Abstract
Intracellular bacteria have been shown to cause autophagy, which impacts infectious outcomes, whereas extracellular bacteria have not been reported to activate autophagy. Here, we demonstrate that Pseudomonas aeruginosa, a Gram-negative extracellular bacterium, activates autophagy with considerably increased LC3 punctation in both an alveolar macrophage cell line (MH-S) and primary alveolar macrophages. Using the LC3 Gly120 mutant, we successfully demonstrated a hallmark of autophagy, conjugation of LC3 to phosphatidylethanolamine (PE). The accumulation of typical autophagosomes with double membranes was identified morphologically by transmission electron microscopy (TEM). Furthermore, the increase of PE-conjugated LC3 was indeed induced by infection rather than inhibition of lysosome degradation. P. aeruginosa induced autophagy through the classical beclin-1–Atg7–Atg5 pathway as determined by specific siRNA analysis. Rapamycin and IFN-γ (autophagy inducers) augmented bacterial clearance, whereas beclin-1 and Atg5 knockdown reduced intracellular bacteria. Thus, P. aeruginosa-induced autophagy represents a host protective mechanism, providing new insight into the pathogenesis of this infection.
Key words: Extracellular bacteria, Punctation, Autophagolysosome, Host defense, Respiratory immunity
Introduction
Pseudomonas aeruginosa accounts for 25% of the Gram-negative bacteria isolated from hospital environments. P. aeruginosa frequently infects immunodeficient individuals who are afflicted with tuberculosis, cancer and particularly cystic fibrosis. However, traditional antibiotic therapies are inadequate because P. aeruginosa has become increasingly resistant to antibiotics (Chastre and Fagon, 2002). P. aeruginosa was classified as an extracellular pathogen with a spectrum of virulence factors against host clearance (Sadikot et al., 2005). Alveolar macrophages are the first line of host defense in the lung and also perform various other functions, but their role in fighting off this pathogen remains to be fully defined. Thus, elucidating the macrophage–pathogen interaction will improve our knowledge of host defense against this pathogen, ultimately leading to new therapeutic targets.
Autophagy is an intracellular process that delivers cytoplasmic components to the autophagosome and lysosome for degradation, a crucial homeostasis mechanism involved in many physiological and pathological conditions (Cuervo, 2004; Klionsky, 2005). During this process, cytosolic components, such as organelles and long-lived proteins, are sequestered into a double-membrane autophagosome (an autophagic vacuole). The classical intracellular signaling mechanism of this process relies on two ubiquitin-like conjugation systems involving autophagy-related genes: Atg7–Atg12–Atg5 or Atg4–Atg7–Atg8 (Atg8 is also known as LC3 in mammals) (Ohsumi and Mizushima, 2004). However, both these systems depend on Atg6 (beclin-1 in mammals), which is crucial in forming an early complex containing class III phosphoinositide 3-kinase (PI3K; also known as VPS34), and eventually forming the autophagosome.
Recently, cumulative publications indicate a essential role of autophagy in immune response in many diseases including viral and bacterial infection (Colombo, 2007; Ogawa et al., 2005; Rioux et al., 2007; Shintani and Klionsky, 2004). Viruses and bacteria are capable of escaping from phagosomes and entering autophagosomes for survival and replication (Campoy and Colombo, 2009; Dorn et al., 2002). Conversely, autophagy potentially captures bacteria that have escaped from phagosomes into the cytoplasm, thereby delivering the bacteria into autophagosomes and autolysosomes where they are destroyed (Campoy and Colombo, 2009). The outcome of autophagy is pathogen specific, indicating that subtle and varied mechanisms exist to counter intracellular bacteria (Ogawa et al., 2005). For example, mycobacterium tuberculosis and group A streptococci (GAS) infection also induce autophagy, in the end benefiting host defense (Nakagawa et al., 2004). However, most studies of bacterial autophagy only involve intracellular pathogens (Deretic, 2011). Up to now, whether autophagy is a part of P. aeruginosa pathogenesis has been completely unknown. We have studied autophagy in P. aeruginosa-infected MH-S cells, and for the first time reveal the induction of autophagy by P. aeruginosa through the beclin-1–Atg7–Atg5 canonical pathway. This observation could provide useful information for further understanding of the role of autophagy in airway P. aeruginosa infection.
Results
P. aeruginosa infection induced LC3 punctation
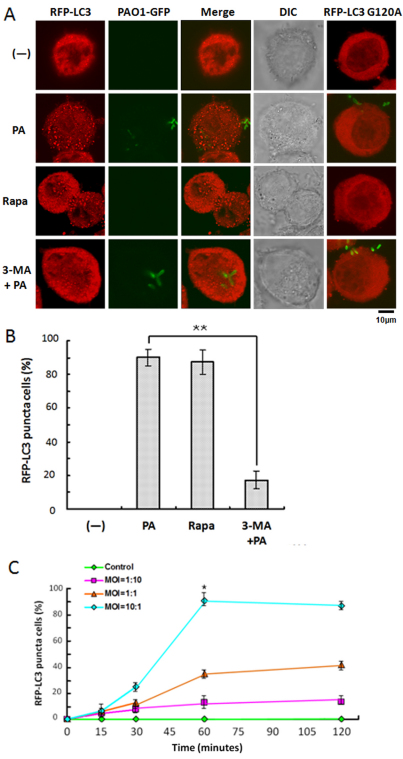
To determine whether infection by P. aeruginosa can induce autophagy, MH-S cells were transfected with RFP-LC3 plasmids. After confirming successful transfection, the MH-S cells were infected with a genome-sequenced strain of P. aeruginosa, PAO1, in a dose- and time-dependent manner. We observed that PAO1 infection with a bacteria:cell multiplicity of infection (MOI) of 10:1 induced the most significant LC3 punctation in MH-S cells (Fig. 1A,B). To exclude the possibility that autophagy induction by PAO1 was caused by infection stress, such as induction of cell death, the levels of cellular apoptosis and necrosis were also examined. Punctation began after infection, reached its peak at 1 hour (no appreciable cell death was observed at this time point; supplementary material Fig. S1), and thereafter gradually decreased, probably because of increased cell death caused by infection (Fig. 1C, P<0.05). To confirm LC3 punctation as a specific autophagic alteration, a mutant of RFP-LC3, RFP-LC3 G120A, was used to account for the possibility of non-specific induction of LC3 punctation by PAO1 infection. Gly120 is evolutionarily conserved in mammalian cells and is essential for LC3 cleavage by Atg4. Thus, nascent LC3 G120A mutant cannot be processed into LC3-I, which, in turn, means it cannot be conjugated to phosphatidylethanolamine (PE) (Ohsumi and Mizushima, 2004). As expected, LC3 punctation was abolished by introducing RFP-LC3 G120A into MH-S cells, which serves as a gold standard for identifying autophagy (Levine et al., 2011). Transfected MH-S cells were also treated with the autophagy activator rapamycin and inhibitor 3-MA as positive and negative controls, respectively (Fig. 1A). The confocal microscopy images were then used to semi-quantitatively measure the percentage of cells with significant LC3 punctation staining (100 cells/sample). The threshold for positive expression was set to 10 visible LC3 punctae. This analysis confirmed that PAO1 infection specifically induced LC3 punctation in MH-S cells (P<0.05). Although P. aeruginosa was considered as an extracellular bacterium, it could be phagocytosed by macrophages. Thus, it could induce autophagy through intracellular pathways. However, many cells were found to show significant LC3 punctation without PAO1 internalization, indicating that autophagy induction does not require intracellular bacteria (supplementary material Fig. S2). Moreover, to determine whether PAO1 could also induce autophagy in vivo, we isolated primary alveolar macrophages from C57/BL6 mice. Similarly, the primary alveolar macrophages showed a substantial increase in LC3 punctation upon PAO1 infection, whereas cells treated with 3-MA largely exhibited reduction in LC3 punctation (Fig. 2). To further confirm these data, we transfected primary alveolar macrophages with the RFP-LC3 plasmid and examined autophagy following infection with PAO1-GFP. Indeed, punctate staining of LC-3 was induced following infection (supplementary material Fig. S2). Furthermore, inhibition with 3-MA reduced the autophagy in primary alveolar macrophages (data not shown). Our results demonstrate that PAO1 infection induced autophagy in primary alveolar macrophages.
Fig. 1.
P. aeruginosa infection induced RFP-LC3 punctation in a dose- and time-dependent manner. (A) MH-S cells were transfected with RFP-LC3 and RFP-LC3 G120A plasmids for 24 hours. Then, the cells were infected with PAO1-GFP for 1 hour (MOI=10:1). Before infection, the cells were also treated with rapamycin (3 μM, 12 hours) and 3-MA (3 mM, 3 hours). (B) The puncta in each cell were counted and cells with more than 10 punctae were considered as LC3-RFP puncta cells. Values are from 100 cells/sample. (C) MH-S cells were infected at different times and with different MOIs. The percentage of LC3-RFP puncta cells was determined (one-way ANOVA; Tukey's post-hoc test, **P<0.01). Data are representative of three experiments with similar results.
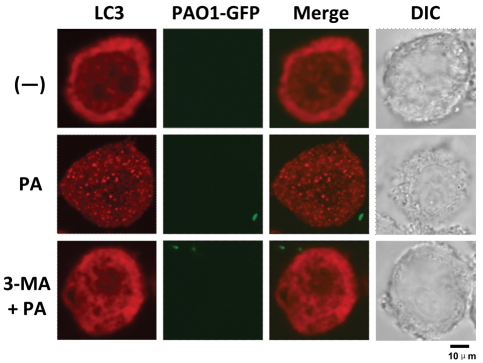
Fig. 2.
P. aeruginosa infection induced LC3 punctation in mouse primary alveolar macrophages. Mouse primary alveolar macrophages were isolated using bronchoalveolar lavage. The cells were infected with PAO1-GFP for 1 hour (MOI=10:1). Before infection, the cells were treated with 3-MA (3 mM, 3 hours). After infection, the cells were fixed and stained with anti-LC3 antibody. The puncta in each cell were counted and cells with more than 10 punctae were considered as LC3 puncta cells. Representative cells from each group are shown.
To determine whether autophagy is a general phenomenon in P. aeruginosa infection, other cell types were also investigated. We also noted that PAO1 infection induced LC3 punctation in several cell types, such as normal mouse alveolar epithelial MLE-12 cells, human alveolar epithelial adenocarcinoma A549 cells and murine macrophage RAW264.7 cells (supplementary material Fig. S3). Interestingly, cancerous A549 cells appeared to exhibit more autophagy than normal epithelial MLE-12 cells and normal RAW264.7 macrophages, which could be due to the intrinsic characteristics of these cells.
P. aeruginosa infection increased autophagosome formation
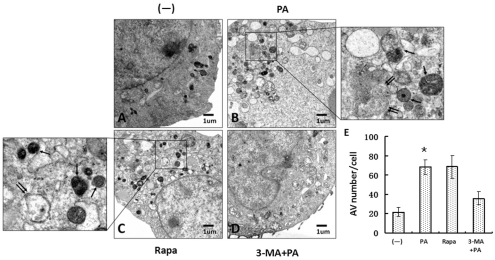
Although we have shown above that PAO1 infection could specifically induce LC3 punctation, there might still be a possibility that the punctation flux was caused by transient overexpression of RFP-LC3 proteins. To determine changes in autophagosome formation, we detected newly formed autophagosomes using transmission electron microscopy (TEM) on PAO1-infected MH-S cells. According to the typical autophagosomes with double membranes and cellular contents, vacuole-containing autophagosomes were identified by TEM (Fig. 3A–D). We found that autophagosomes were significantly increased in PAO1-infected MH-S cells as compared with untreated MH-S cells (Fig. 3E, P<0.05). Similarly, rapamycin, as a positive control, also induced double membrane autophagosomes (Fig. 3C). Finally, the induction of autophagosomes by PAO1 infection was blocked by 3-MA treatment (Fig. 3D). Thus, the morphological evidence obtained by TEM confirms that autophagosome formation is consistent with the accumulation of LC3 punctation.
Fig. 3.
P. aeruginosa infection resulted in elevated autophagosome formation. MH-S cells were transfected with a RFP-LC3 plasmid for 24 hours. The cells were infected with PAO1 for 1 hour (MOI=10:1). Before infection, the cells were also treated with rapamycin (3 μM, 12 hours) and 3-MA (3 mM, 3 hours). After infection, cells were processed and examined by TEM. (A) Untreated cells (–). (B) Cells infected with PAO1 (PA). (C) Cells treated with rapamycin. (D) Cells treated with 3-MA and then infected with PAO1. Boxed areas in B and C were further enlarged. Single arrows indicate autolysosomes and double arrows indicate autophagosomes. (E) The number of autophagic vesicles (AV) in each cells was determined with 20 cells in each sample, respectively (one-way ANOVA; Tukey's post-hoc test, *P<0.05). Data are representative of three experiments with similar results.
P. aeruginosa infection promoted autophagic degradation
Normally, a specific increase of autophagsome formation revealed by TEM is recognized as solid proof of autophagy induction. However, this accumulation of autophagosomes might be caused by the blockage of autophagosome degradation rather than induction of autophagosome formation. Thus, we performed a series of biochemical experiments to examine autophagy activation at the molecular level (Levine et al., 2011).
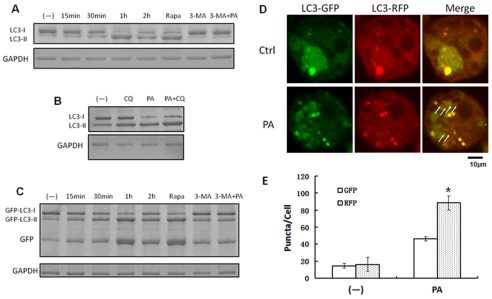
Endogenous LC3 transformation into PE-conjugated LC3-II was dramatically increased by PAO1 infection in a time-dependent manner (Fig. 4A). This lipidation increase was also detected in rapamycin-treated MH-S cells (without infection). Moreover, the transformation was inhibited by 3-MA treatment. To determine whether the accumulation of LC3-II resulted from the blockage of degradation, we utilized a lysosome degradation inhibitor, chloroquine. The inhibition of lysosome degradation by chloroquine reduced the degradation of LC3-II, thus enhancing LC3-II accumulation (Fig. 4B). We also studied this transformation through expression of exogenous GFP-LC3. Consistent with the induction of RFP-LC3 punctation by PAO1 infection, GFP-LC3 showed an increase in lipidation. More importantly, because of the resistance of GFP to lysosome hydrolysis, we also detected a time-dependent increase in the GFP moiety in PAO1-infected MH-S cells, again indicating that GFP-LC3 was efficiently degraded by lysosomes (Fig. 4C).
Fig. 4.
P. aeruginosa infection upregulated autophagic degradation. (A) MH-S cells were infected with PAO1 at different times. Before infection, the cells were also treated with rapamycin (3 μM, 12 hours) and 3-MA (3 mM, 3 hours). (B) Western blotting of LC3 was performed. MH-S cells were treated with chloroquine (CQ; 40 μM, 6 hours) and then infected with PAO1 for 1 hour (MOI=10:1). Western blotting of LC3 was performed. (C) MH-S cells were transfected with GFP-LC3 plasmids for 24 hours, and then treated as in A. Western blotting of GFP was performed. GAPDH was used as a loading control in A–C. (D) MH-S cells were transfected with tandem GFP-RFP-LC3 plasmids for 24 hours. Then the cells were infected with PAO1 for 1 hour (MOI=10:1). Arrows indicate LC3 punctae, which could only be detected in RFP channel. (E) Puncta numbers in each cell was determined. The data are representative of 100 cells for each channel (one-way ANOVA; Tukey's post-hoc test, *P<0.05). Data are representative of three experiments with similar results.
Next, we utilized a recently developed tool, the tandem RFP–GFP–LC3 construct, to further confirm autophagy induction by PAO1 infection (Fig. 4D,E, P<0.05) (Kimura et al., 2007). This construct was designed to differentiate two major autophagic vesicles, the autophagosome and the autolysosome. Similar to the RFP–LC3 construct, the tandem RFP–GFP–LC3 construct can form punctae that represent autophagosome formation. When an autophagosome fuses with a lysosome, the GFP moiety degrades from the tandem protein, but RFP–LC3 maintains the puncta, which then tracks the autolysosomes. After transfection with the tandem construct, we evaluated the successful introduction of the plasmid showing both fluorescent proteins. Following PAO1 infection, we again demonstrated an increase of LC3 punctation in both green and red channels. However, there were markedly more red puncta in infected cells than in the control cells, which reassuringly confirmed the induction of autolysosome formation. Taken together, these findings firmly establish that the infection by PAO1 can specifically induce autophagy in MH-S cells.
P. aeruginosa infection induced autophagy in MH-S cells through the classical autophagy pathway
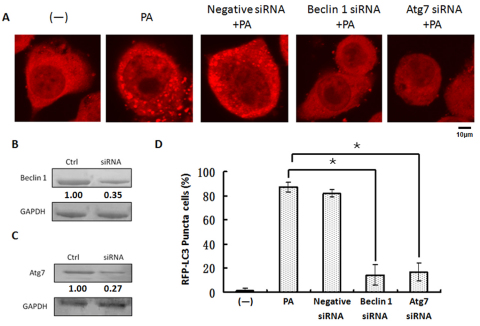
Having confirmed autophagy induction, we sought to characterize the underlying pathways. We first examined the involvement of several canonical autophagic proteins (Fig. 5A,D). Consistent with this established model, we found that beclin-1, an upstream regulator of autophagy, is involved in initiating the autophagy. We demonstrated that PAO1 infection-induced autophagy was largely blocked by beclin-1-specific siRNA (Fig. 5A,B). To further dissect this pathway, we examined a major downstream autophagy-related protein, Atg7. As with beclin-1, Atg7-specific siRNA also resulted in a substantial downregulation of autophagy induced by PAO1 infection (Fig. 5A,C). To determine the involvement of Atg5 in this pathway, we used Atg5-specific siRNA as well, and showed that Atg5 is also required for induction of autophagy with PAO1 (supplementary material Fig. S4). Thus, our data identified that PAO1 infection induces the classical beclin-1–Atg7–Atg5 autophagy pathway in MH-S cells.
Fig. 5.
P. aeruginosa infection induced autophagy through a classical pathway. (A) MH-S cells were transfected with LC3-RFP plasmids for 24 hours and then infected with PAO1 for 1 hour. Before infection, the cells were also treated with negative control siRNA, beclin-1 siRNA and Atg7 siRNA. Confocal images show LC3 puncta induction. (B) Western blotting of beclin-1. (C) Western blotting of Atg7. GAPDH was probed as a loading control in B and C. (D) Puncta numbers in each cell was determined and cells with more than 10 punctae were considered as LC3-RFP puncta cells. The data are representative of 100 cells (one-way ANOVA; Tukey's post-hoc test, *P<0.05). Data are representative of three experiments with similar results.
Autophagy regulates P. aeruginosa clearance in MH-S cells
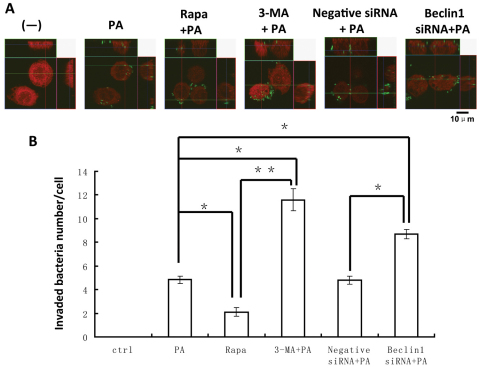
P. aeruginosa has long been considered as an extracellular pathogen, and most autophagy studies thus far have only involved intracellular pathogens. To define the role of autophagy, we infected MH-S cells with PAO1-GFP to monitor internalization and bacterial clearance. After P. aeruginosa infection and LC3 immunostaining, we utilized Z-stack confocal images to count the invading bacteria inside the cells. This approach showed bacterial internalization as well as the extent of autophagy (Fig. 6A). To determine the role of autophagy in regulating phagocytosis of P. aeruginosa and clearance, MH-S cells were pre-treated with rapamycin, 3-MA, negative siRNA or beclin-1 siRNA before infection. The intracellular PAO1-GFP count decreased in rapamycin-treated MH-S cells, indicating that induction of autophagy can increase host defense against this pathogen. By contrast, the number of intracellular bacteria was increased by blocking autophagy with 3-MA or beclin-1 siRNA in MH-S cells. In particular, 3-MA treatment resulted in a marked increase in intracellular bacteria (P<0.01; Fig. 6B). The data indicate that blocking autophagy with 3-MA or beclin-1 siRNA reduced P. aeruginosa bacterial clearance. An autophagy activator, rapamycin, however, produced the opposite effect, improving bacterial clearance. To further confirm these results, we examined the effects of another biological autophagy inducer, IFN-γ, which was hypothesized to provide resistance to the pathogen. As expected, treatment with IFN-γ also resulted in better bacterial clearance following infection in MH-S cells (supplementary material Fig. S5). Furthermore, we examined the role of the exoenzyme-S- and pili-deficient strains and found that whereas the pili-deficient strain did not induce autophagy, the exoenzyme-S-deficient strain induced autophagy (supplementary material Fig. S5). Our data suggest that virulence factors in this bacterium have differential roles in inducing autophagy. Collectively, our observations indicate that autophagy might be a major benefit to host defenses by augmenting bacterial clearance.
Fig. 6.
Autophagy enhanced P. aeruginosa clearance. (A) MH-S cells were transfected with LC3-RFP plasmid for 24 hours and then infected with PAO1-GFP for 1 hour (MOI=10:1). Before infection, the cells were also treated with rapamycin (3 μM, 12 hours), 3-MA (3 mM, 3 hours), negative siRNA or beclin-1 siRNA. The confocal Z-stack images are displayed as orthogonal views to confirm the internalization of bacteria. (B) The number of internalized bacteria per cell. The data are representative of 100 cells (one-way ANOVA; Tukey's post-hoc test, *P<0.05, **P<0.01). Data are representative of three experiments with similar results.
Discussion
This study has demonstrated that P. aeruginosa infection specifically induces autophagy in alveolar macrophages. Importantly, our observations showed that the autophagy was induced through the classical beclin-1–Atg7–Atg5 autophagy pathway. Previous studies have shown that several different bacteria could induce autophagy, although the pathways involved and the impact on infection outcomes vary with intracellular bacteria. P. aeruginosa is traditionally considered as an extracellular pathogen, and many virulence factors, such as biofilm and type III secretion systems, might contribute to the extracellular pathogenic features of the bacterium (Cornelis, 2000; Høiby et al., 2011). To our knowledge, prior to the current study it was not known whether P. aeruginosa infection can induce autophagy. This study is the first to demonstrate the induction of autophagy by an extracellular bacterium as well as its physiological significance in relation to improved bacterial clearance.
Nascent LC3 is processed at its C-terminus by Atg4 and becomes LC3-I. Most of the endogenous LC3 proteins maintain the status of LC3-I and distribute homogenously in the cytoplasm. However, under starvation or infection, LC3 can conjugate with PE to form LC3-II (LC3-PE) by ubiquitinylation-like reactions (Ichimura et al., 2004). In contrast to the cytoplasmic localization of LC3-I, LC3-II associates with both the outer and inner membranes of the autophagosome, thereby being a typical marker of autophagy formation. We identified LC3 punctation in PAO1-infected MH-S cells. To account for transient LC3 overexpression, a dominant-negative mutant of LC3 (LC3 G120A) (Gao et al., 2010; Ichimura et al., 2004) was used and confirmed the increased autophagosome formation in PAO1-infected MH-S cells. The elevated autophagosome formation was also shown to be a result of autophagy induction rather than of blocked lysosome degradation. Moreover, blocking the crucial upstream autophagy regulator beclin-1 led to blockade of autophagy. Rapamycin, a widely used inducer for autophagy induction, caused similar autophagy. However, 3-MA, a typical autophagy inhibitor, reduced autophagy following P. aeruginosa infection.
The classical intracellular signaling mechanism of autophagy relies on two ubiquitin-like conjugation systems (Deretic and Levine, 2009; Levine and Deretic, 2007). Atg12 is activated by Atg7 (a ubiquitin-E1 enzyme) in an ATP-dependent manner. Atg12 is transferred to Atg10 (an E2 enzyme) and is next delivered to Atg5 (Mizushima et al., 1998) to form a multimeric complex with Atg16 (Suzuki et al., 2001). Another system, involving Atg8 (LC3), is first cleaved by Atg4 to expose its C-terminal Gly residue. Similar to the first system, processed Atg8 is activated by Atg7 and then transferred to Atg3 (a ubiquitin-2-like protein). Interestingly, Atg8 forms a final conjugate not with a protein, but with PE, an abundant membrane phospholipid (Ichimura et al., 2000). To evaluate a key downstream autophagy-related protein Atg7, siRNA was used, which abolished autophagy induction by P. aeruginosa. Similarly, Atg5 siRNA transfection abolished autophagy induced by PAO1 infection. Taken together, our studies indicate that P. aeruginosa infection specifically induces autophagy and that this induction depends on the classical autophagy pathway.
Recently, the role of autophagy in intracellular bacterial infections has garnered increasing interest (Deretic, 2010), and it has been shown to play crucial roles in host defense, especially in immunological cells. Moreover, autophagy has a direct impact on immunity and inflammatory response within the whole organism. For example, autophagy might participate in the elimination of invasive bacteria through autolysosome degradation (Deretic and Levine, 2009). Autophagy can also serve as an effector and regulator during immune response against pathogen invasion, for instance by functioning as downstream factors of pattern recognition receptors (PPRs, such as TLRs) (Saitoh et al., 2008) and pathogen-associated molecular patterns (PAMPs) (Delgado et al., 2008). We also noted that P. aeruginosa-induced autophagy probably needs the participation of TLR-4 (supplementary material Fig. S5). Blockade of TLR-4 abrogated the autophagy (supplementary material Fig. S5), whereas the ligand of TLR-4 (LPS) enhanced autophagy (data not shown). However, LPS-induced autophagy was not as strong as that induced by P. aeruginosa. Further assessment of TLRs in autophagy is currently underway. Because alveolar macrophages play a vital role in innate and adaptive immunity against bacterial infection, autophagy in macrophage cells could impact the fate of P. aeruginosa infection. To study whether this autophagy only occurs in macrophages, we investigated P. aeruginosa infection in alveolar epithelial A549 and MLE-12 cells. Similar autophagy induction was observed, despite the inductions being much greater in A549 cells (supplementary material Fig. S3). Furthermore, we demonstrated that P. aeruginosa infection also induced autophagy in primary human alveolar macrophages, suggesting that autophagy helps in immune defense in the alveolar space.
Autophagy can function as an effector of Th1 and Th2 cytokines in very different ways. Th1 cytokines, such as IFN-γ, induce autophagy to eliminate intracellular pathogens, whereas Th2 cytokines, such as IL-4 and IL-13, inhibit autophagy (Levine and Deretic, 2007). Because IFN-γ is implicated in immunity against P. aeruginosa infection (Hazlett et al., 2002; Yamaguchi et al., 2000), we tested the effect of IFN-γ on autophagy and found increased autophagy by pre-treating cells with IFN-γ, leading to reduced bacterial survival. Even after blocking the function of autocrine IFN-γ in macrophages, the induction of autophagy was still observed. Previous studies have shown that polymyxin B might be effective in killing P. aeruginosa (Giamarellou and Poulakou, 2009), and we found that polymyxin B induced autophagy in MH-S cells (supplementary material Fig. S5). These results indicate that autophagy could be a crucial mechanism against P. aeruginosa infection. To further determine virulence factors in inducting autophagy, we used the exoenzyme-S- and pili-deficient strains and found that whereas the pili-deficient strain failed to induce autophagy, the exoenzyme-S-deficient strain did induce autophagy (supplementary material Fig. S5). Exoenzyme S shows anti-phagocytic activity by inhibiting the small GTPases by ADP ribosylation and GTPase activity, thereby preventing activation of key regulators of the actin cytoskeleton (Maresso et al., 2004). Pili are bacterial structures associated with the inhibition of the initial immune defense of the host (Zolfaghar et al., 2003). Our observations indicate that particular virulence factors might involve autophagy induction. Finally, this bacterially induced autophagy might depend on the expression of p62 (also known as SQSTM1; supplementary material Fig. S5), consistent with previous studies showing that p62 can deliver cytosolic components to autolysosome in mycobacterial infection (Ponpuak et al., 2010). These data suggest that autophagy could also contribute to extracellular bacterial clearance by facilitating the delivery of bacterial components to lysosomes.
Having successfully demonstrated that P. aeruginosa infection caused autophagy in MH-S cells and primary alveolar macrophages, we addressed another important question: what specific role does autophagy play in the immune response? Again, we demonstrated that autophagy plays a regulatory role in bacterial clearance. Autophagy has been found to function during the bacterial clearance process rather than the early internalization process (Levine and Deretic, 2007). However, autophagy could play completely opposite roles with different pathogens or different cell types (Ogawa et al., 2005). Previous work has focused on autophagy for intracellular bacterial pathogenesis (Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005), with the exception of a recent report indicating that extracellular bacteria such as H. pylori might be involved in autophagy in dendritic cells (Wang et al., 2010). Our studies herein represent the first compelling evidence for autophagy induction by an extracellular bacterium. This suggests that autophagy, as an ancient defense mechanism, plays a role against extracellular or quasi-extracellular pathogens. Thus, P. aeruginosa could serve as a model pathogen for investigating the impact of autophagy on crucial cellular events such as bacterial entry and clearance.
Materials and Methods
Cells
MH-S, RAW264.7, A549 and MLE-12 cells were obtained from ATCC and maintained following the supplier's instructions (Kannan et al., 2008; Kannan et al., 2009; Wu et al., 2011; Wu et al., 2001b). Isolation of alveolar macrophages and functional assessment was described previously (Wu et al., 2001a).
Bacteria strains
P. aeruginosa strain PAO1 wild-type (WT) was a gift from Stephen Lory (Harvard Medical School, Boston, MA). PAO1-GFP and PAK pili-deficient strains were obtained from Gerald Pier (Channing Laboratory, Harvard Medical School, Boston, MA) (Kannan et al., 2009). PAO1 exoenzyme S and T (two key exoenzymes associated with invasion by targeting ADP ribosylation and actin polymerization) deletion mutant (ΔExoS and ΔExoT) strains were obtained from Joseph Barbieri (Medical College of Wisconsin, Milwaukee, WI).
Infection experiments
Bacteria were grown overnight in Luria–Bertani (LB) broth at 37°C with vigorous shaking. The next day, the bacteria were pelleted by centrifugation at 8000 g and resuspended in 10 ml of fresh LB broth, in which they were allowed to grow until the mid-logarithmic phase (Kannan et al., 2009). Thereafter, the optical density (OD) at 600 nm was measured, and the density was adjusted to ~0.25 OD (0.1 OD=1×108 cells/ml). Cells were washed once with PBS after overnight culture in serum-containing medium and changed to serum-free and antibiotic-free medium immediately before infection (Kannan et al., 2006a). Bacterial clearance was determined using the colony forming units (CFU) assay after treating the infected macrophages with 100 μg/ml polymyxin B (Kannan et al., 2006a).
Cell transfection
MH-S cells were transfected with RFP-LC3, GFP-LC3, RFP-LC3 G120A and RFP-GFP-LC3 plasmids using Lipofectamine 2000 reagent (Invitrogen) in serum-free RPMI 1640 medium (Thermofisher Scientific) following the manufacturer's instructions (Wu et al., 2009). the RFP-LC3 G120A construct was made by replacing Gly120 of RFP-LC3 with alanine and was kindly provided by Xiao-min Yin (Indiana University) (Gao et al., 2010). The tandem RFP-GFP-LC3 plasmid was created and kindly provided by Tamotsu Yoshimori of Osaka University, Japan (Kimura et al., 2007).
Cell death assay
Cell death (apoptosis and necrosis) was evaluated using the Vybrant assay (Invitrogen) following the manufacturer's instructions. Apoptosis and necrosis were calculated according to the intensity of YO-PRO1 (green; apoptosis) and propidium iodide (red; necrosis) staining, respectively.
Western blotting
Rabbit polyclonal Abs against MAP LC3β and goat polyclonal antibodies against beclin-1 were obtained from Santa Cruz Biotechnology. Rabbit monoclonal antibody against GAPDH was obtained from Cell Signaling Technology. The samples from cells were lysed and quantified. The lysates were boiled for 5 minutes, and protease inhibitor cocktail added. The supernatants were collected and 30 μg of each sample were loaded onto 10% SDS-polyacrylamide mini-gels and electrophoresed for protein resolution (Wu et al., 1995). The proteins were then transferred to polyvinylidine difluoride membranes (Pierce Biotechnology) and blocked for 2 hours at room temperature using 5% non-fat milk blocking buffer. Membranes were incubated overnight at 4°C with appropriate first antibodies diluted at 1:1000 in 5% bovine serum albumin (BSA) western antibody buffer. After washing three times with washingsolution, the membranes were incubated for 45 minutes at room-temperature with horseradish peroxidase-conjugated secondary antibody (Rockland Immunochemicals, Gilbertsville, PA) diluted 1:2000 (Wu et al., 2002). Signals were visualized using an enhanced chemiluminescence detection kit (SuperSignal West Pico; Pierce).
Confocal microscopy and indirect immunofluorescence staining
Cells were grown either on coverslips in a 24-well plate or in glass-bottomed dishes (MatTek, Ashland, MA). For immunostaining, the cells were fixed in 3.7% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS and blocked with blocking buffer for 30 minutes (Kannan et al., 2008). Cells were incubated with primary antibodies at 1/500 dilution in blocking buffer for 1 hour and washed three times with wash buffer. After incubation with appropriate fluorophore-conjugated secondary antibodies, the coverslips were mounted on slides with Vectashield mounting medium. The images were captured using an LSM 510 Meta confocal microscope (Carl Zeiss MicroImaging), and processed using the software provided by the manufacturer (Kannan et al., 2006b).
Transmission electron microscopy (TEM)
TEM was employed for identifying autophagosomes using modified Karnovsky's fixative (Karnovsky, 1965). Images were taken and analyzed according to our previous published methods (Teiken et al., 2008; Wu et al., 2003; Wu et al., 2005).
Statistical analysis
All experiments were performed in triplicate and repeated at least three times. Data are presented as percentage changes compared with the controls ± s.d. from the three independent experiments. Group means were compared by one-way ANOVA (post-hoc), using SPSS software, and differences were accepted as significant at P<0.05 (Wu et al., 2009).
Supplementary Material
Acknowledgements
We thank Sarah Rolling of the University of North Dakota imaging core for help with confocal imaging.
Footnotes
Funding
This project was supported by National Institutes of Health [grant numbers 5R03 ES014690 to M.W., 5R01HL092905-04 to H.G., 3R01HL092905-02S1 to H.G.]; the National 973 Basic Research Program of China [2012CB518900]; and an American Heart Association Scientist Development Grant [grant number 535010N to M.W.). Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.094573/-/DC1
References
- Campoy E., Colombo M. I. (2009). Autophagy in intracellular bacterial infection. Biochim. Biophys. Acta 1793, 1465-1477 [DOI] [PubMed] [Google Scholar]
- Chastre J., Fagon J. Y. (2002). Ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 165, 867-903 [DOI] [PubMed] [Google Scholar]
- Colombo M. I. (2007). Autophagy: a pathogen driven process. IUBMB Life 59, 238-242 [DOI] [PubMed] [Google Scholar]
- Cornelis G. R. (2000). Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 681-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M. (2004). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70-77 [DOI] [PubMed] [Google Scholar]
- Delgado M. A., Elmaoued R. A., Davis A. S., Kyei G., Deretic V. (2008). Toll-like receptors control autophagy. EMBO J. 27, 1110-1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. (2010). Autophagy in infection. Curr. Opin. Cell Biol. 22, 252-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. (2011). Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Levine B. (2009). Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn B. R., Dunn W. A. J., Progulske-Fox A. (2002). Bacterial interactions with the autophagic pathway. Cell. Microbiol. 4, 1-10 [DOI] [PubMed] [Google Scholar]
- Gao W., Kang J. H., Liao Y., Ding W. X., Gambotto A. A., Watkins S. C., Liu Y. J., Stolz D. B., Yin X. M. (2010). Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. J. Biol. Chem. 285, 1371-1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellou H., Poulakou G. (2009). Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs 69, 1879-1901 [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753-766 [DOI] [PubMed] [Google Scholar]
- Hazlett L. D., Rudner X. L., McClellan S. A., Barrett R. P., Lighvani S. (2002). Role of IL-12 and IFN-gamma in Pseudomonas aeruginosa corneal infection. Invest. Ophthalmol. Vis. Sci. 43, 419-424 [PubMed] [Google Scholar]
- Høiby N., Ciofu O., Johansen H. K., Song Z. J., Moser C., Jensen P. Ø., Molin S., Givskov M., Tolker-Nielsen T., Bjarnsholt T. (2011). The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 3, 55-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Imamura Y., Emoto K., Umeda M., Noda T., Ohsumi Y. (2004). In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279, 40584-40592 [DOI] [PubMed] [Google Scholar]
- Kannan S., Audet A., Knittel J., Mullegama S., Gao G. F., Wu M. (2006a). Src kinase Lyn is crucial for Pseudomonas aeruginosa internalization into lung cells. Eur. J. Immunol. 36, 1739-1752 [DOI] [PubMed] [Google Scholar]
- Kannan S., Pang H., Foster D., Rao Z., Wu M. (2006b). Human 8-oxoguanine DNA glycosylase links MAPK activation to resistance to hyperoxia in lung epithelial cells. Cell Death Differ. 13, 311-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S., Audet A., Huang H., Chen L. J., Wu M. (2008). Cholesterol-rich membrane rafts and lyn are involved in phagocytosis during Pseudomonas aeruginosa infection. J. Immunol. 180, 2396-2408 [DOI] [PubMed] [Google Scholar]
- Kannan S., Huang H., Seeger D., Audet A., Chen Y., Huang C., Gao H., Li S., Wu M. (2009). Alveolar epithelial type II cells activate alveolar macrophages and mitigate P. Aeruginosa infection. PLoS ONE 4, e4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky M. J. (1965). A formaldehyde-gltuaradldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27, 117-137 [Google Scholar]
- Kimura S., Noda T., Yoshimori T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452-460 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. (2005). The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Deretic V. (2007). Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7, 767-777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso A. W., Baldwin M. R., Barbieri J. T. (2004). Ezrin/radixin/moesin proteins are high affinity targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. J. Biol. Chem. 279, 38402-38408 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. (1998). A protein conjugation system essential for autophagy. Nature 395, 395-398 [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., et al. (2004). Autophagy defends cells against invading group A Streptococcus. Science 306, 1037-1040 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. (2005). Escape of intracellular Shigella from autophagy. Science 307, 727-731 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y., Mizushima N. (2004). Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 15, 231-236 [DOI] [PubMed] [Google Scholar]
- Ponpuak M., Davis A. S., Roberts E. A., Delgado M. A., Dinkins C., Zhao Z., Virgin H. W. T., Kyei G. B., Johansen T., Vergne I., et al. (2010). Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux J. D., Xavier R. J., Taylor K. D., Silverberg M. S., Goyette P., Huett A., Green T., Kuballa P., Barmada M. M., Datta L. W., et al. (2007). Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264-268 [DOI] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. (2004). Autophagy in health and disease: a double-edged sword. Science 306, 990-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971-5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teiken J. M., Audettey J. L., Laturnus D. I., Zheng S., Epstein P. N., Carlson E. C. (2008). Podocyte loss in aging OVE26 diabetic mice. Anat. Rec. 291, 114-121 [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Gorvel J. P., Chu Y. T., Wu J. J., Lei H. Y. (2010). Helicobacter pylori impairs murine dendritic cell responses to infection. PLoS ONE 5, e10844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Brown W. L., Stockley P. G. (1995). Cell-specific delivery of bacteriophage-encapsidated ricin A chain. Bioconjug. Chem. 6, 587-595 [DOI] [PubMed] [Google Scholar]
- Wu M., Hussain S., He H. Y., Pasula R., Smith P. A., Martin W. J., II (2001a). Genetically engineered macrophages expressing IFN-γ restore alveolar immune function in scid mice. Proc. Natl. Acad. Sci. USA 98, 14589-14594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Kelley M. R., Hansen W. K., Martin W. J., II (2001b). Reduction of BCNU toxicity to lung cells by high-level expression of O6-methylguanine-DNA methyltransferease. Am. J. Physiol. Lung Cell. Mol. Physiol. 280, L755-L761 [DOI] [PubMed] [Google Scholar]
- Wu M., Stockley P. G., Martin W. J., II (2002). An improved Western blotting effectively reduces the background. Electrophoresis 23, 2373-2376 [DOI] [PubMed] [Google Scholar]
- Wu M., Pasula R., Smith P. A., Martin W. J., II (2003). Mapping alveolar binding sites in vivo using phage display peptide libraries. Gene Ther. 10, 1429-1436 [DOI] [PubMed] [Google Scholar]
- Wu M., Sherwin T., Brown W. L., Stockley P. G. (2005). Delivery of antisense oligonucleotides to leukaemia cells by RNA bacteriophage capsids. Nanomedicine 1, 67-76 [DOI] [PubMed] [Google Scholar]
- Wu M., Audet A., Cusic J., Seeger D., Cochran R., Ghribi O. (2009). Broad DNA repair responses in neural injury are associated with activation of the IL-6 pathway in cholesterol-fed rabbits. J. Neurochem. 111, 1011-1021 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu M., Huang H., Zhang W., Kannan S., Weaver A., Mckibben M., Herington D., Zeng H., Gao H. (2011). Host DNA repair proteins in response to P. aeruginosa in lung epithelial cells and in mice. Infect. Immun. 79, 75-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Hirakata Y., Izumikawa K., Miyazaki Y., Maesaki S., Tomono K., Yamada Y., Kohno S., Kamihira S. (2000). Prolonged survival of mice with Pseudomonas aeruginosa-induced sepsis by rIL-12 modulation of IL-10 and interferon-gamma. J. Med. Microbiol. 49, 701-707 [DOI] [PubMed] [Google Scholar]
- Zolfaghar I., Evans D. J., Fleiszig S. M. (2003). Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71, 5389-5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.