Autophagosome biogenesis and maturation in primary neurons is a constitutive process that is spatially and temporally regulated along the axon.
Abstract
Autophagy is an essential cellular degradation pathway in neurons; defects in autophagy are sufficient to induce neurodegeneration. In this paper, we investigate autophagosome dynamics in primary dorsal root ganglion neurons. Autophagosome biogenesis occurs distally in a constitutive process at the neurite tip. Autophagosomes initially move bidirectionally and then switch to unidirectional, processive movement toward the cell soma driven by dynein. Autophagosomes copurify with anterograde and retrograde motors, suggesting that the activity of bound kinesin motors is effectively down-regulated to yield robust retrograde motility driven by dynein. Both organelle and soluble cargoes are internalized into autophagosomes, including mitochondria and ubiquitin. As autophagosomes move distally to proximally, they undergo maturation and become increasingly acidified, consistent with the formation of an autolysosomal compartment that may more efficiently degrade cargo. This maturation is accompanied by a switch to bidirectional motility characteristic of lysosomes. Together, autophagosome biogenesis and maturation in primary neurons is a constitutive process that is spatially and temporally regulated along the axon.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a lysosomal degradation process that removes and degrades damaged proteins and organelles. Autophagy is initiated when a portion of the cytoplasm is enclosed within a double-membrane organelle, termed an autophagosome (Xie and Klionsky, 2007). Autophagosomes then fuse with degradative compartments in the endosomal–lysosomal pathway. Autophagy occurs at a basal, constitutive level and functions as a quality control system that can be up-regulated in response to cellular stresses, such as starvation (Mizushima et al., 2008).
Autophagy is an essential pathway in postmitotic cells, such as neurons, cells that are particularly susceptible to the accumulation of defective proteins and organelles. Neuron-specific disruption of autophagy results in neurodegeneration (Hara et al., 2006; Komatsu et al., 2006). Defects in autophagy have been observed in multiple models of neurodegenerative disease, including Alzheimer’s, Huntington’s, and amyotrophic lateral sclerosis (ALS; Rubinsztein et al., 2005; Ventruti and Cuervo, 2007; Mariño et al., 2011). However, the biogenesis, maturation, and dynamics of autophagosomes in neurons are only poorly understood. Most studies to date have focused on model systems that lack the extended and highly polarized processes that characterize neurons (Jahreiss et al., 2008; Kimura et al., 2008).
Here, we track the dynamics of autophagosomes in real time along the axons of primary neurons. We find that autophagosome initiation is a constitutive and spatially restricted process in the distal axon. Autophagosomes engulf both cytosolic and organelle cargo and are driven by dynein and kinesin motors. Once formed, autophagosomes undergo a spatially defined maturation as they move processively along the axon toward the cell body. We find that autophagosome dynamics remain robust in a mouse model of neurodegenerative disease, but autophagosome flux is not increased even as protein aggregates accumulate along the axon. Thus, in primary neurons, the dynamics of the autophagy pathway are robust and spatially specific but may not be efficiently up-regulated in response to increased protein aggregation.
Results and discussion
Distal initiation followed by robust retrograde transport of autophagosomes in primary neurons
To monitor the dynamics of autophagosomes in primary neurons, we isolated dorsal root ganglion (DRG) neurons from transgenic mice expressing the autophagosome marker GFP-LC3 (Mizushima et al., 2004). After 2 d in vitro, DRG neurons extend neurites ∼1,000 µm in length that are tau positive (Perlson et al., 2009). Microtubules in the neurite are uniformly polarized with plus ends outward as indicated by the directionality of the plus-tip protein EB3 (Fig. S1 A). The linear separation provided by the extended length of the processes allows sufficient spatial resolution to examine the steps of autophagosome formation and maturation along the axon using live-cell imaging.
Cytosolic GFP-LC3 is found throughout the neuron but becomes lipidated and incorporated into autophagosomes upon their formation (Xie and Klionsky, 2007). Imaging the distal tips of neurons revealed the initiation of autophagosome formation. Within bulbous regions near the distal end of the axon, ∼1 mm from the cell soma, we observed the incorporation of GFP-LC3 into ringlike structures ∼800 nm in diameter (Fig. 1, A and B). Often, multiple rings were observed to accumulate within a single bulbous region (Fig. 1 B). We identified these ringlike structures as autophagosomes based on their strong LC3-positive signals and the similarity in morphology to the LC3-positive structures seen in vivo in transgenic GFP-LC3 mice (Mizushima et al., 2004).
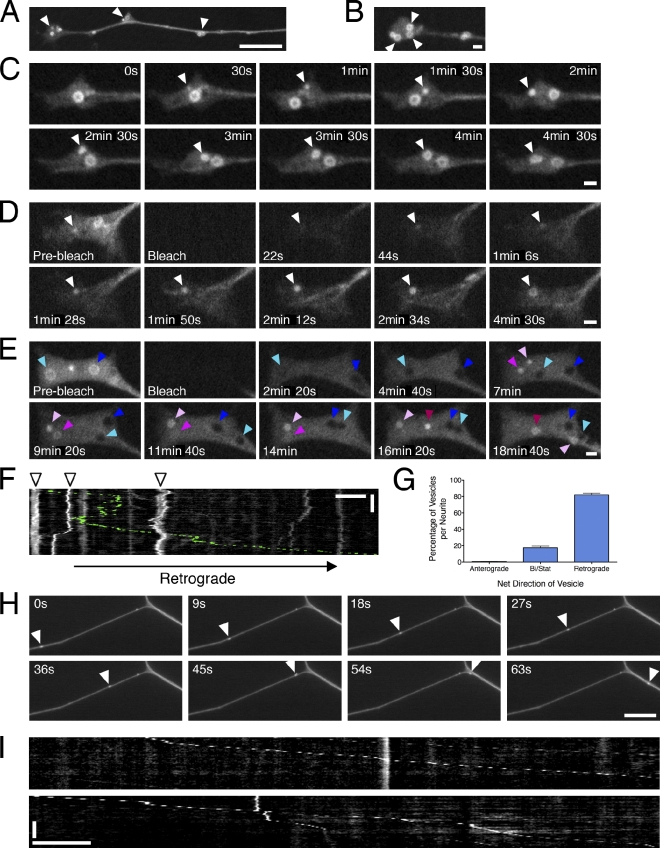
Figure 1.
Autophagosomes initiate distally and undergo retrograde movement toward the cell soma in primary neurons. (A) GFP-LC3 localization at the distal end of DRG neurons. Arrowheads denote the accumulation of GFP-LC3–positive puncta. (B) Autophagosomes initiate in the distal tip of the neurite, where pronounced ringlike structures accumulate (arrowheads). (C) Time series of autophagosome biogenesis; arrowheads denote the appearance and growth of GFP-LC3–positive puncta into a ring (Video 1). (D and E) FRAP analysis of the distal neurite. Open arrowheads denote recovering puncta and growth into a ring, cyan and dark blue arrowheads denote two different examples of black holes of bleached preexisting rings, three different pink-shaded arrowheads denote the appearance of three different GFP-LC3–positive puncta that grow into rings. (F) Kymograph of autophagosome motility in the distal neurite showing bidirectional movement within a constrained region (arrowheads). Occasional autophagosomes escape from the tip and move processively toward the cell soma (retrograde track is pseudocolored green; Video 2). (G) Percentage of retrograde, anterograde, or bidirectional/stationary (Bi/Stat) vesicles (means ± SEM; n = 91 neurites). (H and I) Times series and corresponding kymographs showing processive movement of GFP-LC3–positive puncta along the axon (Video 3). Arrowheads denote an autophagosome traveling along the axon toward the cell soma. Retrograde motility is toward the right in all figures. Horizontal bars: (B–E) 1 µm; (A, F, H, and I) 10 µm. Vertical bars: (F) 2 min; (I) 1 min.
Real-time analysis of distal neurites revealed direct evidence for autophagosome biogenesis. GFP-LC3–positive puncta appeared in distal bulbs and grew progressively into ring structures (Fig. 1, C–E; and Video 1). This growth was further resolved using FRAP. Within ∼20 s of photobleaching, puncta began to recover their fluorescence and continued to grow into a ring, indicating continued recruitment of unbleached GFP-LC3 into the expanding organelle (Fig. 1 D). This process was rapid, taking 4–6 min for recovering puncta to develop into a ring. In contrast, preexisting rings did not recover their fluorescence after photobleaching, appearing as black holes within the GFP-LC3–labeled cytoplasm (Fig. 1 E). The lack of recovery suggests that GFP-LC3 incorporated into the autophagosome ring is relatively stable and not rapidly exchanged with the cytosolic pool. Autophagosome biogenesis appears to be spatially restricted to the distal neurite, as we performed FRAP along the axon and did not observe either formation or growth of autophagosomes; also, preexisting autophagosomes along the axon showed no recovery (Fig. S1 B).
Thus, in primary neurons, autophagosome precursors are generated distally. We have not observed biogenesis along the axon but cannot rule out initiation in the cell body caused by high levels of autofluorescence that limit our resolution. This spatial regulation of autophagosome biogenesis in primary neurons is distinct from observations in smaller, less polarized cells in which autophagosomes form throughout the cytosol (Jahreiss et al., 2008). The origin of the membrane that forms the distal LC3-positive rings remains to be determined. Possible sources include endocytosed plasma membrane, ER, mitochondria, or Golgi membranes (Hollenbeck, 1993; Hayashi-Nishino et al., 2009; Hailey et al., 2010; Ravikumar et al., 2010; van der Vaart and Reggiori, 2010; Yen et al., 2010).
GFP-LC3–positive rings in the distal process either remained stationary or displayed saltatory bidirectional motility in a constrained region within ∼150–200 µm of the neurite tip (Fig. 1 F). Occasionally, autophagosomes could be seen exiting this bidirectional pool to initiate travel toward the cell body (Fig. 1 F and Video 2). Along the axon shaft, autophagosomes exhibited a different type of movement. Within the axon (50–500 µm from the cell body), autophagosomes displayed robust and primarily unidirectional motility (Fig. 1, G–I; and Video 3), with 82 ± 2% (±SEM) moving a net distance of ≥10 µm in the retrograde direction. Anterograde movement was rarely observed (<1%), whereas <20% of autophagosomes exhibited nonprocessive bidirectional motility or remained stationary.
To further characterize autophagosome dynamics, we tracked the motility of individual GFP-LC3–positive autophagosomes that moved a net distance of ≥10 µm along extended neurites. The observed instantaneous velocities were strongly biased in the retrograde direction (Fig. 2 A). Autophagosomes moved at a mean speed of 0.45 µm/s (Fig. S1 C), with most exhibiting few reversals in direction (Fig. 2 B). The few anterograde movements represent short reversals in direction for vesicles that were moving in the net retrograde direction. Pauses represent ∼12% of the total time tracked (median value; Fig. S1 D).
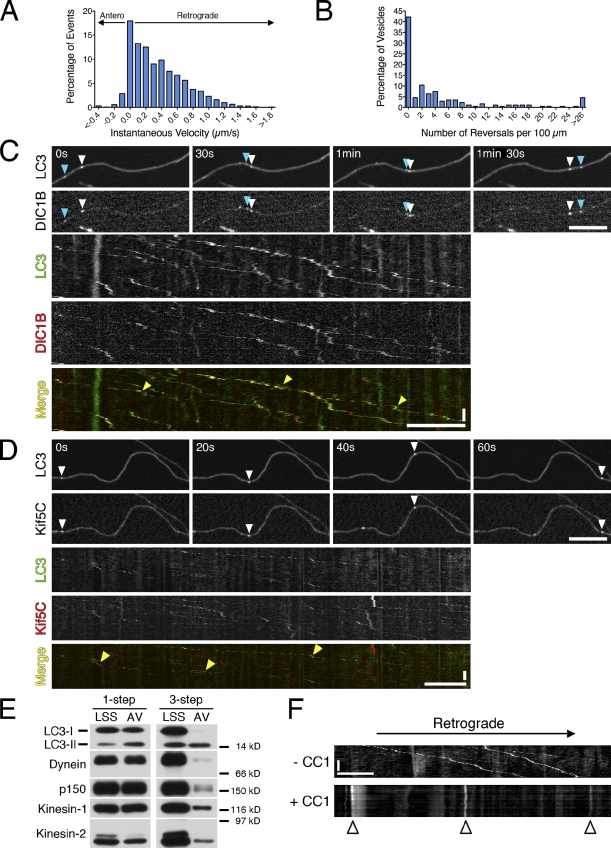
Figure 2.
Autophagosome dynamics in primary neurons are driven primarily by cytoplasmic dynein. (A) Individual GFP-LC3 puncta along the axon were tracked to yield a histogram of instantaneous velocities (n = 8,872 events). Shown are pooled data for 173 vesicles from 59 neurons from 12 mice analyzed in 12 separate experiments. (B) Reversals within 100 µm for each vesicle (median is 1.6 reversals/100 µm, n = 173 vesicles analyzed as in A). Median reversal length is 0.3 µm. (C) Time series and corresponding kymograph of GFP-LC3 and DIC1B-mCherry motility along the axon (Video 4). Open and cyan arrowheads denote two different examples of vesicles positive for both markers. Yellow arrowheads in the kymograph denote multiple examples of vesicles positive for both markers. The red signal may occasionally precede the green signal because of delays in acquiring consecutive images. (D) Time series and corresponding kymograph of GFP-LC3 and mCherry-Kif5C tail motility along the axon. Open arrowhead denotes a vesicle positive for both markers. Yellow arrowheads in the kymograph denote multiple examples of vesicles positive for both markers. (E) Fractions enriched for autophagosomes were prepared using a one-step gradient (Morvan et al., 2009) or a three-step gradient (Strømhaug et al., 1998) that more effectively selects for the lipidated form of LC3, LC3-II. Equal total protein from the low speed supernatant (LSS) and autophagosome-enriched fraction (AVs) were analyzed by immunoblotting. Autophagosome membrane-associated LC3-II is distinguished from cytosolic LC3-I by molecular mass (shown is endogenous LC3). Motors may not be quantitatively retained on autophagosome membranes through the three-step gradient. (F) Kymograph of GFP-LC3 motility from DRG neurons transfected with the dominant-negative CC1, which disrupts dynein/dynactin function. Arrested autophagosomes are marked with arrowheads. Horizontal bars, 10 µm. Vertical bars, 1 min.
Once formed, autophagosomes accumulate and undergo spatially restricted bidirectional motility; release from this bidirectional pool may be triggered by changes in signaling or maturation. After this release, autophagosomes initiate robust, primarily unidirectional transport to the cell body. Autophagosome transport in neurons is also distinct from motility in nonneuronal cells. In both normal rat kidney and PC12 cells, LC3 motility is primarily bidirectional (Jahreiss et al., 2008; Yang et al., 2008, 2011), unlike the characteristic robust retrograde transport we have visualized along the axons of primary neurons (this study; Lee et al., 2011).
The pronounced retrograde movement of autophagosomes along the axon suggests that this motility is likely dependent on the microtubule minus end–directed motor dynein and its activator dynactin. To investigate this possibility, we expressed a fluorescently tagged subunit of dynein, the neuron-specific intermediate chain isoform DIC1B (Ha et al., 2008), in neurons expressing GFP-LC3. Dual-color imaging revealed the co-migration of dynein with autophagosomes (Fig. 2 C and Video 4). Retrograde autophagosomes were also positive for the ubiquitously expressed dynein intermediate chain DIC2C-mCherry (Fig. S2 A and Video 5). Expression of the dominant-negative dynein inhibitor CC1, which blocks the dynein–dynactin interaction (Quintyne et al., 1999), arrested autophagosome motility (Fig. 2 F). We also observed co-migration of autophagosomes with anterograde motors kinesin-1 and kinesin-2 (Fig. 2 D and Fig. S2 B). In the case of kinesin-1, overexpression of the dominant-negative kinesin-1 tail was sufficient to label motile autophagosomes yet did not disrupt transport.
To further define the complement of associated motors, we analyzed isolated autophagosomes by immunoblotting. We saw copurification of dynein and dynactin as well as anterograde motors kinesin-1 and kinesin-2 with autophagosomes (Fig. 2 E). Dynein is the major retrograde motor for vesicular transport and has been implicated in autophagosome dynamics (Jahreiss et al., 2008; Kimura et al., 2008; Cai et al., 2010; Katsumata et al., 2010), consistent with our observations. More surprising is the copurification of both kinesin-1 and kinesin-2 motors with isolated autophagosomes. Although we cannot exclude the possibility that some fraction of motors is internalized in the autophagosome as it forms, the effects of CC1 expression demonstrate that motors are bound to the cytosolic face of the autophagosome. However, the primarily unidirectional motility of autophagosomes suggests that the activities of these anterograde motors are likely to be tightly regulated on these organelles.
Constitutive transport of enveloped cargo along the axon
To determine whether autophagosomes in the axon contain cargo, we expressed DsRed2-mito, a marker for mitochondria, in neurons expressing GFP-LC3. Using dual-color imaging, we observed fragments of mitochondria that co-migrated with autophagosomes along the axon (Fig. 3 A). These mitochondrial fragments were distinct from the large tubular mitochondria that exhibited primarily stationary/bidirectional motility. The frequency of double labeling was relatively low (∼10–20% of autophagosomes along the axon), suggesting that the majority of mitochondria in the axon are healthy and not targeted for degradation.
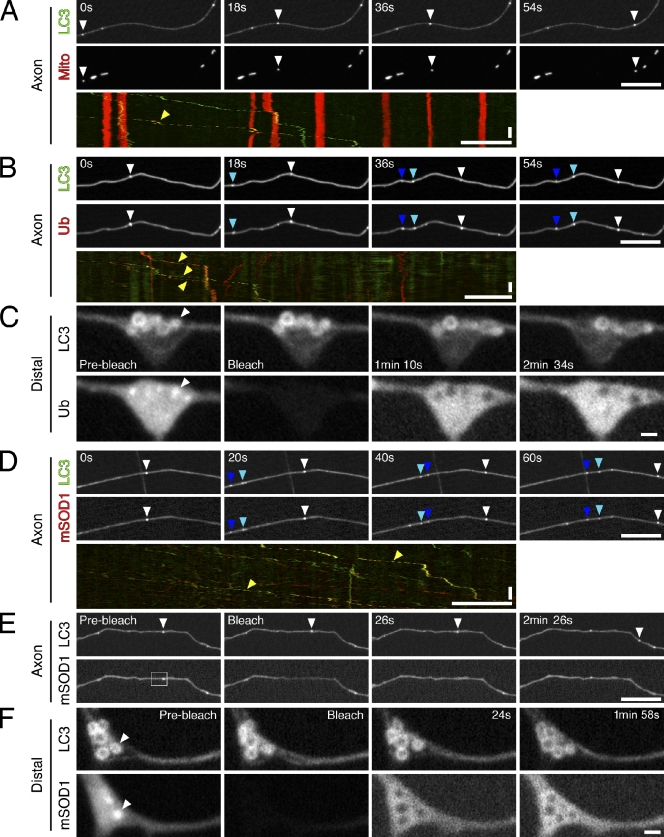
Figure 3.
Autophagosomes contain engulfed cargo. (A) Time series and corresponding kymograph of GFP-LC3 and DsRed2-mito motility along the axon. Open arrowheads in times series and yellow arrowhead in kymograph denote a vesicle positive for both markers. (B) Time-lapse images and corresponding kymograph of GFP-LC3 and RFP-Ub motility along the axon. Open, cyan, and dark blue arrowheads denote three different examples of vesicles positive for both markers. Yellow arrowheads in kymograph denote multiple examples of vesicles positive for both markers. (C) FRAP analysis of RFP-Ub fluorescence inside autophagosome rings at the distal end of the neurite. Arrowheads denote enrichment of RFP-Ub inside autophagosome ring. (D) Time series and corresponding kymograph of GFP-LC3 and mCherry-SOD1G93A motility along the axon. Open, cyan, and dark blue arrowheads denote three different examples of vesicles positive for both markers. Yellow arrowheads in kymograph denote two different examples of vesicles positive for both markers. (E) FRAP analysis of mCherry-SOD1G93A fluorescence inside an autophagosome along the axon. Box denotes photobleached region, and arrowheads denote bleached vesicle. (F) FRAP analysis of mCherry-SOD1G93A fluorescence inside autophagosomes at the neurite tip. Arrowheads denote enrichment of mCherry-SOD1G93A inside the autophagosome ring. For distal FRAP images, the contrast enhancement applied to prebleach and bleach images was different from that applied to recovery images. Horizontal bars: (axon images) 10 µm; (distal images) 1 µm. Vertical bars, 1 min.
Nearly all of the autophagosomes traveling along the axon were positive for RFP-Ubiquitin (Ub; Fig. 3 B). Distally, autophagosome rings were also positive for RFP-Ub, but only a few contained a concentrated signal (Fig. 3 C). To confirm internalization of RFP-Ub, we performed FRAP analysis. As the cytosol regained fluorescence, the lumen of the autophagosome remained bleached, indicating that the internalized RFP-Ub was not exchangeable with the cytosolic pool.
We also tested for the uptake of SOD1G93A, an ALS-linked mutation that forms aggregates in vivo (Fig. S3 A). When expressed in DRG neurons, mCherry-SOD1G93A was predominantly cytosolic with motile puncta that may represent aggregates (Fig. 3 D). Nearly all autophagosomes in the axon were positive for mCherry-SOD1G93A (Fig. 3 D). Again, we used FRAP analysis and found that autophagosomes both distally and along the axon failed to recover mCherry-SOD1G93A fluorescence (Fig. 3, E and F). Thus, mCherry-SOD1G93A is internalized into autophagosomes.
Collectively, these data provide evidence for cargo accumulation inside retrogradely moving autophagosomes. RFP-Ub and mCherry-SOD1G93A are likely engulfed as the autophagosome forms, consistent with their presence in distal as well as axonal autophagosomes. Incorporation of cytosolic proteins appears to be a constitutive process, as we also noted that mCherry alone is engulfed into autophagosomes. In contrast, mitochondria have been shown to be specifically targeted for degradation by autophagy in response to stress or insult (Narendra et al., 2008). The lower frequency of autophagosomes positive for mitochondrial fragments is consistent with a low basal level of organelle autophagy in this model. We also did not see incorporation of a Golgi marker GPP130 (Fig. S2 C).
We also observed that the formation of aggregated SOD1G93A along the axon does not alter autophagosome dynamics. We crossed GFP-LC3 transgenic mice with mice overexpressing SOD1G93A (mouse model of familial ALS [fALS]; Gurney et al., 1994) and performed live-cell imaging of DRG neurons isolated at early onset of disease (3 mo of age) versus late-stage disease (4 mo of age). Despite the formation of SOD1G93A aggregates along the axon (Fig. S3 A), autophagosomes displayed robust and directed retrograde transport in neurons, even at late-stage disease (Fig. S3, B–E). We did not observe any significant changes in autophagosome formation, density, or flux along the axon (Fig. S3, F and G). Based on these data, autophagosome transport is a constitutive and robust pathway that is not altered in this model of neurodegenerative disease.
Maturation of autophagosomes moving distally to proximally along the axon
For engulfed cargoes to be efficiently degraded, autophagosomes must fuse with lysosomes (Xie and Klionsky, 2007). To look for evidence of organelle fusion, we expressed the late endosome/lysosome marker lysosomal-associated membrane protein 1 (LAMP1)–RFP in GFP-LC3–positive neurons. At the distal neurite, both markers were enriched within the same bulbous regions, although they did not colocalize extensively (∼10%; Fig. 4 A). Imaging along the axon, however, showed colocalization of GFP-LC3 and LAMP1-RFP in nearly all retrogradely moving autophagosomes (∼97%; Fig. 4 B). These data suggest that as autophagosomes exit the distal tip, they acquire markers for late endosomes/lysosomes, consistent with autophagosome–lysosome fusion occurring distally.
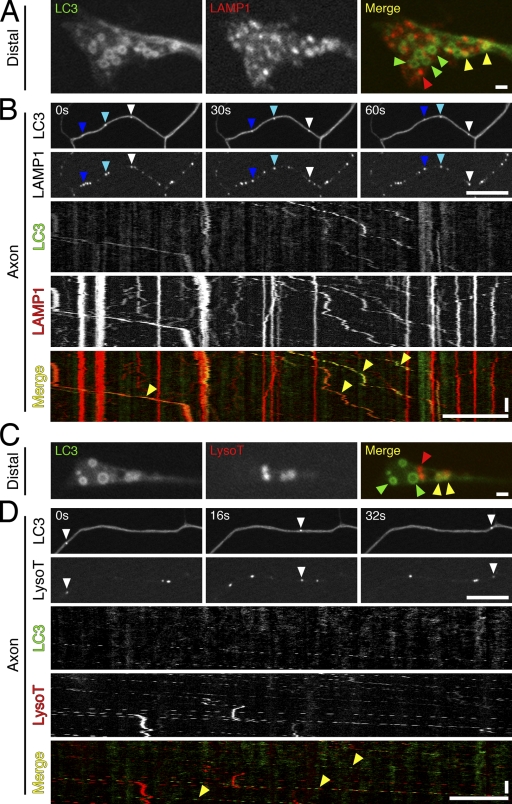
Figure 4.
Autophagosomes along the axon have fused with late endosomes or lysosomes. (A) GFP-LC3 and LAMP1-RFP distribution at the distal end of the neurite. Yellow, green, and red arrowheads designate vesicles positive for both markers, GFP-LC3 only, or LAMP1-RFP only, respectively. (B) Time series and corresponding kymograph of GFP-LC3 and LAMP1-RFP motility along the axon. Open, cyan, and dark blue arrowheads denote three different examples of vesicles positive for both markers. Yellow arrowheads in kymograph denote multiple examples of vesicles positive for both markers. (C) GFP-LC3 and LysoTracker (LysoT) red localization at the neurite tip. Yellow, green, and red arrowheads designate vesicles positive for both markers, GFP-LC3 only, or LysoTracker red only, respectively. (D) Time series and corresponding kymograph of GFP-LC3 and LysoTracker red motility along the axon. Open arrowheads denote a vesicle positive for both markers. Yellow arrowheads in kymograph denote multiple examples of vesicles positive for both markers. Horizontal bars: (distal images) 1 µm; (axon images) 10 µm. Vertical bars, 1 min.
Consistent with this observation, GFP-LC3–positive organelles along the axon were stained by LysoTracker red, a fluorescent dye that preferentially labels acidic organelles, colocalizing with Rab7 (late endosomes/lysosomes) but not Rab5 (early endosomes; Fig. S2, D and E). Although some autophagosome rings in the distal neurite were positive for LysoTracker red (∼30–40%), the majority were negative (Fig. 4 C). In contrast, nearly all of the autophagosomes along the axon were positive for LysoTracker red (∼96%; Fig. 4 D). These data suggest that upon exit from the distal neurite, autophagosomes become acidified, consistent with effective fusion with late endosomes or lysosomes (Jahreiss et al., 2008; Kimura et al., 2008; Lee et al., 2011).
To further examine autophagosome maturation, we used a dual-color fluorescent LC3 reporter, mCherry-EGFP-LC3 (Pankiv et al., 2007). Before acidification, this protein appears yellow in merged images because of the fluorescence of both mCherry and GFP. Once the lumen of the organelle is acidified, the GFP moiety is preferentially quenched, and only the red fluorescence persists (Pankiv et al., 2007). We find that GFP fluorescence of the dual-color probe is more sensitive to quenching at a low pH (Fig. 5) than the singly tagged GFP-LC3 expressed in neurons from the transgenic mouse, as signal from the transgenic GFP-LC3 reporter persists in LysoTracker-positive compartments (Fig. 4 D).
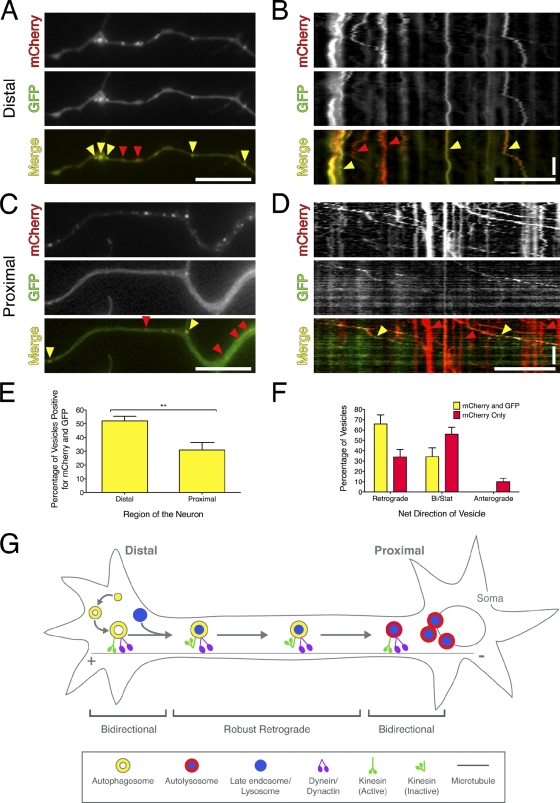
Figure 5.
Autophagosomes mature as they move distally to proximally along the axon. (A) Wild-type neurons transfected with mCherry-EGFP-LC3. In acidic environments, the GFP moiety is preferentially quenched, and only the red fluorescence persists (yellow arrowheads show LC3 puncta positive for mCherry and EGFP; red arrowheads show LC3 puncta positive for mCherry only). (B) Corresponding kymograph of mCherry-EGFP-LC3 motility at the distal tip. (C and D) Images from live-cell analysis and corresponding kymographs from regions proximal to the cell soma. (E) Quantitation of LC3 puncta positive for mCherry and GFP in the distal versus proximal regions of the axon (means ± SEM; n = 9 neurites; **, P = 0.0043, t test). (F) Percentage of retrograde, anterograde, or bidirectional/stationary vesicles (means ± SEM; n = 9 neurites). (G) Model for autophagosome biogenesis and maturation along the axon in primary neurons. Horizontal bars, 10 µm. Vertical bars, 1 min.
In wild-type DRG neurons transfected with mCherry-EGFP-LC3, we observed a decreasing gradient along the axon of autophagosomes positive for both mCherry and GFP (Fig. 5, A, C, and E). At the distal end of the axon, the majority of LC3-positive puncta were positive for both mCherry and GFP (Fig. 5, A, B, and E). In contrast, proximal to the cell body, we observed a decrease in the number of autophagosomes positive for both mCherry and GFP, with ∼70% of LC3 puncta positive for mCherry only (Fig. 5, C–E). This indicates that most autophagosomes within 200 µm of the cell body have fully acidified.
This change in the apparent acidity of the compartment was accompanied by a change in motility. Within the proximal axon, organelles positive for both GFP and mCherry showed robust retrograde motility (Fig. 5, D and F). Organelles positive for mCherry only, however, showed less biased retrograde motility with a corresponding increase in bidirectional/stationary motility that more closely resembles that of lysosomes (Fig. 5, D and F; Hendricks et al., 2010).
These observations suggest that autophagosomes undergo maturation as they move distally to proximally along the axon (Fig. 5 G). Exit from the distal tip is accompanied by fusion with late endosomes and/or lysosomes. This is supported by our observations that although distal autophagosomes are largely negative for LAMP1 and LysoTracker red, autophagosomes along the axon are positive for both markers. As they approach the cell soma, autophagosomes become increasingly acidified, as supported by our experiments with the dual-color LC3 reporter. This maturation is accompanied by a change in motility. Fully acidified autophagosomes exhibit bidirectional motility.
Collectively, our observations support a maturation model, in which autophagosomes initiate distally in a constitutive mechanism (Fig. 5 G). These early autophagosomes move bidirectionally along microtubules driven by bound kinesin and dynein motors. At some point after formation, autophagosomes escape from this distal pool. There is an apparent down-regulation of the associated kinesin motors, allowing for robust retrograde motility driven by dynein. However, these plus end–directed motors remain stably associated with cargo. Escape from this distal pool may be coincident with fusion with late endosomes or lysosomes; fusion may initiate robust retrograde transport along the axon. Autophagosomes move processively toward the cell body carrying engulfed cargo, such as mitochondria and Ub. As they approach the cell soma, LC3-positive organelles become increasingly acidified, consistent with the formation of an autolysosomal compartment that may catalyze a more efficient degradation of engulfed contents (Jahreiss et al., 2008; Kimura et al., 2008; Lee et al., 2011). Degradation of cargo near the cell soma, the primary site of protein synthesis, may facilitate efficient recycling of amino acids and lipids. Once fully acidified, the compartment reverts to bidirectional motility resembling that of lysosomes. This switch in motility may involve activation of associated kinesin motors, suggesting that organelle motility may be regulated by changes in the activity of bound motors rather than recruitment or dissociation of additional motors from a cytosolic pool. Together, our observations of autophagosome dynamics in primary neurons establish a linear pathway of distal initiation followed by maturation during retrograde transport toward the cell soma.
Materials and methods
Reagents
GFP-LC3 transgenic mice, strain name B6.Cg-Tg(CAG-EGFP/LC3)53Nmi/NmiRbrc, were developed by N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan; Mizushima et al., 2004) and deposited into the RIKEN BioResource Center in Japan. Transgenic mice overexpressing SOD1G93A, strain name B6SJL-Tg(SOD*G93A)1Gur/J, were purchased from The Jackson Laboratory. Constructs include DsRed2-mito (gift from T. Schwarz, Harvard Medical School, Boston, MA), monomeric RFP-Ub (Addgene), LAMP1-RFP (Addgene), mCherry-EB3 (gift from I. Kaverina, Vanderbilt University Medical Center, Nashville, TN), GFP-Rab5 (gift from M. Zerial, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany), GFP-Rab7 (Addgene), GPP130-mCherry (gift from A. Linstedt, Carnegie Mellon University, Pittsburgh, PA), and mCherry-EGFP-LC3 (gift from T. Johansen, University of Tromsø, Tromsø, Norway; Pankiv et al., 2007). SOD1G93A, DIC1B, and DIC2C (gifts from K. Pfister, University of Virginia, Charlottesville, VA), Kif5C tail (gift from M. Setou, Hamamatsu University School of Medicine, Shizuoka, Japan), Kif3A (gift from K. Kaibuchi, Nagoya University Graduate School of Medicine, Nagoya, Japan), and the CC1 domain (E216-Q550) of p150Glued were recloned into pmCherry (Takara Bio Inc.). Antibodies include a polyclonal antibody against LC3B (Abcam) and monoclonal antibodies against dynein intermediate chain (clone 74.1; Millipore), p150Glued (BD), kinesin-1 heavy chain (clone H2; Millipore), kinesin-2 (clone K2.4; Abcam), and SOD1 (Sigma-Aldrich).
GFP-LC3 × SOD1G93A cross
GFP-LC3 transgenic mice were crossed with SOD1G93A transgenic mice in our animal facility. DRG neurons were isolated at early stage disease (84 d) and late-stage disease (mean of 125 d). Late-stage disease was determined based on the criteria established by The Jackson Laboratory (score of 3 in the neurological scoring system). Age-matched littermates served as controls. The Institutional Animal Care and Use Committee at the University of Pennsylvania approved all animal protocols.
Live-cell imaging of DRG neurons
DRG neurons were isolated according to Perlson et al. (2009) and maintained in F-12 media (Invitrogen) with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. For live-cell analysis, DRG neurons were plated on glass-bottom dishes (World Precision Instruments, Inc.) and cultured for 2 d at 37°C in a 5% CO2 incubator. If necessary, before plating, neurons were transfected with 0.5 µg plasmid DNA using a Nucleofector (Lonza) according to the manufacturer’s specifications. Imaging was performed in low fluorescence nutrient medium (Hibernate A; BrainBits) with 2% B27 and 2 mM GlutaMAX. For LysoTracker red labeling, DRGs were incubated in 100 nM LysoTracker red (Invitrogen) in F-12 culture medium for 30 min at 37°C in a 5% CO2 incubator and washed twice in culture medium before imaging.
For GFP-LC3 motility and particle tracking analysis (Fig. 1, F–I; Fig. 2, A, B, and F; Fig. S1, C and D; and Fig. S3) as well as mCherry-EGFP-LC3 analysis (Fig. 5), imaging was performed on an inverted epifluorescence microscope (DMI6000B; Leica) using an Apochromat 63×, 1.4 NA oil immersion objective (Leica) in an environmental chamber at 37°C. Digital images were acquired with a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics) using LAS-AF software (Leica). Images were taken once every 3 s for a total of 3 min. For dual-color videos, images were taken consecutively, with green followed by red.
For the biogenesis experiments (Fig. 1, A–E; and Fig. S1 B) and colocalization experiments (Fig. 2, C and D; Fig. 3; Fig. 4; Fig. S1 A; and Fig. S2), imaging was performed on a spinning-disk confocal (UltraVIEW VoX; PerkinElmer) with a microscope (Eclipse Ti; Nikon) with the Perfect Focus System using an Apochromat 100×, 1.49 NA oil immersion objective (Nikon) in an environmental chamber at 37°C. Digital images were acquired with an EM charge-coupled device camera (C9100; Hamamatsu Photonics) using Volocity software (PerkinElmer). For biogenesis experiments, images were taken once every 2, 3, or 5 s for 10–20 min. Photobleaching of GFP-LC3 was achieved using a 488-nm laser at 100% power for 15 iterations. The GFP-LC3 signal was bleached completely throughout the depth of the neurite. For colocalization experiments, images were taken once every 2 s for 5 min, with green followed by red. Photobleaching of mCherry-SOD1G93A and RFP-Ub was achieved using a 561-nm laser at 100% power for 30 iterations. Recovery images were taken once every 2 s for 5 min. Images were assembled using ImageJ (National Institutes of Health) and Photoshop (Adobe).
Image analysis
GFP-LC3–positive puncta that moved a net distance ≥10 µm were manually tracked frame to frame for the duration of the entire video (3 min) using the particle tracking function in MetaMorph (Molecular Devices). A mean vesicle velocity was calculated by averaging all frame to frame instantaneous velocities (excluding paused values) for a single puncta. A pause was defined as a single or consecutive instantaneous velocity value of <0.067 µm/s, empirically determined to be the resolution of our system. A reversal was defined as a single or consecutive instantaneous velocity value of >0.067 µm/s in the opposite direction as compared with the net displacement of the vesicle. The number of reversals within 100 µm was determined based on the net distance a vesicle traveled during the 3 min.
Kymographs were generated using MetaMorph from neurites having at least one GFP-LC3–positive puncta that traveled a net distance of ≥10 µm. From each kymograph, the percentage of autophagosomes moving in the net retrograde direction (≥10 µm) versus net anterograde direction (≥10 µm) was determined. Nonprocessive vesicles that did not move a net distance of 10 µm exhibited bidirectional and stationary motility. From these kymographs, the total number of vesicles was determined and normalized by kymograph length (micrometers). Flux (number of vesicles moving within 100 µm/min) was determined by the sum of retrograde and anterograde vesicles (excluding bidirectional/stationary vesicles) and normalized by kymograph and video length. For mCherry-EGFP-LC3 analysis, the number of LC3 puncta positive for both GFP and mCherry fluorescence was counted based on kymographs. The proximal region of the neurite was defined as being within ∼200 µm of the cell body, and the distal region was within ∼100 µm of the end of the neurite.
Biochemistry
Fractions enriched for autophagosomes were prepared from brains of GFP-LC3 transgenic mice using protocols adapted from Morvan et al. (2009) and Strømhaug et al. (1998). Two brains were homogenized in 10 ml of 250 mM sucrose in 10 mM Hepes-KOH, pH 7.4 (with 1 mM EDTA for three-step gradient protocol) using a 30-ml homogenizer with a round-bottom Teflon pestle. Volumes of the gradient steps were scaled proportionately for a rotor (SW41; Beckman Coulter). The final gradient of the three-step fractionation protocol (Strømhaug et al., 1998) was spun in a rotor (TLS-55; Beckman Coulter). Equal total protein of low speed supernatant and the autophagosome-enriched fraction was separated by SDS-PAGE and subjected to immunoblot analysis.
Immunofluorescence
For immunofluorescence analysis, DRG neurons were plated on coverslips and cultured for 2 d. Cells were washed once in PBS (150 mM NaCl in 50 mM NaPO4, pH 7.4) and fixed in 3% PFA in PBS for 15 min at room temperature. Cells were washed twice in PBS and blocked and permeabilized in 2% (wt/vol) BSA and 0.1% (wt/vol) saponin in PBS for 1 h. All subsequent steps were performed in blocking/permeabilization buffer. Samples were incubated in primary antibody for 1 h, washed 3 × 5 min, incubated in secondary antibody for 1 h, washed 3 × 5 min, and mounted with ProLong gold.
Online supplemental material
Fig. S1 shows the anterograde movement of EB3 in DRG axons, FRAP of GFP-LC3 along the axon, and distributions of mean vesicle velocities and percentage of pausing for GFP-LC3 puncta along the axon. Fig. S2 shows that autophagosomes along the axon are positive for DIC2C-mCherry and mCherry-Kif3A but not the Golgi marker GPP130-mCherry. Fig. S2 also shows that LysoTracker red–positive compartments are positive for the late endosomal marker Rab7 but are largely negative for the early endosomal marker Rab5. In Fig. S3, we present data showing that despite the accumulation of SOD1G93A aggregates along the axon, autophagosome motility, direction, velocity, density, and flux are unaffected in a mouse model of fALS. Video 1 shows the appearance and growth of GFP-LC3–positive puncta in the neurite tip that grow into ringlike structures characteristic of autophagosomes. Video 2 shows an autophagosome escaping from the bidirectional pool at the neurite tip and moving processively toward the cell soma. Video 3 shows the robust retrograde motility of autophagosomes along the axon, and Videos 4 and 5 show that dynein co-migrates with these autophagosomes transfected with DIC1B-mCherry (Video 4) or DIC2C-mCherry (Video 5). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106120/DC1.
Acknowledgments
The authors gratefully acknowledge the technical assistance of Mariko Tokito and support from National Institutes of Health grant NS060698 to E.L.F. Holzbaur.
Footnotes
Abbreviations used in this paper:
- ALS
- amyotrophic lateral sclerosis
- DRG
- dorsal root ganglion
- fALS
- familial ALS
- LAMP1
- lysosomal-associated membrane protein 1
- Ub
- Ubiquitin
References
- Cai Q., Lu L., Tian J.H., Zhu Y.B., Qiao H., Sheng Z.H. 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 68:73–86 10.1016/j.neuron.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. 1994. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 264:1772–1775 10.1126/science.8209258 [DOI] [PubMed] [Google Scholar]
- Ha J., Lo K.W., Myers K.R., Carr T.M., Humsi M.K., Rasoul B.A., Segal R.A., Pfister K.K. 2008. A neuron-specific cytoplasmic dynein isoform preferentially transports TrkB signaling endosomes. J. Cell Biol. 181:1027–1039 10.1083/jcb.200803150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Hendricks A.G., Perlson E., Ross J.L., Schroeder H.W., III, Tokito M., Holzbaur E.L. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20:697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121:305–315 10.1083/jcb.121.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L., Menzies F.M., Rubinsztein D.C. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 9:574–587 10.1111/j.1600-0854.2008.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K., Nishiyama J., Inoue T., Mizushima N., Takeda J., Yuzaki M. 2010. Dynein- and activity-dependent retrograde transport of autophagosomes in neuronal axons. Autophagy. 6:378–385 10.4161/auto.6.3.11262 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2008. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33:109–122 10.1247/csf.08005 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Lee S., Sato Y., Nixon R.A. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci. 31:7817–7830 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Madeo F., Kroemer G. 2011. Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 23:198–206 10.1016/j.ceb.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 15:1101–1111 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morvan J., Köchl R., Watson R., Collinson L.M., Jefferies H.B., Tooze S.A. 2009. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy. 5:676–689 10.4161/auto.5.5.8378 [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., Youle R.J. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Perlson E., Jeong G.-B., Ross J.L., Dixit R., Wallace K.E., Kalb R.G., Holzbaur E.L.F. 2009. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J. Neurosci. 29:9903–9917 10.1523/JNEUROSCI.0813-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Gill S.R., Eckley D.M., Crego C.L., Compton D.A., Schroer T.A. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321–334 10.1083/jcb.147.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C., DiFiglia M., Heintz N., Nixon R.A., Qin Z.H., Ravikumar B., Stefanis L., Tolkovsky A. 2005. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 1:11–22 10.4161/auto.1.1.1513 [DOI] [PubMed] [Google Scholar]
- Strømhaug P.E., Berg T.O., Fengsrud M., Seglen P.O. 1998. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 335:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A., Reggiori F. 2010. The Golgi complex as a source for yeast autophagosomal membranes. Autophagy. 6:800–801 10.4161/auto.6.6.12575 [DOI] [PubMed] [Google Scholar]
- Ventruti A., Cuervo A.M. 2007. Autophagy and neurodegeneration. Curr. Neurol. Neurosci. Rep. 7:443–451 10.1007/s11910-007-0068-5 [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Yang Y., Xu K., Koike T., Zheng X. 2008. Transport of autophagosomes in neurites of PC12 cells during serum deprivation. Autophagy. 4:243–245 [DOI] [PubMed] [Google Scholar]
- Yang Y., Feng L.Q., Zheng X.X. 2011. Microtubule and kinesin/dynein-dependent, bi-directional transport of autolysosomes in neurites of PC12 cells. Int. J. Biochem. Cell Biol. 43:1147–1156 10.1016/j.biocel.2011.04.007 [DOI] [PubMed] [Google Scholar]
- Yen W.L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z., Hughson F.M., Baba M., Klionsky D.J. 2010. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188:101–114 10.1083/jcb.200904075 [DOI] [PMC free article] [PubMed] [Google Scholar]