Alternative NF-κB signaling modulates the activity of PGC-1β to promote oxidative metabolism in skeletal muscle.
Abstract
Although the physiological basis of canonical or classical IκB kinase β (IKKβ)–nuclear factor κB (NF-κB) signaling pathway is well established, how alternative NF-κB signaling functions beyond its role in lymphoid development remains unclear. In particular, alternative NF-κB signaling has been linked with cellular metabolism, but this relationship is poorly understood. In this study, we show that mice deleted for the alternative NF-κB components IKKα or RelB have reduced mitochondrial content and function. Conversely, expressing alternative, but not classical, NF-κB pathway components in skeletal muscle stimulates mitochondrial biogenesis and specifies slow twitch fibers, suggesting that oxidative metabolism in muscle is selectively controlled by the alternative pathway. The alternative NF-κB pathway mediates this specificity by direct transcriptional activation of the mitochondrial regulator PPAR-γ coactivator 1β (PGC-1β) but not PGC-1α. Regulation of PGC-1β by IKKα/RelB also is mammalian target of rapamycin (mTOR) dependent, highlighting a cross talk between mTOR and NF-κB in muscle metabolism. Together, these data provide insight on PGC-1β regulation during skeletal myogenesis and reveal a unique function of alternative NF-κB signaling in promoting an oxidative metabolic phenotype.
Introduction
Skeletal muscle is a dynamic tissue and a major site for energy storage that adapts to a variety of metabolic demands, including growth, exercise, and starvation. Such regulation is managed by signaling pathways acting through transcription factors that control expression of specialized genes and their protein products to control the differentiation as well as bioenergetic capacity of skeletal muscle cells. Of these signaling pathways, increasing attention has been placed on nuclear factor κB (NF-κB), which is emerging as a major regulator of skeletal muscle both in development and disease (Peterson et al., 2011).
Skeletal muscle differentiation or myogenesis orchestrates the fusion of proliferating myoblasts into cell cycle–arrested multinucleated contractile myotubes (Sabourin and Rudnicki, 2000; Tedesco et al., 2010). Evidence indicates that during myogenesis, two distinct signaling pathways direct NF-κB function. In the more familiar canonical or classical pathway, NF-κB is activated through the IκB kinase (IKK) complex comprised of the IKK2/IKKβ subunit whose catalytic activity, in coordination with IKKγ/NEMO, induces the phosphorylation and subsequent degradation of the inhibitor IκB protein, thus allowing the predominant p65–p50 heterodimer complex to translocate to the nucleus and activate gene expression (Hayden and Ghosh, 2008). Studies support that classical NF-κB is constitutively active (CA) in proliferating myoblasts and functions to inhibit differentiation (Guttridge et al., 1999, 2000; Dogra et al., 2006; Bakkar et al., 2008; Wang et al., 2008). This activity is mediated by several mechanisms that either reduce the rate at which myoblasts exit from the cell cycle or limit the expression of required myogenic regulators and myofibrillar genes (Guttridge et al., 1999, 2000; Dogra et al., 2006; Wang et al., 2007). Chronic stimulation of classical NF-κB also contributes to multiple muscle disorders such as cachexia (Guttridge et al., 2000; Cai et al., 2004; Mourkioti et al., 2006), muscular dystrophy (Acharyya et al., 2007), and rhabdomyosarcoma (Wang et al., 2008).
In comparison, noncanonical or alternative NF-κB signaling is regulated by an IKK1–IKKα homodimer complex responsible for phosphorylating the p100 precursor subunit of NF-κB and in turn promoting partial p100 proteolysis to produce the p52 subunit (Hayden and Ghosh, 2008). This subunit forms into a RelB–p52 heterodimer complex that translocates to the nucleus to activate transcription of a selected set of genes considered distinct from the classical pathway (Dejardin et al., 2002; Bonizzi et al., 2004). In addition, unlike the CA antimyogenic activity of classical NF-κB, alternative signaling is induced relatively late in the myogenic program, only after myotubes begin to form (Bakkar et al., 2008). Evidence from cultured muscle cells also indicates that activation of alternative NF-κB is not required for myotube formation but may instead promote mitochondrial biogenesis and maintain myotube homeostasis (Bakkar et al., 2008).
Similar to skeletal myogenesis, mitochondrial biogenesis and oxidative respiration in muscle cells are tightly regulated by gene products that function to replicate mitochondrial DNA, assemble electron transport chain complexes, and regulate the fission of newly formed organelles (Scarpulla, 2008; Shaw and Winge, 2009). Mitochondrial induction causes a metabolic shift in muscle cells, switching from glycolysis in proliferating myoblasts to oxidative phosphorylation in postmitotic myotubes. The basis for this switch is presumably to accommodate the energy demands for a newly formed contractile myofiber (Kraft et al., 2006). Mitochondrial biogenesis is regulated by a set of transcription factors that include NRF (nuclear respiratory factor) 1 and 2, mtTFA (mitochondrial transcription factor A), the PPARs (peroxisome proliferator-activated receptors), and PPAR-γ coactivator 1α (PGC-1α) and PGC-1β transcriptional coactivators (Kelly and Scarpulla, 2004; Spiegelman, 2007). PGC-1 proteins additionally serve as mediators between various external physiological stress conditions that enhance mitochondrial activity as well as drive the formation of slow type I myofibers and their associated oxidative metabolic phenotype (Arany, 2008). Relatively little is known about what controls PGC-1 gene expression or other mitochondrial regulators, although some myogenic factors have been identified (Czubryt et al., 2003; Chang et al., 2006). The nutrient sensor, mammalian target of rapamycin (mTOR), was also shown to promote mitochondrial oxidative function (Schieke et al., 2006) through transcriptional control of PGC-1α (Cunningham et al., 2007).
Although previous links have been made with NF-κB and mitochondria, these associations have been primarily limited to classical signaling (Bottero et al., 2001; Cogswell et al., 2003; Guseva et al., 2004), and even with some evidence implicating the alternative pathway (Bakkar et al., 2008), the physiological relevance and mechanism by which alternative signaling regulates mitochondria remain unknown. In this study, we demonstrate by genetic means that mitochondrial biogenesis during muscle differentiation is specific to alternative NF-κB, and such regulation is mediated by direct transcriptional regulation of PGC-1β but not PGC-1α. Furthermore, we reveal that activation of the alternative pathway is controlled by mTOR and that mTOR itself promotes mitochondrial biogenesis through NF-κB. Collectively, these findings offer a fresh perspective on the contrasting roles of NF-κB signaling pathways and identify a connection between PGC-1β and alternative NF-κB required for the production of mitochondria and the resulting oxidative respiration phenotype in differentiating muscle cells.
Results
IKKα and RelB are regulators of mitochondria and oxidative muscle metabolism
Previous results using murine immortalized C2C12 myoblast cells implicated alternative NF-κB as a regulator of mitochondria during muscle cell differentiation (Bakkar et al., 2008), but how this occurs and whether this regulation is relevant in vivo are not known. To make these determinations, we used mice deficient in alternative signaling components IKKα and RelB. Although IKKα−/− mice are perinatally lethal because of complications in skin development (Hu et al., 1999; Li et al., 1999; Takeda et al., 1999), limb muscles in these mice, including fiber number in soleus, which was measureable at this development stage, were comparable with wild type (657 ± 31 in IKKα+/+ vs. 688 ± 61 in IKKα−/−). We were also unable to detect differences in general muscle gene expression and myogenesis by myotube formation or troponin I–luciferase reporter activity between multiple pairs of IKKα+/+ and IKKα−/− primary myoblast lines (Fig. S1, A–C). However, upon assessing mitochondrial content, both RNA and protein markers of mitochondria were reproducibly reduced (n = 8 litters) in IKKα−/− limb muscles (Fig. 1, A and B). Similar findings were observed from primary myotube cultures (Fig. S1 D), indicating that changes observed in limb muscles likely derived from muscle cells themselves rather than other cell types localized within the muscle fascicle. In addition, mitochondrial DNA copy number was significantly decreased in IKKα−/− limbs (Fig. 1 C). This reduction was accompanied by increases in the phosphorylation of the energy sensor AMP-activated protein kinase (Fig. 1 B), which in response to high AMP to ATP ratios becomes activated to stimulate skeletal muscle fatty acid oxidation and glucose uptake (Winder, 2001). Therefore, AMP-activated protein kinase activation in IKKα−/− muscles was indicative of a state of energy deprivation, possibly resulting from reduced mitochondrial content.
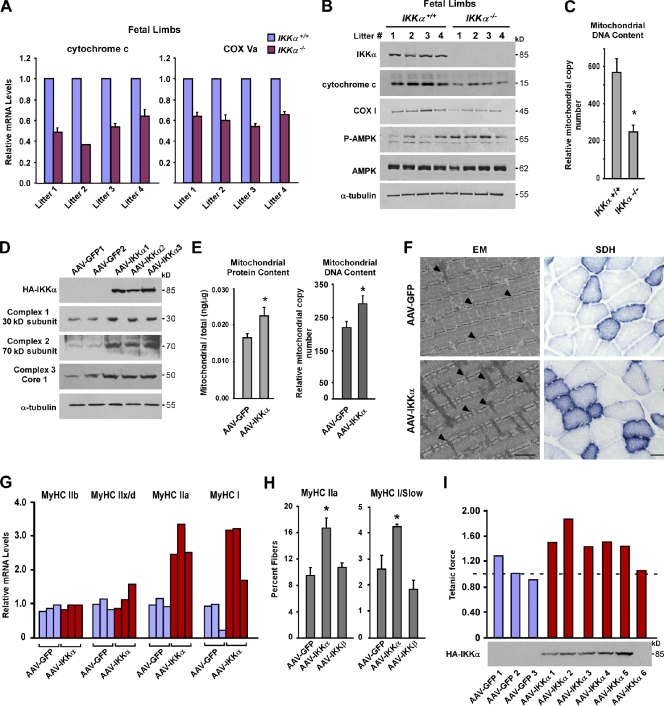
Figure 1.
IKKα regulates mitochondrial biogenesis and fiber type specification in vivo. (A) Hind limbs from four independent litters of IKKα+/+ and IKKα−/− embryos (total n = 8) were isolated at E20. Cytochrome c and COX Va were measured by real-time quantitative RT-PCR. (B) Additional tissues from E20 limbs from A were homogenized and probed by Western blotting. (C) Total DNA was extracted from E20 limbs, and real-time RT-PCR analysis was performed to quantitate mitochondrial and nuclear (GAPDH) genome copy number. Results are means of five mice per genotype and calculated as fold mitochondrial over genomic DNA. P = 0.004. (D) AAV-GFP and AAV-IKKα viruses were injected into the TA. Mice were sacrificed after 3–4 mo, and protein lysates were prepared and analyzed by Western blotting. Shown is a representative blot from five independent experiments. (E and F) After similar viral injections as described in D, mitochondria were fractionated, and protein concentrations or DNA content was determined and normalized to total protein/DNA content (E; n = 5 per group; P < 0.005) or TA muscles were analyzed by EM (F). Bar, 1 µm. Parallel muscles were stained for SDH. Bar, 200 µm. Arrowheads point to individual mitochondria. (G) TA muscles were isolated from AAV-GFP– and AAV-IKKα–injected mice (n = 3 per group, repeated five times), and myosin isoforms were measured by real-time RT-PCR. The data shown are from a single representative experiment out of three replicates. (H) Sections from AAV-GFP– and AAV-IKKα–injected mice (n = 4) were immunostained with MyHC IIa and I forms, and positive fibers were expressed as a percentage of total fibers in each section (n = 3 sections per muscle per mouse). (I) EDL muscles were isolated from AAV-GFP (n = 3)– or AAV-IKKα (n = 6)–injected mice, and force measurements were made after isometric contractions. Values are expressed as a ratio of the force generated over values from uninjected contralateral muscles. The dotted line shows levels of 1. Values shown are from a single experiment. Asterisks denote significance. Error bars are means ± SEM. P, phospho; AMPK, AMP-activated protein kinase.
To further test the in vivo function of IKKα, we used an adeno-associated virus (AAV) delivery system, analogous to other systems studying skeletal muscle metabolism (Lustig et al., 2011). Consistent with IKKα−/− results, overexpression of IKKα in tibialis anterior (TA) and extensor digitorum longus (EDL) muscles enhanced total mitochondrial proteins and genomes as well as increased expression of mitochondrial gene products and oxidative respiratory complexes (Fig. 1, D and E; and Fig. S1 E). Histologically, no effects on muscle architecture were apparent even after 6 mo of IKKα expression, as observed in 29 out of 30 mice carrying the transgene (Fig. S1 F). In sharp contrast, viral delivery in limb muscles of the NF-κB classical pathway component IKKβ (Fig. S1 G) caused a runted phenotype, which in turn led to either early lethality (8/28 cases), complete clearance of the IKKβ transgene (15/28 cases), or severe muscle inflammation (1/28 cases). Importantly, of the remaining cases (4/28), no associated changes in mitochondrial genes were detected as observed with IKKα (unpublished data). Although transgenic expression of a CA form of IKKβ in skeletal muscle was shown to cause extensive muscle atrophy (Cai et al., 2004), we were unable to observe a similar phenotype using the wild-type version of IKKβ, which likely derives from differences in expression systems and isoforms of IKKβ used in these respective experiments. Nevertheless, our data are consistent with a previous study demonstrating that up-regulated IKKβ has a negative outcome on skeletal muscle (Cai et al. 2004). In addition, in our hands, exogenous addition of IKKα CA induced p65 phosphorylation and concomitantly repressed myogenic activity (Fig. S1, H and I), which are two characteristic features of classical signaling activity in muscle cells (Bakkar et al., 2008). Because activation of classical NF-κB is favored by an IKKα–IKKβ heterodimer complex (Hayden and Ghosh, 2008), we suspect that overexpression of IKKα CA would induce muscle atrophy as was previously described (Van Gammeren et al., 2009). For these reasons, we restricted our analysis to the wild-type form of IKKα.
Next, ultrastructural and histological analyses were performed. By EM, muscle expression of IKKα caused a substantial 40% (P = 0.0008) increase in the number and elongation of mitochondria, (Fig. 1 F and Fig. S1 J, high magnification images), which is consistent with the increases found in mitochondrial proteins and genome numbers (Fig. 1, D and E). In addition, immunohistochemical staining of succinate dehydrogenase (SDH), as a measure of oxidative capacity, showed a similar 45% (P = 0.05) increase in both the number and staining intensity of SDH-positive fibers (Fig. 1 F). Together, these results support that IKKα regulates mitochondrial biogenesis in vivo, and this activity is distinct from classical IKKβ.
Because mitochondrial density varies among muscle fiber types, with higher levels contained in slow twitch, oxidative type I and intermediate type IIa myofibers compared with fast twitch, glycolytic type IIb fibers (Bassel-Duby and Olson, 2006), we sought to determine whether IKKα had a corresponding effect on myosin gene expression. Results showed that IKKα expression in TA and EDL muscles substantially increased levels of MyHC I and IIa subtypes but did not affect fast MyHC IIb or the glycolytic intermediate MyHC IIx/d (Fig. 1 G and Fig. S1, K and L). Consistent with these findings, in predominantly fast-type glycolytic TA muscles, IKKα also led to an impressive >75% increase in type IIa fibers (P = 0.008) with a less robust, but still significant, enhancement of type I slow fibers (P = 0.01; Fig. 1 H and Fig. S1 M). In line with our other analyses, similar changes in slow fiber types were not observed with IKKβ-expressing muscles, implicating a specificity of IKKα signaling in regulating fiber type specification.
Next, we reasoned that because IKKα increased myosin content and the fast twitch intermediate IIa fibers (Bassel-Duby and Olson, 2006), such characteristics should translate to overall contractile muscle force. To test this notion, maximal isometric tension was measured with EDL muscles isolated from mice injected with AAV-IKKα or AAV-GFP viruses and normalized to uninjected contralateral muscles. Compared with GFP, muscles expressing IKKα (n = 5) exhibited a 50% increase in contractile strength, whereas EDL negative for IKKα generated a peak contractile force that was similar to controls (1.03 ± 0.18 for GFP vs. 1.53 ± 0.19 for IKKα; P = 0.005; Fig. 1 I and Fig. S1 N). In contrast, IKKβ expression produced a 40% force deficit compared with control (unpublished data).
To determine whether regulation on mitochondria by IKKα was associated with alternative NF-κB, we repeated a similar analysis in RelB−/− mice (Wu et al., 1998). As observed with IKKα−/− muscles, those lacking RelB also contained reduced levels of mitochondrial gene products in both adult and neonatal limb muscles (Fig. 2 A and Fig. S2). To determine whether changes in these markers represented a more generalized effect on mitochondria, we performed global transcriptional profiling. Hierarchical clustering results revealed that a large number of genes involved in mitochondrial metabolism were down-regulated in soleus from RelB−/− compared with RelB+/+ littermates (n = 3; Fig. 2 B and Table S1). Gene ontology analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery) algorithm further showed that out of a total of 649 repressed genes, 271 were classified as mitochondrial or oxidative (Fig. 2 C). In agreement with transcriptional changes, soleus muscles from RelB−/− mice were remarkably pale in comparison to typical dark red oxidative wild-type muscles (Fig. 2 D). Furthermore, mitochondrial content and function declined in the absence of RelB (Fig. 2, E and F). This general loss in oxidative phenotype was tightly correlated with decreases in MyHC IIa, whereas no changes in MyHC I were observed (Fig. 2 G). In striking contrast, levels of MyHC IIb in RelB−/− soleus were pronouncedly elevated. This switch in myosin expression was suggestive of a metabolic shift from oxidative phosphorylation to glycolysis. In support of this notion, lactate production in RelB−/− TA was significantly increased (P = 0.008; Fig. 2 H). Moreover, oxygen consumption monitored over 3 d using a CLAMS (Comprehensive Laboratory Animal Monitoring System) revealed that RelB−/− mice exhibited a higher respiratory exchange ratio (RER) during total light–dark cycles (P = 0.05; Fig. 2 I), which is consistent with a shift toward glycolysis. In particular, RER levels were significantly more glycolytic during the nonfeeding day hours, a phenotype that was not caused by changes in mice ambulatory activity or food consumption (Fig. 2 I). Collectively, these data argue strongly that mitochondrial synthesis and an accompanying oxidative respiratory phenotype in skeletal muscles are regulated by alternative NF-κB signaling components IKKα and RelB.
Figure 2.
The alternative NF-κB subunit RelB regulates mitochondria and a glycolytic phenotype. (A) RNA was extracted from TA or soleus of 5-wk-old mice (n = 4 each), and mitochondrial genes were quantitated by real-time PCR. Asterisks denote significance for COX IVi in TA (P = 0.04) and soleus (P = 0.0006) and COX Va in TA (P = 0.03) and soleus (P = 0.003). (B) Microarray analysis from soleus of adult RelB+/+ and RelB−/− littermates. Mitochondrial genes that are most down-regulated are shown. Green and red colors indicate lower and higher RNA expression, respectively. (C) Gene ontology classification of down-regulated genes in RelB−/− soleus muscles (n = 3). Shown in parentheses is the number of genes that changes in each category. (D) Representative soleus from RelB+/+ and RelB−/− mice. (E) Total DNA was extracted from hind-limb muscles of 5-wk-old RelB+/+ and RelB−/− mice (n = 3 each), and relative mitochondrial to nuclear copy numbers were calculated for TA (P = 0.04), soleus (P = 0.001), and gastrocnemius (Gas; P = 0.02). (F) TA muscles from 4-wk-old mice were harvested and stained for SDH. Bar, 200 µm. (G) RNA was prepared from soleus of RelB+/+ and RelB−/− adult mice as in A, and levels of myosin isotypes were determined by real-time RT-PCR. Asterisk denotes significance for MyHC IIa (P = 0.02) and MyHC IIb (P = 0.01). (H) TA homogenates from RelB+/+ and RelB−/− mice (n = 4) were prepared and measured for lactate normalized to micrograms of muscle proteins. Asterisk denotes significance. (I) 4-wk-old RelB+/+ and RelB−/− mice were housed in individual metabolic cages for 72 h, and RER, total activity, and food intake were measured. Results shown are the mean of four mice per genotype, monitored during light and dark hours. *, P < 0.05. Error bars are means ± SEM.
NF-κB alternative signaling regulates PGC-1β
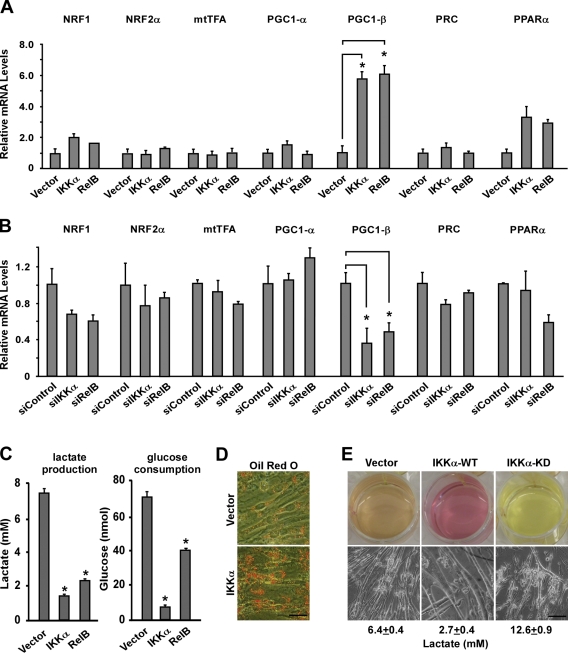
Next, we probed into the mechanism by which alternative NF-κB controls oxidative metabolism in muscle cells in vivo. Because of the large number of mitochondrial genes that were seen affected by NF-κB in our transcriptional profiling analysis, we hypothesized that such regulation might be controlled by one of the master regulators of mitochondrial biogenesis. Therefore, a targeted screen was initiated in differentiating C2C12 myoblasts to examine transcription factors NRF1, NRF2, mtTFA, and PPARα as well as the transcriptional coactivators PGC-1α, PGC-1β, and PRC under conditions in which IKKα and RelB were either overexpressed or silenced by siRNA. Using these criteria, only PGC-1β was significantly increased by over sixfold with addition of IKKα (P = 0.0003) and RelB (P = 0.0007) or suppressed by over twofold with depletion of this kinase (P = 0.007) or NF-κB subunit (P = 0.004; Fig. 3, A and B). In keeping with other studies (Lelliott et al., 2006; Fang et al., 2011; Sahin et al., 2011), this analysis was restricted to steady-state levels of RNA because a reliable commercial PGC-1β antibody could not be obtained. Interestingly, regulation of PGC-1β did not extend to other PGC family members PGC-1α or PRC. This regulation was also pathway specific because a similar analysis with classical IKKβ and p65 instead negatively impacted PGC-1β expression (Fig. S3 A). In addition, in line with the notion that alternative signaling promotes an oxidative phenotype, overexpression of IKKα or RelB led to reduced lactate production and glucose uptake in C2C12 myotubes (Fig. 3 C) while increasing fatty acid accumulation for β oxidation, as indicated by Oil red O–positive staining (Fig. 3 D). Reduction in lactic acid in IKKα-expressing myotubes correlated well with changes in culture media color and integrity of muscle cells compared with vector control cells. In contrast, cells expressing a kinase-dead (KD) version of IKKα increased lactate production while compromising the integrity of myotubes and mitochondrial gene expression (Fig. 3 E and Fig. S3 B).
Figure 3.
PGC-1β is regulated by components of the NF-κB alternative pathway. (A) C2C12 myoblasts were transfected with an empty vector, wild-type IKKα, or RelB expression constructs and differentiated for 3 d. RNA was isolated, and real-time RT-PCR analysis was performed probing for the indicated mitochondrial regulators. (B) C2C12 myoblasts were transfected with control siRNA (siControl) or siRNA oligonucleotides targeting IKKα or RelB. Cells were differentiated and analyzed as in A. (C) C2C12 myoblasts were transfected as in A, and after differentiation, media were collected and tested for lactate production and glucose content. Glucose consumption was calculated as total glucose present minus glucose that remained in the media. (D) Cells were transfected as in A, differentiated, and stained for lipid content with Oil red O. (E) C2C12 myoblasts were transfected with wild type or a KD form of IKKα, and after 5 d of differentiation without medium replenishment, cultured cells was photographed to highlight the change in the media color (top) or integrity of myotubes (bottom). Lactate values are representative of three independent experiments. *, P < 0.05. Error bars are means ± SEM. Bars, 100 µm.
To address the physiological significance of PGC-1β regulation, we examined muscles injected with AAV-IKKα. Consistent with in vitro results, muscles overexpressing IKKα repeatedly led to corresponding increases in PGC-1β (P = 0.03; Fig. 4 A). Conversely, in vivo siRNA knockdown of IKKα in limb muscles reduced the levels of mitochondrial genes, as well as PGC-1β, but not PGC-1α (Fig. 4 B and Fig. S4, A and B). It is noteworthy that in mice that did not exhibit efficient knockdown of the kinase (siIKKα 3), no effects on PGC-1β or mitochondrial genes were observed, supporting that PGC-1β regulation was directly mediated by IKKα. Likewise, siRNA knockdown of RelB in hind-limb muscles was associated with reduced levels of mitochondrial products, as well as PGC-1β, but not PGC-1α (Fig. S4, A–C).
Figure 4.
IKKα regulates PGC-1β in vivo. (A) TA muscles injected with AAV-GFP or AAV-IKKα (n = 4 mice per group) were harvested and probed for PGC-1β. Results are compared with the mean expression of PGC-1β from AAV-GFP–injected muscles, set to a value of 1. *, P = 0.001. (B) siRNA oligonucleotides were injected into TA. For each mouse, siRNA for IKKα was injected on one side, whereas control siRNA was administered to the contralateral side, and expression of PGC-1β was determined by real-time RT-PCR. Shown are data from a single experiment out of two replicates. (C) Skeletal muscles from E20 IKKα+/+ and IKKα−/− embryos from eight separate litters were isolated, and expression for PGC-1α and PGC-1β was examined by real-time RT-PCR. For each litter, values were normalized to the wild-type controls. The asterisk denotes significance. (D) Limbs from 3–8-d-old RelB+/+ and RelB−/− pups from six separate litters were isolated, and relative PGC-1 levels were measured, normalized to wild-type controls. Similarly, limb muscles were isolated from 3–8-d-old p65+/+ and p65−/− pups from seven separate litters to determine PGC-1 levels. (E) Soleus and TA were harvested from RelB+/+ and RelB−/− adult mice (n = 7), and PGC-1 levels were determined by real-time PCR analysis as described in A. *, P < 0.05. Error bars are means ± SEM.
To substantiate these results, we compared PGC-1 from IKKα and RelB knockout mice. From eight litters examined, a consistent down-regulation of PGC-1β, but not PGC-1α, was observed in IKKα−/− as compared with wild-type littermates (n = 14; P < 0.001; Fig. 4 C). Similarly, in RelB−/− neonatal hind limbs (n = 9) or adult soleus and TA (n = 8), PGC-1β, but not PGC-1α, was significantly reduced compared with RelB+/+ muscles (Fig. 4, D and E), and analogous to C2C12 myotubes, this regulation was selective to alternative NF-κB because no change in PGC-1β was observed from age-matched p65−/− mouse muscles (Fig. 4 D). Although PGC cofactors play prominent roles in regulating mitochondria in liver tissue, we were unable to observe a general reduction of PGC-1β in livers of either IKKα−/− or RelB−/− mice (Fig. S4 D), suggesting that regulation of PGC-1β by alternative NF-κB is tissue restrictive. It is also notable that in RelB−/− muscles, a compensatory increase in PGC-1α was observed both by real-time RT-PCR and microarray analyses (Fig. 4 E and not depicted), findings that are consistent with previous observations described in PGC-1β−/− mice (Sonoda et al., 2007). Collectively, results confirm that PGC-1β regulation by alternative NF-κB occurs in vivo.
PGC-1β is transcriptionally regulated by alternative NF-κB
To gain further insight into the regulatory mechanism of PGC-1β, we asked whether the PGC-1β gene might itself be a transcriptional target of alternative NF-κB. Use of the sequence comparative analysis program rVista 2.0 identified three putative NF-κB binding sites within the PGC-1β locus that closely matched the consensus sequence GGGRNNYYCC and, in addition, were conserved across species (Fig. 5 A). The first site (5′-GGGGGTGCCC-3′; 95% conserved), which we designated κB-S1, is located 200 base pairs upstream of the putative transcription start site (TSS). Two other predicted sites, κB-S2 (5′-GGGAAATTCC-3′; 95% conserved) and κB-S3 (5′-GGGGGTTCCC-3′; 100% conserved), are located within a large 78-kb first intron that occupies most of the 107-kb PGC-1β gene (Fig. 5 A). To test the efficiency of NF-κB binding, electrophoretic mobility shift assays (EMSAs) were performed with nuclear extracts from C2C12 cells transfected with classical or alternative NF-κB complexes. Although binding occurred on all three sites, RelB–p52 binding on κB-S2 and κB-S3 was noticeably stronger as compared with p65–p50 (Fig. 5 B). This activity appeared specific because complex formation was decreased with κB-S2 and κB-S3 oligonucleotides containing mutations in NF-κB consensus sites (Fig. 5 B, WT vs. Mut). Consistent with these findings, chromatin immunoprecipitation (ChIP) analysis revealed preferential binding for RelB on κB-S2 and κB-S3 (Fig. 5 C). RNA polymerase II (Pol II) also bound to regions overlapping NF-κB binding sites, supporting earlier findings that PGC-1β is transcriptionally active in skeletal muscle cells (Saunders et al., 2006). Significantly, RelB binding to κB-S2 and κB-S3 was confirmed in adult skeletal muscle, which again was preferentially bound in comparison to p65 (Fig. 5 D), thus demonstrating that RelB binding occurs in vivo. Although IKKα can in some cell types reside in the nucleus and associate with chromatin (Anest et al., 2003; Yamamoto et al., 2003), nuclear localization was not detectable, nor could we find evidence by ChIP of IKKα binding to PGC-1β in regions encompassing sites κB-S2 and κB-S3 (unpublished data). Together, results suggest that in differentiating muscle, NF-κB binds to PGC-1β within intron 1 and that such binding is favored by an alternative complex.
Figure 5.
PGC-1β is a direct transcriptional target of NF-κB. (A) Schematic of PGC-1β displaying intron–exon (Ex) locations and conserved NF-κB binding sites. (B) Oligonucleotides corresponding to wild-type (WT) or mutant (Mut) versions of the NF-κB binding sites within PGC-1β were used in EMSA from C2C12 myoblasts transfected with classical and alternative NF-κB dimer complexes. (C) C2C12 myoblasts were transfected with HA-RelB and p52 and differentiated, and ChIPs were performed using antibodies against HA, Pol II, or IgG. DNA fragments were amplified with primers surrounding κB-S1, κB-S2, and κB-S3 sites, and values were represented as fold enrichment over IgG controls. All ChIP data shown in C, D, H, and I are from a single representative experiment repeated in triplicate. (D) TA from adult wild-type muscles was isolated, and ChIPs were performed as in C. DNA fragments around κB-S2 and κB-S3 were amplified. (E) NF-κB–PGC-1β luciferase reporter activities were monitored in myoblasts and myotubes. (F) Myoblasts were transfected with reporter constructs described in E along with siRNA oligonucleotides directed against a nonspecific control or components of classical or alternative NF-κB signaling. Cells were differentiated for 3 d, and luciferase values were subsequently determined. *, P < 0.05. (G) Wild-type or mutant sequences corresponding to κB-S2 and κB-S3 fused to a luciferase reporter were transfected into C2C12 myoblasts, and activities were determined in myoblasts and myotubes. (H and I) C2C12 myoblasts were transfected with IKKα or RelB expression plasmids or siRNA oligonucleotides targeting IKKα, and ChIPs were performed on myotubes to detect for H3K4 in a region of PGC-1β ∼200 base pairs upstream of exon 1, or ChIPs were performed with Pol II on the κB-S2 site. Binding was normalized to IgG used as a control. Error bars are means ± SEM. RLU, relative luciferase unit; meH3K4, methylated H3K4.
To address the functional relevance of NF-κB binding, luciferase reporter assays were performed with constructs containing a minimally acting promoter and ∼400 base pairs of genomic sequence surrounding each of the κB-S1, κB-S2, and κB-S3 sites. In line with EMSAs, we found that the highest transcriptional activity occurred with constructs containing κB-S2 and κB-S3 (Fig. 5 E). These reporters were also greatly induced during muscle differentiation, which mirrored the endogenous induction of PGC-1β during myogenesis (Fig. S5 A).
To further investigate the specificity of PGC-1β transcription with respect to classical and alternative signaling, reporters were tested along with siRNA targeted against both NF-κB pathways. Consistent with EMSAs and ChIPs, knockdown of classical or alternative components had no significant effect on reporter activity containing κB-S1 (Fig. 5 F). This confirmed that κB-S1 is not functional and therefore was no longer considered in subsequent analyses. In contrast, depletion of IKKα or RelB led to a pronounced reduction in PGC-1β transcriptional activity containing sites κB-S2 and κB-S3, whereas only a small, but nonsignificant, effect occurred with knockdown of classical NF-κB (Fig. 5 F). Moreover, mutating κB-S2 and κB-S3 sites substantially reduced PGC-1β transcriptional activity (Fig. 5 G), which together argue that κB-S2 and κB-S3 sites function as PGC-1β enhancer elements during skeletal muscle differentiation.
Next, we used published genomic ChIP sequencing analysis to identify chromatin marks of active transcription within the TSS and first intron of PGC-1β (Mikkelsen et al., 2007). We reasoned that such marks could be used as a guide to further assess the relevance of the NF-κB regulatory sites with respect to PGC-1β transcription. In line with the aforementioned results, the methylated form of H3K4 was profoundly elevated in a region upstream of the TSS in IKKα-expressing differentiated muscle cells (Fig. 5 H), which was also tightly associated with increases in PGC-1β (Fig. S5 A). Notably, a similar enhancement of H3K4 methylation occurred in myoblasts expressing RelB or IKKα wild type but not IKKα KD (Fig. 5 H). Furthermore, IKKα or RelB was sufficient to recruit Pol II on κB-S2 under differentiating conditions (Fig. 5 I). This recruitment was specific because noticeably less Pol II binding occurred in cells expressing IKKα KD or depleted of IKKα by siRNA. Collectively, these data strongly support that PGC-1β is a direct transcriptional target of NF-κB alternative signaling in differentiating muscle cells.
Alternative NF-κB regulation of mitochondria occurs through PGC-1β
Given the link that we established between alternative NF-κB and PGC-1β, we subsequently asked whether regulation of mitochondrial biogenesis by alternative signaling occurred through PGC-1β. Therefore, induction of mitochondrial genes by IKKα was examined under PGC-1β knockdown conditions. Results showed that, whereas IKKα robustly increased PGC-1β, mitochondrial genes, and ATP production in myotubes, PGC-1β silencing completely abolished this regulation (Fig. 6, A–C). Similar findings occurred with RelB expression with and without PGC-1β siRNA. Control experiments confirmed that PGC-1β knockdown was specific because these effects occurred in the absence of any changes in PGC-1α (Fig. 6 A), and in line with the view that PGC-1β serves as a vital regulator of mitochondria (Lin et al., 2002a; Arany et al., 2007), PGC-1β knockdown alone in differentiating myoblasts was sufficient to decrease the levels of mitochondrial genes and the cellular ATP content (Fig. 6, A–C). These data are consistent with the notion that IKKα signaling to RelB in skeletal muscle cells modulates mitochondria predominantly through PGC-1β.
Figure 6.
IKKα regulation of mitochondrial biogenesis is mediated through PGC-1β. (A) C2C12 myoblasts were transfected with IKKα and RelB expression constructs, along with siRNA oligonucleotides directed against a nonspecific control or PGC-1β. After differentiation, levels of the PGC-1 coactivators as well as mitochondrial genes were examined by real-time RT-PCR. Vect, vector. (B) C2C12 myoblasts were transfected and differentiated as in A and harvested to monitor for ATP levels normalized to total cellular protein. *, P < 0.05. (C) C2C12 myoblasts were transfected with IKKα or RelB, along with siRNA oligonucleotides targeted against either a control or PGC-1β, and after differentiation, Western blotting was performed. Error bars are means ± SEM.
mTOR activates alternative signaling to regulate PGC-1β in myogenesis
To ascertain what lies upstream of IKKα to regulate PGC-1β during muscle differentiation, we investigated how alternative signaling was itself regulated during myogenesis. Activation of alternative signaling in lymphoid cells occurs through a receptor-mediated mechanism that causes proteolysis of TRAF3, leading to NF-κB–inducing kinase (NIK) stability and subsequent IKK phosphorylation (Vallabhapurapu et al., 2008; Zarnegar et al., 2008). However, in differentiating muscle, although we were able to capture alternative NF-κB activation through co-translational p100 to p52 processing, regulation on NIK or TRAF3 was not detected, even after repeated attempts (Fig. 7 A). This suggested that activation of alternative NF-κB in muscle cells does not occur through an analogous mechanism as previously described in maturing lymphoid cells (Liao et al., 2004).
Figure 7.
mTOR mediates PGC-1β induction through alternative NF-κB. (A) Whole-cell extracts were prepared from C2C12 cells under proliferating (myoblast) or differentiated (myotube) conditions, and Western blotting was performed (ns denotes a nonspecific band). (B) C2C12 myotubes were treated with rapamycin for 24 h, and PGC-1β levels were monitored. ChIP was performed for the methylated H3K4 (meH3K4) chromatin mark in the PGC-1β promoter. (C) Cell extracts were prepared as in A. mTOR or IgG as a control was individually immunoprecipitated (IP), and lysates were subsequently probed (IB, immunoblot) for IKKα. (D) C2C12 cell extracts from myoblasts or myotubes were prepared, and IKK kinase activity (KA) was assessed in the absence (−) or presence (+) of 20 nM rapamycin. (E) Myoblasts were transfected with the κB-S2–luciferase reporter, and after differentiation and treatment, cells were harvested for luciferase activity. (F) C2C12 myoblasts were differentiated, and ChIP analysis for endogenous RelB binding to the κB-S2 site of PGC-1β was performed in the absence or presence of 20 nM rapamycin. (G) C2C12 myoblasts were transfected with RelB, and after differentiation, cells were either untreated (−) or treated (+) with rapamycin and subsequently monitored for PGC-1β by real-time RT-PCR. Asterisks denote significance. Error bars are means ± SEM. CMV, cytomegalovirus; RLU, relative luciferase unit.
Because mTOR and the rapamycin-sensitive TORC1 complex have independently been shown to promote mitochondrial oxidative function (Cunningham et al., 2007; Bentzinger et al., 2008; Risson et al., 2009) as well as regulate NF-κB in prostate cells (Dan et al., 2008), we considered the possibility that mTOR might act upstream of IKKα to regulate PGC-1β and the oxidative capacity of differentiating muscle. To test this hypothesis, we initially determined whether PGC-1β expression during myogenesis is mTOR dependent. Results showed that PGC-1β was indeed decreased in the presence of rapamycin and further that this decrease was reflected in dramatically lower H3K4 methylation on the PGC-1β promoter (Fig. 7 B). These results supported the idea that mTOR regulates PGC-1β at the transcriptional level. To explore the potential link with NF-κB, coimmunoprecipitations were performed to analyze the interaction of mTOR with IKK. An endogenous interaction between IKKα and mTOR was detected in muscle cells (Fig. 7 C), which curiously was unaltered upon differentiation even though IKK activity clearly increased during this process (Fig. 7, C and D; Bakkar et al., 2008). Nevertheless, the interaction between mTOR and IKKα in muscle cells appears relevant because inhibition of mTOR was sufficient to diminish IKK activity (Fig. 7 D). To then address whether induction of PGC-1β by alternative NF-κB was controlled by mTOR, we treated myotubes with rapamycin and investigated PGC-1β transcriptional activity as well as recruitment of RelB on κB-S2. Inhibition of mTOR resulted in lower κB-S2 activity and lower levels of RelB binding to PGC-1β (Fig. 7, E and F). Furthermore, addition of RelB markedly rescued the inhibitory effect of rapamycin on PGC-1β expression (P = 0.005; Fig. 7 H), thus indicating that RelB is downstream of mTOR. Collectively, these results are highly suggestive that mTOR, in complex with IKKα, activates an alternative NF-κB pathway during myogenesis to stimulate PGC-1β and oxidative metabolism.
Discussion
Skeletal myogenesis involves a series of tightly regulated events that together culminate in the assembly of multinucleated myotubes. This process also involves a shift in the metabolic profile mediated by activation of mitochondrial biogenesis, presumably to meet the energy demands of a newly contracting myofiber (Kraft et al., 2006). Numerous studies have focused on identifying the regulators of mitochondrial biogenesis in skeletal muscle in response to exercise or cold exposure as reporter earlier in this paper. In contrast, little is known regarding the signaling pathways that couple the generation of mitochondria with myogenic differentiation. In this current study, we showed that the alternative NF-κB pathway is one such signaling cascade regulating mitochondrial content and oxidative phosphorylation during myogenesis. We further revealed that this mechanism is controlled by PGC-1β, which we propose as a newly identified transcriptional target of NF-κB. Thus, results support that oxidative metabolism in developing myotubes is regulated by alternative NF-κB–dependent induction of PGC-1β.
Past and more recent studies established a link between NF-κB and mitochondrial metabolism (Cogswell et al., 2003; Guseva et al., 2004; Suliman et al., 2010; Johnson et al., 2011; Mauro et al., 2011; Schilling et al., 2011). However, these findings almost exclusively were restricted to the classical pathway. In comparison, our current findings implicate a regulation specific to alternative signaling. This specificity was shown genetically by demonstrating that IKKα−/− and RelB−/− muscles are defective in mitochondrial content and oxidative function. In addition, in vivo expression of alternative and not classical signaling components was shown to be sufficient to induce mitochondrial genes and genomes as well as slow fiber myosin isoforms, characteristic of a mitochondrial oxidative phenotype.
Cis-acting factors that control the transcription of PGC-1β during skeletal myogenesis have not been well defined. Our results highlighted two NF-κB regulatory sites within the first long intron of PGC-1β that favored RelB binding over p65 and preferentially function through IKKα rather than IKKβ. IKKα and RelB were also required for Pol II recruitment and H3K4 methylation on the PGC-1β promoter, which suggest that NF-κB can mediate epigenetic regulation on myogenic genes. Previous results indicate that this regulation can occur indirectly, through p65-mediated expression of YY1 that cooperates with the polycomb complex to epigenetically silence transcription of miR-29 (Wang et al., 2008) or directly as shown here when RelB binding promotes H3K4 methylation in the PGC-1β promoter. This latter mechanism is in line with reported functions of NF-κB on distal enhancer elements (Ghisletti et al., 2010; Goh et al., 2010). Such data bring a new understanding for how PGC-1β is regulated during muscle differentiation and support a mechanism whereby RelB binding at a distal site signals to upstream TSS chromatin remodeling modulators and the transcriptional machinery to promote PGC-1β expression.
The contrast in PGC-1β regulation between alternative and classical NF-κB is intriguing because, from the standpoint of NF-κB binding sites, both κB-S2 and κB-S3 conform perfectly to the consensus sequence and therefore, in principle, should be competent to bind both classical and alternative complexes. Only a handful of genes expressed in the developing spleen have been shown to contain binding sites specific to an alternative NF-κB complex (Bonizzi et al., 2004). This specificity derives from a single nucleotide modification in the 3′ end position of the decamer sequence. The absence of this variance in sites κB-S2 and κB-S3 of PGC-1β suggests that binding to the classical subunit p65 remains possible, and although only a minor fraction of p65 binding was detected by EMSA, we suspect that in different cell types or under different physiological conditions, classical NF-κB may yet contribute to PGC-1β transcriptional control.
Results from a series of analysis in our study are consistent with the notion that alternative NF-κB regulation of PGC-1β mediates oxidative respiration relevant to myogenesis and fiber type specification. This notion is supported by phenotypes from PGC-1 transgenic and knockout models. Although muscle-specific transgenic expression of PGC-1α or PGC-1β induces an oxidative phenotype and concomitant fiber type specification in both mice (Lin et al., 2002b; Arany et al., 2007) and cultured cells (Mortensen et al., 2006), conditional deletion of these PGC-1 genes in muscles reduces expression of types I and IIa fibers (Handschin et al., 2007).
Given the functional similarities between PGC-1α and PGC-1β, it was enlightening to find in our study that NF-κB regulation appears specific to the β isoform. Unlike NF-κB binding sites in PGC-1β, similar sites are not present in PGC-1α (unpublished data). This is consistent with our inability to detect a similar regulation on PGC-1α from IKKα−/− and RelB−/− mice or to induce or suppress PGC-1α levels in cells or tissues in response to alternative NF-κB components. Because expression of PGC-1 isoforms is stimulated during myogenesis, we do not suspect that this is when the selectivity with alternative NF-κB comes into play. Rather, we favor that this relates to specific conditions in which the two homologues differentially regulate mitochondria. For instance, although both PGC-1α and PGC-1β induce MyHC-I, only PGC-1β increases type IIa (Mortensen et al., 2006). In addition, although PGC-1α is predominantly responsible for mitochondrial uncoupling, PGC-1β mainly controls mitochondrial volume (Lin et al., 2002a). In line with these distinct properties of PGC-1 isoforms, our results implicate IKKα as a positive regulator of type IIa myofibers and of mitochondrial volume. Control of mitochondrial size is mediated by PGC-1β through stimulation of ERRα, leading to expression of mitofusin 2, a mammalian mitochondrial fusion regulator (Liesa et al., 2008). Our results revealed that muscles expressing PGC-1β contained elongated mitochondria that correlated with increases in mitofusin 2 (Fig. S5 B). Therefore, it is possible that alternative NF-κB signaling is involved in stimulating mitochondrial fusion indirectly through PGC-1β–mediated transcriptional regulation of mitofusin 2.
In the final part of our study, we addressed the mechanism linking NF-κB to PGC-1β regulation during myogenesis. We showed that this link was independent of TRAF3 and NIK stabilization but rather was under control of mTOR and the TORC1 complex. Based on these findings, we propose that mTOR represents an important node of NF-κB signaling in maturing muscle cells. A study dedicated to elucidating mTOR function in myogenesis has shown that expression and activity are rapidly stimulated after a switch in differentiation (Erbay and Chen, 2001). This early activation is required for myogenesis because inhibition of TORC1 prohibits myotube formation. However, a second function of mTOR was implicated in differentiation that only until recently was shown to involve myoblast fusion (Sun et al., 2010). Based on our findings, we propose that mTOR possesses an additional function to facilitate alternative NF-κB activation. The interaction of mTOR with IKKα likely regulates IKK activity that in turn leads to RelB–p52 binding to κB-S2 and κB-S3 sites in PGC-1β. This binding promotes chromatin remodeling and PGC-1β expression, resulting in subsequent generation of mitochondria critical for myogenesis and muscle energy metabolism.
A concluding point that should not be overlooked is that various muscle and nonmuscle diseases are associated with mitochondrial mutations and metabolic disorders (Arany, 2008; Shaw and Winge, 2009). Stimulating mitochondrial biogenesis by manipulating the production or activities of mitochondrial regulators has been touted as an attractive therapeutic strategy. In general, the focus to design pharmacological agents to suppress IKK activity remains firmly centered on the classical pathway. However, given the connections made in this study, careful consideration is warranted to ensure that such compounds are indeed selective and do not impinge on alternative NF-κB signaling that might otherwise contribute to compromised mitochondrial function.
Materials and methods
Antibodies and materials
Antibodies against IKKα (IMG-136) and IKKβ (IMG-159A) were obtained from Imgenex, and HA (MMS-101P) was purchased from Covance. MyHC IIb (MY-32), MyHC I (slow; NOQ7.5.4D), and α-tubulin (B-5-1-2) were purchased from Sigma-Aldrich, cytochrome c and COX IV were purchased from BD (556433), RelB (sc-226), TRAF3 (M20), p100 (K27), and Pol II (sc-899X) were purchased from Santa Cruz Biotechnology, Inc., myosin forms MyHC IIa (2F7) and MyHC IIx (6H1) were purchased from the Developmental Hybridoma Bank, NIK (4994) was obtained from Cell Signaling Technology, anti–mono/di/trimethyl-histone H3 (Lys4; 05–791) and p65 (C terminus; 06–418) were obtained from Millipore, and anti–histone H3 (NB-100) was obtained from Novus Biologicals. Antibodies against the oxidative phosphorylation complex 1, 30-kD subunit (459130), complex 2, 70-kD subunit (459200), and complex 3 core, 1 subunit (459140), were obtained from Invitrogen. Rapamycin was obtained from EMD, secondary antibodies for immunofluorescence were obtained from Invitrogen, and materials for immunohistochemical analysis were obtained from Vector Laboratories.
Mice and genotyping
Animals were housed in the animal facility at The Ohio State University Comprehensive Cancer Center, fed a standard diet, and kept at constant temperature and humidity. Treatment of mice was in accordance with the institutional guidelines of the Institutional Animal Care and Use Committee. Wild-type postnatal day 2 (P2) mice were injected once with AAV with an intramuscular dose of 3 × 1010 infectious particles in the frontal area of TA muscles. Transgene expression was monitored by immunoblotting. For in vivo siRNA knockdown experiments, 5 µM siRNA oligonucleotides were preincubated with Lipofectamine for 15 min before injection into the TA of P2 mice in a total volume of 30 µl Opti-MEM. Injections were repeated every other day, and mice were sacrificed at P8. IKKα−/− mice were generously obtained from I. Verma (The Salk institute, La Jolla, CA) and lack a 3-kb genomic region covering the promoter and exon 1 as previously described (Li et al., 1999; Takeda et al., 1999). TNF−/−;p65+/+ and TNF−/−;p65−/− mice were previously described in which the first exon of p65 is deleted (Bakkar et al., 2008). These strains were used, representing postnatal p65+/+ and p65−/− mice in this current study. RelB mice were obtained from Bristol-Myers Squibb.
Mitochondrial DNA and respiratory markers
Protein quantification of mitochondria was made through differential centrifugation in 10 mM Tris-MOPS, 0.1 mM EGTA/Tris, and 200 mM sucrose solution (Frezza et al., 2007), and spectrophotometric readings were performed at 280 nm using a spectrophotometer (ND-1000; Nanodrop) normalized to total protein. ATP was measured as recommended by the manufacturer using a luminescent assay kit (CellTiter-Glo; Promega). Lactate and glucose concentrations were determined using assays obtained from BioVision. Glucose consumption was calculated from glucose levels in media at days 2 and 4 after differentiation. For mitochondrial genome quantification, total DNA from tissues was extracted and used to amplify a nuclear gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the MTCO1 (mitochondrial cytochrome c oxidase subunit 1) by real-time PCR analysis. Relative mitochondrial to nuclear DNA copy numbers were determined normalized to the nuclear gene copy numbers. For whole-body metabolic experiments, mice were individually housed in CLAMS (Columbus Instruments) and maintained at thermoneutrality (28°C) for 72 h with normal 12-h light–dark cycles. Mice were allowed to acclimate for 12 h, and rates of oxygen consumption and CO2 release were determined at 8-min intervals, normalized to body mass, and used to calculate the RER. Ambulations were simultaneously recorded in all three x, y, and z planes to determine total activity.
ChIP analysis
ChIP assays were performed as recommended by the manufacturer (Millipore) with only minor modifications. Cells were lysed in Pipes nuclei isolation buffer (20 mM Pipes, 85 mM KCl, and 0.5% NP-40) before lysis and sonication. Samples were precleared and incubated overnight with 2 or 4 µg (RelB) antibody. Protein–DNA complexes were then collected, washed, and reverse cross-linked to release DNA fragments. DNA was precipitated overnight and purified with PCR purification kits (QIAGEN). Samples were resuspended in 25–40 µl TE (Tris-EDTA), and real-time PCR amplification was performed with 3–5 µl of reaction volume. Values were normalized to respective input and computed as fold enrichment over IgG controls. For murine muscles, fresh tissues were fixed in formaldehyde, homogenized, and lysed in Pipes buffer as described in this paragraph. All assays shown from a representative experiment were performed in triplicate.
Muscle functional measurements
AAV-injected mice were euthanized, and EDL muscles were rapidly excised, weighed, and placed in oxygenated Krebs-Henseleit solution at 30°C in a circulating bath as previously described (Rodino-Klapac et al., 2007). In brief, one end of the muscle was tied to a force transducer, and the other was tied to a motor. Stimulation was delivered via two parallel electrodes. Isometric tetanic contractions were recorded and normalized to muscle length and/or cross-sectional area.
Plasmids
Troponin I– and MyHC IIb–luciferase reporter plasmids containing muscle-specific enhancers in the troponin I or MyHC IIB genes fused to luciferase as well as the pcDNA3 RelB, p65, p50, and p52 expression plasmids were previously described (Guttridge et al., 1999; Acharyya et al., 2004; Bakkar et al., 2008). IKK constructs were obtained from J. Didonato (Cleveland Clinic, Cleveland, OH). AAV1 pseudotyped viruses were generated by the Viral Vector Core facility at Nationwide Children’s Research Institute (Columbus, OH). PGC-1β–NF-κB reporter plasmids were generated by subcloning ∼400 bp of genomic DNA surrounding each of the NF-κB binding sites in the mouse PGC-1β gene into Nhe1–BglII cloning sites of pGL3-control (Promega). For PGC-1β mutant reporter plasmids, mutagenesis (QuikChange XL; Agilent Technologies) was used to delete NF-κB binding sites.
Histology, EM, and immunohistochemistry
For histology, tissues were sectioned at 10 µm on a cryostat (CM 3050S; Leica) and stained with hematoxylin and eosin or processed for staining. SDH staining was performed for 15 min at 37°C using succinate as a substrate and nitro blue tetrazolium as a redox indicator. Sections were then washed in acetone, mounted in PermaSlip, and visualized using a microscope (BX51; Olympus). Pictures were taken using a camera (ALTRA20; Olympus) and the corresponding Microsuite FIVE pathology software. Values were recorded from three to five injected animals per group. The darkest staining fibers were counted and expressed as the percentage of total fibers/section. Immunohistochemistry for myosin subtypes was performed at 1:500 dilutions as described previously (Acharyya et al., 2007). Ultrastructural analysis was performed on fixed tissues, sectioned using a microtome (EM UC6; Leica) at 70 nm, and visualized using a transmission electron microscope (Spirit Tecnai; FEI) at the Ohio State University Medical Center Microscopy and Imaging Facility. Images were captured using a camera (XR60; Advanced Microscopy Techniques). Mitochondria were counted in multiple sections (10–12 per mouse; two mice per group) and expressed as numbers per cross-sectional area.
EMSA, IKK assay, and gene expression analysis
EMSA were performed as previously described (Guttridge et al., 1999). In brief, nuclear extracts were prepared and incubated with 20,000 cpm of radio-labeled probes. Complexes were then resolved on 5% polyacrylamide gels. For IKK activity assays, IKK was immunoprecipitated and incubated with a GST-IκB fusion protein in the presence of γ-[32P]ATP as previously described (Bakkar et al., 2008). For quantitative RNA expression, total RNA was prepared with TRIZOL (Invitrogen). cDNA was synthesized, and real-time RT-PCR was performed using SYBR green master mix (Bio-Rad Laboratories). For gene transcription profile, RNA was extracted using the RNeasy kit (QIAGEN). Hybridization and scanning of the murine 430 2.0 chip (GeneChip; Affymetrix) were performed by the Microarray Shared Resource at the Ohio State University Comprehensive Cancer Center.
Cell culture, transfections, and luciferase assays
C2C12 myoblasts were cultured and transfected in 12-well plates for reporter assays using transfection reagent (SuperFect; QIAGEN) and 0.25 µg of total DNA per well as previously described (Guttridge et al., 2000). Primary myoblasts were prepared from embryonic day 20 (E20) embryos for IKKα−/− or 2-d-old neonates for all other genotypes (Bakkar et al., 2008). siRNA for IKKα, IKKβ, RelB, p65, and PGC-1β was obtained from Thermo Fisher Scientific, and transfections were performed using Lipofectamine 2000 (Invitrogen).
Bioinformatic and statistical analysis
Microarray gene expression estimates were obtained by the Robust Multichip Average method with quantile normalization using R software. These expression data were analyzed using the class comparison algorithm of Biometric Research Branch–ArrayTools with random variance model (Simon et al., 2007). Significant lists of genes were obtained with P < 0.01, fold change >1.25, and with controlling the false discovery rate at 0.15 to adjust for multiple comparisons. Gene ontology clustering classification was performed on genes down-regulated >1.25-fold (P < 0.01) using DAVID functional annotation bioinformatics microarray analysis (Huang et al., 2009). Promoter sequences were retrieved from the University of California, Santa Cruz Genome Bioinformatics database. Mouse and human sequences for PGC-1α and PGC-1β were aligned, and prediction of conserved transcription factor binding sites was performed using rVista. All quantitative data are represented as means ± SEM. Analysis was performed between different groups using a two-tailed t test. Statistical significance was set at P < 0.05.
Online supplemental material
Fig. S1 shows that IKKα is not required for myogenic differentiation. Fig. S2 shows that RelB is not required for this myogenic differentiation process. Fig. S3 shows that activation of classical NF-κB signaling inhibits the regulators of mitochondria. Fig. S4 demonstrates that PGC-1α is not regulated by alternative NF-κB signaling. Fig. S5 shows that PGC-1β is induced during skeletal muscle differentiation and mitofusin 2 levels. For Table S1, the list indicates mitochondria-related genes repressed in RelB−/− muscles. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201108118/DC1.
Acknowledgments
We are grateful to I. Verma for the use of IKKα−/− mice. We also thank J. Didonato for expression plasmids and H. Wang, B. Kaspar, V. Jin, and members of the Ostrowski laboratory for technical advice as well as M. Ostrowski, D. Perrotti, and P. Reiser for helpful discussions. We are also thankful to members of the Guttridge laboratory for their support throughout the course of this study.
This work was supported by National Institutes of Health grant AR052787 to D.C. Guttridge.
Footnotes
Abbreviations used in this paper:
- AAV
- adeno-associated virus
- CA
- constitutively active
- ChIP
- chromatin immunoprecipitation
- EDL
- extensor digitorum longus
- EMSA
- electrophoretic mobility shift assay
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- IKK
- IκB kinase
- KD
- kinase dead
- mTOR
- mammalian target of rapamycin
- NF-κB
- nuclear factor κB
- NIK
- NF-κB–inducing kinase
- PGC
- PPAR-γ coactivator
- Pol II
- polymerase II
- RER
- respiratory exchange ratio
- SDH
- succinate dehydrogenase
- TA
- tibialis anterior
- TSS
- transcription start site
References
- Acharyya S., Ladner K.J., Nelsen L.L., Damrauer J., Reiser P.J., Swoap S., Guttridge D.C. 2004. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J. Clin. Invest. 114:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharyya S., Villalta S.A., Bakkar N., Bupha-Intr T., Janssen P.M., Carathers M., Li Z.W., Beg A.A., Ghosh S., Sahenk Z., et al. 2007. Interplay of IKK/NF-kappaB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J. Clin. Invest. 117:889–901 10.1172/JCI30556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anest V., Hanson J.L., Cogswell P.C., Steinbrecher K.A., Strahl B.D., Baldwin A.S. 2003. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 423:659–663 10.1038/nature01648 [DOI] [PubMed] [Google Scholar]
- Arany Z. 2008. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr. Opin. Genet. Dev. 18:426–434 10.1016/j.gde.2008.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B.M. 2007. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 5:35–46 10.1016/j.cmet.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Bakkar N., Wang J., Ladner K.J., Wang H., Dahlman J.M., Carathers M., Acharyya S., Rudnicki M.A., Hollenbach A.D., Guttridge D.C. 2008. IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 180:787–802 10.1083/jcb.200707179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R., Olson E.N. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37 10.1146/annurev.biochem.75.103004.142622 [DOI] [PubMed] [Google Scholar]
- Bentzinger C.F., Romanino K., Cloëtta D., Lin S., Mascarenhas J.B., Oliveri F., Xia J., Casanova E., Costa C.F., Brink M., et al. 2008. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8:411–424 10.1016/j.cmet.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Bonizzi G., Bebien M., Otero D.C., Johnson-Vroom K.E., Cao Y., Vu D., Jegga A.G., Aronow B.J., Ghosh G., Rickert R.C., Karin M. 2004. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 23:4202–4210 10.1038/sj.emboj.7600391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottero V., Rossi F., Samson M., Mari M., Hofman P., Peyron J.F. 2001. Ikappa b-alpha, the NF-kappa B inhibitory subunit, interacts with ANT, the mitochondrial ATP/ADP translocator. J. Biol. Chem. 276:21317–21324 10.1074/jbc.M005850200 [DOI] [PubMed] [Google Scholar]
- Cai D., Frantz J.D., Tawa N.E., Jr, Melendez P.A., Oh B.-C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Shoelson S.E. 2004. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 119:285–298 10.1016/j.cell.2004.09.027 [DOI] [PubMed] [Google Scholar]
- Chang J.H., Lin K.H., Shih C.H., Chang Y.J., Chi H.C., Chen S.L. 2006. Myogenic basic helix-loop-helix proteins regulate the expression of peroxisomal proliferator activated receptor-gamma coactivator-1alpha. Endocrinology. 147:3093–3106 10.1210/en.2005-1317 [DOI] [PubMed] [Google Scholar]
- Cogswell P.C., Kashatus D.F., Keifer J.A., Guttridge D.C., Reuther J.Y., Bristow C., Roy S., Nicholson D.W., Baldwin A.S.J., Jr 2003. NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J. Biol. Chem. 278:2963–2968 10.1074/jbc.M209995200 [DOI] [PubMed] [Google Scholar]
- Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 450:736–740 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- Czubryt M.P., McAnally J., Fishman G.I., Olson E.N. 2003. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA. 100:1711–1716 10.1073/pnas.0337639100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H.C., Cooper M.J., Cogswell P.C., Duncan J.A., Ting J.P., Baldwin A.S. 2008. Akt-dependent regulation of NF-kappaB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 22:1490–1500 10.1101/gad.1662308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E., Droin N.M., Delhase M., Haas E., Cao Y., Makris C., Li Z.W., Karin M., Ware C.F., Green D.R. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 17:525–535 10.1016/S1074-7613(02)00423-5 [DOI] [PubMed] [Google Scholar]
- Dogra C., Changotra H., Mohan S., Kumar A. 2006. Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-kappaB and degradation of MyoD protein. J. Biol. Chem. 281:10327–10336 10.1074/jbc.M511131200 [DOI] [PubMed] [Google Scholar]
- Erbay E., Chen J. 2001. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 276:36079–36082 10.1074/jbc.C100406200 [DOI] [PubMed] [Google Scholar]
- Fang S., Suh J.M., Atkins A.R., Hong S.H., Leblanc M., Nofsinger R.R., Yu R.T., Downes M., Evans R.M. 2011. Corepressor SMRT promotes oxidative phosphorylation in adipose tissue and protects against diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. USA. 108:3412–3417 10.1073/pnas.1017707108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Scorrano L. 2007. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2:287–295 10.1038/nprot.2006.478 [DOI] [PubMed] [Google Scholar]
- Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C.L., et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 32:317–328 10.1016/j.immuni.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Goh F.G., Thomson S.J., Krausgruber T., Lanfrancotti A., Copley R.R., Udalova I.A. 2010. Beyond the enhanceosome: cluster of novel κB sites downstream of the human IFN-β gene is essential for lipopolysaccharide-induced gene activation. Blood. 116:5580–5588 10.1182/blood-2010-05-282285 [DOI] [PubMed] [Google Scholar]
- Guseva N.V., Taghiyev A.F., Sturm M.T., Rokhlin O.W., Cohen M.B. 2004. Tumor necrosis factor-related apoptosis-inducing ligand-mediated activation of mitochondria-associated nuclear factor-kappaB in prostatic carcinoma cell lines. Mol. Cancer Res. 2:574–584 [PubMed] [Google Scholar]
- Guttridge D.C., Albanese C., Reuther J.Y., Pestell R.G., Baldwin A.S., Jr 1999. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge D.C., Mayo M.W., Madrid L.V., Wang C.-Y., Baldwin A.S., Jr 2000. NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 289:2363–2366 10.1126/science.289.5488.2363 [DOI] [PubMed] [Google Scholar]
- Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N.K., Yan Z., Spiegelman B.M. 2007. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J. Biol. Chem. 282:30014–30021 10.1074/jbc.M704817200 [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell. 132:344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Hu Y., Baud V., Delhase M., Zhang P., Deerinck T., Ellisman M., Johnson R., Karin M. 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 284:316–320 10.1126/science.284.5412.316 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Johnson R.F., Witzel I.I., Perkins N.D. 2011. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-κB. Cancer Res. 71:5588–5597 10.1158/0008-5472.CAN-10-4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D.P., Scarpulla R.C. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357–368 10.1101/gad.1177604 [DOI] [PubMed] [Google Scholar]
- Kraft C.S., LeMoine C.M., Lyons C.N., Michaud D., Mueller C.R., Moyes C.D. 2006. Control of mitochondrial biogenesis during myogenesis. Am. J. Physiol. Cell Physiol. 290:C1119–C1127 10.1152/ajpcell.00463.2005 [DOI] [PubMed] [Google Scholar]
- Lelliott C.J., Medina-Gomez G., Petrovic N., Kis A., Feldmann H.M., Bjursell M., Parker N., Curtis K., Campbell M., Hu P., et al. 2006. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 4:e369 10.1371/journal.pbio.0040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lu Q., Hwang J.Y., Büscher D., Lee K.F., Izpisua-Belmonte J.C., Verma I.M. 1999. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 13:1322–1328 10.1101/gad.13.10.1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G., Zhang M., Harhaj E.W., Sun S.C. 2004. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279:26243–26250 10.1074/jbc.M403286200 [DOI] [PubMed] [Google Scholar]
- Liesa M., Borda-d’Agua B., Medina-Gómez G., Lelliott C.J., Paz J.C., Rojo M., Palacín M., Vidal-Puig A., Zorzano A. 2008. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS ONE. 3:e3613–e3624 10.1371/journal.pone.0003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. 2002a. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 277:1645–1648 10.1074/jbc.C100631200 [DOI] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. 2002b. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 418:797–801 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- Lustig Y., Ruas J.L., Estall J.L., Lo J.C., Devarakonda S., Laznik D., Choi J.H., Ono H., Olsen J.V., Spiegelman B.M. 2011. Separation of the gluconeogenic and mitochondrial functions of PGC-1alpha through S6 kinase. Genes Dev. 25:1232–1244 10.1101/gad.2054711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C., Leow S.C., Anso E., Rocha S., Thotakura A.K., Tornatore L., Moretti M., De Smaele E., Beg A.A., Tergaonkar V., et al. 2011. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 13:1272–1279 10.1038/ncb2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 448:553–560 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen O.H., Frandsen L., Schjerling P., Nishimura E., Grunnet N. 2006. PGC-1alpha and PGC-1beta have both similar and distinct effects on myofiber switching toward an oxidative phenotype. Am. J. Physiol. Endocrinol. Metab. 291:E807–E816 10.1152/ajpendo.00591.2005 [DOI] [PubMed] [Google Scholar]
- Mourkioti F., Kratsios P., Luedde T., Song Y.H., Delafontaine P., Adami R., Parente V., Bottinelli R., Pasparakis M., Rosenthal N. 2006. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J. Clin. Invest. 116:2945–2954 10.1172/JCI28721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J.M., Bakkar N., Guttridge D.C. 2011. NF-κB signaling in skeletal muscle health and disease. Curr. Top. Dev. Biol. 96:85–119 [DOI] [PubMed] [Google Scholar]
- Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., et al. 2009. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187:859–874 10.1083/jcb.200903131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac L.R., Janssen P.M., Montgomery C.L., Coley B.D., Chicoine L.G., Clark K.R., Mendell J.R. 2007. A translational approach for limb vascular delivery of the micro-dystrophin gene without high volume or high pressure for treatment of Duchenne muscular dystrophy. J. Transl. Med. 5:45–56 10.1186/1479-5876-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin L.A., Rudnicki M.A. 2000. The molecular regulation of myogenesis. Clin. Genet. 57:16–25 10.1034/j.1399-0004.2000.570103.x [DOI] [PubMed] [Google Scholar]
- Sahin E., Colla S., Liesa M., Moslehi J., Müller F.L., Guo M., Cooper M., Kotton D., Fabian A.J., Walkey C., et al. 2011. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 470:359–365 10.1038/nature09787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557–567 10.1038/nrm1981 [DOI] [PubMed] [Google Scholar]
- Scarpulla R.C. 2008. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 88:611–638 10.1152/physrev.00025.2007 [DOI] [PubMed] [Google Scholar]
- Schieke S.M., Phillips D., McCoy J.P.J., Jr, Aponte A.M., Shen R.F., Balaban R.S., Finkel T. 2006. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281:27643–27652 10.1074/jbc.M603536200 [DOI] [PubMed] [Google Scholar]
- Schilling J., Lai L., Sambandam N., Dey C.E., Leone T.C., Kelly D.P. 2011. Toll-like receptor-mediated inflammatory signaling reprograms cardiac energy metabolism by repressing peroxisome proliferator-activated receptor γ coactivator-1 signaling. Circ Heart Fail. 4:474–482 10.1161/CIRCHEARTFAILURE.110.959833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.M., Winge D.R. 2009. Shaping the mitochondrion: mitochondrial biogenesis, dynamics and dysfunction. Conference on Mitochondrial Assembly and Dynamics in Health and Disease. EMBO Rep. 10:1301–1305 10.1038/embor.2009.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Lam A., Li M.C., Ngan M., Menenzes S., Zhao Y. 2007. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 3:11–17 [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., Mehl I.R., Chong L.W., Nofsinger R.R., Evans R.M. 2007. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. USA. 104:5223–5228 10.1073/pnas.0611623104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman B.M. 2007. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found. Symp. 287:60–63 10.1002/9780470725207.ch5 [DOI] [PubMed] [Google Scholar]
- Suliman H.B., Sweeney T.E., Withers C.M., Piantadosi C.A. 2010. Co-regulation of nuclear respiratory factor-1 by NFkappaB and CREB links LPS-induced inflammation to mitochondrial biogenesis. J. Cell Sci. 123:2565–2575 10.1242/jcs.064089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ge Y., Drnevich J., Zhao Y., Band M., Chen J. 2010. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 189:1157–1169 10.1083/jcb.200912093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Takeuchi O., Tsujimura T., Itami S., Adachi O., Kawai T., Sanjo H., Yoshikawa K., Terada N., Akira S. 1999. Limb and skin abnormalities in mice lacking IKKalpha. Science. 284:313–316 10.1126/science.284.5412.313 [DOI] [PubMed] [Google Scholar]
- Tedesco F.S., Dellavalle A., Diaz-Manera J., Messina G., Cossu G. 2010. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 120:11–19 10.1172/JCI40373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P.H., Keats J.J., Wang H., Vignali D.A., Bergsagel P.L., Karin M. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 9:1364–1370 10.1038/ni.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gammeren D., Damrauer J.S., Jackman R.W., Kandarian S.C. 2009. The IkappaB kinases IKKalpha and IKKbeta are necessary and sufficient for skeletal muscle atrophy. FASEB J. 23:362–370 10.1096/fj.08-114249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hertlein E., Bakkar N., Sun H., Acharyya S., Wang J., Carathers M., Davuluri R., Guttridge D.C. 2007. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 27:4374–4387 10.1128/MCB.02020-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Garzon R., Sun H., Ladner K.J., Singh R., Dahlman J., Cheng A., Hall B.M., Qualman S.J., Chandler D.S., et al. 2008. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 14:369–381 10.1016/j.ccr.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder W.W. 2001. Energy-sensing and signaling by AMP-activated protein kinase in skeletal muscle. J. Appl. Physiol. 91:1017–1028 [DOI] [PubMed] [Google Scholar]
- Wu L., D’Amico A., Winkel K.D., Suter M., Lo D., Shortman K. 1998. RelB is essential for the development of myeloid-related CD8alpha- dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 9:839–847 10.1016/S1074-7613(00)80649-4 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Verma U.N., Prajapati S., Kwak Y.T., Gaynor R.B. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 423:655–659 10.1038/nature01576 [DOI] [PubMed] [Google Scholar]
- Zarnegar B.J., Wang Y., Mahoney D.J., Dempsey P.W., Cheung H.H., He J., Shiba T., Yang X., Yeh W.C., Mak T.W., et al. 2008. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378 10.1038/ni.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]