Abstract
Background
Mupirocin has been used for the treatment of skin infections and eradication of nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA). The increased use of this antibiotic has been accompanied by outbreaks of MRSA that are resistant to mupirocin.
Objective
This study aims to determine the prevalence, genotype and antimicrobial susceptibility of mupirocin-resistant MRSA from 4 Korean hospitals.
Methods
A total 193 MRSA clinical isolates were collected from four university hospitals. Antimicrobial susceptibility tests, including mupirocin, and pulsed-field gel electrophoresis (PFGE) pattern analysis were performed.
Results
Overall, 27 of the 193 (14.1%) MRSA isolates were resistant to mupirocin. All of the (A) hospital isolates showed high-level (HL) mupirocin resistance and the low-level (LL) mupirocin resistant strains were from three other hospitals. The PFGE patterns of 16 mupirocin-resistant isolates were divided into 5 clusters (1-5), and the nine HL mupirocin-resistant isolates belonged to cluster 1. Both the HL and LL mupirocin-resistant MRSA isolates were susceptible to vancomycin and rifampin, but they were resistant to ciprofloxacin, clindamycin and tetracycline. The erythromycin and fusidic acid resistance rates were different between the HL and LL resistant isolates.
Conclusion
HL mupirocin-resistant isolates that could transfer this resistance to other bacteria were detected and these isolates were clonally related. The emergence of mupirocin resistant isolates emphasizes the importance of using antibiotics judiciously and carefully monitoring the prevalence of mupirocin resistance.
Keywords: Antimicrobial susceptibility, MRSA, Mupirocin, Pulsed field gel electrophoresis
INTRODUCTION
Mupirocin is a topical antibiotic that was originally isolated from Pseudomonas fluorescens, and it inhibits bacterial protein synthesis by competitively binding to isoleucyl-tRNA synthetase (IleS). This drug is particularly effective against streptococci and staphylococci, including methicillin-resistant Staphylococcus aureus (MRSA). Mupirocin is one of the most popular topical agents used for the treatment of skin infection and eradication of nasal carriage of MRSA1. The increased use of this antibiotic has been accompanied by outbreaks of MRSA that are resistant to mupirocin2.
The mupirocin resistant strains are divided into two groups depending on the minimum inhibitory concentration (MIC) against mupirocin: low-level resistance (MIC=8~256µg/ml) and high-level resistance (MIC>256µg/ml). High-level mupirocin resistant strains are important in the clinical field because they are able to transfer resistance genes to other bacteria by plasmid conjugation, consequently protecting these bacteria from being eradicated with mupirocin3.
The use of mupirocin ointment has been increasing in Korea, but there are only a few studies about mupirocin resistance in MRSA4-6. Therefore, we measured the prevalence, genotype and antimicrobial susceptibility of mupirocin-resistant MRSA from four Korean university hospitals.
MATERIALS AND METHODS
Bacterial isolates
A total of 193 MRSA isolates were collected from the clinical cultures taken from patients at four Korean university hospitals (A~D) from April 2008 to July 2009. The specimens originated from the skin, pus, blood, central venous catheter tip, sputum, tracheal tip, urine, and wound. Of these, 103 strains were from (A) hospital and 30 strains were from the other 3 hospitals, respectively. All the MRSA isolates were randomly collected from each of the hospitals.
Antimicrobial susceptibility test
The susceptibility to antimicrobial agents was determined by agar dilution tests, according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI). Eight antimicrobial agents were tested: ciprofloxacin, clindamycin, erythromycin, fusidic acid, mupirocin, rifampin, tetracycline and vancomycin. S. aureus ATCC 33591 was used as a control strain.
Briefly, serial two-fold dilutions of the antibiotics were prepared in Muller-Hinton agar (Difco Laboratories, Detroit, MI, USA). The strains were subcultured on tryptic soy agar (Difco Laboratories), suspended in tryptic soy broth (Difco Laboratories) and adjusted to the turbidity of a 0.5 McFarland standard. Next, the suspension was diluted 1:10 and inoculated on each plate coated with antibiotic containing media by a Steer replicator. The inoculated plates were incubated at 35℃ for 24 hr. The lowest concentration of antibiotic that inhibits the visible growth of an organism was regarded as the MIC, and growth of one isolate was ignored. The MIC50 and MIC90 were the MICs that were required to inhibit 50% and 90% of the organisms, respectively. The MIC determination was evaluated according to the CLSI guidelines7.
Detection of the femA, mecA and mupA genes by polymerase chain reaction (PCR)
All the isolates were confirmed to be S. aurues and MRSA by detection of the femA and mecA genes using a PCR assay. The high level mupirocin resistant strains were also confirmed by detection of the mupA gene (Fig. 1~4). Three primer pairs were used for the detection of the femA, mecA and mupA genes. For femA gene detection, forward primer (5'-CATGATGGCGAGATTACAGG-3') and reverse primer (5'-CGCTAAAGGTACTAACACACGG-3') with a fragment of 372 bp were used8. For mecA gene detection, forward primer (5'-ATGAGATTAGGCATCGTTCC-3') and reverse primer (5'-TGGATGACAGTACCTGAGCC-3') with a fragment of 554 bp were used9. For mupA gene detection, forward primer (5'-CCCATGGCTTACCAGTTGA-3') and reverse primer (5'-CCATGGAGCACTATCCGAA-3') with a fragment of 1.6 kb were used10.
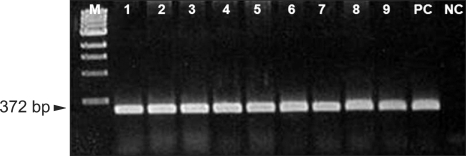
Fig. 1.
Agarose gel electrophoresis of the amplified 372-bp DNA fragments of the femA gene. The samples are lane M: a 100 bp size marker, lanes 1-9: mupirocin resistant MRSA, PC: femA-positive control (S. aureus ATCC 33591) and NC: femA-negative control (S. epidermidis ATCC 2228). MRSA: methicillin-resistant Staphylococcus aureus.
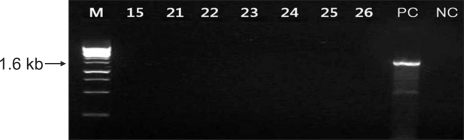
Fig. 4.
Agarose gel electrophoresis of the amplified 1.6 kb DNA fragments of the mupA gene. The samples are lane M: a 1-kb size marker, lanes 15, 21-26: low-level mupirocin resistant MRSA, PC: mupA-positive control (KCKC 6129) and NC: mupA-negative control (ATCC 25923). MRSA: methicillin-resistant Staphylococcus aureus.
Prior to DNA extraction, frozen bacteria were subcultured twice onto Mueller-Hinton agar plates. For extraction, one to five bacterial colonies were suspended in 50µl of cell lysis buffer (Genotek Co., Daejeon, Korea) and this was heated at 100℃ for 10 min. After centrifugation at 12,000 rpm for 10 min, 2µl of the supernatant was used for DNA extraction. The PCR reactions were performed using a Primer Mix Kit (Genotek Co.). The thermal cycler (GeneAmp PCR system 9700, Perkin-Elmer Cetus, Foster City, CA, USA) was programmed with the following parameters: pre-denaturation at 95℃ for 15 min, 30 cycles of denaturation at 94℃ for 30 s, annealing at 68℃ for 90 s, extension at 72℃ for 90 s and a final extension at 72℃ for 10 min. The amplified products were run on a 2% agarose gel for 20 min at 200 volts and visualized with ethidium bromide.
Pulsed-field gel electrophoresis (PFGE)
A total of 16 strains of mupirocin resistant MRSA (10 isolates with high-level resistance and 6 isolates with low-level resistance) were used for PFGE11. The high-level resistant strains were from (A) hospital. For the low-level strains, 2 strains were from (B) hospital, 3 strains were from (C) hospital and one strain was from (D) hospital.
Each isolate was inoculated into 3 ml Luria-Vertani broth (Difco Laboratories) and incubated at 37℃ overnight. The concentration of the cell suspensions were adjusted with saline using a spectrophotometer to an absorbance of 0.3~0.25 at 610 nm. After adjustment, 1 ml of the cell suspension was centrifuged at 14,000 rpm for 5 min and the supernatant was aspirated. The pellet was washed once in 1 ml of Tris-EDTA (TE) buffer (10 mM Tris HCl, 1 mM EDTA [pH 8]) and centrifuged again. The washed cells were resuspended in 50µl of lysis II buffer (6 mM Tris-HCl, pH 8.0, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroylsarcosine) and equilibrated at 37℃ for 15 min. Fifty microliters of 2% low-melting-pint agarose and 2µl of 100 units lysostaphin were added to the cell suspension, gently mixed and dispensed into a plug mold. The plugs were allowed to solidify in the refrigerator at 4℃ for 5 min. The plugs were placed into a tube containing 250µl of lysis I buffer (50 mM Tris-HCl, pH 7.4, 100 unit lysostapin, lysozyme 1 mg/ml) and the cells in the plug were lysed for 1 h at 37℃. Then the lysis I buffer were removed and replaced with 250µl of lysis II buffer at 37℃ for 1 h. The lysis II buffer was removed and 250µl of proteinase K buffer (0.5 M EDTA, 25 unit/ml proteinase K) was added and this was incubated at 50℃ overnight. The TE washings were repeated 3 more times.
For electrophoresis, the plug was cut into small slices (2 by 5 mm) and these were placed in 125µl of a total restriction enzyme mixture that contained 20 U of SmaI (Sib Enzyme Ltd, Novosibirk, Russia). After 2 h incubation at 25℃ with shaking at 140 rpm, the chromosomal restriction fragment patterns were analyzed by loading the trimmed slices of the plug into a well of SeaKem 1% agarose gel. The running gel was prepared in 0.5×TBE buffer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Electrophoresis was performed with a CHEP-DR III (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The running parameters were an initial pulse of 5 s, a final pulse of 40 s, 200 V, 20 hrs and 12~14℃. After the electrophoresis run was completed, the gel was stained with 0.5µg/ml ethidium bromide for 20 min and destained in fresh distilled water for 20 min.
The gels were photographed and digitalized with a GelDoc (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The images were stored electronically as TIFF files. The restriction pattern was analyzed with the GelCompar II program (version 4.5; Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient, and cluster analysis of the similarity matrices was performed by the unweighted pair group method using the arithmetic average with 0.75% of tolerance12,13. The similarity cut-off value was 99%.
RESULTS
Prevalence of mupirocin resistant MRSA
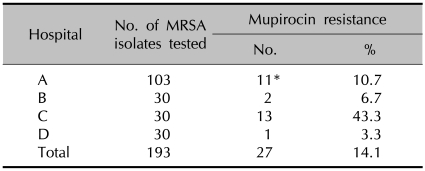
Of the total 193 clinical isolates of MRSA from the four university hospitals, 27 (14.1%) were mupirocin resistant (Table 1). Of these, 11 (5.7%) were found to have high-level resistance to mupirocin and all of them were from (A) hospital. The low-level strains were composed of 2, 13 and 1 isolates from (B), (C) and (D) hospitals, respectively.
Table 1.
Rates of mupirocin resistant MRSA isolates in four Korean hospitals
MRSA: methicillin-resistant Staphylococcus aureus. *High level mupirocin resistant MRSA isolates.
Antimicrobial susceptibility tests of the mupirocin resistant MRSA
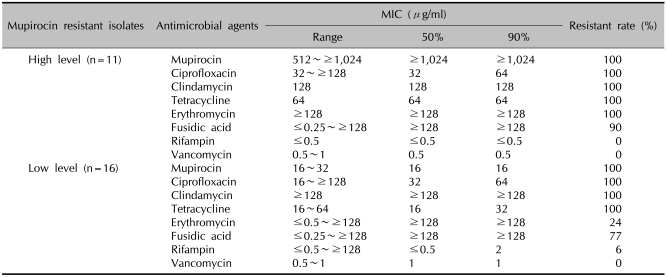
The antimicrobial susceptibility test results for the mupirocin resistant MRSA isolates are summarized in Table 2. The MICs of the high-level mupirocin resistant MRSA ranged from 512 to ≥1,024µg/ml, and the MIC50 and MIC90 were ≥1,024µg/ml, respectively. But for the low-level mupirocin resistant strains, the MICs ranged from 16 to 32, and the MIC50 and MIC90 was 16µg/ml in both cases. Compared with the MIC90 among the high-level resistant strains, rifampin and vancomycin showed high antimicrobial activity and the MICs were ≤0.5µg/ml and 0.5µg/ml, respectively. The MICs of ciprofloxacin and tetracycline were 64µg/ml, respectively and those for erythromycin and fusidic acid were ≥128µg/ml, respectively. For the low-level mupirocin resistant MRSA strains, vancomycin showed the highest antimicrobial activity (MIC=1µg/ml) and this was followed by rifampin (2µg/ml), tetracycline (32µg/ml) and ciprofloxacin (64µg/ml). The MICs of clindamycin, erythromycin and fusidic acid were ≥128µg/ml, respectively.
Table 2.
Antimicrobial susceptibilities of the 11 high-level and 16 low-level mupirocin resistant MRSA isolates
MRSA: methicillin-resistant Staphylococcus aureus, MIC: minimum inhibitory concentration.
The high-level mupirocin resistant MRSA strains were susceptible to rifampin and vancomycin, but 90% were resistant to fusidic acid, and 100% were resistant to ciprofloxacin, clindamycin, tetracycline and erythromycin. While the low-level mupirocin resistant MRSA strains were susceptible to vancomycin, 6% were resistant to rifampin, 24% were resistant to erythromycin, 77% were resistant to fusidic acid and 100% were resistant to ciprofloxacin, clindamycin and tetracycline.
Genotyping the mupirocin resistant MRSA using the PFGE patterns
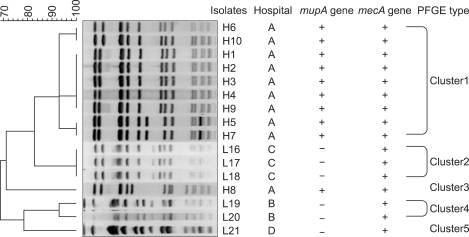
The PFGE patterns of the genomic DNA of the 16 strains of mupirocin resistant MRSA were classified into five clusters (Fig. 5). Of the 10 high-level resistant strains from (A) hospital, 9 (H1-H7, H9-H10) were classified in cluster 1 with 92% similarity. Among these, H6 and H10, H1-4 and H9, and H5 and H7 were the same clones. The H8 strain was in cluster 4 with 72% similarity. Of the 6 low-level resistant strains, L16-L18 from (B) hospital were classified in cluster 2 and they were the same clone with 100% similarity. The L19 and L20 from (A) hospital were in cluster 4 with 93% similarity, and L21 from (D) hospital was in cluster 5 with 79% similarity.
Fig. 5.
A dendrogram generated with the Gelcompar II program showing the five PFGE types (1-5) of SmaI-restricted chromosome DNA of the 16 mupirocin-resistant MRSA isolates. H1-H10: isolates with high-level mupirocin resistance, L16-21: isolates with low-level mupirocin resistance. PFGE: pulsed-field gel electrophoresis, MRSA: methicillin-resistant Staphylococcus aureus.
DISCUSSION
Mupirocin is a topical antibiotic that shows a high level of activity against streptococci and staphylococci, and certain gram-negative bacteria, including Haemophilus influenzae and Neisseria gonorrhoae, but it is much less active against most gram-negative bacilli and anaerobes1. Therefore, it has been used to treat skin infection and to eradicate MRSA. Mupirocin was introduced into clinical practice in 1985 and the use of mupirocin ointment has been progressively increasing worldwide.
Clinical isolates resistant to mupirocin were first reported in 1987, and the resistance rate has been increasing14. Prolonged use and frequent use of mupirocin were thought to be the most important factors associated with the increase of mupirocin resistance3,15. A Canadian study by Simor et al.2 reported an increase in mupirocin resistance among MRSA over time. From 1995 to 1999, the proportions of MRSA strains with high- and low-level mupirocin resistance were 1.6% and 6.4%, respectively, whereas from 2000 to 2004, the resistant rates were 7.0% and 10% respectively. Pérez-Fontán et al.16 reported the emergence of mupirocin-resistant S. aureus in peritoneal dialysis patients who applied mupirocin over 10 years. From 1990 to 1996, there were no mupirocin resistant strains, but they increased to 8.3% from 1997 to 1998, and to 12.4% from 1999 to 2000. According to Wise and Johnson17, the mupirocin resistance rate was 8.3% in a dermatological hospital, while it was 0.2% in the nearby general hospitals over the same period.
In Korea, topical mupirocin has been used since 1994 to eradicate staphylococcal infection in hospitals and the use of mupirocin has been dramatically increasing. However, there has been very little awareness and research about mupirocin resistance. The study conducted in Korea up to 1999 failed to detect mupirocin resistant strains18. Yet Yun et al.4 first identified 16 high-level mupirocin resistant isolates in 2003 and the prevalence of mupirocin resistance was 5%. In a study from long-term-care facilities in Korea, the rate of mupirocin resistance was 11.3% (6.1% high-level mupirocin resistance and 5.2% low-level mupirocin resistance)5. Another study carried out at intensive care units reported that the mupirocin resistance rate was 25.3% and all the isolates showed low-level resistance. In our study, the prevalence of mupirocin resistance was 14.1%. The 5.7% of the isolates were high-level resistant strains (11/193) and all of them were from (A) hospital. The low-level resistant strains were from the other three hospitals. Also, the prevalence of mupirocin resistance of (C) hospital was 43.3%, which is much higher than that of the other hospitals, and the reason for this was thought to be there were more dermatological samples from (C) hospital.
Mupirocin resistance is divided into two groups: low-level resistance (MIC=8~256µg/ml) and high-level resistance (MIC>256µg/ml). Low-level mupirocin resistance is due to point mutation in the chromosomally encoded native IleS gene, whereas high-level resistance is related to the acquisition of a plasmid containing a modified additional IleS-2 gene mupA3,4. However, the mupA gene was detected in the low-level mupirocin resistant strains by western blot analysis in a small case study10. It is also possible that high- and low-level mupirocin resistance are mediated by different, but closely related genes that hybridize with the mupA probe, so further studies are needed to resolve this issue.
Low-level mupirocin resistant strains are not considered to have clinical significance because the concentration of mupirocin in the 2% ointment (20,000µg/ml) exceeds the MICs of the low-level mupirocin resistant strains, so low-level mupirocin resistant strains can be treated by topical mupirocin19. On the contrary, high-level mupirocin resistant strains that can not be treated by mupirocin are more clinically important. Fortunately, low-level strains have so far been more frequently isolated than high-level strains. In our study, the prevalence of low-level mupirocin resistance (8.3%) was higher than the prevalence of high-level resistance (5.7%)2,10,20. However, high-level mupirocin resistant strains have been increasing in prevalence recently. Also, the mupirocin resistance rates were higher for the methicillin-resistant isolates than for the methicillin-susceptible isolates4,21. Chaves et al.21 detected a much higher percentage of mupirocin resistance among the isolates of MRSA (14.8%) than among the isolates of methicillin-susceptible S. aureus (0.6%). Therefore, the evaluation of mupirocin resistance, and especially high-level resistance, is very important in MRSA isolates.
High-level mupirocin resistant strains have an additional mupA gene in a plasmid that can be transferred to other strains by plasmid conjugation22,23. Bastos et al.23 showed that mupirocin resistance is transferred from mupirocin resistant strains of S. aureus to mupirocin susceptible S. aureus via a conjugative plasmid. Furthermore, this plasmid was transferred between S. aureus and Staphylococcus epidermidis. This result indicates the possibility of horizontal transfer of the conjugative plasmid among Staphylococcus species and this suggests that Staphylococcus epidermidis could be a reservoir of this plasmid. This enables the wide spread of mupirocin resistance. Further, the mupA gene may co-transfer with other antibacterial resistance, such as that against triclosan, tetracycline and trimethoprim3.
PFGE is the most widely used genotyping or genetic fingerprinting tool, although it requires technical skill, a long processing time and expensive instruments. It is commonly considered the gold standard method for epidemiologic typing and the determination of genetic relatedness11,24. Yoo et al.5 showed 20 of 25 high-level and 20 of 21 low-level mupirocin resistant S. aureus from eight long-term-care facilities belonged to the same PFGE groups. A study by Chaves et al.21 also identified that 13 of 14 high-level mupirocin resistant strains showed the same PFGE band patterns. These results indicate that clonal transmission of mupirocin-resistant S. aureus occurred in hospitals. In our study, 9 of 10 high-level mupirocin resistant MRSA isolates from (A) hospital and 5 of 6 low-level resistant isolates (2 from (A) hospital, 3 from (B) hospital) belonged to the same PFGE group.
The antimicrobial susceptibility of the mupirocin resistant strains can vary depending on different studies. A study conducted on 4980 MRSA strains showed mupirocin-resistant strains were more likely to be resistant to fusidic acid and to be susceptible to tetracycline, trimethoprim-sulfamethoxazole and ciprofloxacin as compared to that of the mupirocin-susceptible strains2. Yoo et al.5 showed all the mupirocin resistant S. aureus were resistant to oxacillin, penicillin and erythromycin and they were susceptible to ampicillin and vancomycin. In our study, the mupirocin resistant MRSA isolates were susceptible to vancomycin and rifampin, and they were resistant to ciprofloxacin, clindamycin and tetracycline. Compared to the low-level mupirocin resistant strains, the high-level mupirocin resistant strains were more resistant to erythromycin and fusidic acid.
Several agents have been shown to have activity against mupirocin resistant S. aureus3. These include azelaic acid, nitrofurazone and silver sulphadiazine. Maple et al.25 showed that the concentrations of azelaic acid, nitrofurazone and silver sulphadiazine close to the MIC were bactericidal, but mupirocin was only bactericidal at concentrations substantially greater than the MIC.
Prolonged use and frequent usage of mupirocin appear to be the most important factors for increasing mupirocin resistance. Riley et al.26 recommended less than 10 days treatment and at least 1 month between treatment.
The increasing number of reports of high-level mupirocin resistance could mean the potential loss of one of the major treatment methods for controlling MRSA. Therefore, mupirocin treatment should be used cautiously and judiciously. Careful monitoring of mupirocin usage and testing of S. aureus, including MRSA, for mupirocin resistance seems to be indicated.
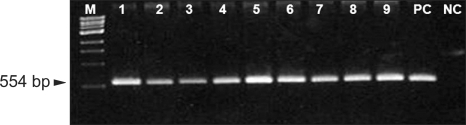
Fig. 2.
Agarose gel electrophoresis of the amplified 554-bp DNA fragments of the mecA gene. The samples are lane M: a 100 bp size marker, lanes 1-9: mupirocin resistant MRSA, PC: mecA-positive control (ATCC 33591) and NC: mecA-negative control (ATCC 25923). MRSA: methicillin-resistant Staphylococcus aureus.
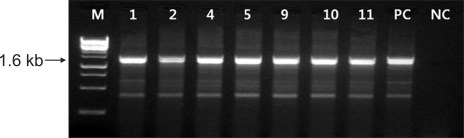
Fig. 3.
Agarose gel electrophoresis of the amplified 1.6 kb DNA fragments of the mupA gene. The samples are lane M: a 1-kb size marker, lanes 1-2, 4-5, 9-11: high-level mupirocin resistant MRSA, PC: mupA-positive control (KCKC 6129), and NC: mupA-negative control (ATCC 25923). MRSA: methicillin-resistant Staphylococcus aureus.
References
- 1.Sutherland R, Boon RJ, Griffin KE, Masters PJ, Slocombe B, White AR. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob Agents Chemother. 1985;27:495–498. doi: 10.1128/aac.27.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simor AE, Stuart TL, Louie L, Watt C, Ofner-Agostini M, Gravel D, et al. Canadian Nosocomial Infection Surveillance Program. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob Agents Chemother. 2007;51:3880–3886. doi: 10.1128/AAC.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cookson BD. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J Antimicrob Chemother. 1998;41:11–18. doi: 10.1093/jac/41.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Yun HJ, Lee SW, Yoon GM, Kim SY, Choi S, Lee YS, et al. Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. J Antimicrob Chemother. 2003;51:619–623. doi: 10.1093/jac/dkg140. [DOI] [PubMed] [Google Scholar]
- 5.Yoo JI, Shin ES, Cha JO, Lee JK, Jung YH, Lee KM, et al. Clonal dissemination and mupA gene polymorphism of mupirocin-resistant Staphylococcus aureus isolates from long-term-care facilities in South Korea. Antimicrob Agents Chemother. 2006;50:365–367. doi: 10.1128/AAC.50.1.365-367.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang JA, Park DW, Sohn JW, Yang IS, Kim KH, Kim MJ. Molecular analysis of isoleucyl-tRNA synthetase mutations in clinical isolates of methicillin-resistant Staphylococcus aureus with low-level mupirocin resistance. J Korean Med Sci. 2006;21:827–832. doi: 10.3346/jkms.2006.21.5.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. eighth edition. Wayne: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 8.Lee HK, Lee EJ, Pahk YJ, Kim BK, Kang MW, Shim SI. Relationship between the level of methicillin resistance and mecA, mecI, femA genes genes in Staphylococci. Korean J Infect Dis. 1998;30:36–44. [Google Scholar]
- 9.Yokoyama T. Study on mec gene in methicillin-resistant staphylococci. Kansenshogaku Zasshi. 1993;67:1203–1210. doi: 10.11150/kansenshogakuzasshi1970.67.1203. [DOI] [PubMed] [Google Scholar]
- 10.Ramsey MA, Bradley SF, Kauffman CA, Morton TM. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob Agents Chemother. 1996;40:2820–2823. doi: 10.1128/aac.40.12.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41:1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyndrickx M, Vandemeulebroecke K, Scheldeman P, Kersters K, de Vos P, Logan NA, et al. A polyphasic reassessment of the genus Paenibacillus, reclassification of Bacillus lautus (Nakamura 1984) as Paenibacillus lautus comb. nov., and of Bacillus peoriae (Montefusco et al. 1993) as Paenibacillus peoriae comb. nov., and emended descriptions of P. lautus and of P. peoriae. Int J Syst Bacteriol. 1996;46:988–1003. doi: 10.1099/00207713-46-4-988. [DOI] [PubMed] [Google Scholar]
- 13.Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. University of Kansas Scientific Bulletin. 1958;28:1409–1438. [Google Scholar]
- 14.Rahman M, Noble WC, Cookson B, Baird D, Coia J. Mupirocinresistant staphylococcus aureus. Lancet. 1987;330:387–388. [Google Scholar]
- 15.Conly JM, Vas S. Increasing mupirocin resistance of Staphylococcus aureus in CAPD--should it continue to be used as prophylaxis? Perit Dial Int. 2002;22:649–652. [PubMed] [Google Scholar]
- 16.Pérez-Fontán M, Rosales M, Rodríguez-Carmona A, Falcón TG, Valdés F. Mupirocin resistance after long-term use for Staphylococcus aureus colonization in patients undergoing chronic peritoneal dialysis. Am J Kidney Dis. 2002;39:337–341. doi: 10.1053/ajkd.2002.30553. [DOI] [PubMed] [Google Scholar]
- 17.Wise R, Johnson J. Mupirocin resistance. Lancet. 1991;338(8766):578. doi: 10.1016/0140-6736(91)91148-n. [DOI] [PubMed] [Google Scholar]
- 18.Lee HJ, Suh JT, Kim YS, Lenz W, Bierbaum G, Schaal KP. Typing and antimicrobial susceptibilities of methicillin resistant Staphylococcus aureus (MRSA) strains isolated in a hospital in Korea. J Korean Med Sci. 2001;16:381–385. doi: 10.3346/jkms.2001.16.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redhead RJ, Lamb YJ, Rowsell RB. The efficacy of calcium mupirocin in the eradication of nasal Staphylococcus aureus carriage. Br J Clin Pract. 1991;45:252–254. [PubMed] [Google Scholar]
- 20.Janssen DA, Zarins LT, Schaberg DR, Bradley SF, Terpenning MS, Kauffman CA. Detection and characterization of mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:2003–2006. doi: 10.1128/aac.37.9.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaves F, García-Martínez J, de Miguel S, Otero JR. Molecular characterization of resistance to mupirocin in methicillin-susceptible and -resistant isolates of Staphylococcus aureus from nasal samples. J Clin Microbiol. 2004;42:822–824. doi: 10.1128/JCM.42.2.822-824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas DG, Wilson JM, Day MJ, Russell AD. Mupirocin resistance in staphylococci: development and transfer of isoleucyl-tRNA synthetase-mediated resistance in vitro. J Appl Microbiol. 1999;86:715–722. doi: 10.1046/j.1365-2672.1999.00718.x. [DOI] [PubMed] [Google Scholar]
- 23.Bastos MC, Mondino PJ, Azevedo ML, Santos KR, Giambiagi-deMarval M. Molecular characterization and transfer among Staphylococcus strains of a plasmid conferring high-level resistance to mupirocin. Eur J Clin Microbiol Infect Dis. 1999;18:393–398. doi: 10.1007/s100960050306. [DOI] [PubMed] [Google Scholar]
- 24.Kim SM, Lee DC, Park SD, Kim BS, Kim JK, Choi MR, et al. Genotype, coagulase type and antimicrobial susceptibility of methicillin-resistant staphylococcus aureus isolated from dermatology patients and Healthy Individuals in Korea. J Bacteriol Virol. 2009;39:307–316. [Google Scholar]
- 25.Maple PA, Hamilton-Miller JM, Brumfitt W. Comparison of the in-vitro activities of the topical antimicrobials azelaic acid, nitrofurazone, silver sulphadiazine and mupirocin against methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1992;29:661–668. doi: 10.1093/jac/29.6.661. [DOI] [PubMed] [Google Scholar]
- 26.Riley TV, Carson CF, Bowman RA, Mulgrave L, Golledge CL, Pearman JW, et al. Mupirocin-resistant methicillin-resistant Staphylococcus aureus in Western Australia. Med J Aust. 1994;161:397–398. doi: 10.5694/j.1326-5377.1994.tb127503.x. [DOI] [PubMed] [Google Scholar]