Abstract
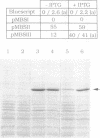
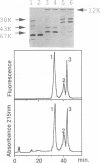
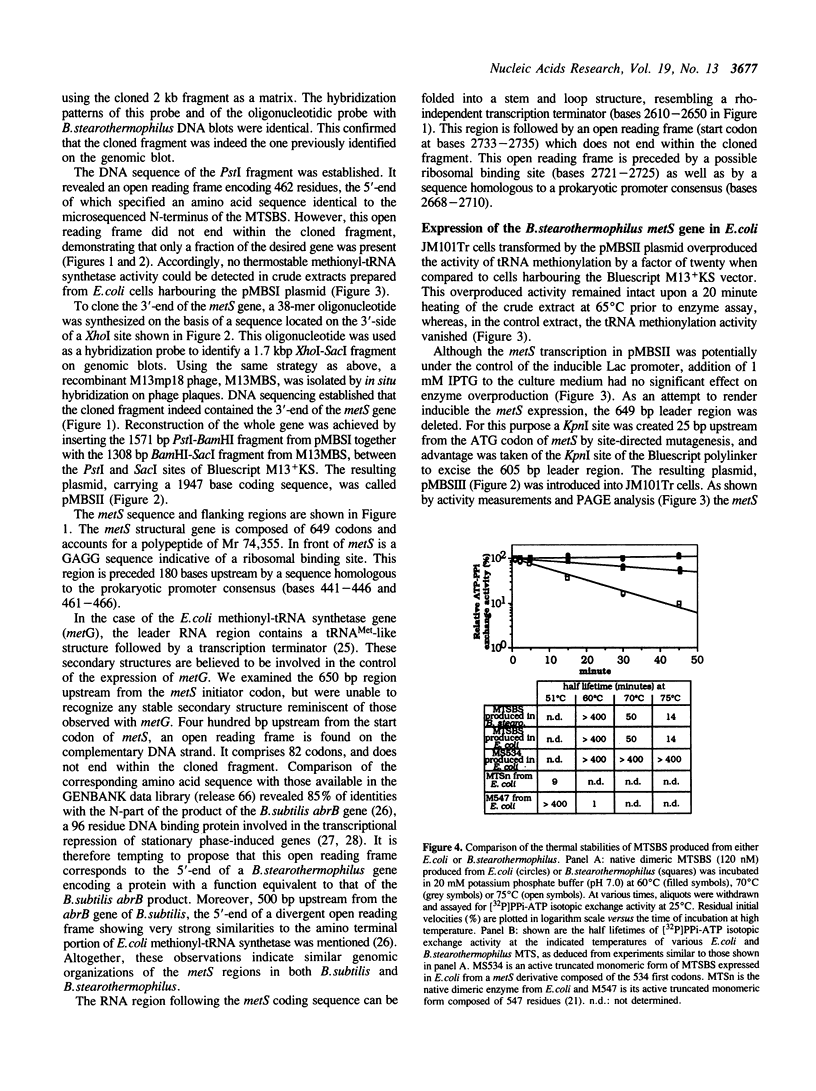
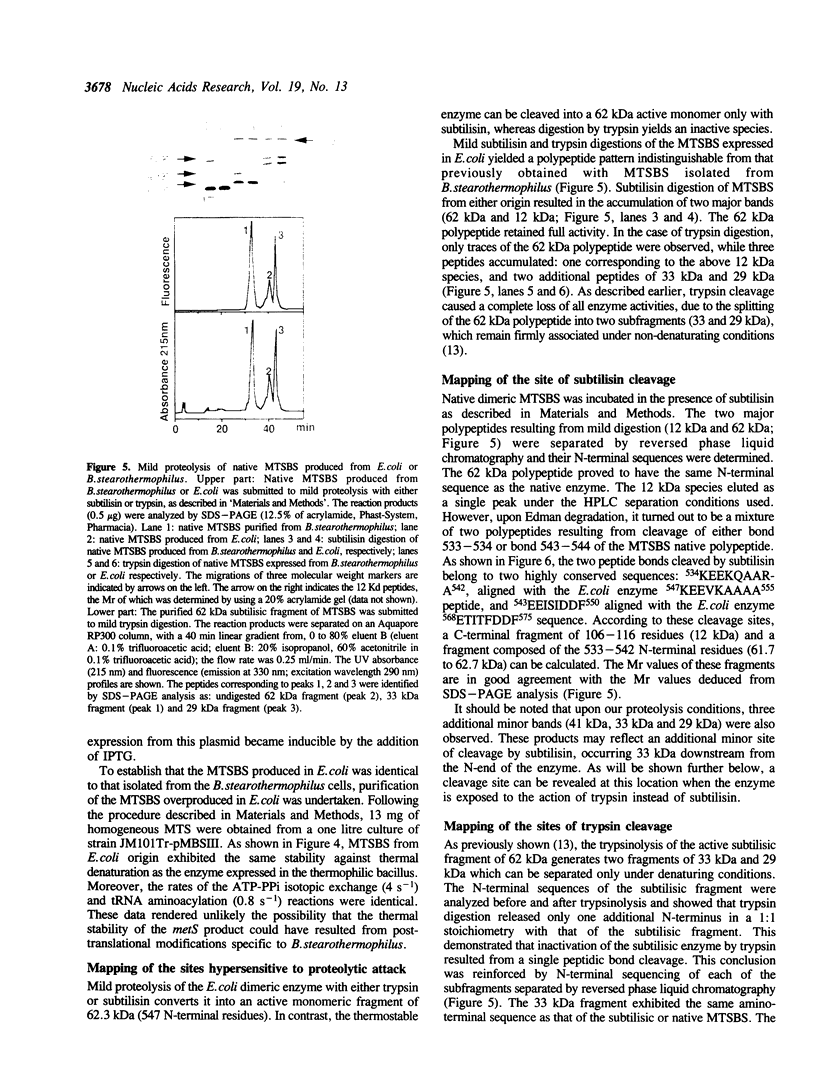
The metS gene encoding homodimeric methionyl-tRNA synthetase from Bacillus stearothermophilus has been cloned and a 2880 base pair sequence solved. Comparison of the deduced enzyme protomer sequence (Mr 74,355) with that of the E. coli methionyl-tRNA synthetase protomer (Mr 76,124) revealed a relatively low level (32%) of identities, although both enzymes have very similar biochemical properties (Kalogerakos, T., Dessen, P., Fayat, G. and Blanquet, S. (1980) Biochemistry 19, 3712-3723). However, all the sequence patterns whose functional significance have been probed in the case of the E. coli enzyme are found in the thermostable enzyme sequence. In particular, a stretch of 16 amino acids corresponding to the CAU anticodon binding site in the E. coli synthetase structure is highly conserved in the metS sequence. The metS product could be expressed in E. coli and purified. It showed structure-function relationships identical to those of the enzyme extracted from B. stearothermophilus cells. In particular, the patterns of mild proteolysis were the same. Subtilisin converted the native dimer into a fully active monomeric species (62 kDa), while trypsin digestion yielded an inactive form because of an additional cleavage of the 62 kDa polypeptide into two subfragments capable however of remaining firmly associated. The subtilisin cleavage site was mapped on the enzyme polypeptide, and a gene encoding the active monomer was constructed and expressed in E. coli. Finally, trypsin attack was demonstrated to cleave a peptidic bond within the KMSKS sequence common to E. coli and B. stearothermophilus methionyl-tRNA synthetases. This sequence has been shown, in the case of the E. coli enzyme, to have an essential role for the catalysis of methionyl-adenylate formation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauvallet C., Hountondji C., Schmitter J. M. Analytical strategy for determination of active site sequences in aminoacyl-tRNA synthetases. J Chromatogr. 1988 Apr 22;438(2):347–357. doi: 10.1016/s0021-9673(00)90266-8. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Fayat G., Waller J. P., Iwatsubo M. The mechanism of reaction of methionyl-tRNA synthetase from Escherichia coli. Interaction of the enzyme with ligands of the amino-acid-activation reaction. Eur J Biochem. 1972 Jan 21;24(3):461–469. doi: 10.1111/j.1432-1033.1972.tb19707.x. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Assembly of a class I tRNA synthetase from products of an artificially split gene. Biochemistry. 1991 Jan 15;30(2):319–324. doi: 10.1021/bi00216a002. [DOI] [PubMed] [Google Scholar]
- Cassio D., Waller J. P. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971 May 28;20(2):283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Dardel F., Panvert M., Fayat G. Transcription and regulation of expression of the Escherichia coli methionyl-tRNA synthetase gene. Mol Gen Genet. 1990 Aug;223(1):121–133. doi: 10.1007/BF00315804. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987 Dec 15;26(25):8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Pelka H., Schulman L. H. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990 Mar 6;29(9):2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Elkana Y., Ehrlich S. D., Lederberg J. Electrophoretic separation of Bacillus subtilis genes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2207–2211. doi: 10.1073/pnas.72.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel P. H., Lévêque F., Mellot P., Dardel F., Panvert M., Mechulam Y., Fayat G. Genetic engineering of methionyl-tRNA synthetase: in vitro regeneration of an active synthetase by proteolytic cleavage of a methionyl-tRNA synthetase--beta-galactosidase chimeric protein. Biochimie. 1988 Jun;70(6):773–782. doi: 10.1016/0300-9084(88)90107-1. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Blanquet S., Lederer F. Methionyl-tRNA synthetase from Escherichia coli: primary structure at the binding site for the 3'-end of tRNAfMet. Biochemistry. 1985 Feb 26;24(5):1175–1180. doi: 10.1021/bi00326a018. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Schmitter J. M., Beauvallet C., Blanquet S. Mapping of the active site of Escherichia coli methionyl-tRNA synthetase: identification of amino acid residues labeled by periodate-oxidized tRNA(fMet) molecules having modified lengths at the 3'-acceptor end. Biochemistry. 1990 Sep 4;29(35):8190–8198. doi: 10.1021/bi00487a029. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Schmitter J. M., Fukui T., Tagaya M., Blanquet S. Affinity labeling of aminoacyl-tRNA synthetases with adenosine triphosphopyridoxal: probing the Lys-Met-Ser-Lys-Ser signature sequence as the ATP-binding site in Escherichia coli methionyl-and valyl-tRNA synthetases. Biochemistry. 1990 Dec 25;29(51):11266–11273. doi: 10.1021/bi00503a016. [DOI] [PubMed] [Google Scholar]
- Kalogerakos T., Dessen P., Fayat G., Blanquet S. Proteolytic cleavage of methionyl transfer ribonucleic acid synthetase from Bacillus stearothermophilus: effects on activity and structure. Biochemistry. 1980 Aug 5;19(16):3712–3723. doi: 10.1021/bi00557a012. [DOI] [PubMed] [Google Scholar]
- Kohda D., Yokoyama S., Miyazawa T. Functions of isolated domains of methionyl-tRNA synthetase from an extreme thermophile, Thermus thermophilus HB8. J Biol Chem. 1987 Jan 15;262(2):558–563. [PubMed] [Google Scholar]
- Mechulam Y., Dardel F., Le Corre D., Blanquet S., Fayat G. Lysine 335, part of the KMSKS signature sequence, plays a crucial role in the amino acid activation catalysed by the methionyl-tRNA synthetase from Escherichia coli. J Mol Biol. 1991 Feb 5;217(3):465–475. doi: 10.1016/0022-2836(91)90750-z. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Dardel F., Schmitter J. M., Hountondji C., Brunie S., Dessen P., Fayat G., Blanquet S. Methionyl-tRNA synthetase from E. coli--a review. Biochimie. 1990 Aug;72(8):625–632. doi: 10.1016/0300-9084(90)90126-2. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B., Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988 Nov;2(6):689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Robertson J. B., Gocht M., Marahiel M. A., Zuber P. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8457–8461. doi: 10.1073/pnas.86.21.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe J. J., Goldberg I. D., Amelunxen R. E. Development of defined and minimal media for the growth of Bacillus stearothermophilus. J Bacteriol. 1975 Oct;124(1):279–284. doi: 10.1128/jb.124.1.279-284.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schmitter J. M., Mechulam Y., Fayat G., Anselme M. Rapid purification of DNA fragments by high-performance size-exclusion chromatography. J Chromatogr. 1986 Jun 13;378(2):462–466. doi: 10.1016/s0378-4347(00)80743-4. [DOI] [PubMed] [Google Scholar]
- Strauch M. A., Spiegelman G. B., Perego M., Johnson W. C., Burbulys D., Hoch J. A. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989 May;8(5):1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Vambutas A., Akai A. Characterization of MSM1, the structural gene for yeast mitochondrial methionyl-tRNA synthetase. Eur J Biochem. 1989 Feb 1;179(2):365–371. doi: 10.1111/j.1432-1033.1989.tb14562.x. [DOI] [PubMed] [Google Scholar]
- Walter P., Gangloff J., Bonnet J., Boulanger Y., Ebel J. P., Fasiolo F. Primary structure of the Saccharomyces cerevisiae gene for methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 May;80(9):2437–2441. doi: 10.1073/pnas.80.9.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]