
Constructs for protein expression were prepared using PCR and verified by sequencing. See Table S1 in the Supporting Material for the list of primers used to generate the constructs. Verified PCR products were introduced into the pET-28a(+) vector (Novagen, Darmstadt, Germany), which encodes for a 6x histidine tag at the N-terminus of proteins. The assembled plasmids were introduced into Rosetta (DE3) cells (Novagen) for protein expression. His-tagged recombinant proteins were affinity purified using a nickel column and subsequently desalted using a PD-10 column (GE Healthcare, Piscataway, NJ) and exchanged into BRB80 buffer (80 mM piperazine-1,4-bis(2-ethanesulfonic acid), 1 mM MgCl2, and 1 mM EGTA, pH 6.8). Protein aliquots were flash frozen in liquid nitrogen and stored at −80°C until use.
MT-binding assays
All MTs in this study were assembled in BRB80 buffer using purified bovine tubulin (Cytoskeleton, Denver, CO). The MT-binding assays were conducted by coincubating increasing concentrations of MTs with 1.5 μM of the specified recombinant protein along with 20 μM paclitaxel (Cytoskeleton) at 25°C for 30 min. The samples were then centrifuged at 39,000 × g for 20 min at 25°C to sediment the MTs. The resultant supernatant and pellet fractions were analyzed by SDS/PAGE and densitometry to calculate the bound fraction. The ΔR1 protein comigrates with tubulin, and therefore, we used Western blot analysis with a monoclonal Tetra-His antibody (Qiagen, Valencia, CA) to detect ΔR1 in the supernatant and pellet fractions. Densitometry was carried out using ImageJ. For analysis of MT bundling using taxol-stabilized MTs, 1 μM of rhodamine-labeled and taxol-stabilized MTs was coincubated with 1 μM of the specified recombinant protein at 25°C for 30 min and then visualized using fluorescence microscopy.
Reconstitution experiments with dynamic MTs
The in vitro reconstitution assay was developed based on our previously described method (20). Briefly, flow chambers of ∼20 μl volume were prepared using silanized coverslips attached to slides with double-sided sticky tape. The flow cell was coated with 20% monoclonal anti-biotin antibody (clone BN-34, Sigma, St. Louis, MO) and then blocked with 5% pluronic F-127 (Sigma). About 150 nM rhodamine-labeled and biotinylated guanosine 5′-(α,β-methylene)triphosphate (GMPCPP) MT seeds were then introduced into the flow cell. MT growth and bundling was initiated by introducing 20 μM 1:40 rhodamine-labeled bovine tubulin in BRB80 buffer and the specified MAP65-1 protein along with 0.15% methylcellulose, 100 mM DTT, an oxygen scavenging system consisting of 250 μg/ml glucose oxidase, 35 μg/ml catalase, and 4.5 mg/ml glucose, and 2 mM GTP. The samples were excited with 488-nm (at 10 mW output) and 561-nm (at 4 mW output) diode-pumped solid-state lasers (Melles Griot, Albuquerque, NM) to visualize MAP65-1-GFP and rhodamine-labeled MTs, respectively. Time-lapse images were captured with a back-illuminated electron-multiplying CCD camera (Hamamatsu, Bridgewater, NJ, ImageEM) and GFP (500–550 nm emission) and rhodamine (582–636 nm emission) filter sets. The polarity of growing MTs was assigned based on the difference in growth velocity between the plus and minus end. Kymograph analysis was conducted using Slidebook 5.0 (Intelligent Imaging Innovations, Denver, CO). Curve-fitting and statistical analysis were conducted using KaleidaGraph (Synergy Software, Reading, PA).
Single-molecule imaging
For photobleaching assays, 1 nM MAP65-1-GFP bound to rhodamine-labeled and taxol-stabilized MTs was imaged at the higher laser power (20 mW output from the 488-nm laser), and the fluorescence intensities of individual spots were measured over time to determine the number of bleaching steps. For comparison, 10 nM human kinesin1-GFP bound to rhodamine-labeled and taxol-stabilized MTs in the presence of AMPPNP was analyzed using identical image-acquisition conditions. For dwell-time analysis, reconstitution assays were conducted using 400 nM unlabeled MAP65-1 containing 8 nM MAP65-1-GFP. Kymographs of single and bundled MTs were generated using Slidebook 5.0 and used to measure the dwell times of individual molecules.
Electron microscopy
For negative-stain electron microscopy of MT bundles, 1 μM of taxol-stabilized MTs and 1 μM recombinant protein were coincubated at 25°C for 30 min. The MT suspension was then applied to formvar-coated grids and stained with a 7% (aqueous) solution of uranyl acetate for 2 min. The grids were then blotted dry and examined in an LEO 912 AB energy filter transmission electron microscope operated at 120 kV.
Results
MAP65-1 inherently selects shallow-angle MT encounters for bundling
To study the MT bundling activity of MAP65-1, we purified full-length MAP65-1 expressed in bacteria (Fig. 1 A). In vitro MT binding experiments showed that the equilibrium Kd of MAP65-1 for MTs is 1.03 ± 0.75 μM (Fig. 1 B), which is similar to that of Ase1, PRC1, and tobacco MAP65-1b (19,21,22). Since MAP65-1 is thought to dimerize within MT bundles (15), the measured Kd of MAP65-1 is likely to be a composite of MT binding and MAP65-1 dimerization.
Figure 1.
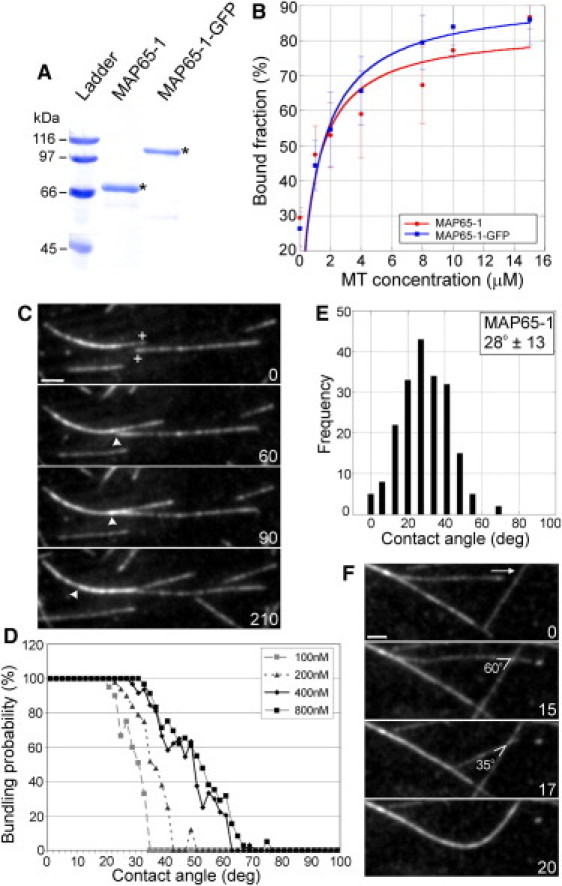
MAP65-1 preferentially bundles antiparallel MTs after shallow-angle encounters. (A) Coomassie-stained gel of purified MAP65-1 and MAP65-1-GFP proteins. The expected protein sizes are marked by asterisks. (B) Binding curves with 1.5 μM MAP65-1 and MAP65-1-GFP proteins at increasing MT concentrations. Each data point represents the mean ± SD from at least three independent experiments. The data were fit to the Michaelis-Menten equation yielding Kd values of 1.03 ± 0.75 μM and 1.27 ± 0.68 μM for MAP65-1 and MAP65-1-GFP, respectively. (C) Montage showing antiparallel MT bundling by 400 nM MAP65-1. The plus-ends of the MTs of interest are indicated in the first frame. Arrowheads mark the position of the plus end within the MT bundle. (D) Plots showing the probability for MT bundling as a function of the encounter angle at various MAP65-1 concentrations. The bundling probability was calculated as a percentage of the number of MT encounters that resulted in MT bundling at a particular angle. The total number of MT encounters observed for 100 nM, 200 nM, 400 nM, and 800 nM of MAP65-1 are 245, 243, 323, and 311, respectively. (E) Distribution of the frequency of MT bundling at various encounter angles in the presence of 400 nM MAP65-1 (N = 199 events). The mean MT bundling angle is 28 ± 13°. (F) MT bundling after a decrease in the crossover angle from 60° to 35°. The arrow indicates the direction of the growing plus end of the MT of interest. Numbers in C and F indicate time in seconds. Scale bars, 2 μm.
Next, we developed an in vitro reconstitution assay consisting of dynamic MTs and purified MAP65-1 to observe MT bundling by MAP65-1 using time-lapse total internal reflection fluorescence microscopy. In these experiments, growing MTs encountered each other along the coverglass surface and we noticed that only a subset of these MT encounters led to bundling. In control experiments lacking MAP65-1, we never observed MT bundling (Fig. S2). Analysis of the polarity of MAP65-1-induced MT bundles demonstrated that ∼90% of the MT bundles consisted of antiparallel MTs (N = 148; Fig. 1 C and Movie S1). Only ∼10% of MT bundles were between parallel MTs (Movie S2). Therefore, MAP65-1 inherently discriminates between parallel and antiparallel MTs.
In addition to the strong preference of MAP65-1 for cross-linking antiparallel MTs, we found that only a narrow range of MT encounter angles yielded MT bundles. Specifically, shallow-angle MT encounters invariably led to MT bundling, whereas steep-angle MT encounters led to MT crossover. This was true for both antiparallel (Fig. 1, D and E) and parallel MT bundling (Fig. S1, A and B). To determine whether the MT bundling angle is a function of the MAP65-1 concentration, we conducted reconstitution experiments at increasing MAP65-1 concentrations. Increasing the MAP65-1 concentration from 100 nM to 400 nM shifted the bundling probability to larger encounter angles (Fig. 1 D). A further increase in MAP65-1 concentration to 800 nM did not significantly affect the probability of MT bundling compared to 400 nM MAP65-1 (Fig. 1 D). Therefore, 400 nM MAP65-1 is sufficient to result in maximal MT bundling under our experimental conditions.
The distribution of bundling angles at 400 nM MAP65-1 (Fig. 1 E) is strikingly similar to the distribution of bundling angles for cortical MTs in living Arabidopsis plants (5). The different types of cortical MT bundling events that have been seen in cells were also observed in our reconstitution experiments. 1), In 64% of the cases, the growing plus end of an MT encountered the sidewall of another MT, which led to reorientation of its growth trajectory and continued polymerization alongside the impeding MT (Movie S3). This scenario has been called plus-end entrainment (1). 2), In 30% of the cases, MTs are observed to progressively coalign along their lengths (Movie S4), which has been called zippering (1,7). 3), In 6% of the cases, individual MTs instantly snapped together to form a bundle (Movie S5), as seen in both wild-type Arabidopsis plants and clasp-1 mutants (1,5). Together, these data suggest that our reconstitution experiments with 400 nM MAP65-1 mimic the physiological conditions in plant cells.
The dependency on the encounter angle for MT bundling was most convincingly demonstrated in cases where an MT initially crossed over another MT (at a steep encounter angle) but later become bundled as the crossover angle decreased to a shallow angle (Fig. 1 F and Movie S6). These examples highlight the inherent ability of MAP65-1 to discriminate between MT encounter angles and to selectively target shallow-angle MT encounters for bundling.
MAP65-1 dynamically tracks regions of MT overlap
To understand how MAP65-1 selectively bundles particular MT configurations, we generated a construct to express full-length MAP65-1 with GFP fused to its C-terminus. In vitro MT binding experiments showed that the Kd of MAP65-1-GFP for MTs is 1.27 ± 0.68 μM, which is statistically indistinguishable from the Kd of untagged MAP65-1 (Fig. 1 B). Initial tests also showed that MAP65-1-GFP is able to bundle taxol-stabilized MTs (Fig. S2). Therefore, the GFP tag does not interfere with the ability of MAP65-1 to bind and cross-link MTs.
Ase1 and PRC1 bind to MTs as dimers (19,23,24). Attempts to determine whether MAP65-1 binds to MTs as a dimer or as a monomer have yielded mixed results (14,15). To directly determine whether our purified MAP65-1 binds to MTs as a monomer or dimer, we performed photobleaching experiments of 1 nM MAP65-1-GFP bound to taxol-stabilized MTs. Analysis of the intensity traces of individual spots revealed that the fluorescence intensity of a majority of MAP65-1-GFP spots decreased to background levels in a single step, indicating the presence of a single GFP molecule that photobleached during the observation period (Fig. 2 A). In contrast, photobleaching analysis of human kinesin1-GFP under identical imaging conditions showed mostly two bleaching steps, consistent with the presence of two GFP molecules in the kinesin-1 dimer (Fig. 2 A). Therefore, our data indicate that MAP65-1-GFP binds to MTs predominantly as a monomer.
Figure 2.
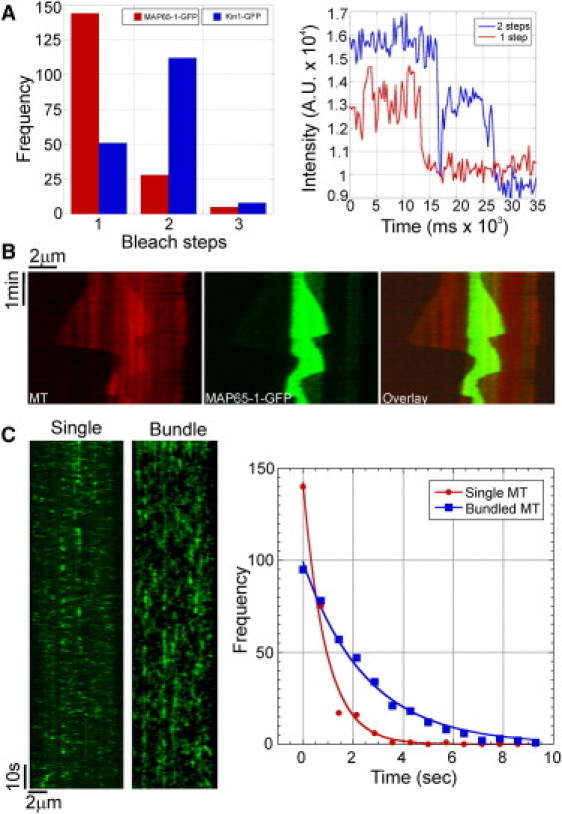
MAP65-1 binds to MTs as a monomer and preferentially localizes to regions of MT overlap. (A) Bar graph of the number of bleaching steps for MAP65-1-GFP and Kinesin1-GFP molecules bound to taxol-stabilized MTs (N = 177 and 171 for MAP65-1-GFP and Kinesin1-GFP, respectively). Examples of fluorescence intensity traces showing one and two bleaching steps are shown at right. (B) Kymograph showing the localization of 400 nM MAP65-1-GFP in an antiparallel MT bundle. MAP65-1-GFP specifically tracks the region of MT overlap and is barely detectable along stretches with a single MT. (C) To the left are kymographs showing the binding of 8 nM MAP65-1-GFP to a single MT and an antiparallel MT bundle. To the right are the distributions of dwell-times of single binding events of MAP65-1-GFP on single MTs (N = 257) and bundled MTs (N = 384). Exponential fits to the data yielded half-times of 0.62 ± 0.07 s and 1.82 ± 0.01 s, respectively.
We next carried out reconstitution experiments using 400 nM MAP65-1-GFP. We found that MAP65-1-GFP specifically accumulated at regions of antiparallel MT bundling after shallow-angle MT encounters (Movie S7). Kymograph analysis of MT bundles showed that MAP65-1-GFP dynamically tracks the regions of MT overlap (Fig. 2 B). In the same experiments, we detected little MAP65-1-GFP accumulation along single MTs and parallel MT bundles. Analysis of the dwell times of individual MAP65-1-GFP spots revealed that the dwell time of MAP65-1-GFP increased approximately threefold on antiparallel MT bundles compared to the dwell time on single MTs (Fig. 2 C) and on parallel MT bundles (Fig. S1 C). These results indicate that a decrease in the MT unbinding rate underlies the ability of MAP65-1 to selectively accumulate at regions of antiparallel MT overlap, similar to that described for Ase1 (23).
The length of the rod domain of MAP65-1 specifies the range of MT bundling angles
To test whether the rod domain of MAP65-1 is involved in specifying MT bundling angles, we generated constructs to express and purify several mutant versions of MAP65-1 with either shorter or longer rod domains compared to wild-type MAP65-1 (Fig. 3, A and B). The mutant proteins were designated ΔR1 (first spectrin repeat deleted), ΔR2 (second spectrin repeat deleted), and R1R4 (entire spectrin-repeat domain duplicated). In vitro MT binding experiments showed that the Kd of the various mutant proteins for MTs is similar to that of wild-type MAP65-1 (Fig. 3 C). Once again, we note that these values represent a convolution of both MT binding and MAP65 protein dimerization, whose individual contributions cannot be discriminated in these binding experiments. The mutant proteins are also able to bundle taxol-stabilized MTs (Fig. S2). Therefore, all of the mutant proteins are able to bind and bundle MTs. We found that 400 nM ΔR1 bundles MTs more weakly compared to the other proteins. Therefore, for our subsequent experiments we increased the protein concentration to 800 nM ΔR1. This increase in ΔR1 protein concentration does not hinder interpretation of data, because the MT bundling angle distributions are similar using either 400 nM or 800 nM of wild-type MAP65-1 protein (Fig. 1 D).
Figure 3.
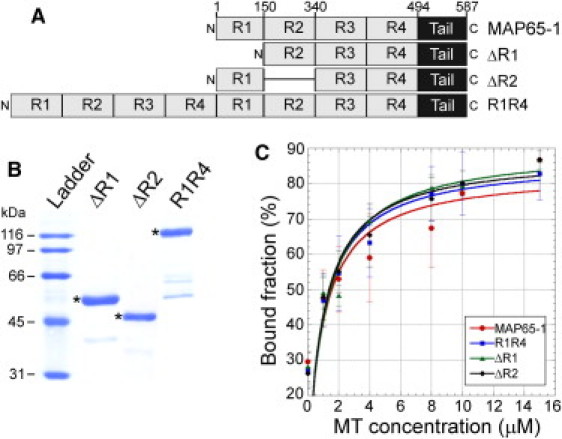
Purification and MT binding of MAP65-1 mutants. (A) Schematic of the domain architecture of MAP65-1 and the various mutants used in this study. The four predicted spectrin repeats are labeled R1–R4. Tail refers to the unstructured domain at the C-terminus of MAP65-1. (B) Coomassie-stained gel of purified ΔR1, ΔR2, and R1R4 proteins. The expected protein sizes are marked by asterisks. (C) Binding curves with 1.5 μM ΔR1, ΔR2, and R1R4 proteins at increasing MT concentrations. Each data point represents the mean ± SD from at least three independent experiments. The data were fit to the Michaelis-Menten equation, yielding Kd values of 1.17 ± 0.73 μM, 1.04 ± 0.61 μM, and 1.04 ± 0.63 μM for ΔR1, ΔR2, and R1R4, respectively. The binding curve for MAP65-1 is reproduced from Fig. 1B for comparison to the mutant proteins.
To confirm that the ΔR1, ΔR2, and R1R4 mutants altered the spacing between bundled MTs, as expected from the predicted lengths of their rod domains, we performed negative-stain electron microscopy of MTs incubated with these proteins. Electron micrographs of MTs bundled by wild-type MAP65-1 showed coaligned MTs separated by an average distance of ∼24 nm (Fig. 4 B). In contrast, the spacings between MTs in bundles induced by ΔR1 and ΔR2 are ∼9 nm and 10 nm, respectively (Fig. 4, C and D). This is consistent with previous measurements of distances separating MTs bundled by ΔR1 and ΔR2 (15). Electron micrographs of MTs bundled by R1R4 showed that the average distance between MTs is increased to ∼37 nm (Fig. 4 E). These results indicate that the ΔR1, ΔR2, and R1R4 mutants indeed produce the expected decrease or increase in inter-MT spacing, as predicted by the number of spectrin repeats in their rod domain.
Figure 4.
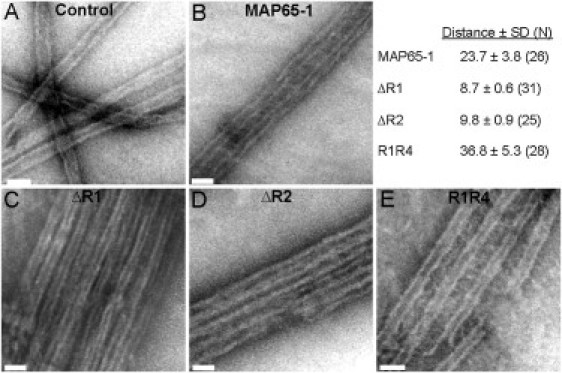
Length of the rod domain of MAP65-1 determines the distance between MTs in a bundle. Negative-stain electron microscopy of 1 μM MTs alone (A) or 1 μM MTs coincubated with 1 μM of MAP65-1 (B), ΔR1 (C), ΔR2 (D), and R1R4 (E), respectively. The mean ± SD of the distance (nm) is shown in the figure. The number of independent measurements between separate MTs is shown in parentheses. Scale bars, 50 nm.
To determine whether the ΔR1, ΔR2, and R1R4 mutants altered the distribution of the MT bundling angles, we conducted reconstitution experiments with 800 nM ΔR1, 400 nM ΔR2, and 400 nM R1R4. We found that both ΔR1 and ΔR2 target only very shallow-angle MT encounters for bundling and generally take several attempts to initiate MT bundling compared to MAP65-1 (Movie S8 and Movie S9). Analysis of the MT bundling angles showed that both ΔR1 and ΔR2 shift the distribution of MT bundling angles to smaller angles compared to MAP65-1 (Fig. 5, A–C). The mean bundling angles are 16° and 18° with ΔR1 and ΔR2, respectively, which are significantly lower than the mean bundling angle of 28° with MAP65-1 (p < 0.0001 using the t-test). In contrast, R1R4 frequently resulted in MT bundling, even after steep-angle MT encounters (Movie S10). Analysis of the MT bundling angles showed that R1R4 dramatically expands the distribution of MT bundling angles to include steep angles (Fig. 5 D). The mean bundling angle with R1R4 is 36°, which is significantly higher than the mean bundling angle with MAP65-1 (p < 0.0001 using the t-test). Analysis of the bundling probability as a function of the contact angle shows a striking leftward shift for ΔR1 and ΔR2, whereas R1R4 shows a striking rightward shift as compared to MAP65-1 (Fig. 5 E). Based on these results, we conclude that the length of the rod domain of MAP65-1 is a major determinant of the MT bundling angle.
Figure 5.
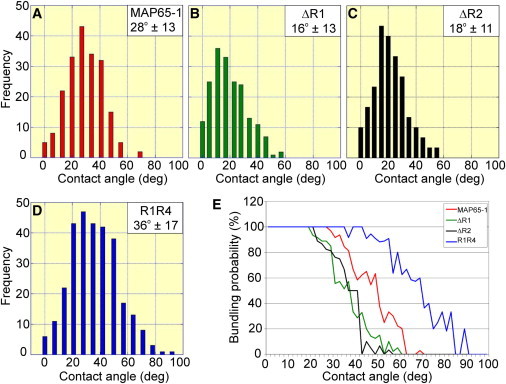
Length of the rod domain of MAP65-1 determines the MT bundling angle. (A–D) Distribution of the frequency of MT bundling at various encounter angles in the presence of 400 nM MAP65-1 (A), 800 nM ΔR1 (B), 400 nM ΔR2 (C), and 400 nM R1R4 (D). The data for MAP65-1 are reproduced from Fig. 1E for comparison to the mutant proteins. N = 199, 189, 231, and 295 for MAP65-1, ΔR1, ΔR2, and R1R4, respectively. The means ± SD of the bundling angle are shown in the figure. (E) Plots showing the probability for MT bundling as a function of the encounter angle in the presence of 400 nM MAP65-1, 800 nM ΔR1, 400 nM ΔR2, and 400 nM R1R4.
Discussion
In this study, we sought to understand the molecular basis for the observation that only shallow-angle encounters between cortical MTs result in bundle formation in plant cells. This feature is an important aspect of cortical MT array organization, because only similarly oriented cortical MTs are allowed to productively interact and form bundles, thus promoting the formation of linearly ordered arrays. Using a minimal system consisting of dynamic MTs and purified MAP65-1, we found that the ability to selectively bundle MTs that interact at a shallow angle is an intrinsic property of MAP65-1 and does not require additional factors. Furthermore, we found that the length of the rod domain of MAP65-1 determines the range of MT bundling angles, thus providing insight into the structural feature of MAP65-1 that is responsible for bundling-angle selection.
We found that increasing the MAP65-1 concentration increases the range of MT bundling angles up to a certain limit. This observation is consistent with the prediction from a theoretical model of cortical MT interactions, which posits that an increase in the concentration of an MT cross-linking protein will increase the probability of MT bundling by increasing the torque necessary to bend an incoming MT along the impeding MT (25). It is worth noting that once the torque exerted by the cross-linking protein exceeds the bending rigidity of the incoming MT, any further increase in the concentration of the cross-linking protein would have little effect, in agreement with our finding. Based on our data, regulation of the intracellular concentration of MAP65 proteins offers cells a mechanism to specify which MT encounters will lead to bundling. This ability may be important during MT array formation, remodeling, and disassembly.
Like other members of the MAP65/Ase1/PRC1 family, we found that MAP65-1 can inherently distinguish between parallel and antiparallel MTs. In our in vitro experiments, ∼90% of the MAP65-1-induced MT bundles consisted of antiparallel MTs. This is comparable to Ase1p and PRC1, which yield antiparallel MT bundles ∼70% (23) and 90% (19) of the time, respectively. Our results are also consistent with previous work showing that MAP65-1 localizes to antiparallel MT bundles both in vitro (15) and in vivo (12). Dwell-time analysis of individual MAP65-1 molecules showed that the off rate on antiparallel MT overlaps was around threefold lower than on single MTs and parallel-MT overlaps. The increased affinity for antiparallel MTs provides a possible explanation for the selective cross-linking of antiparallel MTs by MAP65-1.
In our assays, the constituent MTs within a bundle remain dynamic and MAP65-1 is observed to dynamically track the regions of antiparallel MT overlap, strikingly illustrating the differential binding of MAP65-1 to antiparallel MT overlaps versus single MTs. Fluorescently tagged MAP65-1 is similarly observed to track along bundled segments of cortical MTs in Arabidopsis plants (12). This property of MAP65-1 is similar to that of the mitotic MAP65-4 (26), MAP65-3 (27), Ase1 (28), and PRC1 (19,24) and thus appears to be a conserved feature of the MAP65/Ase1/PRC1 family.
Our measured dwell time of ∼2 s for individual MAP65-1 molecules on antiparallel MTs in vitro is in good agreement with the reported bulk turnover rate of ∼5 s for MAP65-1 on cortical MT bundles in vivo (11,12). Photobleaching analysis of individual MAP65-1 molecules showed that MAP65-1 binds to MTs as a monomer, which is consistent with the results of Gaillard et al. (15), who concluded, based on analytical ultracentrifugation and size-exclusion chromatography experiments, that MAP65-1 is monomeric in solution. Therefore, it is not necessary for MAP65/Ase1/PRC1 homologs to assemble into preformed dimers to be able to bundle MTs.
MT bundling requires the formation of antiparallel dimers from monomeric MAP65-1 subunits bound to separate MTs to form a stable cross-link between encountering MTs. Biochemical evidence indicates that the spectrin repeats in the rod domain of MAP65-1 mediate the formation of an antiparallel dimer as described for muscle α-actinin (15,18). The rod domain of MAP65-1 is also likely to be a flexible structure when bound to a single MT (15,19). The conformational flexibility of the rod domain may allow MAP65-1 monomers at multiple orientations to dimerize, thus increasing the chances for MT bundling. A possible mechanism for why only certain MT encounter angles lead to MT bundling is that these MT orientations position the MAP65-1 monomers in a way that allows them to productively interact and dimerize. Thus, MT orientations that allow MAP65-1 monomers to dimerize will lead to bundling, whereas other MT orientations that are not conducive for MAP65-1 dimerization will fail to produce MT bundles (Fig. 6, A–C).
Figure 6.
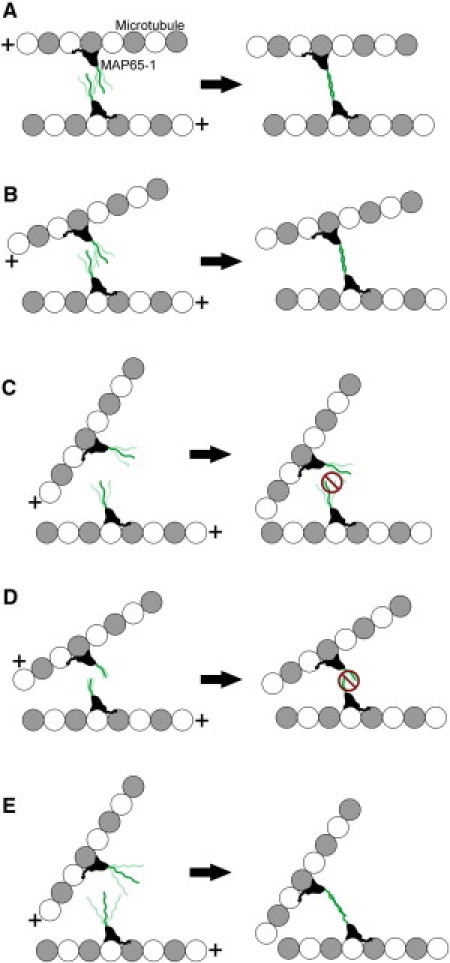
Model for encounter-angle-dependent MT bundling by MAP65-1. MAP65-1 monomers are shown bound to MTs (a single MT protofilament is shown for simplicity). The plus sign indicates the MT plus end. The N-terminal rod domain of MAP65-1 is shown projecting from the MT surface, and its conformational flexibility is represented by its multiple positions. If two MTs encounter each other in a nearly parallel orientation (A) or at a shallow-angle (B), the MAP65-1 monomers are able to dimerize and form a stable cross-link, thus resulting in MT bundling. In contrast, if two MTs encounter each other at a steep angle, the MAP65-1 monomers are unable to dimerize, because their rod domains cannot interact productively at these angles (C). Consequently, these MTs do not bundle. In the case of the ΔR1 and ΔR2 mutants, their shorter rod domains are probably stiffer, thus requiring even shallower encounter angles for dimer formation (D). In contrast, the R1R4 mutant has a longer rod domain that is likely to be more flexible than the rod domain of wild-type MAP65-1, which allows dimer formation and MT bundling even at steep encounter angles (E).
In our in vitro reconstitution experiments, the length of the rod domain had a strong effect on the MT bundling angle. Shortening the rod domain by deleting a spectrin repeat constrained the bundling angles to smaller values, whereas lengthening the rod domain by including additional spectrin repeats greatly expanded the range of bundling angles to include larger values, as compared to wild-type MAP65-1. Deletion of either the first or the second spectrin repeat resulted in a similar shift in the distribution of bundling angles, indicating that the length of the rod domain, and not a particular sequence, is the key determinant of the MT bundling angle. The length of the rod domain of MAP65-1 may impact the MT bundling angle in at least two ways that are not mutually exclusive: 1) it might affect the efficiency and/or strength of dimer formation based on the extent of overlap that would be possible between the rod domains of MAP65-1 monomers; and 2), it might affect the range of the angular sector that the rod domain explores given its conformational flexibility. In particular, the shorter rod domains of the ΔR1 and ΔR2 mutants might be stiffer, thus allowing their dimerization and consequent MT bundling only at very shallow encounter angles (Fig. 6 D). In contrast, the longer rod domain of the R1R4 mutant is envisioned to be more flexible than the rod domain of wild-type MAP65-1, which would allow the R1R4 mutant to dimerize and bundle MTs at even higher encounter angles (Fig. 6 E). Besides affecting the MT bundling angle, we found that the length of the rod domain of MAP65-1 also acts as a spacer that determines the distance between MTs within a bundle.
A similar mechanism for selectively bundling shallow-angle MT interactions is probably applicable to MAP65 homologs that bind to MTs as dimers. Ase1 and PRC1 bind to MTs as dimers, and both specifically bundle MTs that interact at shallow angles (19,28). It has been proposed that the flexibility of the rod domain of PRC1 dimers allows contact with a second MT within a certain range of MT orientations, thus determining the acceptable MT bundling angles (19). It is interesting that the distribution of MT bundling angles for Ase1 is very similar to that of the ΔR1 and ΔR2 mutants (28), and the distance between Ase1-induced MT bundles is ∼6 nm (21), which is in the range of the MT spacing by the ΔR1 and ΔR2 mutants. Therefore, the length of the rod domain is likely to be an important determinant of the MT bundling angle even for dimeric MAP65 homologs.
Acknowledgments
The authors thank Dr. Howard Berg at the Donald Danforth Plant Science Center for the electron microscopy of microtubule bundles.
This work was partially supported by funds from I-CARES, Washington University in St. Louis.
Supporting Material
References
- 1.Wasteneys G.O., Ambrose J.C. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 2009;19:62–71. doi: 10.1016/j.tcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Shaw S.L., Kamyar R., Ehrhardt D.W. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 3.Chan J., Sambade A., Lloyd C. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell. 2009;21:2298–2306. doi: 10.1105/tpc.109.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura M., Ehrhardt D.W., Hashimoto T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 2010;12:1064–1070. doi: 10.1038/ncb2110. [DOI] [PubMed] [Google Scholar]
- 5.Ambrose J.C., Wasteneys G.O. CLASP modulates microtubule-cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell. 2008;19:4730–4737. doi: 10.1091/mbc.E08-06-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton D.A., Vantard M., Overall R.L. Analysis of cortical arrays from Tradescantia virginiana at high resolution reveals discrete microtubule subpopulations and demonstrates that confocal images of arrays can be misleading. Plant Cell. 2008;20:982–994. doi: 10.1105/tpc.108.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit R., Cyr R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell. 2004;16:3274–3284. doi: 10.1105/tpc.104.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eren E.C., Dixit R., Gautam N. A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol. Biol. Cell. 2010;21:2674–2684. doi: 10.1091/mbc.E10-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussey P.J., Hawkins T.J., Smertenko A. The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 2002;50:915–924. doi: 10.1023/a:1021236307508. [DOI] [PubMed] [Google Scholar]
- 10.Van Damme D., Bouget F.Y., Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 2004;40:386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- 11.Smertenko A.P., Kaloriti D., Hussey P.J. The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell. 2008;20:3346–3358. doi: 10.1105/tpc.108.063362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas J.R., Courtney S., Shaw S.L. Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell. 2011;23:1889–1903. doi: 10.1105/tpc.111.084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasabe M., Kosetsu K., Machida Y. Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal. Behav. 2011;6:743–747. doi: 10.4161/psb.6.5.15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smertenko A.P., Chang H.Y., Hussey P.J. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell. 2004;16:2035–2047. doi: 10.1105/tpc.104.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard J., Neumann E., Vantard M. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell. 2008;19:4534–4544. doi: 10.1091/mbc.E08-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardham A.R., Gunning B.E. Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 1978;77:14–34. doi: 10.1083/jcb.77.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J., Jensen C.G., Lloyd C.W. The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA. 1999;96:14931–14936. doi: 10.1073/pnas.96.26.14931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Mao T., Yuan M. The AtMAP65-1 cross-bridge between microtubules is formed by one dimer. Plant Cell Physiol. 2007;48:866–874. doi: 10.1093/pcp/pcm059. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian R., Wilson-Kubalek E.M., Kapoor T.M. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixit R., Ross J.L. Elsevier; Philadelphia, PA: 2010. Studying Plus-End Tracking at Single Molecule Resolution Using TIRF Microscopy. [DOI] [PubMed] [Google Scholar]
- 21.Schuyler S.C., Liu J.Y., Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wicker-Planquart C., Stoppin-Mellet V., Vantard M. Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J. 2004;39:126–134. doi: 10.1111/j.1365-313X.2004.02115.x. [DOI] [PubMed] [Google Scholar]
- 23.Kapitein L.C., Janson M.E., Peterman E.J. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr. Biol. 2008;18:1713–1717. doi: 10.1016/j.cub.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 24.Bieling P., Telley I.A., Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 25.Allard J.F., Ambrose J.C., Cytrynbaum E.N. A mechanochemical model explains interactions between cortical microtubules in plants. Biophys. J. 2010;99:1082–1090. doi: 10.1016/j.bpj.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fache V., Gaillard J., Vantard M. Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell. 2010;22:3804–3815. doi: 10.1105/tpc.110.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho C.M., Hotta T., Liu B. Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell. 2011;23:2909–2923. doi: 10.1105/tpc.110.078204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janson M.E., Loughlin R., Tran P.T. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


