Neuropeptide signaling, which is known to affect synaptic function in neural communication, also promotes neuromuscular junction growth in Drosophila.
Abstract
Neuropeptide signaling is integral to many aspects of neural communication, particularly modulation of membrane excitability and synaptic transmission. However, neuropeptides have not been clearly implicated in synaptic growth and development. Here, we demonstrate that cholecystokinin-like receptor (CCKLR) and drosulfakinin (DSK), its predicted ligand, are strong positive growth regulators of the Drosophila melanogaster larval neuromuscular junction (NMJ). Mutations of CCKLR or dsk produced severe NMJ undergrowth, whereas overexpression of CCKLR caused overgrowth. Presynaptic expression of CCKLR was necessary and sufficient for regulating NMJ growth. CCKLR and dsk mutants also reduced synaptic function in parallel with decreased NMJ size. Analysis of double mutants revealed that DSK/CCKLR regulation of NMJ growth occurs through the cyclic adenosine monophosphate (cAMP)–protein kinase A (PKA)–cAMP response element binding protein (CREB) pathway. Our results demonstrate a novel role for neuropeptide signaling in synaptic development. Moreover, because the cAMP–PKA–CREB pathway is required for structural synaptic plasticity in learning and memory, DSK/CCKLR signaling may also contribute to these mechanisms.
Introduction
Proper synaptic growth is essential for normal development of the nervous system and its function in mediating complex behaviors such as learning and memory. The Drosophila melanogaster larval neuromuscular junction (NMJ) has become a powerful system for studying the molecular mechanisms underlying synaptic development and plasticity (Collins and DiAntonio, 2007), and many of the key synaptic proteins are evolutionarily conserved.
Genetic and molecular analysis in Drosophila has uncovered numerous molecules and pathways that regulate NMJ growth, including proteins required for cell adhesion, endocytosis, cytoskeletal organization, and signal transduction via TGF-β, Wingless, JNK, cAMP, and other signaling molecules (Featherstone and Broadie, 2000; Collins and DiAntonio, 2007). For example, previous studies revealed that increased cAMP levels led to a down-regulation of the cell adhesion protein FasII at synapses, and the activation of the cAMP response element binding protein (CREB) transcription factor to achieve long-lasting changes in synaptic structure and function (Davis et al., 1996; Schuster et al., 1996a,b). Despite the identification and characterization of these various positive and negative regulators, our understanding of the networks that govern synaptic growth is still incomplete, with many of the key components and mechanisms yet to be uncovered and analyzed.
To search for new regulators of synaptic growth, we conducted a forward genetic screen for mutants exhibiting altered NMJ morphology. In this screen, we discovered a new mutation that exhibits strikingly undergrown NMJs, which indicates disruption of a positive regulator of NMJ growth. We have identified the affected protein as cholecystokinin (CCK)-like receptor (CCKLR), a putative neuropeptide receptor that belongs to the family of G protein–coupled receptors (GPCRs) sharing a uniform topology with seven transmembrane domains. When activated by their ligands, neuropeptide GPCRs affect levels of second messengers such as cAMP, diacylglycerol, inositol trisphosphate, and intracellular calcium (Nässel, 2002). Through activation of their cognate receptors, secreted neuropeptides mediate communication among various sets of neurons as well as other cell types to regulate several physiological activities, including feeding and growth, molting, cuticle tanning, circadian rhythm, sleep, and learning and memory (Nässel and Winther, 2010). In general, neuropeptides act by modulating neuronal activity through both short-term and long-term effects. Short-term effects include modifications of ion channel activity and alterations in release of or response to neurotransmitters. Long-term effects include changes in gene expression through activation of transcription factors and protein synthesis. In contrast with the well-known effects of neuropeptide signaling on neuronal activity and the strength of synaptic transmission, regulation of synaptic growth and development by neuropeptides has not previously been clearly established.
Here, we demonstrate that CCKLR is required presynaptically to promote NMJ growth. Moreover, mutations of drosulfakinin (dsk), which encodes the predicted ligand of CCKLR, cause similar NMJ undergrowth and interact genetically with CCKLR mutations, indicating that DSK and CCKLR are components of a common signaling mechanism that regulates NMJ growth. In addition to the morphological phenotypes caused by mutations of CCKLR and dsk, mutant larvae also exhibit deficits in synaptic function. Through phenotypic analysis of double mutant combinations, we show that DSK/CCKLR signals through the cAMP–PKA–CREB pathway to regulate NMJ growth. Our results suggest a novel role for neuropeptide signaling in regulation of synaptic development. Moreover, because the cAMP-PKA-CREB pathway is required for structural plasticity of synapses in learning and memory, DSK/CCKLR signaling may also contribute to these mechanisms.
Results
Loss of CCKLR leads to NMJ undergrowth
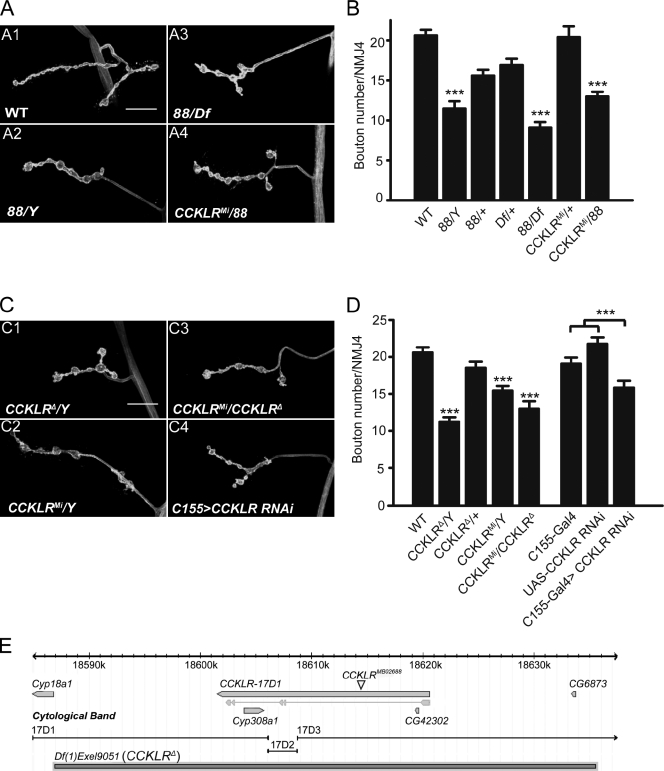
To identify mutations that perturb synaptic growth and development, we used ethyl methanesulfonate for mutagenesis and screened directly for viable, X-linked mutants with aberrant larval NMJ morphology. One such mutant (referred to by its original isolation number, 88) exhibits striking NMJ undergrowth, with a 50% reduction in bouton number. In addition, the length of the NMJ branches is significantly reduced, and the boutons are enlarged (Fig. 1, A2 and B). Although we focused on the analysis of NMJ4 because of its relative simplicity, we observed similar phenotypes at NMJ6/7. The 88 mutation is recessive and localizes by deletion mapping to a small region (17D1-17D3) on the X chromosome (Fig. 1 E).
Figure 1.
CCKLR positively regulates NMJ growth in Drosophila. (A and C) Confocal images of NMJ4 labeled with FITC–anti-HRP. (A) 88/Y, 88/Df, and 88/CCKLRMi larvae all exhibit similar NMJ undergrowth compared with control. (B) Quantification of bouton numbers at NMJ4 for genotypes shown in A. Df shown in A and B is Df(1)fu-A7. (C) CCKLRΔ/Y, CCKLRMi/Y, CCKLRMi/CCKLR-17D1Δ, and C155-Gal4>CCKLR RNAi all exhibit similar NMJ undergrowth. Bars, 20 µM. (D) Quantification of bouton numbers at NMJ4 for genotypes shown in C. CCKLRMi is CCKLR-17D1MB02688. Df(1)Exel9051 is designated as CCKLRΔ. The genotype of C155>CCKLR RNAi larvae is C155-Gal4 UAS-Dcr2/Y; UAS-CCKLR-RNAi7231/+. Error bars denote SEM. ***, P < 0.001 (compared with WT control unless indicated otherwise). (E) Gene structure and genomic view of CCKLR-17D1 (modified from FlyBase.org).
Df(1)Exel9051, which uncovers the mutant phenotype, removes ∼40 kb and spans only four genes. The biggest gene in this interval is CCKLR-17D1, which is predicted to encode a CCK-like neuropeptide receptor. The deficiency is sufficiently small that it is viable when hemizygous or homozygous, enabling us to examine NMJs of larvae completely lacking CCKLR-17D1. These larvae exhibit NMJ undergrowth very similar to that of 88 hemizygotes or homozygotes (Fig. 1, C1 and D), which indicates that 88 is a strong hypomorphic or null allele. Furthermore, we examined an insertional allele, CCKLR-17D1MB02688 (referred to as CCKLRMi for short), which carries a transposon inserted in the first intron (Fig. 1 E), and is thus presumed to be a weak hypomorph. Nonetheless, it failed to complement 88 (Fig. 1, A4 and B) and causes NMJ undergrowth when hemizygous (Fig. 1, C2, C3 and D). The undergrowth phenotype is stronger in CCKLRMi/Df(1)Exel9051 larvae, which is consistent with the prediction that CCKLRMi is a hypomorphic allele (Fig. 1, C3 and D). We sequenced the entire CCKLR-17D1 coding region in the 88 mutant but did not find any lesion. Thus, 88 most likely contains a regulatory mutation that interferes with expression of this gene. Because Df(1)Exel9051 homozygotes are definitely null for CCKLR-17D1, we used these larvae (designated as CCKLR-17D1Δ/Y or CCKLR-17D1Δ/CCKLR-17D1Δ) in subsequent experiments.
Because the Drosophila genome contains two CCKLR genes, CCKLR-17D1 and CCKLR-17D3, separated by only ∼40 kb, we sought to determine whether CCKLR-17D3 also affects NMJ growth. To answer this question, we used two different chromosomal duplications, Dp(1;3)DC351 (designated as Dp(17D1-D5)) and Dp(1;3)DC352 (designated as Dp(17D3-E1)), to rescue the NMJ phenotype of CCKLR-17D1Δ/Y. Dp(17D1-D5), which contains a wild-type (WT) copy of both CCKLR-17D1 and CCKLR-17D3, completely rescues NMJ undergrowth in CCKLR-17D1Δ/Y, as expected (Fig. S1, A3 and B). In contrast, Dp(17D3-E1), which contains only a WT copy of CCKLR-17D3, fails to rescue CCKLR-17D1Δ/Y (Fig. S1, A4 and B); this suggests that CCKLR-17D3 cannot substitute for CCKLR-17D1 in regulating NMJ growth. To test whether there is genetic interaction between these two CCKLRs, we used a deficiency, Df(1)ED7413 (designated as Df(17D1-F1)), which removes both CCKLR-17D1 and CCKLR-17D3. The NMJ phenotypes of Df(17D1-F1)/+ and CCKLR-17D1Δ/+ larvae do not differ (Fig. S1, C1 and D), which indicates that removing one copy of CCKLR-17D3 does not further enhance the phenotype of CCKLR-17D1Δ/+ heterozygotes. Moreover, even in CCKLR-17D1Δ/Df(17D1-F1) larvae, where one copy of CCKLR-17D3 is removed in a CCKLR-17D1-null background, the NMJ phenotype is unchanged compared with CCKLR-17D1–null homozygotes (Fig. S1, C2 and D). We also examined an insertional allele of CCKLR-17D3 (CCKLR-17D3KG), and found no effect on NMJ growth in CCKLR-17D3KG/Y or CCKLR-17D3KG/Df(17D1-F1) larvae (Fig. S1, C3, C4, and D). On the basis of these results, we conclude that CCKLR-17D1 but not CCKLR-17D3 is involved in regulating NMJ growth. We thus focus on CCKLR-17D1, hereafter referred to as CCKLR for short.
CCKLR functions presynaptically to regulate NMJ growth
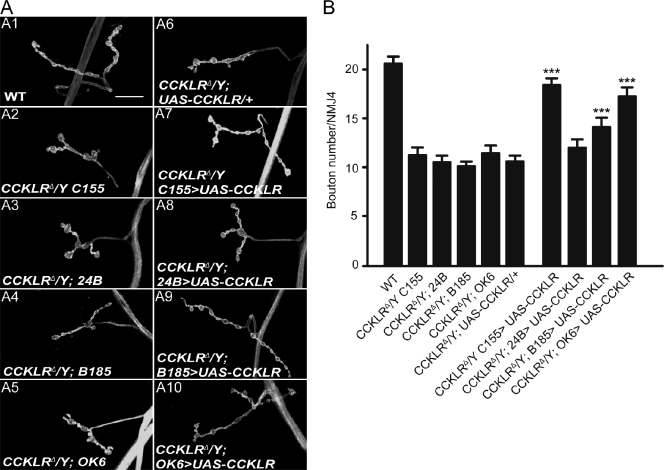
Because neuropeptide signaling has not previously been characterized in synaptic development, it was of interest to determine where CCKLR function is required for proper NMJ growth. Neural-specific expression of RNAi directed against CCKLR using C155-Gal4 resulted in NMJ undergrowth similar to the original mutant (Fig. 1, C4 and D). Conversely, rescue experiments using a series of different Gal4 drivers demonstrated that presynaptic expression of CCKLR was sufficient to rescue the mutant phenotypes: Presynaptic expression of upstream activating sequence (UAS)-CCKLR using C155-Gal4 substantially rescues the mutant phenotype of CCKLRΔ/Y both for bouton number and overall NMJ morphology (17.9 ± 0.8 vs. 11.2 ± 0.8 boutons; Fig. 2, A7 and B), whereas postsynaptic expression via 24B-Gal4 has no significant effect on these phenotypes (12.0 ± 0.8 vs. 10.5 ± 0.7 boutons; Fig. 2, A8 and B). When we use B185-Gal4 to drive UAS-CCKLR expression in all muscles and motor neurons (Davis et al., 1997), phenotypic rescue was not further increased beyond that observed with presynaptic expression alone (14.2 ± 0.9 vs. 10.1 ± 0.5 boutons; Fig. 2, A9 and B). The slight decrement in rescue with B185-Gal4 compared with C155-Gal4 is most likely caused by differences in the level of expression associated with the different drivers. We were also able to rescue CCKLRΔ/Y by driving UAS-CCKLR with OK6-Gal4, which is expressed preferentially in motor neurons (Fig. 2, A10 and B). Collectively, these data indicate that presynaptic CCKLR function is both necessary and sufficient to regulate NMJ growth.
Figure 2.
CCKLR functions presynaptically to regulate NMJ growth. (A) Confocal images of NMJ4 labeled with FITC–anti-HRP. Neuronal expression of UAS-CCKLR by C155-Gal4 (A7) or OK6-Gal4 (A10) in CCKLRΔ/Y background produces significant rescue of the NMJ undergrowth phenotype. Similar but less complete rescue is achieved when UAS-CCKLR is expressed under the control of B185-Gal4 (A9), which drives expression both in motor neurons and in muscles. No rescue is seen when 24B-Gal4 is used to drive UAS-CCKLR in muscle (A8). Bar, 20 µm. (B) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown. The genotype of CCKLRΔ/Y C155>UAS-CCKLR is C155-Gal4 CCKLRΔ/Y; UAS-CCKLR/+. CCKLRΔ/Y; OK6 >UAS-CCKLR is CCKLRΔ/Y; OK6-Gal4/UAS-CCKLR. CCKLRΔ/Y; 24B>UAS-CCKLR is CCKLRΔ/Y; UAS-CCKLR/+; 24B-Gal4/+. CCKLRΔ/Y; B185 >UAS-CCKLR is CCKLRΔ/Y; B185-Gal4/UAS-CCKLR. Error bars denote SEM. ***, P < 0.001 (compared with corresponding CCKLRΔ/Y; Gal4/+ and CCKLRΔ/Y; UAS-CCKLR/+ controls).
Our efforts to raise a specific antibody against CCKLR to investigate its cellular localization were unsuccessful. As an alternative, we generated a tagged UAS-CCKLR–mCherry construct to examine the subcellular localization of CCKLR in neurons. Although GAL4-driven expression of this construct is not equivalent to the endogenous pattern of CCKLR expression, we found that it successfully rescued CCKLRΔ/Y when its expression was driven presynaptically by C155-Gal4 (Fig. S2, A and B), which indicates that expression, subcellular localization, and function of the tagged protein overlap sufficiently well with that of endogenous CCKLR to confer a nearly normal phenotype. In these rescued larvae, CCKLR–mCherry is strongly expressed in cell bodies of motor neurons in the ventral ganglion (Fig. S2 C). Because C155-Gal4 drives transcription pan-neuronally (unpublished data), the apparent enrichment of CCKLR–mCherry in motor neurons compared with other neurons in the ventral ganglion suggests that there are regulatory mechanisms that promote the translation or stabilization of CCKLR preferentially in motor neurons. We also detected CCKLR–mCherry in presynaptic boutons at NMJs, but the intensity is much lower than in cell bodies (Fig. S2 D). These results further suggest that CCKLR functions within motor neurons to regulate NMJ development.
dsk interacts with CCKLR to regulate NMJ growth
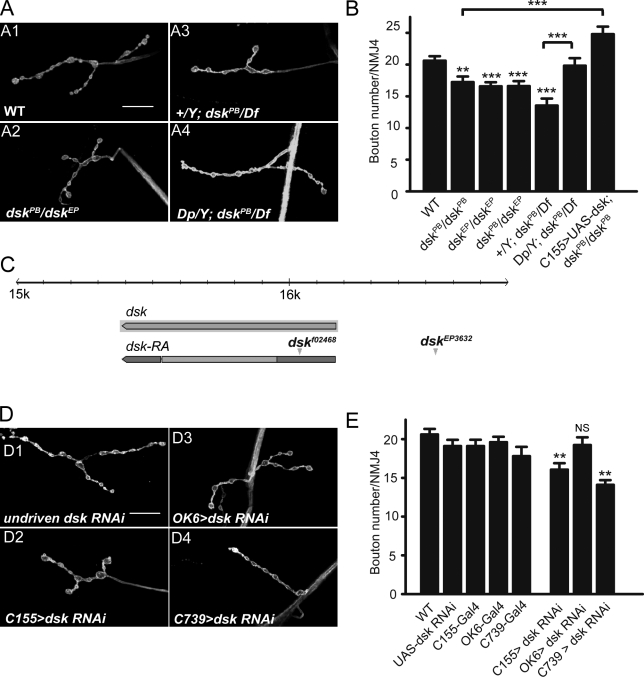
Because neuropeptide receptors are typically activated in response to binding of specific neuropeptide ligands, we wished to determine if CCKLR’s role in regulating NMJ growth was also ligand dependent. On the basis of sequence comparisons, the Drosophila orthologue of CCK is DSK, encoded by the dsk gene. Consequently, we examined the NMJ phenotype of two P-element insertion alleles, dskf02468 (referred to as dskPB) and dskEP3632 (referred to as dskEP; Fig. 3 C). Although neither mutation disrupts the coding region of dsk, both exhibited mild NMJ undergrowth as homozygotes or in heterozygous combination with each other (Fig. 3, A2 and B). Hemizygous dskPB/Df(3R)2-2 larvae exhibit a more pronounced undergrowth phenotype, as expected, if the insertion is a weak hypomorphic allele (Fig. 3, A3 and B). The undergrowth phenotypes of dskPB/Df(3R)2-2 or dskPB/dskPB are rescued by an X chromosome that carries an insertional duplication of a small segment of 3R that includes dsk+ (Fig. 3, A4 and B) or by C155-Gal4–driven expression of UAS-dsk (Fig. 3 B and Fig. 4 A6). Conversely, expression of dsk RNAi using the C155-Gal4 driver results in NMJ undergrowth (Fig. 3, D2 and E). These results demonstrate that dsk is also a positive regulator of NMJ growth. Moreover, the NMJ undergrowth phenotype of dsk mutants is reminiscent of that observed in CCKLR mutants, which is consistent with the idea that DSK is the CCKLR ligand.
Figure 3.
dsk is a positive regulator of NMJ growth. (A and D) Confocal images of NMJ4 labeled with FITC–anti-HRP. (A) dskPB/dskEP double heterozygotes (A2) and dskPB/Df larvae (A3) show NMJ undergrowth similar to CCKLR mutants. A duplicated genomic segment containing dsk+ inserted on the x chromosome rescues the phenotype of dskPB/Df (A4). (B) Quantification of bouton numbers at NMJ4 in dsk mutant larvae and rescued larvae. dskPB is dskf02468. dskEP is dskEP3632. Df is Df(3R)2-2. Dp is Dp(3;1)2-2. (C) Genomic organization of dsk showing sites of transposon insertion for two dsk mutations (modified from Flybase.org). (D) dsk RNAi driven by C155-Gal4 (D2) and C739-Gal4 (D4), but not OK6-Gal4 (D3), leads to NMJ undergrowth. (E) Quantification of bouton numbers at NMJ4 for larvae shown in D. The genotype of C155>dsk RNAi larvae is C155-Gal4 UAS-Dcr2/+; UAS-dsk-RNAi14201/+. OK6>dsk RNAi is OK6-Gal4/+; UAS-dsk-RNAi14201/+. C739>dsk RNAi is C739-Gal4/+; UAS-dsk-RNAi14201/+. Error bars denote SEM. ***, P < 0.001; **, P < 0.01 (compared with WT control unless indicated otherwise). Bars, 20 µm.
Figure 4.
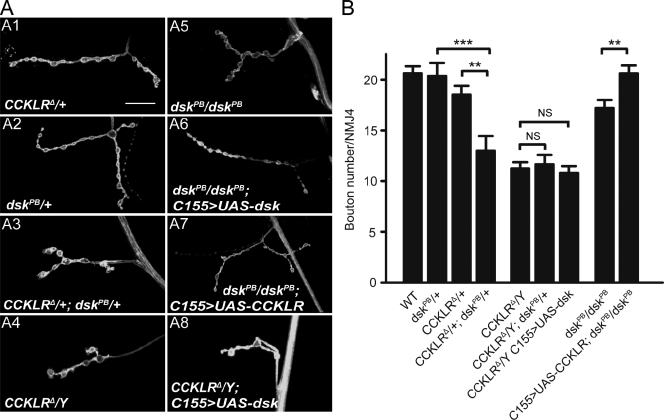
Genetic interactions between CCKLR and dsk. (A) Confocal images of NMJ4 labeled with FITC–anti-HRP. CCKLRΔ/+; dskPB/+ double heterozygotes (A3) exhibit significantly enhanced NMJ undergrowth compared with either single heterozygote alone (A1 and A2). C155-Gal4–driven UAS-dsk rescues dskPB/dskPB (A6) but not CCKLRΔ/Y (A8). C155-Gal4–driven UAS-CCKLR overexpression also rescues dskPB/dskPB (A7). Bar, 20 µm. (B) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown in A. Error bars denote SEM. ***, P < 0.001; **, P < 0.01.
To determine where DSK is required for normal NMJ growth, we used different Gal4 drivers to express UAS-dsk-RNAi. OK6-Gal4–driven dsk RNAi in motor neurons does not cause the undergrowth phenotype that we observed with C155-Gal4 (Fig. 3, D3 and E). In contrast, UAS-dsk-RNAi driven by C739-Gal4 did result in significant NMJ undergrowth (Fig. 3, D4 and E). Although C739-Gal4 is commonly used as a mushroom body driver (Tettamanti et al., 1997), it also drives expression in two longitudinal stripes in the ventral ganglion (Manseau et al., 1997) as well as in a group of neurosecretory cells, as revealed by C739-Gal4–driven UAS-CD8–GFP (Fig. S3). Compared with earlier immunohistochemical studies by Nichols (1992, 2003), it appears that C739-Gal4 expression overlaps with DSK-positive cells (Fig. S3). Although at this point we do not know which specific groups of cells are required for DSK production, the results of these RNAi experiments suggest that DSK is not secreted by motor neurons, but instead is most likely secreted by neurosecretory cells in the central nervous system (CNS).
If DSK is the ligand for CCKLR and both of them affect the same pathway regulating NMJ growth, we might expect to observe phenotypic interactions between mutations that partially impair both dsk and CCKLR. In fact, whereas CCKLRΔ/+ alone (18.5 ± 0.8 boutons) or dskPB/+ heterozygotes alone (20.2 ± 1.3 boutons) do not significantly affect NMJ growth, the CCKLRΔ/+; dskPB/+ double heterozygotes exhibit a strong reduction in bouton number (13.0 ± 1.4; Fig. 4, A3 and B). In contrast, heterozygosity for dskPB does not further enhance the NMJ undergrowth phenotype of CCKLRΔ/Y-null mutants (Fig. 4 B), and C155-Gal4–driven overexpression of UAS-dsk is unable to rescue the CCKLRΔ/Y-null phenotype (Fig. 4, A8 and B). However, the NMJ undergrowth of homozygotes for dskPB, a hypomorphic dsk allele, is rescued by C155-Gal4–driven overexpression of UAS-CCKLR (Fig. 4, A7 and B). These results all support the idea that DSK and CCKLR are components of a common regulatory pathway affecting NMJ growth and that DSK is the ligand of CCKLR.
CCKLR acts upstream of the cAMP pathway
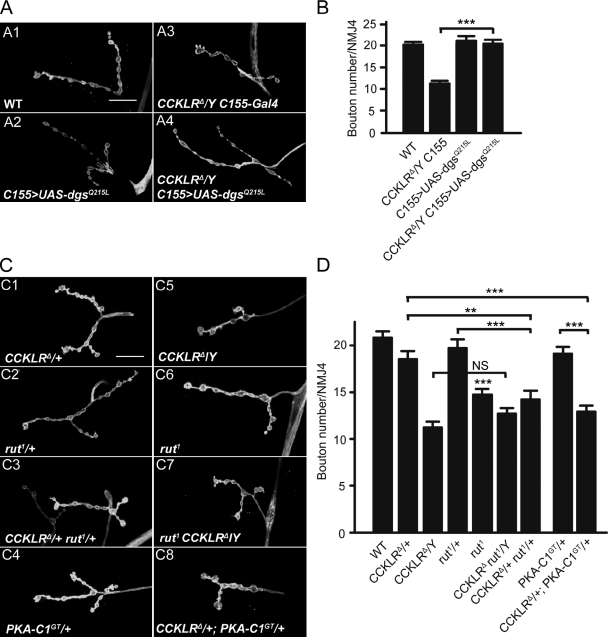
When activated, GPCRs undergo a conformational change that allows the receptor to promote the exchange of GDP for GTP on the Gα subunit of heterotrimeric G proteins (McCudden et al., 2005). This exchange triggers the dissociation of the Gα subunit from the Gβγ dimer, and both Gα-GTP and Gβγ can then activate different second messenger pathways such as the cAMP signal pathway through stimulation of adenylyl cyclase (AC; Gilman, 1987; Neves et al., 2002). Previous studies have shown that a loss-of-function mutation in the gene encoding the stimulatory G protein α subunit (Gsα) in Drosophila (dgsM19) causes NMJ undergrowth (Wolfgang et al., 2004). If dgs functions downstream of CCKLR, we hypothesized that overexpression of UAS-dgsQ215L, a constitutively active Gsα construct (Renden and Broadie, 2003), could suppress the CCKLR mutant phenotype. When driven by C155-Gal4, overexpression of UAS-dgsQ215L in a WT background did not alter NMJ morphology (Fig. 5, A2 and B). However, overexpression of UAS-dgsQ215L completely suppresses the NMJ undergrowth phenotype of CCKLRΔ-null mutants (20.4 ± 0.9 vs. 11.2 ± 0.6 boutons; Fig. 5, A4 and B). Thus, NMJ undergrowth in CCKLR mutants appears to entail a decrease in signaling through Gsα.
Figure 5.
CCKLR genetically interacts with components of the cAMP–PKA pathway. (A and C) Confocal images of NMJ4 labeled with FITC–anti-HRP. (A) Overexpression of dgsQ215L, a gain-of-function allele, suppresses NMJ undergrowth of CCKLRΔ/Y. (B) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown in A. (C) rut1/+ CCKLRΔ/+ double heterozygotes (C3) show significantly enhanced undergrowth compared with either single heterozygote (C1 and C2). Similarly, CCKLRΔ/+; PKA-C1GT/+ double heterozygotes (C8) exhibit significantly enhanced undergrowth compared with either single heterozygote (C1 and C4). rut1 homozygotes show NMJ undergrowth (C6). rut1 CCKLRΔ double mutants (C7) do not show further undergrowth compared with CCKLRΔ/Y (C5). Bars, 20 µm. (D) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown in C. PKA-C1GT is PKA-C1BG02142. C155>UAS-dgsQ215L is C155/Y; UAS-dgsQ215L/+. Error bars denote SEM. ***, P < 0.001; **, P < 0.01 (compared with WT control unless indicated otherwise).
A major signal transduction pathway downstream of GPCRs and stimulatory G proteins is the cAMP pathway via activation of AC, one of which is encoded by the rutabaga (rut) locus in Drosophila (Levin et al., 1992). Therefore, we examined rut1 mutations for their effect on NMJ growth and interaction with CCKLR mutations. As previously reported (Zhong et al., 1992), rut1 causes a mild undergrowth phenotype (Fig. 5, C6 and D). Heterozygous rut1/+ (19.7 ± 0.9 boutons) and heterozygous CCKLRΔ/+ (18.5 ± 0.8 boutons) larvae have NMJs that do not differ from WT (Fig. 5, C1 and C2). However, heterozygosity for rut1 in a CCKLRΔ/+ background results in a significant reduction in NMJ growth (14.2 ± 0.9 boutons; Fig. 5, C3 and D), which suggests impairments in a common pathway. We generated a rut1 CCKLRΔ double mutant by recombination and found that there was no further reduction in bouton number in double mutant homozygotes compared with CCKLRΔ single mutants (Fig. 5, C7 and D). Collectively, these results point toward CCKLR and rut being involved in a common pathway.
Similarly, we found that the undergrowth phenotype of CCKLRΔ/+ heterozygotes is significantly enhanced by heterozygosity for Df(2L)N22-14, which uncovers PKA-C1, a cAMP-dependent protein kinase (data not shown), or for a mutation in this gene (PKA-C1BG02142, referred to as PKA-C1GT; 12.9 ± 0.6 boutons for CCKLRΔ/+; PKA-C1GT/+ vs. 18.5 ± 0.8 boutons for CCKLRΔ/+ and 19.1 ± 0.7 boutons for PKA-C1GT/+; Fig. 5, C8 and D). These results strongly support the idea that CCKLR regulates NMJ growth via activation of the cAMP pathway.
dCreb2 positively regulates NMJ growth and is epistatic to CCKLR
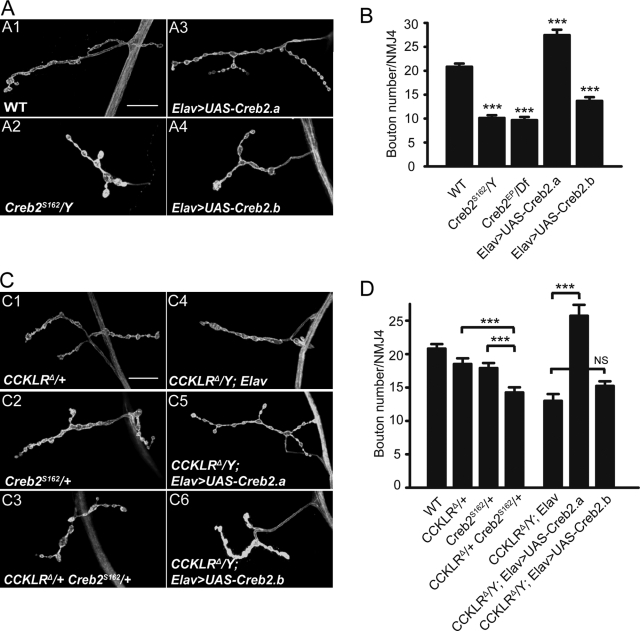
A major downstream regulator of the cAMP signaling pathway is CREB. Activation of CREB in response to elevated levels of cAMP leads to transcription of genes required for modifications in synaptic structure and function that are involved in long-term plasticity (Davis et al., 1996; Alberini, 2009). In Drosophila, although dCreb2 has been shown to regulate activity-dependent functional plasticity at larval NMJs, its role in regulating NMJ growth during development has not been previously established (Davis et al., 1996; Koon et al., 2011). Having found that the cAMP pathway is required for CCKLR-dependent NMJ growth, we sought to determine if this phenotype is mediated through dCreb2. We observed a significant decrease in bouton number in dCreb2 mutants dCreb2S162 and dCreb2EP/Df (Fig. 6, A2 and B), which is comparable to that of CCKLR mutants. Similarly, overexpression of a dominant-negative (transcriptional blocker) dCreb2 transgene (UAS-Creb2.b) in otherwise WT neurons also causes NMJ undergrowth (Fig. 6, A4 and B). In contrast, overexpression of UAS-Creb2.a, a WT form (transcriptional activator) of dCreb2, causes a mild but significant increase in bouton number (Fig. 6, A3 and B).
Figure 6.
dCreb2 positively regulates NMJ growth and is epistatic to CCKLR. (A and C) Confocal images of NMJ4 labeled with FITC–anti-HRP. (A) Creb2S162 hemizygotes exhibit strong NMJ undergrowth similar to CCKLRΔ/Y (A2). Neuronal overexpression of Creb2.a by Elav-Gal4 (Elav>UAS-Creb2.a) results in NMJ overgrowth (A3), whereas neuronal overexpression of Creb2.b (Elav>UAS-Creb2.b) leads to NMJ undergrowth (A4). (B) Quantification of bouton numbers at NMJ4 in Creb2 mutant larvae and larvae with Creb2 misexpression. CrebEP/Df is CrebEP1378/Df (1)osUE69. (C) Genetic interactions between CCKLR and Creb2. Creb2S162/+ CCKLRΔ/+ double heterozygotes (C3) exhibit significantly enhanced NMJ undergrowth compared with either single heterozygote (C1 and C2). Neuronal overexpression of Creb2.a suppresses NMJ undergrowth of CCKLRΔ/Y and leads to overgrowth (C5), whereas overexpression of Creb2.b does not (C6). Bars, 20 µm. (D) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown in C. Error bars denote SEM. ***, P < 0.001 (compared with WT control unless indicated otherwise). Elav>UAS-Creb2.a or -b is Elav-Gal4/UAS-Creb2.a or -b.
Loss of one copy of dCreb2 in an otherwise WT background has no effect on NMJ morphology but significantly enhances NMJ undergrowth in CCKLRΔ/+ heterozygotes (14.3 ± 0.8 vs. 18.5 ± 0.8 boutons; Fig. 6, C3 and D). Remarkably, overexpression of UAS-dCreb2.a by Elav-Gal4 not only fully suppresses the NMJ undergrowth of CCKLR mutants but even produces the same mild overgrowth phenotype as in a WT background (26.4 ± 1.0 vs. 13.2 ± 1.2 boutons; Fig. 6, C5 and D). In contrast, overexpression of UAS-dCreb2.b neither suppresses nor enhances CCKLR phenotypes (15.4 ± 0.8 vs. 13.2 ± 1.2 boutons; Fig. 6, C6 and D). These results demonstrate that in addition to its previously described role in functional synaptic plasticity (Davis et al., 1996), dCREB2 strongly regulates NMJ growth during development and is a major downstream target through which CCKLR promotes NMJ growth.
Presynaptic overexpression of CCKLR leads to NMJ overgrowth
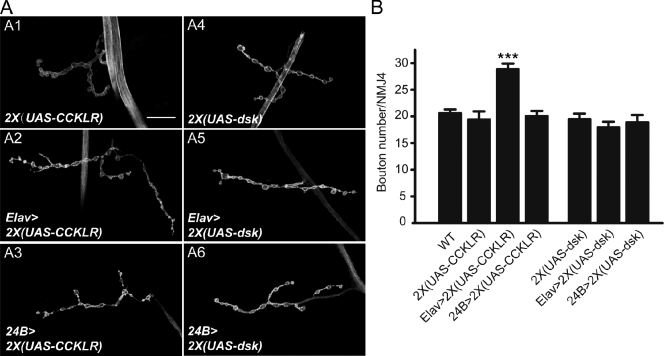
Our results thus far indicate that CCKLR is a positive regulator of NMJ growth. If the CCKLR pathway does stimulate NMJ growth, enhancement of the activity of this pathway could lead to NMJ overgrowth. To investigate this possibility, we overexpressed the UAS-CCKLR transgene in an otherwise WT background. Overexpression of a single copy of UAS-CCKLR either in neurons (Elav-Gal4) or in muscles (24B-Gal4) has no effect on NMJ morphology (unpublished data). However, when two copies of UAS-CCKLR are driven by Elav-Gal4 (Gal4/+; UAS/UAS, designated as Gal4>2X(UAS)), there is a significant increase in bouton number (28.9 ± 1.0 vs. 19.4 ± 1.5; Fig. 7, A2 and B). This overgrowth phenotype is very similar to that caused by overexpression of UAS-Creb2.a (Fig. 6, A3 and B). To avoid possible genetic background effects associated with homozygosity for UAS-CCKLR, we repeated the 2X(UAS-CCKLR) neuronal overexpression using a heterozygous combination of two independent UAS-CCKLR transgenes and observed the same overgrowth phenotype (unpublished data). In contrast, overexpression of two copies of UAS-CCKLR in muscle still had no effect on NMJ morphology (Fig. 7, A3 and B). These results provide additional support for the conclusion that CCKLR functions presynaptically as a positive regulator of NMJ growth.
Figure 7.
Presynaptic overexpression of CCKLR leads to NMJ overgrowth. (A) Confocal images of NMJ4 labeled with FITC–anti-HRP. Neuronal overexpression of two copies of CCKLR by Elav-Gal4 (Elav>2X(UAS-CCKLR)) results in a significant increase in NMJ growth (A2). Overexpression in muscle (24B>2X(UAS-CCKLR)) does not alter NMJ growth (A3). Overexpression of 2X(UAS-dsk) by either Elav-Gal4 (A5) or 24B-Gal4 (A6) does not affect NMJ growth. Bar, 20 µm. (B) Quantification of bouton numbers at NMJ4 in larvae of the genotypes shown in A. The genotype of Gal4>2X(UAS) is Gal4/+; UAS /UAS. Error bars denote SEM. ***, P < 0.001 (compared with WT control).
We also examined overexpression of 2X(UAS-dsk) in either neurons or muscles in an otherwise WT background. In neither instance did we observe a significant effect on NMJ morphology (Fig. 7, A5, A6, and B). Thus, increasing the level of dsk expression in WT larvae does not perturb NMJ growth, which suggests that it is the endogenous receptor rather than the neuropeptide ligand that is normally present in limiting amounts.
DSK/CCKLR signaling is required for normal synaptic function
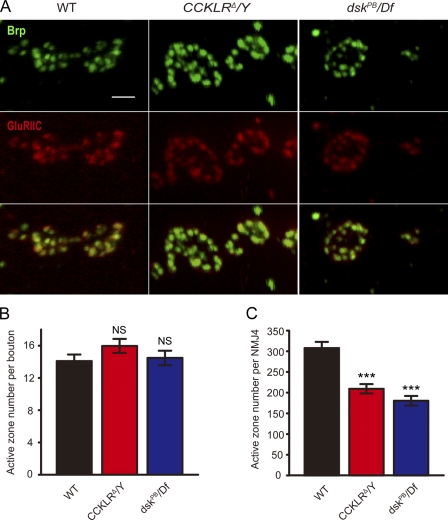
Although both CCKLR and dsk mutants have reductions in bouton number, the mutant boutons appear generally larger than WT (especially in CCKLRΔ/Y larvae), raising the question of whether there could be some type of structural compensation for the deficit in bouton number. One way the NMJ could compensate for having fewer boutons would be to increase the density of active zones within the remaining boutons. To determine if there was any effect on the number or distribution of active zones, we stained with anti-Bruchpilot (anti-Brp) and anti-GluRIIC antibodies to label both pre- and postsynaptic components. The number of active zones per bouton is slightly higher in CCKLRΔ/Y mutants compared with WT, but this difference is not significant (Fig. 8, A and B). Similarly, the number of active zones per bouton is normal in dsk mutants (Fig. 8, A and B). Thus, as a result of the reduction in bouton number, the total number of active zones per NMJ is significantly reduced in both CCKLR (209 ± 11) and dsk mutants (180 ± 12) compared with WT (308 ± 15; Fig. 8 C). The apposition of pre- and postsynaptic active zones, stained with anti-Brp and anti-GluRIIC antibodies, respectively, appear normal in both dsk and CCKLR mutants, as do the levels of these proteins (Fig. 8 A). Similarly, the levels of GluRIIB in the mutants do not differ from control larvae (Fig. S4).
Figure 8.
Active zone density per bouton is normal in CCKLR and dsk mutants but the total number per NMJ is reduced. (A) Confocal images of NMJ4 boutons labeled with anti-Brp (green) and anti-GluRIIC (red). Bar, 2 µm. (B) Quantification of active zones per bouton for larvae of the indicated genotypes. (C) Quantification of total active zones per NMJ4 for larvae of the indicated genotypes (WT, n = 10; CCKLRΔ/Y, n = 10; dskPB/Df, n = 12. n is the number of NMJs scored). Error bars denote SEM. ***, P < 0.001(compared with WT control). NS, not significant compared with WT control.
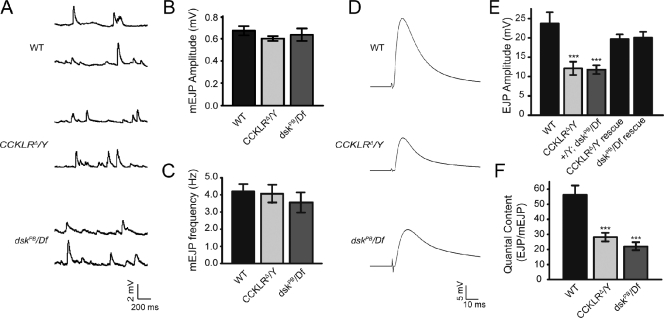
To determine if the reduction in active zone number is associated with a reduction in synaptic transmission, we recorded excitatory junction potentials (EJPs) from muscle 6. We did not detect any significant difference in the size or frequency of miniature EJPs (mEJPs) in CCKLR or dsk mutants compared with WT controls (Fig. 9, A–C). However, the size of EJPs in both CCKLR (12.12 ± 1.71 mV) and dsk mutants (11.78 ± 1.13 mV) is reduced by 50% compared with controls (23.73 ± 2.90 mV; Fig. 9, D and E). Thus, there is a significant reduction in the quantal content (number of vesicles released per stimulus) in CCKLR (27.15 ± 2.90) and dsk mutants (21.68 ± 2.44) compared with WT (55.98 ± 6.19) that parallels their decrease in number of active zones (Fig. 9 F). The EJP defect in CCKLR and dsk mutants is substantially rescued by expressing a UAS-CCKLR transgene under the direction of C155-Gal4 or by introducing a WT copy of dsk via a chromosome duplication (Fig. 9 E), which is similar to what we previously found for bouton number. Thus, CCKLR and dsk mutants have deficits in synaptic transmission, and there is very good correlation between the reduction in number of active zones and the reduction in EJP amplitude. Nonetheless, it is important to note that the CCKLR–cAMP–CREB signaling pathway could still be affecting transmitter release via a mechanism that is distinct from its effects on NMJ morphology.
Figure 9.
CCKLR and dsk mutants exhibit a reduction in evoked neurotransmission. (A) Representative traces of spontaneous neurotransmitter release recorded in 0.5 mM Ca2+ from muscle 6 in third instar larvae of the indicated genotypes. (B and C) Quantification of the mean mEJP amplitude and frequency of WT (n = 12), CCKLRΔ/Y (n = 12), and dsk PB/Df (n = 11). (D) Representative traces of evoked transmitter release recorded in 0.5 mM Ca2+ from muscle 6. (E) Quantification of the mean EJP amplitude. The mean EJP amplitude is significantly reduced in CCKLRΔ/Y (n = 10) and dskPB/Df (n = 9) mutants compared with WT (n = 7; P < 0.001). C155-Gal4–driven UAS-CCKLR expression (n = 15) and the Dp carrying dsk+ (n = 11) rescue the defects of CCKLRΔ/Y and dskPB/Df, respectively. (F) Quantal content is significantly reduced in CCKLRΔ/Y (n = 12) and dskPB/Df (n = 11) mutants (P < 0.001). Mean quantal content was determined for each recording by dividing the mean super-threshold EJP amplitude by the mean mEJP amplitude. EJP amplitudes were corrected for nonlinear summation before calculation of quantal content. Error bars denote SEM. ***, P < 0.001(compared with WT control).
Discussion
Using an unbiased, forward genetic approach, we discovered that CCKLR, a GPCR, together with its presumptive ligand, DSK, are positive regulators of NMJ growth in Drosophila. In addition, we demonstrate that DSK/CCKLR mediates NMJ development via the cAMP–PKA–CREB pathway, which is also required for learning and memory. These findings reveal a novel role for neuropeptide signaling in synaptic development and suggest a possible mechanistic link between pathways regulating synaptic growth and those regulating long-term synaptic plasticity.
A role for DSK/CCKLR signaling in NMJ growth regulation
Neuropeptides, whose effects have been extensively studied at NMJs, are usually described as neuromodulators because they modify the strength of synaptic transmission. For example, proctolin can potentiate the action of glutamate at certain NMJs in insects (Adams and O’Shea, 1983). However, involvement of neuropeptides in regulating neural development has not been well characterized. Recently, a C-type natriuretic peptide acting through a cGMP signaling cascade was found to be required for sensory axon bifurcation in mice (Schmidt et al., 2009), which suggests that neuropeptides may have a broader role in development than previously appreciated. Our studies demonstrate that DSK and its receptor, CCKLR, are strong positive regulators of NMJ growth in Drosophila.
DSK belongs to the family of FMRFamide-related peptides (FaRPs), which is very broadly distributed across invertebrate and vertebrate phyla. Originally identified in clams (Price and Greenberg, 1977), FaRPs affect heart rate, blood pressure, gut motility, feeding behavior, and reproduction in invertebrates (Li et al., 1999; Nichols, 2006; Audsley and Weaver, 2009). They have been shown to enhance synaptic efficacy at NMJs in locust and to modulate presynaptic Ca2+ channel activity in crustaceans (Robb and Evans, 1994; Rathmayer et al., 2002). In Drosophila, various neuropeptides derived from the FMRFa gene can modulate the strength of muscle contraction when perfused onto standard larval nerve–muscle preparations (Hewes et al., 1998). To these previously described functions of FaRPs, we now add a new role as a positive regulator of NMJ development.
DSK/CCKLR signals through the cAMP–PKA–CREB pathway to regulate NMJ development
Transgenic rescue experiments, RNAi expression, and overexpression of WT CCKLR demonstrate that CCKLR functions presynaptically in motor neurons to promote NMJ growth. Downstream components of this pathway were identified on the basis of known biochemistry of GPCRs and phenotypic interactions in double mutant combinations. GPCRs typically function by activating second messenger pathways via G proteins. Because loss-of-function mutations in dgs, which encodes the Gsα subunit in Drosophila, cause NMJ undergrowth (Wolfgang et al., 2004), we hypothesized that CCKLR signals through Gsα. Consistent with this idea, we found that presynaptic constitutively active dgs overexpression rescues the NMJ undergrowth phenotype of CCKLR mutants. Conversely, we observe dominant dose-dependent interactions between CCKLR-null mutations and mutations of rut, which encodes an AC; or PKA-C1, which encodes a cAMP-dependent protein kinase, resulting in significant reductions in NMJ growth. These data place CCKLR together with the other genes in a common cAMP-dependent signaling pathway that regulates NMJ growth.
It is known that the AC encoded by rut is activated by Gsα, and on the basis of our results, we propose that it is downstream of CCKLR signaling. However, the NMJ undergrowth in rut1, which is a presumptive null mutant, is not as severe as that of a CCKLR-null mutant. This is likely due to the fact that the Drosophila genome contains up to seven different AC-encoding genes, all of which are stimulated by Gsα (Cann and Levin, 1998). Presumably, one or more additional AC-encoding genes share some functional overlap with rut in regulation of NMJ growth. This idea is in good agreement with the results of Wolfgang et al., (2004), who found that the NMJ undergrowth phenotype of rut1 is weaker than that of dgs mutants, which they also interpreted as an indication that multiple ACs are activated by the Gsα encoded by dgs.
The primary effector of this pathway is CREB2, a transcriptional regulatory protein that is activated upon phosphorylation by PKA. Consistent with the idea that CCKLR ultimately acts via activation of CREB, loss-of-function mutations of dCreb2 or neuronal overexpression of a dominant-negative dCreb2 transgene cause NMJ undergrowth similar to that of CCKLR-null mutants. Additionally, loss of one copy of dCreb2 in a CCKLR heterozygous background also causes NMJ undergrowth, and overexpression of WT dCreb2 fully rescues the NMJ undergrowth phenotype of CCKLR null, even leading to NMJ overgrowth. Thus, regulation of NMJ growth through the CCKLR signaling pathway is clearly mediated by dCreb2, whose activity is itself necessary and sufficient for regulating NMJ growth. This conclusion differs from Davis et al. (1996), who suggested that dCreb2 is required for NMJ function but not NMJ growth. One possible explanation for this discrepancy is that a weaker, inducible heat shock–driven transgene was used to express dCreb2 in the earlier work, whereas strong constitutive neuronal drivers were used here. In any case, our results demonstrate that in addition to its known role in NMJ function (Davis et al., 1996), CREB2 is also a strong positive regulator of NMJ growth and is likely to play a greater role in structural plasticity of synapses in learning and memory in Drosophila than previously suggested. This conclusion is consistent with a recent study indicating that sprouting of type II larval NMJs in response to starvation is stimulated by a cAMP/CREB-dependent pathway via activation of an octopamine GPCR (Koon et al., 2011).
DSK/CCKLR signaling is required for normal synaptic function
In addition to being undergrown, NMJs in CCKLR mutant larvae also exhibit a functional deficit. This is perhaps less straightforward than it might seem. Previous analyses of mutations affecting growth of the larval NMJ in Drosophila have shown that there is no simple correlation between the size and complexity of the NMJ and the amplitude of EJPs or amount of neurotransmitter release. These discrepancies arise because of various homeostatic compensatory mechanisms and because some of the affected signaling pathways alter synaptic growth and synaptic function in different ways via distinct downstream targets. For example, a mutation in highwire, which has the most extreme NMJ overgrowth phenotype described, is associated with a decrease in synaptic transmission (Wan et al., 2000). Wallenda mutations have been shown to fully suppress the overgrowth phenotype, but have no effect on the deficit in synaptic transmission (Collins et al., 2006).
In the case of CCKLR mutants, however, there appears to be a very good correspondence between the morphological phenotype and the electrophysiological phenotype: the reduction in the total number of active zones in CCKLR larvae as measured morphologically correlates very well with the reduction in quantal content we observe. In addition, we did not detect any difference in CCKLR mutants in calcium sensitivity of transmitter release or in the size or frequency of spontaneous release events. Thus, the synaptic growth phenotype of CCKLR mutant NMJs is sufficient to account for the functional phenotype. However, we cannot rule out the possibility that DSK/CCKLR signaling also exerts some modulatory effect on NMJ function that is distinct from its effect on NMJ development.
DSK as the CCKLR ligand
DSK is identified as the Drosophila orthologue of CCK, the ligand of CCKLR in vertebrates, on the basis of sequence analysis. Our genetic analysis strongly supports the conclusion that DSK is the ligand of CCKLR at the larval NMJ. First, mutations of dsk and expression of dsk RNAi result in NMJ undergrowth phenotypes similar to that of CCKLR mutants. Second, loss of one copy of both dsk and CCKLR in double heterozygotes results in NMJ undergrowth. Third, heterozygosity for dsk does not further enhance the phenotype of a CCKLR-null mutant as expected if DSK regulates NMJ growth through its action on CCKLR. Fourth, overexpression of UAS-dsk does not rescue the undergrowth phenotype of a CCKLR-null mutant, but CCKLR overexpression can rescue NMJ undergrowth of a dsk hypomorphic mutant.
The discovery of an entirely novel role for neuropeptide signaling in NMJ growth raises several questions about how this signaling is regulated and the biological significance of this mechanism. Although answers to these questions will require much additional work, an immediate question is whether a paracrine or autocrine mechanism is involved. In the case of octopamine-mediated synaptic sprouting in response to starvation, both autocrine and paracrine signaling are involved in the sprouting of type II and type I NMJs, respectively (Koon et al., 2011). In an early immunohistochemical investigation, Nichols (1992) reported that DSK was detected in medial neurosecretory cells in the larval CNS that extended projections anteriorly into the brain and posteriorly to the ventral ganglion (Nichols, 1992). As we were unable to obtain the original DSK antiserum and were unsuccessful in raising our own antiserum, we have not been able to confirm or extend the previous report. Instead, we performed tissue-specific RNAi experiments to examine the spatial requirement for DSK. Pan-neuronal dsk RNAi expression indicates that DSK expression in neurons is required to promote NMJ growth. In addition, C739-Gal4–driven dsk RNAi also causes NMJ undergrowth, whereas OK-Gal4–driven dsk RNAi in motor neurons does not. The expression pattern of C739-Gal4 overlaps with the DSK-positive cells previously identified by immunohistochemistry, which suggests that DSK produced by those neurosecretory cells is required for normal NMJ growth. Thus, from available data, it seems most likely that DSK is acting in paracrine fashion to regulate NMJ growth. However, further investigation will be necessary to determine the exact source of the DSK that promotes NMJ growth to fully understand how this neuropeptide regulates NMJ development.
Linking synaptic growth and synaptic plasticity
Studies have demonstrated a role for CREB in long-term synaptic plasticity—structural changes in synaptic morphology that underlie the formation of long-term memories (Benito and Barco, 2010). Here we show that in addition to CREB’s role in structural modification of synapses in response to experience after development is complete, it is also a key regulator of growth and morphology during development of the larval NMJ. Moreover, although CREB is the transcriptional effector for many GPCRs, the fact that NMJs in CCKLR mutants are as undergrown as those of CREB mutants suggests that DSK/CCKLR signaling is a major input to CREB during NMJ growth. Many of the genes encoding intermediate components of the pathway such as dnc, rut, and PKA also have effects on NMJ growth and development as well as on synaptic plasticity and learning and memory, further emphasizing an overlap between the mechanisms that regulate synaptic growth during development and those that regulate postdevelopmental structural synaptic plasticity. These results raise the possibility that DSK/CCKLR signaling also plays a role in long-term synaptic plasticity and learning as well as in synaptic development.
Materials and methods
Fly stocks
w1118 was used as WT control for genetic background. B185-Gal4 was provided by G. Davis (University of California, San Francisco, San Francisco, CA). rut1 was provided by C.F. Wu (University of Iowa, Iowa City, IA). UAS-Creb2.b was provided by J. Yin (University of Wisconsin-Madison, Madison, WI). C155-Gal4 UAS-Dcr was provided by K. O’Connor-Giles (University of Wisconsin-Madison). The following fly lines were obtained from the Bloomington Stock Center: Df(1)fu-A7, Df(1)Exel9051, Df(1)ED7413, Dp(1;3)DC351, Dp(1;3) DC352, Dp(3;1)2-2, Df(3R)2-2, Df(1)osUE69, Df(2L)N22-14, CCKLRMB02688, CCKLR-17D3KG01373, C155-Gal4, Elav-Gal4, 24B-Gal4, OK6-Gal4, C739-Gal4, dskEP3632, UAS-dgsQ215L, PKA-C1BG02142, dCreb2S162, dCreb2EP1378, and UAS-Creb2.a (Perazzona et al., 2004). UAS-CCKLR-RNAi (no. 7231) and UAS-dsk-RNAi (no. 14201) were obtained from the Vienna Drosophila RNAi Center. dskf02468 was obtained from the Exelixis collection at Harvard Medical School. CCKLRΔ C155-Gal4 was derived by recombination between Df(1)Exel9051 and C155-Gal4 and confirmed by PCR for the CCKLR deletion and C155-Gal4–driven UAS-CD8–GFP expression pattern. rut1 CCKLRΔ was built by recombination between Df(1)Exel9051 and rut1 and confirmed by PCR for the CCKLR deletion and sequencing for the rut1 lesion. All stocks were raised on cornmeal/molasses fly medium and maintained at RT (21–23°C).
Immunohistochemistry
Wandering third instar larvae were dissected in Ca2+-free ice-cold saline and fixed in 4% paraformaldehyde for 20 min. Larvae were incubated in primary antibodies overnight at 4°C or for 2–3 h at room temperature and in secondary antibodies for 1–2 h at room temperature, then mounted in VectaShield (Vector Laboratories) for microscopic analysis. We used the following antibodies: FITC-conjugated or Cy5-conjugeted anti-HRP at 1:100 (Jackson ImmunoResearch Laboratories, Inc.), mouse anti-Elav at 1:500 (Developmental Studies Hybridoma Bank), mouse anti-Brp (nc82) at 1:50 (Developmental Studies Hybridoma Bank), rabbit anti-GluRIIB, and rabbit anti-GluRIIC (DiAntonio laboratory, Washington University, St. Louis, MO). To detect CCKLR–mCherry fusion protein, rabbit anti-DsRed (Takara Bio Inc.) was used at 1:500. Specific secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 633 (Invitrogen) were used at 1:200.
Imaging and quantification
Although we concentrated on NMJ4 because of its relative simplicity, we also observed comparable results at NMJ6/7 as well as other NMJs in the mutant larvae. Quantification of bouton number was performed at NMJ4 (Ib) in segments A2–A4. Boutons were scored by counting each distinct spherical anti-HRP stained varicosity along motor terminals. Bouton numbers for each genotype are summarized in Table S1. The muscle size of all larvae examined was similar. Corresponding controls were processed and quantified simultaneously with samples for each experiment. Statistical analyses were performed via analysis of variance and Student’s t tests for multiple comparisons. Significance levels of <0.05, <0.01, or <0.001 are indicated by one, two, and three asterisks, respectively. Error bars denote SEM. Confocal images were obtained on a microscope (LSM 510; Carl Zeiss) with Plan-Apochromat 20× NA 10.8 and EC Plan-Neofluar 40× NA 1.3 oil objective lenses and the accompanying software. Images were processed in the LSM image browser (Carl Zeiss). Brightness and contrast were adjusted using Photoshop (Adobe). All images were captured at room temperature (21–23°C).
Molecular cloning
UAS-CCKLR comprises a full-length CCKLR cDNA cloned into pUAST. Full-length cDNA was obtained by PCR amplification using pOT2-IP18747 full insert cDNA clone (731–2,022 bp; Berkeley Drosophila Genome Project) and genomic DNA of WT flies (1–730 bp), and ligation of the two fragments. UAS-CCKLR–mCherry was generated by cloning CCKLR cDNA into the pUAS-C5 attB-C-term mCherry vector. UAS-dsk comprises a full-length dsk cDNA cloned into pUAST-attB. cDNA constructs for dsk were generated by amplification of the coding sequence using genomic DNA of WT flies. After germline transformation, UAS-CCKLR, UAS-CCKLR–mCherry, and UAS-dsk were inserted on chromosome 2 in a w1118 background, respectively. Primers flanking the predicted open reading frames incorporated restriction sites to facilitate directional cloning into the pUAST or pUAST-attB vector. The sequences of primers used are provided in Table S2. All receptor constructs were fully sequenced.
Electrophysiology
Intracellular recordings were performed at RT on muscle 6 (segments A2 or A3) or wandering third instar larvae as described previously (Petersen et al., 1997). Larvae were dissected in physiological saline HL-3 (Stewart et al., 1994) containing 0.5 mM Ca2+. Data were considered valid when the resting membrane potential was between −60 and −80 mV, and the input resistance of the muscle was >5 MΩ. The mEJP amplitudes were measured by hand using the cursor option in Clampfit software (Axon Instrument), and mean mEJP amplitudes were calculated from mEJPs. Mean EJP size was calculated by measuring the amplitude of the computer-generated trace average of 50 consecutive EJP traces. Quantal content was estimated by dividing the mean EJP by the mean mEJP, and corrected for nonlinear summation according to Martin (1955).
Online supplemental material
Fig. S1 shows that CCKLR-17D3 does not interact with CCKLR-17D1 and does not affect NMJ growth. Fig. S2 shows that CCKLR–mCherry rescues CCKLRΔ/Y and localizes to motor neurons. Fig. S3 shows that the C739-Gal4 expression pattern overlaps with the previously reported DSK antiserum staining pattern. Fig. S4 shows that GluRIIB staining appears normal in CCKLRΔ/Y and dskPB/Df larvae. Table S1 presents quantification of bouton numbers for each genotype. Table S2 presents sequences of primers used in constructs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109044/DC1.
Acknowledgments
We are grateful to Chun-Fang Wu, Jerry Yin, Graeme Davis, Kate O’Connor-Giles, the Bloomington Stock Center, and the Vienna Drosophila RNAi Center for providing fly stocks used in these experiments. We thank all members of the Ganetzky laboratory, Kate O’Connor-Giles, and Grace Boekhoff-Falk for helpful discussion and critical comments on the manuscript.
This research was supported by a grant from the National Institutes of Health (NS15390) to B. Ganetzky.
Footnotes
Abbreviations used in this paper:
- AC
- adenylyl cyclase
- Brp
- Bruchpilot
- CCK
- cholecystokinin
- CCKLR
- CCK-like receptor
- CNS
- central nervous system
- CREB
- cAMP-response element binding protein
- DSK
- drosulfakinin
- EJP
- excitatory junction potential
- FaRP
- FMRFamide-related peptide
- GPCR
- G protein–coupled receptor
- mEJP
- miniature EJP
- NMJ
- neuromuscular junction
- rut
- rutabaga
- UAS
- upstream activating sequence
- WT
- wild type
References
- Adams M.E., O’Shea M. 1983. Peptide cotransmitter at a neuromuscular junction. Science. 221:286–289 10.1126/science.6134339 [DOI] [PubMed] [Google Scholar]
- Alberini C.M. 2009. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 89:121–145 10.1152/physrev.00017.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audsley N., Weaver R.J. 2009. Neuropeptides associated with the regulation of feeding in insects. Gen. Comp. Endocrinol. 162:93–104 10.1016/j.ygcen.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Benito E., Barco A. 2010. CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 33:230–240 10.1016/j.tins.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Cann M.J., Levin L.R. 1998. Genetic characterization of adenylyl cyclase function. Adv. Second Messenger Phosphoprotein Res. 32:121–135 [DOI] [PubMed] [Google Scholar]
- Collins C.A., DiAntonio A. 2007. Synaptic development: insights from Drosophila. Curr. Opin. Neurobiol. 17:35–42 10.1016/j.conb.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Collins C.A., Wairkar Y.P., Johnson S.L., DiAntonio A. 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 51:57–69 10.1016/j.neuron.2006.05.026 [DOI] [PubMed] [Google Scholar]
- Davis G.W., Schuster C.M., Goodman C.S. 1996. Genetic dissection of structural and functional components of synaptic plasticity. III. CREB is necessary for presynaptic functional plasticity. Neuron. 17:669–679 10.1016/S0896-6273(00)80199-3 [DOI] [PubMed] [Google Scholar]
- Davis G.W., Schuster C.M., Goodman C.S. 1997. Genetic analysis of the mechanisms controlling target selection: target-derived Fasciclin II regulates the pattern of synapse formation. Neuron. 19:561–573 10.1016/S0896-6273(00)80372-4 [DOI] [PubMed] [Google Scholar]
- Featherstone D.E., Broadie K. 2000. Surprises from Drosophila: genetic mechanisms of synaptic development and plasticity. Brain Res. Bull. 53:501–511 10.1016/S0361-9230(00)00383-X [DOI] [PubMed] [Google Scholar]
- Gilman A.G. 1987. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615–649 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- Hewes R.S., Snowdeal E.C., III, Saitoe M., Taghert P.H. 1998. Functional redundancy of FMRFamide-related peptides at the Drosophila larval neuromuscular junction. J. Neurosci. 18:7138–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon A.C., Ashley J., Barria R., DasGupta S., Brain R., Waddell S., Alkema M.J., Budnik V. 2011. Autoregulatory and paracrine control of synaptic and behavioral plasticity by octopaminergic signaling. Nat. Neurosci. 14:190–199 10.1038/nn.2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin L.R., Han P.L., Hwang P.M., Feinstein P.G., Davis R.L., Reed R.R. 1992. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 68:479–489 10.1016/0092-8674(92)90185-F [DOI] [PubMed] [Google Scholar]
- Li C., Kim K., Nelson L.S. 1999. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res. 848:26–34 10.1016/S0006-8993(99)01972-1 [DOI] [PubMed] [Google Scholar]
- Manseau L., Baradaran A., Brower D., Budhu A., Elefant F., Phan H., Philp A.V., Yang M., Glover D., Kaiser K., et al. 1997. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209:310–322 [DOI] [PubMed] [Google Scholar]
- Martin A.R. 1955. A further study of the statistical composition on the end-plate potential. J. Physiol. 130:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCudden C.R., Hains M.D., Kimple R.J., Siderovski D.P., Willard F.S. 2005. G-protein signaling: back to the future. Cell. Mol. Life Sci. 62:551–577 10.1007/s00018-004-4462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D.R. 2002. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 68:1–84 10.1016/S0301-0082(02)00057-6 [DOI] [PubMed] [Google Scholar]
- Nässel D.R., Winther A.M. 2010. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92:42–104 10.1016/j.pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Neves S.R., Ram P.T., Iyengar R. 2002. G protein pathways. Science. 296:1636–1639 10.1126/science.1071550 [DOI] [PubMed] [Google Scholar]
- Nichols R. 1992. Isolation and expression of the Drosophila drosulfakinin neural peptide gene product, DSK-I. Mol. Cell. Neurosci. 3:342–347 10.1016/1044-7431(92)90031-V [DOI] [PubMed] [Google Scholar]
- Nichols R. 2003. Signaling pathways and physiological functions of Drosophila melanogaster FMRFamide-related peptides. Annu. Rev. Entomol. 48:485–503 10.1146/annurev.ento.48.091801.112525 [DOI] [PubMed] [Google Scholar]
- Nichols R. 2006. FMRFamide-related peptides and serotonin regulate Drosophila melanogaster heart rate: mechanisms and structure requirements. Peptides. 27:1130–1137 10.1016/j.peptides.2005.07.032 [DOI] [PubMed] [Google Scholar]
- Perazzona B., Isabel G., Preat T., Davis R.L. 2004. The role of cAMP response element-binding protein in Drosophila long-term memory. J. Neurosci. 24:8823–8828 10.1523/JNEUROSCI.4542-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S.A., Fetter R.D., Noordermeer J.N., Goodman C.S., DiAntonio A. 1997. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 19:1237–1248 10.1016/S0896-6273(00)80415-8 [DOI] [PubMed] [Google Scholar]
- Price D.A., Greenberg M.J. 1977. Structure of a molluscan cardioexcitatory neuropeptide. Science. 197:670–671 10.1126/science.877582 [DOI] [PubMed] [Google Scholar]
- Rathmayer W., Djokaj S., Gaydukov A., Kreissl S. 2002. The neuromuscular junctions of the slow and the fast excitatory axon in the closer of the crab Eriphia spinifrons are endowed with different Ca2+ channel types and allow neuron-specific modulation of transmitter release by two neuropeptides. J. Neurosci. 22:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renden R.B., Broadie K. 2003. Mutation and activation of Galpha s similarly alters pre- and postsynaptic mechanisms modulating neurotransmission. J. Neurophysiol. 89:2620–2638 10.1152/jn.01072.2002 [DOI] [PubMed] [Google Scholar]
- Robb S., Evans P. 1994. The modulatory effect of SchistoFLRFmide on heart and skeletal muscle in the locust Schistocerca gregaria. J. Exp. Biol. 197:437–442 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Stonkute A., Jüttner R., Koesling D., Friebe A., Rathjen F.G. 2009. C-type natriuretic peptide (CNP) is a bifurcation factor for sensory neurons. Proc. Natl. Acad. Sci. USA. 106:16847–16852 10.1073/pnas.0906571106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C.M., Davis G.W., Fetter R.D., Goodman C.S. 1996a. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 17:641–654 10.1016/S0896-6273(00)80197-X [DOI] [PubMed] [Google Scholar]
- Schuster C.M., Davis G.W., Fetter R.D., Goodman C.S. 1996b. Genetic dissection of structural and functional components of synaptic plasticity. II. Fasciclin II controls presynaptic structural plasticity. Neuron. 17:655–667 10.1016/S0896-6273(00)80198-1 [DOI] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wang J., Wu C.F. 1994. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 175:179–191 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
- Tettamanti M., Armstrong J.D., Endo K., Yang M.Y., Furukubo-Tokunaga K., Kaiser K., Reichert H. 1997. Early development of the Drosophila mushroom bodies, brain centres for associative learning and memory. Dev. Genes Evol. 207:242–252 10.1007/s004270050112 [DOI] [PubMed] [Google Scholar]
- Wan H.I., DiAntonio A., Fetter R.D., Bergstrom K., Strauss R., Goodman C.S. 2000. Highwire regulates synaptic growth in Drosophila. Neuron. 26:313–329 10.1016/S0896-6273(00)81166-6 [DOI] [PubMed] [Google Scholar]
- Wolfgang W.J., Clay C., Parker J., Delgado R., Labarca P., Kidokoro Y., Forte M. 2004. Signaling through Gs alpha is required for the growth and function of neuromuscular synapses in Drosophila. Dev. Biol. 268:295–311 10.1016/j.ydbio.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Budnik V., Wu C.F. 1992. Synaptic plasticity in Drosophila memory and hyperexcitable mutants: role of cAMP cascade. J. Neurosci. 12:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]