Abstract
In this short communication we report a quick, cost-free method of purification of DNA fragments from agarose gel. Unlike those procedures that involve commercial kits, this method uses glass wool or absorbent cotton to filter agarose gel during a quick spinning-down of DNA, thus significantly simplifying the routine practice of many molecular biologists and decreasing the cost.
Keywords: DNA fragments, agarose gel, method of purification.
Introduction
Extraction of DNA fragments from agarose gel has many uses in biomedical research. For instance, we often need to clone or sequence a DNA fragment digested by restriction enzyme or a product of polymerase chain reactions (PCR). For these purposes, we need to fractionize the DNAs in agarose gel, excise the desired band from the gel, and then purify the DNA by removing the agarose. Sometimes, plasmid DNA isolated from bacteria may also need to be purified from agarose gel so as to remove any residual of bacterial endotoxin that may be cytotoxic to eukaryotic cells during transfection of the cells with the plasmids. Traditionally, special low-melting agarose has to be used for the extraction, so that the excised DNA-containing gel can be melted easily by heating, thus releasing the DNA into solution. This low-melting agarose method has now been replaced by quite a few commercial kits. In this paper, however, we introduce a quicker and cost-free method as an alternative to those commercial kits.
Materials
Key materials include a 22G syringe needle, a small centrifuge machine that can run at 5,000-10,000 revolutions per minute (rpm), depending on the size of the DNA fragment, and some glass wool (Cat# 3352; Ohio Valley Specialty, Marietta, OH, USA; www.ovsc.com) or absorbent cotton which in our case was collected from cotton swabs purchased from Dollar Tree, a chain store everywhere in the USA. Additional materials include regular agarose (gel temperature 33±1.5 0C and melt temperature 87±1.5 0C; Cat # CA3510-8, Denville Scientific Inc; www.denvillescientific.com), and 1% solution of ethidium bromide (Cat # BP1302-10, Denville Scientific Inc, supplied by Fisher Biotech; www.denvillescientific.com). A surgical blade of size 22 (Cat# 12-460-440, from Fisher Scientific; www.fishersci.com), a 500-µl centrifuge tube and a 1.7-ml Eppendorf tube were also needed. For control, the UltraClean Gel DNA Extraction Kit (Cat # G-1210-0300, IscBioExpress; www.bioexpress.com) was used.
Results and Discussion
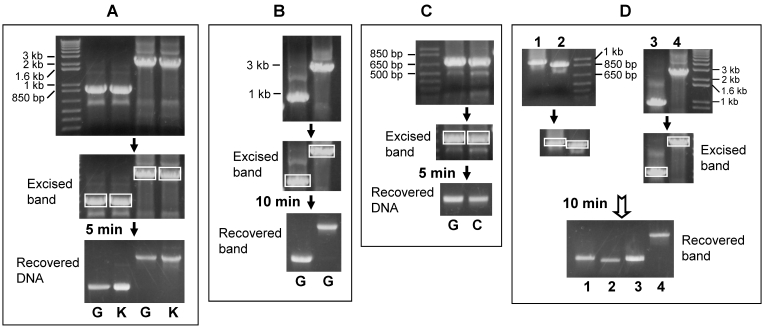
The syringe needle was used to make a hole in the bottom of a 500-µl centrifuge tube. The hole should be made from inside of the tube to the outside, not the other away around (Fig 1). Some glass wool was put to the bottom of the tube, as a roughly 4-mm cushion filter. The 1% agarose gel was pre-added with ethidium bromide (1 µl of 1% ethidium bromide solution in 15 ml of 1% agarose). The desired DNA band of PCR product fractioned in the gel was visualized under ultraviolet light and excised from the gel with a surgical blade. The excised gel should be as small as possible to avoid diluting the recovered DNA. The piece of DNA-containing gel was laid on the cushion filter in the tube. The tube was capped and put into a 1.7-ml Eppendorf tube. The uncapped Eppendorf tube was then put in a centrifuge machine as illustrated in figure 1 and centrifuged at 5,000-10,000 rpm for 5-10 minutes, depending on the length of DNA fragment. A DNA fragment smaller than 1 kilo base-pairs (1 kb) can be recovered by centrifugation at 5,000 rpm for 5 minutes, a 1-2 kb fragment at 10,000 rpm for 5 minutes, and a 2-3 kb fragment at 10,000 rpm for 10 minutes or less. We have not yet tested fragments larger than 3 kb because there is less need for such fragments. Centrifugation extracts the DNA, along with aqueous components, from the gel, and spins them down into the Eppendorf tube through the hole. After centrifugation, the 500-µl tube should be discarded properly because it contains hazardous ethidium bromide residual. The DNA-containing aqueous fraction in the Eppendorf tube should be collected for future use. Usually, the recovered volume is about 15-30 µl, depending on the amount of agarose gel excised. The recovery rate varies between 30-60% of the amount of DNA in the gel piece (Fig 2A, 2B and 2C), with larger fragments at the lower end. The recovery rate is sometimes better, but hardly ever reaches 80%.
Fig 1.
Illustration of cushion-filter preparation. A 22G needle is used to make a hole at the bottom of a 500-µl tube. Some glass wool or cotton is then put into the tube to make a cushion in the bottom of the tube. The tube is capped and put into a 1.7-ml Eppendorf tube. The uncapped Eppendorf tube is put into a centrifuge machine.
Fig 2.
Representative results of purifying DNA from gel by centrifugation at 10,000 rpm for different time periods. A: PCR products were fractioned in 1% agarose gel (top panel). The 3-kb and 1-kb bands (boxed in the middle panel) were excised out and then purified with glass wool (G) or commercial kit (K) by centrifugation for 5 minutes. The recovered DNA was loaded into a new 1% agarose gel shown in the bottom panel. Comparison of the bottom with the top panels reveals that the DNA recovery rates vary between 30-40% by our estimation, with a very slightly better recovery from the commercial kit. B: The same 1-kb and 3-kb DNA fragments shown in A were PCR-amplified again, fractionized in a 1% gel and purified with glass wool (G) by centrifugation for 10 minutes, resulting in about 50% recovery of the 1-kb and 40% recovery of the 3-kb fragment, respectively. C: A DNA fragment of about 800-bp was PCR amplified and then loaded into two wells of a gel (top panel). The fractionized bands (boxed) were cut out and purified with glass wool (G) or absorbent cotton (C) by centrifugation for 5 minutes. The recovery rates are not significantly different between the two methods, considering that the amount of DNA used for the glass wool method is also slightly more than that used for the cotton method (top panel). D: Four DNA fragments of different lengths were excised (boxed bands) and purified with cotton filter by centrifugation for 10 minutes. The recovery rates vary between 30-70% by our estimation.
To compare the efficiency with that resulting from using a commercial kit, the same amount of PCR products was loaded into 1% gel in duplicate. One of the two excised bands as illustrated in figure 2A was run through the above described procedure while the other was run with the UltraClean Gel DNA Extraction Kit by following the manual. The commercial kit sometimes results in a very slightly better recovery (Fig. 2A), in part because we are still more experienced in using the kit. The actual merit of the commercial kit is that the recovered DNA is in water solution, making it ready for various uses. In contrast, the aqueous solution directly spun down from the gel in our method is gel-making buffer, Tris-Borate-EDTA (TBE) buffer in our case. Therefore, further purification or concentration by ethanol or isopropanal precipitation and then resuspension of the DNA in a small volume of water may be needed, depending on what the DNA is later used for.
We have also established an alternative method by replacing the glass wool with absorbent cotton. The cotton cushion is made exactly as described above with glass wool. We compare the DNA recovery rates in both methods and find that cotton works as efficiently as glass wool (Fig 2C and 2D).
A few technical aspects may affect the quality and quantity of the recovered DNA. In both methods, a thicker cushion may retain more DNA, leading to a lower yield, whereas a thinner cushion may have a less filterability to retain all the agarose in the 500-µl tube. A longer time of centrifugation may help recover more DNA or a larger fragment of DNA but, in the meantime, may also risk spinning down some agarose into the Eppendorf tube, thus lowering the quality.
If very high quality and quantity of recovered DNA are needed, experimental optimizations are suggested to harmonize the thickness of cushion filter, the size of excised agarose gel, and the length of DNA fragment with the centrifugation time and speed. We now more often use the cotton method since commercial kits consume more time but are not much better.
Acknowledgments
This work was supported in part by a grant from National Science Foundation of China (#81060200) to Dr. D-Z Luo and by a grant (CON25943) from the Department of Defense of USA to Dr. DJ Liao. We would like to thank Fred Bogott, M.D., Ph.D., at Austin Medical Center, Austin of Minnesota, for his excellent English editing of this manuscript.