Abstract
Early detection of disease plays a crucial role in successful therapy. Early diagnosis and management reduces the severity and possible complications of the disease process. To overcome this challenge, medical researchers are devoted to finding molecular disease biomarkers that reveal a hidden lethal threat before the disease becomes complicated. Saliva, an important physiologic fluid, containing a highly complex mixture of substances, is rapidly gaining popularity as a diagnostic tool. Periodontal disease is a chronic disease of the oral cavity comprising a group of inflammatory conditions affecting the supporting structures of the dentition. In the field of periodontology, traditional clinical criteria are often insufficient for determining sites of active disease, for monitoring the response to therapy, or for measuring the degree of susceptibility to future disease progression. Saliva, as a mirror of oral and systemic health, is a valuable source for clinically relevant information because it contains biomarkers specific for the unique physiologic aspects of periodontal diseases. This review highlights the various potentials of saliva as a diagnostic biomarker for periodontal diseases.
Keywords: Biomarker, diagnostic fluid, periodontal diseases, saliva
INTRODUCTION
Early detection of disease plays a crucial role in successful therapy. In most cases, the earlier the disease is diagnosed, the more likely it is to be successfully cured or well controlled. However, most systemic diseases are not diagnosed until morbid symptoms become apparent in the late phase. To overcome this challenge, medical researchers are devoted to finding molecular disease biomarkers that reveal a hidden lethal threat before the disease becomes complicated.[1]
NEED FOR A NEW BIOMARKER
Currently, three major limitations have prevented people from recognizing the full potential of disease detection, and have seriously hampered the development of clinical diagnostics, namely:
Lack of definitive molecular biomarkers for specific diseases;
Lack of an easy and inexpensive sampling method with minimal discomfort; and
Lack of an accurate, easy-to-use, and portable platform to facilitate early disease detection.
Saliva, an oral fluid that contains an abundance of proteins and genetic molecules and is readily accessible via a totally noninvasive approach, has long been recognized as the potential solution to these limitations.[1]
Saliva provides an easily available, noninvasive diagnostic medium for a rapidly widening range of diseases and clinical situations.[2]
In the field of periodontology, traditional clinical criteria are often insufficient for determining sites of active disease, for monitoring the response to therapy or for measuring the degree of susceptibility to future disease progression. Saliva as a mirror of oral and systemic health is a valuable source for clinically relevant information because it contains biomarkers specific for the unique physiological aspects of periodontal diseases.
PERIODONTAL DISEASE
Periodontal disease is a chronic disease of the oral cavity comprising a group of inflammatory conditions affecting the supporting structures of the dentition.[3,4] The natural history of periodontitis follows a discontinuous pattern of exacerbation and remission characterized by disease activity and inactivity.[5,6] Periodontitis is a multifactorial disease which is affected by both genetic and environmental factors.[7]
Clinical parameters such as probing depth, attachment level, bleeding on probing, plaque index, and radiographic assessment of alveolar bone loss provide information on the severity of periodontitis but they do not measure disease activity, whereas microbiological tests, analysis of host response, and genetic analyses have been proposed in an effort to monitor and identify patients at increased risk for periodontitis.[6,8]
SALIVA AS A DIAGNOSTIC FLUID IN PERIODONTAL DISEASES
Proposed salivary diagnostic markers for periodontal diseases have included serum and salivary molecules such as immunoglobulins, enzymes constituents of gingival crevicular fluid, bacterial components or products, volatile compounds, and phenotypic markers, such as epithelial keratins.[2,8]
COLLECTION OF SALIVA
The fluid mostly collected for salivary diagnostic purpose is expectorated whole saliva, a mix composed largely of the secretions from the major salivary glands along with the modest contributions from the minor salivary glands and gingival crevicular fluid.
Unstimulated or resting saliva is usually collected by passive drooling into a graduated tube or preweighed vial so that flow rate per unit time can be measured.[2]
When volume measurement is not required, saliva can be collected on cotton swabs, cotton rolls, gauze or filter paper strips, then eluted or centrifuged or aspirated directly from the floor of the mouth with plastic pipettes.[2]
When large volumes of saliva are required for analytical purposes, saliva is stimulated by a masticatory or gustatory stimulus, expectorated and handled in a similar manner as the unstimulated fluid. Softened paraffin wax or a washed rubber band are the usual masticatory stimuli and 2% citric acid applied directly to the tongue is the standard gustatory stimulus.[2]
Many a times secretions from individual glands are preferred and this can be accomplished in a noninvasive manner with suitable collecting devices. Parotid saliva is best collected with plastic modifications of a single cup first introduced by Carlson and Crittenden in 1910. Now disposable and individualized collectors have been introduced for this purpose. Submandibular–sublingual saliva can be collected by customization of a basic plastic collector or by aspiration from the duct openings with micropipettes [Figure 1].[2]
Figure 1.
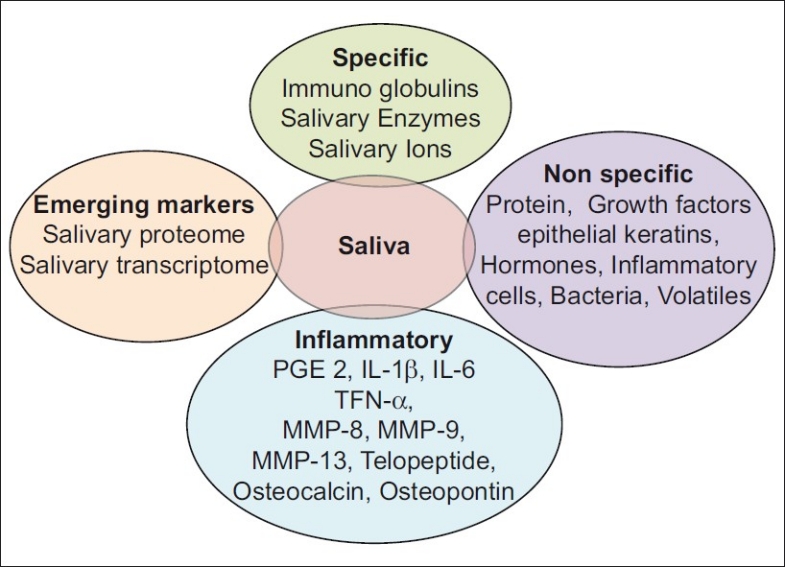
Biomarkers seen in saliva
SALIVARY MARKERS OF PERIODONTAL DISEASES
The various salivary biomarkers are as follows.
Markers affecting the dental biofilm
Specific markers
Immunoglobulins (Ig) are important specific defense factors of saliva. The predominant immunoglobulin in saliva is secretory IgA (sIgA), which is derived from plasma cells in the salivary glands. Lesser amount of IgG and IgM are also found in saliva. IgA, IgG, and IgM influence the oral microbiota by interfering with the bacterial adherence or by inhibiting bacterial metabolism.[3,9] There are two subclasses of IgA: IgA1 and IgA2. IgA1 is predominant in serum while IgA2 is found in higher concentrations in external secretions, that is, tears, saliva, and milk.[10]
Many studies have been attempted to determine a relationship between salivary levels of sIgA and various forms of periodontal diseases. It was found that there was a positive correlation between the severity of inflammation and IgA concentration.[11,12]
Specific immunoglobulins in saliva directed toward periodontal pathogens have also been examined for their diagnostic potential. Eggert et al.[13] reported that saliva from treated periodontitis patients had higher IgA and IgG levels to periodontal pathogens porphyromonas. gingivalis and Treponema. denticola than as compared to saliva from control subjects. Sandholm et al.[14] found increased concentrations of salivary IgG to Aggregatibacter. actinomycetemcomitans in patients of aggressive periodontitis.
Salivary enzymes
Salivary enzymes can be produced by salivary glands, oral microorganisms, polymorphonuclear leukocytes, oral epithelial cells derived from gingival crevicular fluid (GCF).
Lysozyme is an antimicrobial enzyme with the ability to cleave chemical bonds in the bacterial cell wall. It can lyse some bacterial species by hydrolyzing glycosidic linkages in the cell wall peptidoglycan. It may also cause lysis of bacterial cells by interacting with monovalent anions and with proteases found in saliva. This combination leads to destabilization of the cell membrane, probably as a result of the activation and deregulation of endogenous bacterial autolysins. Patients with low levels of lysozyme in saliva are more susceptible to plaque accumulation, which is considered a risk factor for periodontal disease.[15]
Peroxidase is a salivary enzyme produced by acinar cells in the salivary glands. This enzyme removes toxic hydrogen peroxide produced by oral microorganisms and reduces acid production in the dental biofilm, thereby decreasing plaque accumulation and the establishment of gingivitis and caries. Patients with periodontal disease have demonstrated high levels of this enzyme in saliva.[16]
Salivary ions
Calcium (Ca) is the ion that has been most intensely studied as a potential marker for periodontal disease in saliva. Sewon et al.[17] reported a higher concentration of Ca detected in whole stimulated saliva from the periodontitis patients. The authors concluded that an elevated Ca concentration in saliva was characteristic of patients with periodontitis.
Nevertheless, the importance of the salivary Ca concentration in relationship to progression of periodontal disease is not defined. Considering the distribution of Ca, this ion appears to hold only limited promise as a marker for periodontal disease.[8]
Nonspecific markers
Other proteins: A number of studies have examined the correlation between nonenzymatic, nonimmunoglobulin proteins in saliva and periodontal disease.
Mucins are glycoproteins produced by submandibular and sublingual salivary glands and numerous minor salivary glands. The physiological functions of the mucins (MG1 and MG2) are cytoprotection, lubrication, protection against dehydration, and maintenance of viscoelasticity in secretions. The mucin, MG2, affects the aggregation and adherence of bacteria and is known to interact with A. actinomycetemcomitans, and a decreased concentration of MG2 in saliva may increase colonization with this periodontopathogen.[3]
Lactoferrin is an iron-binding glycoprotein produced by salivary glands, which inhibits microbial growth by sequestering iron from the environment, thus depriving bacteria of this essential element. Lactoferrin is strongly upregulated in mucosal secretions during gingival inflammation and is detected at a high concentration in saliva of patients with periodontal disease compared with healthy patients.[18]
Histatin is a salivary protein with antimicrobial properties and is secreted from parotid and submandibular glands. It neutralizes the endotoxic lipopolysaccharides located in the membrane of gram-negative bacteria. Histatin is also an inhibitor of host and bacterial enzymes involved in the destruction of the periodontium. In addition to its antimicrobial activities, histatin is involved in the inhibition of the release of histamine from mast cells, affecting their role in oral inflammation.[3,19]
Fibronectin is a glycoprotein that promotes selective adhesion and colonization of certain bacterial species while inhibiting others. It mediates adhesion between cells and is also involved in chemotaxis, migration, inflammation, and wound healing and tissue repair.[8]
Cystatins (cysteine proteinases) are proteolytic enzymes originated from pathogenic bacteria, inflammatory cells, osteoclasts, and fibroblasts. These enzymes have collagenolytic activity, which may cause tissue destruction.[8] Cystatins are physiological inhibitors of cysteine proteinases, and may function by modulating enzyme activity in the periodontium.
Cystatins are present in a variety of tissues and body fluids, including saliva. Cystatins were found in saliva collected from the submandibular and sublingual salivary glands, and to a lesser degree in saliva from the parotid gland.[20]
Platelet activating factor (PAF) is a potent phospholipid mediator of inflammation. Garito et al. found a positive correlation between PAF and periodontal inflammation.[21] Various other studies also showed similar findings but none of the authors discussed the potential diagnostic significance of their findings.[8]
Amino acids Several studies have examined the levels of free amino acids in saliva in relation to periodontal status. It appears that in some patients, elevated levels of certain amino acids, especially proline, may be detected.[22,23] These amino acids probably appear in whole saliva as a result of bacterial metabolism or degradation of salivary proteins rich in proline. In another study, by the same investigators, no differences in amino acid concentrations in saliva were found between patients with progressive periodontitis and controls. The authors concluded that levels of amino acids in oral fluids (including GCF) has no diagnostic significance for periodontal disease.[24]
Growth factors
Epidermal growth factor (EGF) is involved in oral wound healing and functions with hormone-like properties to stimulate epithelial cells. In humans, the parotid gland is the major source of EGF.[8]
Vascular endothelial growth factor (VEGF), also known as vascular permeability factor or vasculotropin, is a multifunctional angiogenic cytokine important in inflammation and wound healing. This cytokine was found to be a component of whole saliva. Higher levels of VEGF were detected in whole saliva from periodontitis patients.[8]
Epithelial keratins
Epithelial cells from the lining of the oral cavity are found in saliva, but the contribution of crevicular or pocket epithelial cells to the total number of salivary epithelial cells is not known.[25] Furthermore, detection of keratins by monoclonal antibodies may have diagnostic value in the detection of epithelial dysplasia, oral cancer, odontogenic cysts, and tumors.[26]
It has been suggested that phenotypic markers for junctional and oral sulcular epithelia might eventually be used as indicators of periodontal disease, however, there are no studies that demonstrate an association between number or type of epithelial cells or specific types of keratins in saliva and progression of periodontitis.[8]
Hormones
Cortisol: Recent studies have suggested that emotional stress is a risk factor for periodontitis.[7,27,28] One mechanism proposed to account for the relationship is that elevated serum cortisol levels associated with emotional stress exert a strong inhibitory effect on the inflammatory process and immune response.[29] The presence of cortisol in saliva has been recognized for more than 40 years.[30] Recently, salivary cortisol levels were used to evaluate the role of emotional stress in periodontal disease. Higher salivary cortisol levels were detected in individuals exhibiting severe periodontitis, a high level of financial strain, and high emotion focused coping, as compared to individuals with little or no periodontal disease, low financial strain, and low levels of emotion-focused coping.[31] While it appears that emotional stress, as reflected by salivary cortisol levels, is a risk factor for periodontal disease, attempts to diagnose periodontal disease severity or activity based on salivary cortisol levels are premature. It can be argued that in addition to changes in the host response (ie, smoking, poor oral hygiene, and poor compliance), stress also leads to behavioral changes, which could have a significant effect on the periodontium. Additional studies are required to determine the diagnostic value of salivary cortisol for periodontal disease.[8]
Inflammatory cells
The number of leukocytes in saliva varies from person to person, and the cell counts vary for an individual during the course of the day. The majority of salivary leukocytes enter the oral cavity via the gingival crevice.[32] Klinkhammer[33] standardized collection and counting of leukocytes in saliva and developed the orogranulocytic migratory rate (OMR). Raeste et al.,[34] in their study, indicated that the OMR reflects the presence of oral inflammation, and suggested that this measure can be used as a laboratory test.
Occult blood in saliva in relation to gingival inflammation has also been examined using a home-screening test.[35]
Bacteria
Specific species of bacteria colonizing the subgingival environment have been implicated in the pathogenesis of periodontal disease.[36–38] It has been suggested that microorganisms in dental plaque can survive in saliva, and can utilize salivary components as a substrate. It was shown that saliva could serve as a growth medium for oral Streptococus species and Actinomyces. viscous.[39]
In a study by De Jong et al.,[40] microorganisms from supragingival plaque were grown on saliva agar. When supragingival plaque was plated on saliva and blood agar plates, the composition of the microflora isolated from the plates were similar. The authors concluded that the supragingival microflora could utilize saliva as a complete nutrient source. Studies have also shown the presence of periodontopathic microorganisms, A. actinomycetemcomitans, P. gingivalis, Prevotella intermedia, and T. denticola in whole saliva.[41,42]
An oral microbial rinse test (Oratest) was described by Rosenberg et al.[43] In this study Oratest was found to be a simple method for estimating oral microbial levels. In a companion study, Oratest results were correlated with plaque index and gingival index scores, and the authors stated that this test provides a reliable estimate of gingival inflammation.[44]
Volatiles
Volatile sulfur compounds, primarily hydrogen sulfide and methylmercaptan, are associated with oral malodor. Salivary volatiles have been suggested as possible diagnostic markers and contributory factors in periodontal disease.
In a study of self-estimation of oral malodor, estimation of malodor based on saliva yielded a significant correlation with objective parameters.[45]
Markers in saliva via GCF
As GCF traverses through inflamed periodontal tissues en route to the sulcus, biological molecular markers are gathered from the surrounding areas and are subsequently eluted into whole saliva.
Markers of periodontal soft tissue inflammation
Proinflammatory cytokines, such as prostaglandin E2 (PGE2), interleukin (IL)-1beta, IL-6, and tumor necrosis factor-alpha are released from cells of the junctional epithelium, connective tissue fibroblasts, macrophages, and polymorphonuclear leukocytes. Enzymes, such as matrix metalloproteinase (MMP)-8, MMP-9, and MMP-13, produced by polymorphonuclear leukocytes and osteoclasts, all lead to the degradation of connective tissue collagen and alveolar bone. Studies have shown that PGE2 acts as a potent vasodilator and increases capillary permeability, which elicits clinical signs of redness and edema. It also stimulates fibroblasts and osteoclasts to increase the production of MMPs.[46]
Markers of alveolar bone loss
Many different biomarkers associated with bone formation, resorption, and turnover, such as alkaline phosphatase, osteocalcin, osteonectin, and collagen telopeptidases, have been evaluated in GCF and saliva.[47] These mediators are associated with local bone metabolism as well as with systemic conditions.
Matrix metalloproteinases (MMP): They are host proteinases responsible for both tissue degradation and remodeling. During progressive periodontal breakdown, gingival and periodontal ligament collagens are cleaved by host cell–derived interstitial collagenases.
MMP-8: It is the most prevalent MMP found in diseased periodontal tissue and GCF. Recently, the level of MMP-8 was demonstrated to be highly elevated in saliva from patients with periodontal disease using a rapid point-of-care microfluidic device.[48] Studies are required to evaluate MMP-8, either alone or in conjunction with other molecular biomarkers, to predict the risk of future disease occurrence and to monitor treatment interventions.
Gelatinase (MMP-9): Another member of the collagenase family, is produced by neutrophils and degrades collagen intercellular ground substance. In their study, Teng et al.[49] found a twofold increase in the mean MMP-9 levels in patients with progressive attachment loss. Given these results, future use of MMP-9 in oral fluid diagnostics may serve as a guide in periodontal treatment monitoring.
Collagenase-3: Also referred to as MMP-13, is another collagenolytic MMP with an exceptionally wide substrate specificity. In the future, MMP-13 may be useful for diagnosing and monitoring the course of periodontal disease as well as for tracking the efficacy of therapy.[50]
Telopeptide: Given the specificity and sensitivity for bone resorption, pyridinoline cross-links, such as pyridinoline cross-linked carboxyterminal telopeptide of type I collagen represent a potentially valuable diagnostic aid for periodontal disease. Several investigations have explored the ability of pyridinoline cross-links to detect bone resorption in periodontitis as well as in response to periodontal therapy.[51,52] These studies assessing the role of GCF carboxyterminal telopeptide of type I collagen levels as a diagnostic marker of periodontal disease activity have produced promising results to date. Carboxyterminal telopeptide of type I collagen has been shown to be a promising predictor of both future alveolar bone and attachment loss.
Controlled human longitudinal trials are needed to establish fully the role of salivary carboxyterminal telopeptide of type I collagen as a predictor of periodontal tissue destruction, disease activity and response to therapy in periodontal patients.
Osteocalcin: Elevated serum osteocalcin levels have been found during periods of rapid bone turnover, such as in osteoporosis and multiple myeloma and during fracture repair. Therefore, studies have investigated the relationship between GCF osteocalcin levels and periodontal disease.
When a combination of the biochemical markers osteocalcin, collagenase, prostaglandin E2, alpha-2 macroglobulin, elastase, and alkaline phosphatase was evaluated, increased diagnostic sensitivity and specificity values of 80% and 91%, respectively, were reported.[53]
Osteopontin: It is a single-chain polypeptide with a molecular weight of approximately 32,600. In bone matrix, osteopontin is highly concentrated at sites where osteoclasts are attached to the underlying mineral surface (ie, the clear zone attachment areas of the plasma membrane). Results from periodontal studies indicated that osteopontin concentrations in GCF increased proportionally with the progression of disease; and when nonsurgical periodontal treatment was provided, the osteopontin levels in GCF were significantly reduced. Although additional long-term prospective studies are needed, at this point osteopontin appears to hold promise as a possible salivary biomarker of periodontal disease progression.[54]
Systemic markers
C-reactive protein is produced by the liver and is stimulated by circulating cytokines, such as tumor necrosis factor-alpha and interleukin-1, from local and/or systemic inflammation such as periodontal inflammation. Circulating C-reactive protein may reach saliva via GCF or the salivary glands. High levels of C-reactive protein have been associated with chronic and aggressive periodontal diseases and with other inflammatory biomarkers.[19] C-reactive protein has recently been shown to be measurable in saliva from periodontal patients using a lab-on-a-chip method.[3]
Oxidative stress marker
Oxidative stress is defined as the result of an imbalance between oxidant factors and protective antioxidant systems; it may occur due to an excess of free radicals, or by the diminishing of the antioxidant systems. Oxidative stress is enhanced during periodontitis.[55] Oxidative stress can result in DNA damage, including oxidation of nucleosides. 8-Hydroxydeoxyguanosine (8-OHdG) is an oxidized nucleoside that is excreted in the bodily fluids with DNA repair. It has been demonstrated that the 8-OHdG in bodily fluids can act as a biomarker of oxidative stress.[56]
Studies have shown that saliva is a biological product that can be easily collected and may be used for the quantification of 8-OHdG as an oxidative stress biomarker in the diagnosis and monitorization of the treatment in periodontitis.[57,58]
Antioxidative capacity of saliva
Physiologically free radical/reactive oxygen species in the mouth are derived mainly from polymorphonuclear neutrophils (PMN), which may also help to control bacterial growth by the well-known “respiratory burst” (RB). Such physiological processes are usually efficiently counteracted by intrinsic antioxidant systems: if such systems fail, tissue damage can result.[59]
Saliva may constitute a first line of defence against free radical-mediated oxidative stress, since the process of mastication promotes a variety of such reactions, including lipid peroxidation.[60] Moreover, during gingival inflammation, gingival crevicular fluid (GCF) flow increases, adding to saliva with products from the inflammatory response. This is why the antioxidant capacity of saliva is of increasing interest.
Saliva is rich in antioxidants, mainly uric acid, with lesser contributions from albumin, ascorbate and glutathione.[61] It has been reported that uric acid is the major antioxidant in saliva, accounting for more than 85% of the total antioxidant activity of resting and stimulated saliva from both healthy and periodontally compromised subjects.[61]
Antioxidants such as albumin, ascorbic acid, glutathione and traces of transferrin, lactoferrin, and caeruloplasmin are also found in saliva.[59]
Emerging salivary diagnostic tools
The use of saliva for translational and clinical application has emerged at the forefront. Most relevant to periodontal diseases are the emerging toolboxes of the salivary proteome and the salivary transcriptome for early detection, disease progression, and therapeutic monitoring. Using these emerging technologies, it has been shown that salivary proteins and RNAs can be used to detect oral cancer[62,63] and Sjogren's syndrome.[64] The stage is now poised to use these technologies for translational and clinical applications in periodontal diseases.
Salivary proteome: The proteome is the protein complement of the genome, and proteomics is analysis of the portion of the genome that is expressed. The proteomes in body fluids are valuable due to their high clinical potential as sources of disease markers. In principle, a global analysis of the human salivary proteomes can provide a comprehensive spectrum of oral and general health. Furthermore, analysis of salivary proteomes over the course of complications may unveil morbidity signatures in the early stage and monitor disease progression. Significant progress has been made in cataloguing human saliva proteins and exploring their post-translational modifications. By using both two-dimensional gel electrophoresis/mass spectrometry and “shotgun” proteomics approaches, Hu et al.[64,65] identified 309 distinct proteins in human whole saliva.
Collectively, 1166 salivary proteins have been identified: 914 from the parotid fluid and 917 from the combined submandibular and sublingual fluids by Denny et al.[66]
Salivary transcriptome: Salivary transcriptome is an emerging concept. In addition to salivary proteome, salivary transcriptomes (RNA molecules) were discovered that are unusually stable in saliva.[67] They included mRNA molecules that cells use to convey the instructions carried by DNA for subsequent protein production. This discovery presented a second diagnostic alphabet in saliva and opened a door to another avenue of salivary transcriptomic diagnostics.[1]
Li et al.[67] found that RNA molecules elevated in oral cancer tissues are also elevated in saliva, which prompted them to examine the scope and complexity of RNA present in human saliva. Other research groups, particularly from forensic sciences, are focusing on multiplex mRNA profiling for the identification of body fluids, including saliva.[68,69]
Possible testing for HIV
To expedite screening and accurately diagnose HIV infection, rapid point-of-care HIV tests have been developed. Majority of these rapid tests utilize whole blood, plasma, and finger stick blood specimens, whereas a few tests employ nonconventional specimens, such as saliva, oral mucosal fluid and urine.[70,71] Although salivary and oral fluid-based tests have been in development for the past 20 years, it is only recently that oral fluid-based, rapid point-of-care tests have increased in popularity.[71] Oral fluid tests are based on a salivary component, the oral mucosal transudate or crevicular fluid, an interstitial transudate rich in IgG antibodies, used for diagnosing HIV infection.[70–72] Although accuracy is very high, oral fluid test results are considered preliminary. Therefore, the results of oral fluid tests require confirmatory testing with conventional tests, such as ELISA and/or western blot.[73]
DISCUSSION
In the field of periodontal diagnosis, there has been a steady growing trend during the last 2 decades to develop tools to monitor periodontitis. Currently, diagnosis of periodontal disease relies primarily on clinical and radiographic parameters. These measures are useful in detecting evidence of past disease, or verifying periodontal health, but provide only limited information about patients and sites at risk for future periodontal breakdown. Numerous markers in saliva have been proposed and used as diagnostic tests for periodontal disease.
Ideally, diagnostic tests should demonstrate high specificity and sensitivity. Given the complex nature of periodontal disease, it is unlikely that a single marker will prove to be both sensitive and specific. A combination of two or more markers may provide a more accurate assessment of the periodontal patient.
Interest in saliva as a diagnostic medium is escalating due to its many advantages over other diagnostic biofluids. Both saliva and blood serum contain similar proteins and RNA, which is why saliva is considered a “mirror to the body.”[74] Saliva is readily available which makes the collection process fairly straightforward, even when multiple samples are needed. Its collection is noninvasive, which makes the procedure more acceptable to patients and more conducive to a stress-free appointment. Many of the hazards associated with blood collection do not apply to saliva. There is no potential for cross-contamination among patients when used improperly and present a danger to health care personnel. Because of the low concentrations of antigens in saliva, HIV, and hepatitis infections are much less of a danger from saliva than from blood.[75] Saliva is also easier to handle because it does not clot. As salivary testing becomes more commonplace, the costs could drop below those currently incurred for urine/blood sampling. However, due to the uniqueness of the technique, the analysis today is still quite expensive.
GCF has several diagnostic advantages, such as contributing inflammatory mediators and tissue-destructive molecules associated with periodontitis that appear, and can be detected, in GCF. However, GCF analyses are time consuming, requiring multiple sampling of individual tooth sites. The procedure is labor intensive and somewhat technically demanding, requiring equipment for calibrating and measuring fluid volumes. Finally, analysis is expensive, requires laboratory-based assays and generally cannot be done chairside. In addition, GCF analyses involve miniscule amounts of fluid, often approximately 1 μL, which has an impact on laboratory analysis, and can be contaminated with blood, saliva, or plaque.[76] Given some of the problems inherent in sampling GCF, the analysis of salivary biomarkers offers some advantages. Acquisition of saliva is easy, noninvasive, rapid, and requires less manpower and materials than GCF. However, in contrast with GCF, whole saliva clearly provides different diagnostic information.[77–79]
Because of the simple and noninvasive method of collection, salivary diagnostic tests appear to hold promise for the future.
Novel technologies such as lab-on-a-chip and microfluidic devices have the potential to manage complex oral fluids, such as saliva and GCF, and to provide a determination of a patient's periodontal disease-risk profile, current disease activity, and response to therapeutic interventions. This approach should accelerate clinical decision-making and monitoring of episodic disease progression in a chronic infectious disease such as periodontitis.[8]
CONCLUSION
Saliva, like blood, contains an abundance of protein and nucleic acid molecules that reflects physiological status; however, unlike other bodily fluids, salivary diagnostics offer an easy, inexpensive, safe, and noninvasive approach for disease detection, and possess a high potential to revolutionize the next generation of diagnostics.
Although challenges remain ahead, the use of saliva-based oral fluid diagnostics appear promising for future application to diagnose periodontal diseases and to prognosticate periodontal treatment outcomes.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Hari Menon, Dr. Abhijit Gurav, and Dr. Abhijeet Shete for thoughtful discussions, and assistance in preparation of the manuscript.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Lee Y, Wong DT. Saliva: An emerging biofluid for early detection of disease. Am J Dent. 2009;22:241–8. [PMC free article] [PubMed] [Google Scholar]
- 2.Mandel ID. The diagnostic uses of saliva. J Oral Pathol Med. 1990;19:119–25. doi: 10.1111/j.1600-0714.1990.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 3.Giannobile WV, Beikler T, Kinney JS, Ramseier CA, Morelli T, Wong DT. Saliva as a diagnostic tool for periodontal disease: Current state and future directions. Periodontol 2000. 2009;50:52–64. doi: 10.1111/j.1600-0757.2008.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Offenbacher S. Periodontal diseases: Pathogenesis. Ann Periodontol. 1996;1:821–78. doi: 10.1902/annals.1996.1.1.821. [DOI] [PubMed] [Google Scholar]
- 6.Sahingur SE, Cohen RE. Analysis of host responses and risk for disease progression. Periodontol 2000. 2004;34:57–83. doi: 10.1046/j.0906-6713.2002.003425.x. [DOI] [PubMed] [Google Scholar]
- 7.Genco RJ. Current view of risk factors for periodontal diseases. J Periodontol. 1996;67(10 Suppl):1041–9. doi: 10.1902/jop.1996.67.10.1041. [DOI] [PubMed] [Google Scholar]
- 8.Kaufman I, Lamster IB. Analysis of saliva for periodontal diagnosis: A review. J Clin Periodontol. 2000;27:453–65. doi: 10.1034/j.1600-051x.2000.027007453.x. [DOI] [PubMed] [Google Scholar]
- 9.Marcotte H, Lavoie MC. Oral microbial ecology and the role of salivary immunoglobulin A. Microbiol Molecular Biol Rev. 1998;62:71–109. doi: 10.1128/mmbr.62.1.71-109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delacroix DL, Dive C, Rambaud JC, Vaerman JP. IgA subclasses in various secretions and in serum. Immunology. 1982;47:383–5. [PMC free article] [PubMed] [Google Scholar]
- 11.Guven O, De Visscher JG. Salivary IgA in periodontal disease. J Periodontol. 1982;53:334–5. doi: 10.1902/jop.1982.53.5.334. [DOI] [PubMed] [Google Scholar]
- 12.Sandholm L, Gronblad E. Salivary immunoglobulins in patients with juvenile periodontitis and their healthy siblings. J Periodontol. 1984;55:9–12. doi: 10.1902/jop.1984.55.1.9. [DOI] [PubMed] [Google Scholar]
- 13.Eggert FM, Maenz L, Tam YC. Measuring the interaction of human secretory glycoproteins to oral bacteria. J Dent Res. 1987;66:610–2. doi: 10.1177/00220345870660024501. [DOI] [PubMed] [Google Scholar]
- 14.Sandholm L, Tolo K, Olsen I. Salivary IgG, a parameter of periodontal disease activity? High responders to actinobacillus actinomycetemcomitans Y4 in juvenile and adult periodontitis. J Clin Periodontol. 1987;14:289–94. doi: 10.1111/j.1600-051x.1987.tb01535.x. [DOI] [PubMed] [Google Scholar]
- 15.Jalil RA, Ashley FP, Wilson RF, Wagaiyu EG. Concentrations of thiocyanate, hypothiocyanite, “free” and “total” lysozyme, lactoferrin and secretory IgA in resting and stimulated whole saliva of children aged 12-14 years and the relationship with plaque accumulation and gingivitis. J Periodont Res. 1993;28:130–6. doi: 10.1111/j.1600-0765.1993.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 16.Guven Y, Satman I, Dinccag N, Alptekin S. Salivary peroxidase activity in whole saliva of patients with insulin-dependent (type-1) diabetes mellitus. J Clin Periodontol. 1996;23:879–81. doi: 10.1111/j.1600-051x.1996.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 17.Sewon L, Karjalainen SM, Sainio M, Seppa O. Calcium and other salivary factors in periodontitis affected subjects prior to treatment. J Clin Periodontol. 1995;22:267–70. doi: 10.1111/j.1600-051x.1995.tb00146.x. [DOI] [PubMed] [Google Scholar]
- 18.Groenink J, Walgreen-Weterings E, Nazmi K, Bolscher JG, Veerman EC, van Winkelhoff AJ, et al. Salivary lactoferrin and low-Mr mucin MG2 in Actinobacillus actinomycetemcomitans-associated periodontitis. J Clin Periodontol. 1999;26:269–75. doi: 10.1034/j.1600-051x.1999.260501.x. [DOI] [PubMed] [Google Scholar]
- 19.Helmerhorst EJ, Oppenheim FG. Saliva: A dynamic proteome. J Dent Res. 2007;86:680–93. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 20.Henskens YM, van der Velden U, Veerman ECI, Nieuw Amerongen AV. Cystatins S and C in human whole saliva and in glandular salivas in periodontal health and disease. J Dent Res. 1994;73:1606–14. doi: 10.1177/00220345940730100501. [DOI] [PubMed] [Google Scholar]
- 21.Garito ML, Prihoda TJ, McManus LM. Salivary PAF levels correlate with the severity of periodontal inflammation. J Dental Res. 1995;74:1048–56. doi: 10.1177/00220345950740040401. [DOI] [PubMed] [Google Scholar]
- 22.Syrjanen S, Piironen P, Markkanen H. Free amino-acid composition of wax-stimulated whole saliva in human subjects with healthy periodontium, severe chronic periodontitis and post-juvenile periodontitis. Arch Oral Bio. 1984;29:735–8. doi: 10.1016/0003-9969(84)90181-x. [DOI] [PubMed] [Google Scholar]
- 23.Syrjanen S, Piironen P, Markkanen H. Free amino-acid content of wax stimulated whole saliva as related to periodontal disease. Arch Oral Biol. 1987;32:607–10. doi: 10.1016/0003-9969(87)90032-x. [DOI] [PubMed] [Google Scholar]
- 24.Syrjanen S, Alakuijala L, Alakuijala P, Markkanen SO, Markkanen H. Free amino acid levels in oral fluid of normal subjects and patients with periodontal disease. Arch Oral Biol. 1990;35:189–93. doi: 10.1016/0003-9969(90)90054-e. [DOI] [PubMed] [Google Scholar]
- 25.Wilton JM, Curtis MA, Gillett IR, Griffiths GS, Maiden MF, Sterne JA, et al. Detection of high-risk groups and individuals for periodontal diseases: Laboratory markers from analysis of saliva. J Clin Periodontol. 1989;16:475–83. doi: 10.1111/j.1600-051x.1989.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgan PR, Shirlaw PJ, Johnson NW, Leigh IM, Lane EB. Potential applications of anti-keratin antibodies in oral diagnosis. J Oral Pathol. 1987;16:212–22. doi: 10.1111/j.1600-0714.1987.tb02068.x. [DOI] [PubMed] [Google Scholar]
- 27.Linden GJ, Mullally BH, Freeman R. Stress and the progression of periodontal disease. J Clin Periodontol. 1996;23:675–80. doi: 10.1111/j.1600-051x.1996.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 28.Axtelius B, Soderfeldt B, Nilsson A, Edwardsson S, Attstrom R. Therapy-resistant periodontitis. Psychosocial characteristics. J Clin Periodontol. 1998;62:482–91. doi: 10.1111/j.1600-051x.1998.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 29.Chrousos GP, Gold PW. The concepts of stress and stress system disorders: Overview of physical and behavioral homeostasis. J Am Med Assoc. 1992;267:1244–52. [PubMed] [Google Scholar]
- 30.Shannon IL, Prigmore JR, Brooks RA, Feller RP. The 17-hydroxycorticosteroids of parotid fluid, serum and urine following intramuscular administration of repository corticotropin. J Clin Endocrinol. 1959;19:1477–80. doi: 10.1210/jcem-19-11-1477. [DOI] [PubMed] [Google Scholar]
- 31.Genco RJ, Ho AW, Kopman J, Grossi SG, Dunford RG, Tedesco LA. Models to evaluate the role of stress in periodontal disease. Ann Periodontol. 1998;3:288–302. doi: 10.1902/annals.1998.3.1.288. [DOI] [PubMed] [Google Scholar]
- 32.Schiott RC, Loe H. The origin and variation in number of leukocytes in the human saliva. J Periodont Res. 1970;5:36–41. doi: 10.1111/j.1600-0765.1970.tb01835.x. [DOI] [PubMed] [Google Scholar]
- 33.Klinkhammer J. Quantitative evaluation of gingivitis and periodontal disease (I): The orogranulocyte migratory rate. Periodontics. 1968;6:207–11. [PubMed] [Google Scholar]
- 34.Raeste AM, Aura A. Rate of migration of oral leukocytes in-patients with periodontitis. Scand J Dent Res. 1978;86:43–51. doi: 10.1111/j.1600-0722.1978.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 35.Kopczyk RA, Graham R, Abrams H, Kaplan A, Matheny J, Jasper SJ. The feasibility and reliability of using a home screening test to detect gingival inflammation. J Periodontol. 1995;66:52–4. doi: 10.1902/jop.1995.66.1.52. [DOI] [PubMed] [Google Scholar]
- 36.Genco RJ, Zambon JJ, Christersson LA. The role of specific bacteria in periodontal disease: The origin of periodontal infections. Adv Dent Res. 1998;2:245–59. doi: 10.1177/08959374880020020901. [DOI] [PubMed] [Google Scholar]
- 37.Socransky SS. Relationship of bacteria to the etiology of periodontal disease. J Dent Res. 1970;49:203–22. doi: 10.1177/00220345700490020401. [DOI] [PubMed] [Google Scholar]
- 38.Slots J. Bacterial specificity in adult periodontitis: A summary of recent work. J Clin Periodontol. 1986;13:912–7. doi: 10.1111/j.1600-051x.1986.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 39.de Jong MH, van der Hoeven JS, van Os JH, Olijve JH. Growth of oral streptococcus species and actinomyces viscosus in human saliva. App Environ Microbiol. 1984;47:901–4. doi: 10.1128/aem.47.5.901-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Jong MH, van der Hoeven JS, van Os JH. Growth of micro-organisms from supragingival dental plaque on saliva agar. J Dent Res. 1986;65:85–8. doi: 10.1177/00220345860650021601. [DOI] [PubMed] [Google Scholar]
- 41.Asikainen S, Alaluusua S, Saxen L. Recovery of A.actinomycetemcomitans from teeth, tongue and saliva. J Periodontol. 1991;62:203–6. doi: 10.1902/jop.1991.62.3.203. [DOI] [PubMed] [Google Scholar]
- 42.Umeda M, Contreras A, Chen C, Bakker I, Slots J. The utility of whole saliva to detect the oral presence of periodontopathic bacteria. J Periodontol. 1998;69:828–33. doi: 10.1902/jop.1998.69.7.828. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg M, Barki M, Portnoy S. A simple method for estimating oral microbial levels. J Microbiol Method. 1989;9:253. [Google Scholar]
- 44.Tal H, Rosenberg M. Estimation of dental plaque levels and gingival inflammation using a simple oral rinse technique. J Periodontol. 1990;61:339–42. doi: 10.1902/jop.1990.61.6.339. [DOI] [PubMed] [Google Scholar]
- 45.Rosenberg M, Kozlovsky A, Gelernter I, Cherniak O, Gabbay J, Baht R, et al. Self-estimation of oral malodor. J Dent Res. 1995;74:1577–82. doi: 10.1177/00220345950740091201. [DOI] [PubMed] [Google Scholar]
- 46.Airila-Mansson S, Soder B, Kari K, Meurman JH. Influence of combinations of bacteria on the levels of prostaglandin E2, interleukin-1beta, and granulocyte elastase in gingival crevicular fluid and on the severity of periodontal disease. J Periodontol. 2006;77:1025–31. doi: 10.1902/jop.2006.050208. [DOI] [PubMed] [Google Scholar]
- 47.Kinney JS, Ramseier CA, Giannobile WV. Oral fluid-based biomarkers of alveolar bone loss in periodontitis. Ann N Y Acad Sci. 2007;1098:230–51. doi: 10.1196/annals.1384.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, et al. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci USA. 2007;104:5268–73. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng YT, Sodek J, McCulloch CA. Gingival crevicular fluid gelatinase and its relationship to periodontal disease in human subjects. J Periodontal Res. 1992;27:544–52. doi: 10.1111/j.1600-0765.1992.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez M, Valenzuela MA, Lopez-Otin C, Alvarez J, Lopez JM, Vernal R, et al. Matrix metalloproteinase- 13 is highly expressed in destructive periodontal disease activity. J Periodontol. 2006;77:863–70. doi: 10.1902/jop.2006.050461. [DOI] [PubMed] [Google Scholar]
- 51.Giannobile WV. C-telopeptide pyridinoline cross-links. Sensitive indicators of periodontal tissue destruction. Ann N Y Acad Sci. 1999;878:404–12. doi: 10.1111/j.1749-6632.1999.tb07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talonpoika JT, Hamalainen MM. Type I collagen carboxyterminal telopeptide in human gingival crevicular fluid in different clinical conditions and after periodontal treatment. J Clin Periodontol. 1994;21:320–6. doi: 10.1111/j.1600-051x.1994.tb00720.x. [DOI] [PubMed] [Google Scholar]
- 53.Nakashima K, Giannopoulou C, Andersen E, Roehrich N, Brochut P, Dubrez B, et al. A longitudinal study of various crevicular fluid components as markers of periodontal disease activity. J Clin Periodontol. 1996;23:832–8. doi: 10.1111/j.1600-051x.1996.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 54.Sharma CG, Pradeep AR. Gingival crevicular fluid osteopontin levels in periodontal health and disease. J Periodontol. 2006;77:1674–80. doi: 10.1902/jop.2006.060016. [DOI] [PubMed] [Google Scholar]
- 55.Sculley DV, Langley-Evans SC. Periodontal disease is associated with lower antioxidant capacity in whole saliva and evidence of increased protein oxidation. Clin Sci (Lond) 2003;105:167–72. doi: 10.1042/CS20030031. [DOI] [PubMed] [Google Scholar]
- 56.Mahmood S, Kawanaka M, Kamei A, Izumi A, Nakata K, Niiyama G, et al. Immunohistochemical evaluation of oxidative stress markers in chronic hepatitis C. Antioxid Redox Signal. 2004;6:19–24. doi: 10.1089/152308604771978318. [DOI] [PubMed] [Google Scholar]
- 57.Badea V, Balaban D, Amariei C, Nuca C, Bucur L. Salivary 8-hidroxy-2-deoxy guanosine as oxidative stress biomarker for the diagnosis of periodontal disease. Farmacia. 2010;58:5. [Google Scholar]
- 58.Takane M, Sugano N, Ezawa T, Uchiyama T, Ito K. A marker of oxidative stress in saliva: Association with periodontally-involved teeth of a hopeless prognosis. J Oral Sci. 2005;47:53–7. doi: 10.2334/josnusd.47.53. [DOI] [PubMed] [Google Scholar]
- 59.Battino M, Ferreiro MS, Gallardo I, Newman HN, Bullon P. The antioxidant capacity of saliva. J Clin Periodontol. 2002;29:189–94. doi: 10.1034/j.1600-051x.2002.290301x.x. [DOI] [PubMed] [Google Scholar]
- 60.Terao J, Nagao A. Antioxidative effect of human saliva on lipid peroxidation. Agr Biol Chem. 1991;55:869–72. [Google Scholar]
- 61.Moore S, Calder KA, Miller NJ, Rice-Evans CA. Antioxidant activity of saliva and periodontal disease. Free Radical Res. 1994;21:417–25. doi: 10.3109/10715769409056594. [DOI] [PubMed] [Google Scholar]
- 62.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–52. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10:8442–50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 64.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum. 2007;56:3588–600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography ⁄ mass spectrometry and two-dimensional gel electrophoresis- mass spectrometry. Proteomics. 2005;5:1714–28. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 66.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell free saliva using microarray technology. J Dent Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- 68.Ballantyne J. Validity of messenger RNA expression analyses of human saliva. Clin Cancer Res. 2007;13:1350. doi: 10.1158/1078-0432.CCR-06-2796. author reply 1351. [DOI] [PubMed] [Google Scholar]
- 69.Nussbaumer C, Gharehbaghi-Schnell E, Korschineck I. Messenger RNA profiling: A novel method for body fluid identification by Real-Time PCR. Forensic Sci Int. 2006;157:181–6. doi: 10.1016/j.forsciint.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 70.Gottfried TD, Mink RW, Phanuphak P. Calypte AWARE HIV-1/2 OMT antibody test using oral fluid: Special challenges of rapid HIV testing in the developing world. Expert Rev Mol Diagn. 2006;6:139–44. doi: 10.1586/14737159.6.2.139. [DOI] [PubMed] [Google Scholar]
- 71.Jones JP. In the culture of now: The arrival of the OraQuick rapid HIV test. Posit Aware. 2004;15:35–6. [PubMed] [Google Scholar]
- 72.Donovan BJ, Rublein JC, Leone PA, Pilcher CD. HIV infection: Point-of-care testing. Ann Pharmacother. 2004;38:670–6. doi: 10.1345/aph.1D314. [DOI] [PubMed] [Google Scholar]
- 73.Reynolds SJ, Muwonga J. OraQuick ADVANCE Rapid HIV-1/2 antibody test. Expert Rev Mol Diagn. 2004;4:587–91. doi: 10.1586/14737159.4.5.587. [DOI] [PubMed] [Google Scholar]
- 74.Schipper R, Loof A, de Groot J, Harthoorn L, Dransfield E, van Heerde W. SELDI-TOF-MS of saliva: Methodology and pre-treatment effects. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:45–53. doi: 10.1016/j.jchromb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Major CV, Read SE, Coates RA, Francis A, McLaughlin BJ, Millson M, et al. Comparison of saliva and blood for human immunodeficiency virus prevalence testing. J Infect Dis. 1991;163:699–702. doi: 10.1093/infdis/163.4.699. [DOI] [PubMed] [Google Scholar]
- 76.Griffiths GS. Formation, collection and significance of gingival crevice fluid. Periodontol 2000. 2003;31:32–42. doi: 10.1034/j.1600-0757.2003.03103.x. [DOI] [PubMed] [Google Scholar]
- 77.Miller CS, Foley JD, Bailey AL, Campell CL, Humphries RL, Christodoulides N, et al. Current developments in salivary diagnostics. Biomark Med. 2010;4:171–89. doi: 10.2217/bmm.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D’Aiuto F, Ready D, Tonetti MS. Periodontal disease and C reactive protein-associated cardiovascular risk. J Periodontal Res. 2004;39:236–41. doi: 10.1111/j.1600-0765.2004.00731.x. [DOI] [PubMed] [Google Scholar]
- 79.Teng YT, Sodek J, McCulloch CA. Gingival crevicular fluid gelatinase and its relationship to periodontal disease in human subjects. J Periodontal Res. 1992;27:544–52. doi: 10.1111/j.1600-0765.1992.tb01830.x. [DOI] [PubMed] [Google Scholar]


