Abstract
Myotonic dystrophy type 1 and type 2 (DM1 and DM2) are genetic diseases in which mutant transcripts containing expanded CUG or CCUG repeats cause cellular dysfunction by altering the processing or metabolism of specific mRNAs and miRNAs. The toxic effects of mutant RNA are mediated partly through effects on proteins that regulate alternative splicing. Here we show that alternative splicing of exon 29 (E29) of CaV1.1, a calcium channel that controls skeletal muscle excitation–contraction coupling, is markedly repressed in DM1 and DM2. The extent of E29 skipping correlated with severity of weakness in tibialis anterior muscle of DM1 patients. Two splicing factors previously implicated in DM1, MBNL1 and CUGBP1, participated in the regulation of E29 splicing. In muscle fibers of wild-type mice, the CaV1.1 channel conductance and voltage sensitivity were increased by splice-shifting oligonucleotides that induce E29 skipping. In contrast to human DM1, expression of CUG-expanded RNA caused only a modest increase in E29 skipping in mice. However, forced skipping of E29 in these mice, to levels approaching those observed in human DM1, aggravated the muscle pathology as evidenced by increased central nucleation. Together, these results indicate that DM-associated splicing defects alter CaV1.1 function, with potential for exacerbation of myopathy.
INTRODUCTION
Myotonic dystrophy (DM) is an autosomal dominant disorder characterized by skeletal myopathy, cardiac arrhythmia, cataracts, hypogonadism, hypersomnolence, insulin resistance and other symptoms (1). The most conspicuous features are myotonia and muscle weakness. Although the true prevalence of DM is unknown, it is one of the most common forms of muscular dystrophy (2). There are two types of DM, both resulting from expansions of simple tandem repeats in non-coding regions of the genome. DM type 1 (DM1) is caused by an expansion of CTG repeats in the 3′-untranslated region of DMPK (dystrophia myotonica protein kinase) (3), whereas DM type 2 (DM2) is caused by an expansion of CCTG repeats within the first intron of ZNF9 (zinc finger 9) (4).
The disease mechanism in DM involves a toxic gain-of-function by RNAs expressed from the mutant DMPK or ZNF9 alleles. The RNAs with expanded CUG (CUGexp) or CCUG (CCUGexp) repeats bind to Muscleblind-like 1 (MBNL1) protein with high affinity, resulting in sequestration of MBNL1 in nuclear foci and a corresponding loss of its activity as a regulator of splicing and miRNA processing (5–8). In DM1, the CUGexp RNA has the additional effect of upregulating CUG-binding protein 1 (CUGBP1) (9–14), but evidence that this also occurs in DM2 is conflicting (15–17). These effects on RNA-binding proteins lead to misregulated alternative splicing and other changes of the muscle transcriptome (18–21). Although an exact animal model of DM1 does not exist, mouse models with ablation of Mbnl1, overexpression of CUGBP1 or expression of CUGexp RNA partially reproduce the transcriptomic and clinical features of the disease (20–24).
DM is associated with misregulated alternative splicing but for most of the affected transcripts the physiological consequences are unknown. There is evidence that myotonia results from misregulated alternative splicing of the CLCN1 chloride ion channel, causing a loss of channel function and involuntary runs of muscle action potentials (25–29). Insulin resistance is also a characteristic feature of DM1, and may result from misregulated alternative splicing of the insulin receptor (12).
Excessive calcium entry has long been considered a key initiator of muscle degeneration in Duchenne muscular dystrophy (30,31). Studies of mice that overexpress TRPC3, a calcium entry channel, indicated that increased calcium influx is sufficient to cause progressive dystrophic changes in skeletal muscle (32). Malignant hyperthermia and central core disease are other hereditary disorders caused by altered calcium regulation in muscle (33,34). However, few studies have suggested calcium influx as a mechanism for DM (35–37) because there is no primary defect of the muscle membrane and no known alteration of calcium entry channels.
Here we show that DM is associated with misregulated alternative splicing of CACNA1S, the gene encoding CaV1.1. CaV1.1 is an L-type calcium channel (DHPRα1S) and voltage sensor that plays a central role in excitation–contraction (EC) coupling (38–41). In DM1 and DM2, there is increased skipping of CACNA1S exon 29 (E29), an exon that is developmentally regulated in skeletal muscle (42). In a prospective cohort, the extent of E29 skipping was correlated with the severity of muscle weakness. When splice-shifting oligonucleotides were used to induce E29 skipping in wild-type (WT) mice, CaV1.1 conductance and voltage sensitivity were increased and a contribution of Ca2+ influx to the electrically evoked myoplasmic Ca2+ transient was observed in single adult muscle fibers, similar to previous observations from expressing E29-skipped CaV1.1 in dysgenic (CaV1.1-null) myotubes (43). Although E29, like several other DM1-affected exons, showed antagonistic regulation by MBNL1 and CUGBP1, the E29 splicing defect in mice that express CUGexp RNA was much less profound than in individuals with DM1. However, when splice shifting oligonucleotides were used to induce E29 skipping in this mouse model, the extent of the myopathy was enhanced, as evidenced by an increased frequency of central nuclei. These results suggest that the combined effects of misregulated splicing of several genes involved in calcium regulation and EC coupling may contribute to the muscle degeneration in DM.
RESULTS
CaV1.1-E29 skipping in DM1 and DM2 and correlation with muscle strength
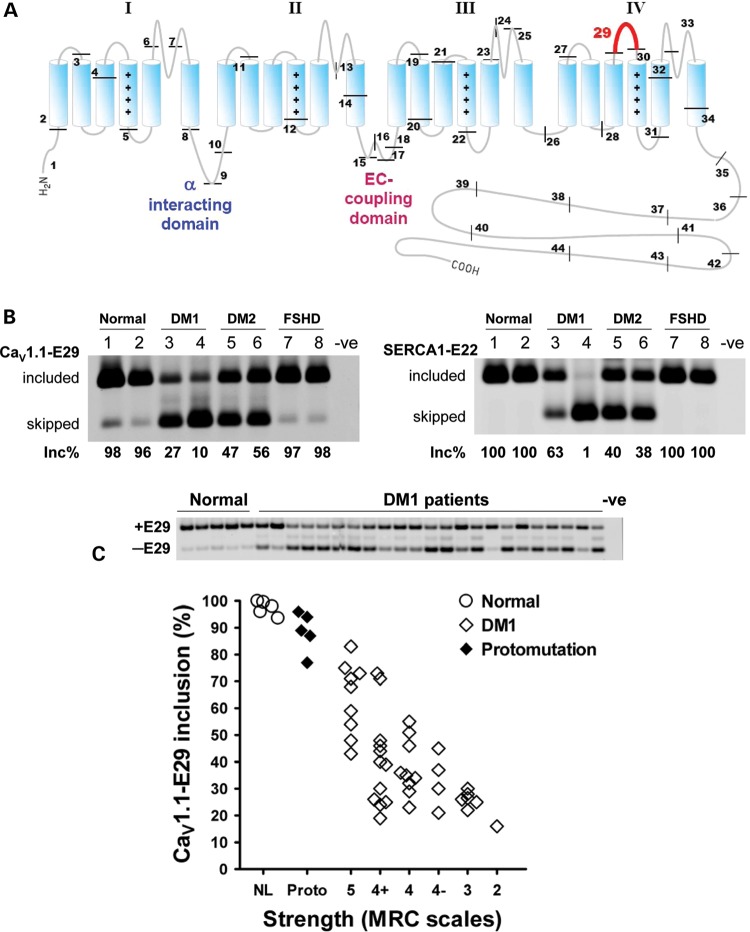
We identified abnormal skipping of CaV1.1-E29 in DM muscle based on all-exon expression profiling of DM1 and DM2 compared with normal and disease controls (Sobczak et al., manuscript in preparation). Skipping of E29 deletes a 19-amino acid fragment within the IVS3–IVS4 extracellular loop, adjacent to the IVS4 voltage sensor (Fig. 1A). Reverse transcriptase polymerase chain reaction (RT-PCR) analysis of DM1 and DM2, compared with disease controls (facioscapulohumeral muscular dystrophy, FSHD) and healthy individuals, confirmed that CaV1.1-E29 splicing was abnormally regulated in both types of DM (Fig. 1B). Compared with the near-complete E29 inclusion in normal and FSHD controls, E29 splicing was markedly repressed in DM1 and DM2 (Fig. 1B, left panel, lanes 3–6). Previous studies have shown that splicing of SERCA1 exon 22 (SERCA1-E22) is also repressed in DM1 patients (15,44). This exon, which is regulated by MBNL1 (15,45), can shift from near-complete inclusion in controls to near-complete skipping in DM1 (Fig. 1B, right; compare lanes 1 and 4). By comparison, the effect on CaV1.1-E29 splicing was nearly as large (Fig. 1B, left; compare lanes 1 and 4).
Figure 1.
Correlation of CaV1.1 E29 skipping and muscle strength in DM. (A) Schematic representation of CaV1.1 topology and exon organization. CaV1.1 is composed of four membrane-spanning repeat domains (I–IV). Each repeat consists of six transmembrane segments (S1–S6). The protein segment encoded by the alternatively spliced E29 (57 nts, highlighted in red) is located in the IVS3–IVS4 extracellular loop. Positive charges in the S4 segments of each repeat are indicated by ‘+’ symbols. Locations of the α interacting domain in the I–II loop, which binds the β1α subunit, and the EC coupling domain in the II–III intracellular loop, are also indicated. (B) RT–PCR assay for alternative splicing of CaV1.1 E29 (CaV1.1-E29, left) and SERCA1 exon 22 (SERCA1-E22, right) in muscle from normal (lanes 1 and 2), DM1 (lanes 3 and 4), DM2 (lanes 5 and 6) and FSHD (lanes 7 and 8) individuals. Inc% denotes the fractional inclusion rate, calculated as the signal intensity of inclusion (upper) band divided by the summed intensities of the corresponding inclusion and exclusion (lower) bands. Negative control (-ve) refers to the absence of template. (C) Upper panel shows representative RT–PCR E29 inclusion results for several normal (lanes 1–5) and DM1-affected (lanes 6–28) individuals. Negative control (-ve) refers to the absence of template. Lower panel shows correlation of ADF strength with fractional E29 inclusion in TA muscles from healthy individuals (n = 5, ‘NL’), DM1 protomutation (n = 5, ‘Proto’) and classical DM1 (n = 41). Strength was determined by standardized manual muscle testing using Medical Research Council scales (46). An MRC scale value of 5 indicates normal strength, 4 indicates moderate weakness, 3 indicates full active range of movement against gravity with no added resistance and 2 indicates inability to dorsiflex fully against gravity. The Spearman rank correlation for E29 inclusion versus strength in classical DM1 was r = 0.6560 (P < 0.0001, excludes normal and protomutation subjects).
Because CaV1.1 plays a key role in EC coupling (41), we examined E29 splicing in relation to muscle strength in individuals with DM1. Ankle dorsiflexion (ADF) is primarily a function of tibialis anterior (TA), a muscle that is preferentially affected in DM1. We used standardized manual muscle testing (46,47) to prospectively assess ADF strength in 41 individuals with genetically proven DM1. The TA muscle was then sampled by needle biopsy. Similar procedures were followed in 5 individuals who carried a DM1 protomutation, defined here as 50–90 CTG repeats, and in 5 healthy controls. The individuals with protomutations had minimal or no symptoms and normal ADF strength; they came to diagnosis through their DM1-affected offspring. RT–PCR analysis of TA muscle RNA showed abnormal CaV1.1-E29 skipping in all individuals with DM1 (<85% inclusion) when compared with 93–100% inclusion in healthy controls (Fig. 1C). Notably, CaV1.1-E29 inclusion was moderately reduced (61 ± 16.8% inclusion, n = 11) in DM1-affected individuals having normal ADF strength (Medical Research Council or MRC grade 5), indicating that splicing of this exon is affected at an early stage of the disease process. Overall, the level of E29 skipping was correlated with weakness of the TA muscle (Spearman non-parametric r = 0.6560, P < 0.0001). All individuals with moderate or severe weakness (MRC grade ≤ 4) had CaV1.1-E29 inclusion levels below 55% (mean value 33 ± 9.6% inclusion, n = 20). E29 splicing did not correlate with length of the CTG expansion in DMPK, as determined by genetic analysis of circulating leucocytes (data not shown). Taken together, these data raised the possibility that misregulated alternative splicing of CaV1.1-E29 could modify CaV1.1 channel function in DM1.
MBNL1 and CUGBP1 participate in the regulation of CaV1.1-E29 alternative splicing
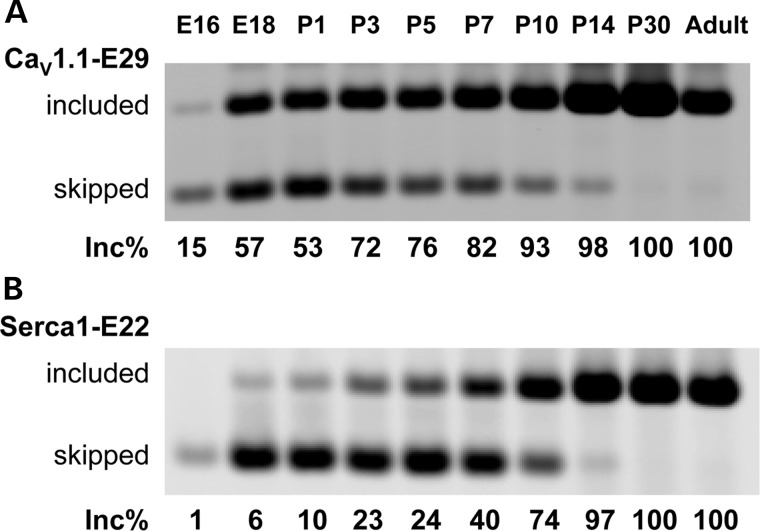
CaV1.1-E29 is predominantly skipped in myoblasts and included in adult muscle (43). Previous studies of SERCA1-E22, fast troponin T (TNNT3-fetal exon) and other MBNL1-dependent exons have shown a coordinate program of splice switching that occurs during late fetal and early postnatal development (15,48). To determine the developmental regulation of CaV1.1 splicing during this period, we analysed E29 inclusion in mouse hindlimb muscle from embryonic day 16 (E16) to postnatal day 30 (P30). The fractional inclusion of E29 was 15% at E16, similar to the lowest levels observed in DM1, rising to 100% at P30. This result was similar to the pattern recently reported by Flucher and Tuluc (42). The postnatal transition from E29 skipping to inclusion occurred during a similar developmental window as SERCA1-E22, from P1 to P14 (Fig. 2). However, different from SERCA1-E22 and TNNT3-fetalEx (15), CaV1.1-E29 exhibited a more prominent early shift towards inclusion between E16 and E18.
Figure 2.
Analysis of CaV1.1 E29 inclusion in mouse hindlimb muscle during late fetal and early postnatal development. (A) The inclusion level of CaV1.1 E29 and (B) Serca1 exon 22 (Serca1-E22) is shown from E16 to P30. Hindlimb muscles from four mice were pooled for RNA extraction at embryonic stages and from two mice for postnatal stages.
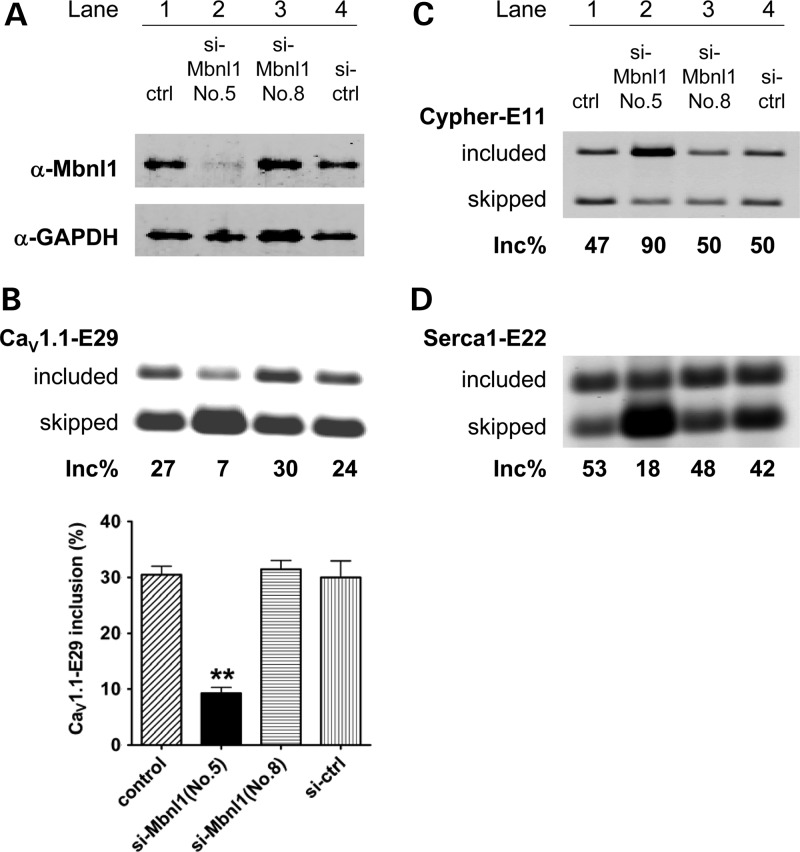
To examine the role of Mbnl1 protein in CaV1.1-E29 splicing regulation, we transfected mouse C2C12 myoblasts with Mbnl1 siRNA. The Mbnl1 protein was knocked down by >80% (Fig. 3A). The basal level of CaV1.1-E29 inclusion (30%) was decreased to 9% by reduction in Mbnl1 (Fig. 3B and C). As positive controls for reduction in Mbnl1 activity, we examined two of its physiological targets, Serca1-E22 and Cypher exon 11 (49), and both responded strongly to Mbnl1 knockdown (Fig. 3C and D). These results indicated that Mbnl1 is a direct or indirect regulator of E29 splicing.
Figure 3.
Mbnl1 knockdown enhances CaV1.1 E29 skipping in C2C12 mouse myoblasts. (A) Immunoblot for Mbnl1 in C2C12 myoblasts treated with mock transfection (lane 1), si-RNA targeting Mbnl1 (lanes 2 and 3) and scrambled si-RNA control (si-ctrl). Efficient knockdown was achieved with si-Mbnl1-No.5 but not si-Mbnl1-No.8. RT–PCR assay for alternative splicing of CaV1.1-E29 (B), Cypher exon 11 (C) and Serca1-E22 (D) (same cells/lanes as labeled in A and C). Quantification of CaV1.1-E29 inclusion for each condition is shown in the lower panel of (B). Mbnl1-knockdown significantly reduced CaV1.1-E29 inclusion (**P < 0.01).
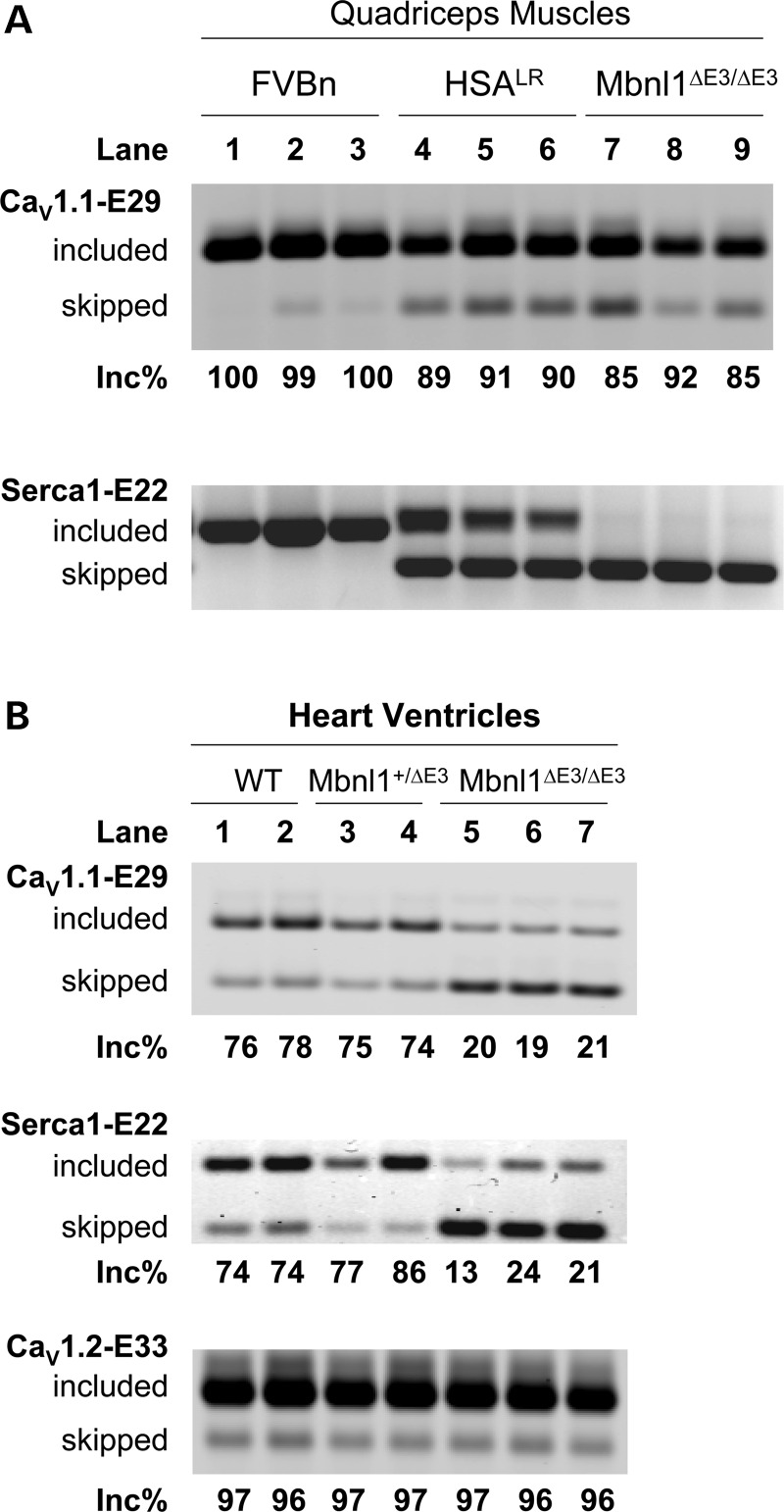
Next, we examined CaV1.1-E29 splicing in two DM mouse models, human skeletal actin-long repeat (HSALR) transgenic mice that express high levels of CUGexp RNA in muscle (22) and Mbnl1▵E3/▵E3 mice that have ablation of Mbnl1 (50). Surprisingly, both models showed only slight repression of E29 in skeletal muscle (Fig. 4A, upper panel, lanes 4–9). This was in sharp contrast to the splicing defect of Serca1-E22, which was strongly repressed in both models (Fig. 4A, lower panel, lanes 4–9). However, a different pattern emerged in cardiac muscle. CaV1.1 expression in heart tissue is much lower than CaV1.2, and CaV1.1 is not known to have a physiological role in cardiomyocytes. Nevertheless, the CaV1.1 transcript was detected and splicing of E29 was markedly repressed in Mbnl1 knockout hearts compared with WT and heterozygous littermates (Fig. 4B, upper panel). A significant repression of Serca1-E22 was also found in Mbnl1▵E3/▵E3 hearts (Fig. 4B, middle panel). In contrast, ablation of Mbnl1 had no effect on splicing of CaV1.2 exon 33, the homologous alternative exon for the L-type calcium channel that predominates in heart (Fig. 4B, lower panel). Taken together, these results support the concept that CaV1.1-E29 is MBNL1 regulable, but the exact role of this protein in sustaining E29 inclusion may depend on the species and cellular context.
Figure 4.
CaV1.1 E29 is only weakly skipped in skeletal muscle of DM1 mouse models, but more strongly repressed in hearts. (A) RT–PCR assay of CaV1.1-E29 (upper) and Serca1-E22 (lower) splicing in quadriceps muscle of FVBn WT, HSALR transgenic and Mbnl1-knockout (Mbnl1ΔE3/ΔE3) mice. (B) RT–PCR assays of CaV1.1-E29 (upper), Serca1-E22 (middle) and CaV1.2 exon 33 (lower) splicing in cardiac ventricular muscle of adult (6 months) male WT, Mbnl1-heterozygote (Mbnl1+/ΔE3) and Mbnl1-knockout (Mbnl1ΔE3/ΔE3) littermates. Note that strong amplification products were observed for CaV1.2-E33 after 25 cycles of RT–PCR, when compared with weaker products for CaV1.1-E29 after 30 cycles, as expected because CaV1.2 is the predominant L-type Ca2+ channel expressed in cardiomyocytes.
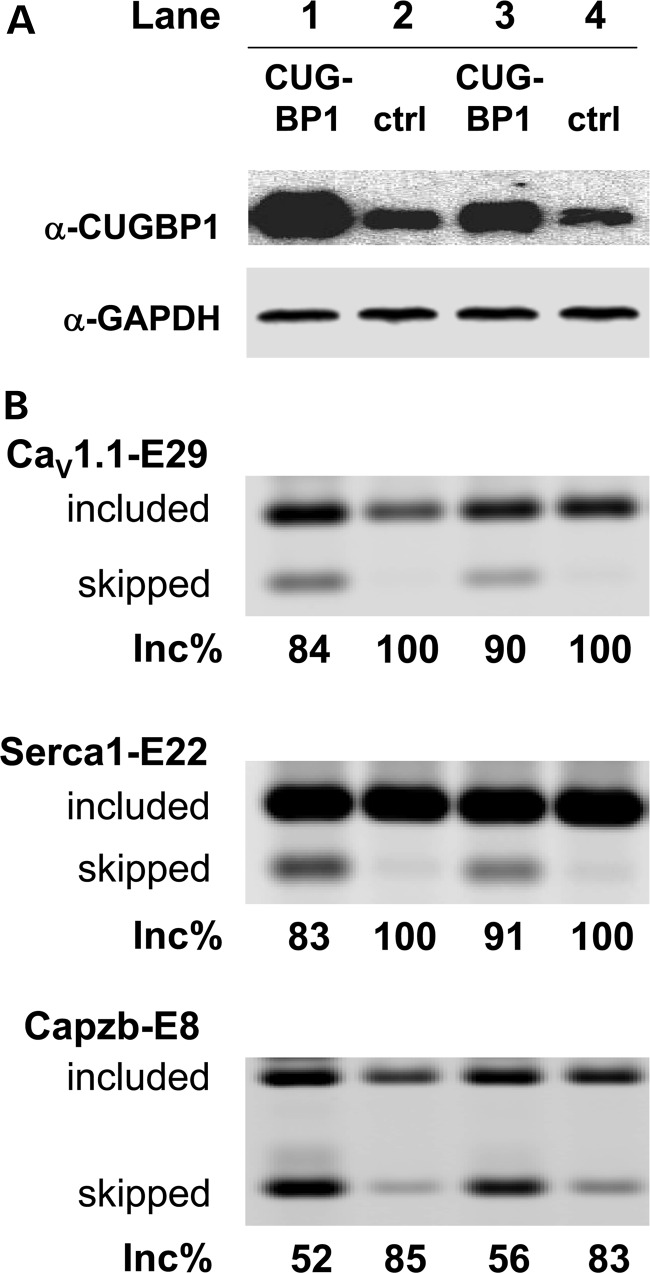
CUGBP1 is another splicing factor implicated in DM pathogenesis (9–14). As discussed above, CUGBP1 is upregulated in DM1 muscle cells, but for unclear reasons this does not occur in HSALR transgenic mice. Because CUGBP1 and MBNL1 have antagonistic effects on splicing of some exons (51), we examined the effect of CUGBP1 overexpression on CaV1.1-E29 splicing in mouse muscle in vivo. A CUGBP1 expression construct was injected and electroporated into TA of WT mice, whereas TA in the opposite hindlimb was electroporated with empty vector. Muscle was harvested 9 days later and overexpression of CUGBP1 was verified by western blot (Fig. 5A, lanes 1 and 3). As expected, CUGBP1 splicing activity was increased, as reflected by repression of its physiological target, exon 8 of Capzb (23,51) (Fig. 5B, lower panel). We found that CUGBP1 overexpression also caused a modest reduction in CaV1.1-E29 splicing (Fig. 5B, upper panel), with the fractional inclusion dropping from 100% in control to 84–90% in TA muscles overexpressing CUGBP1. These results suggest that CaV1.1-E29 is also CUGBP1 responsive, and that upregulation of CUGBP1 may contribute to CaV1.1-E29 skipping in DM. However, we cannot be certain that E29 is directly responsive to CUGBP1 because, as previously reported (23), CUGBP1 overexpression led to muscle regeneration (Supplementary Material, Fig. S1, see Discussion).
Figure 5.
Overexpression of CUGBP1 represses CaV1.1-E29 in mouse TA muscles. (A) Immunoblot for the CUGBP1 protein in mouse TA 9 days after intramuscular injection and electroporation of CUGBP1 expression construct (lanes 1 and 3) or empty vector control (lanes 2 and 4). GAPDH was used as a loading control. (B) RT–PCR assay of CaV1.1 E29 (upper), Serca1-E22 (middle) and Capzb—exon 8 (lower) splicing in mouse TA muscle 9 days after injection/electroporation of CUGBP1 expression construct (lanes 1 and 3) or empty vector control (lanes 2 and 4).
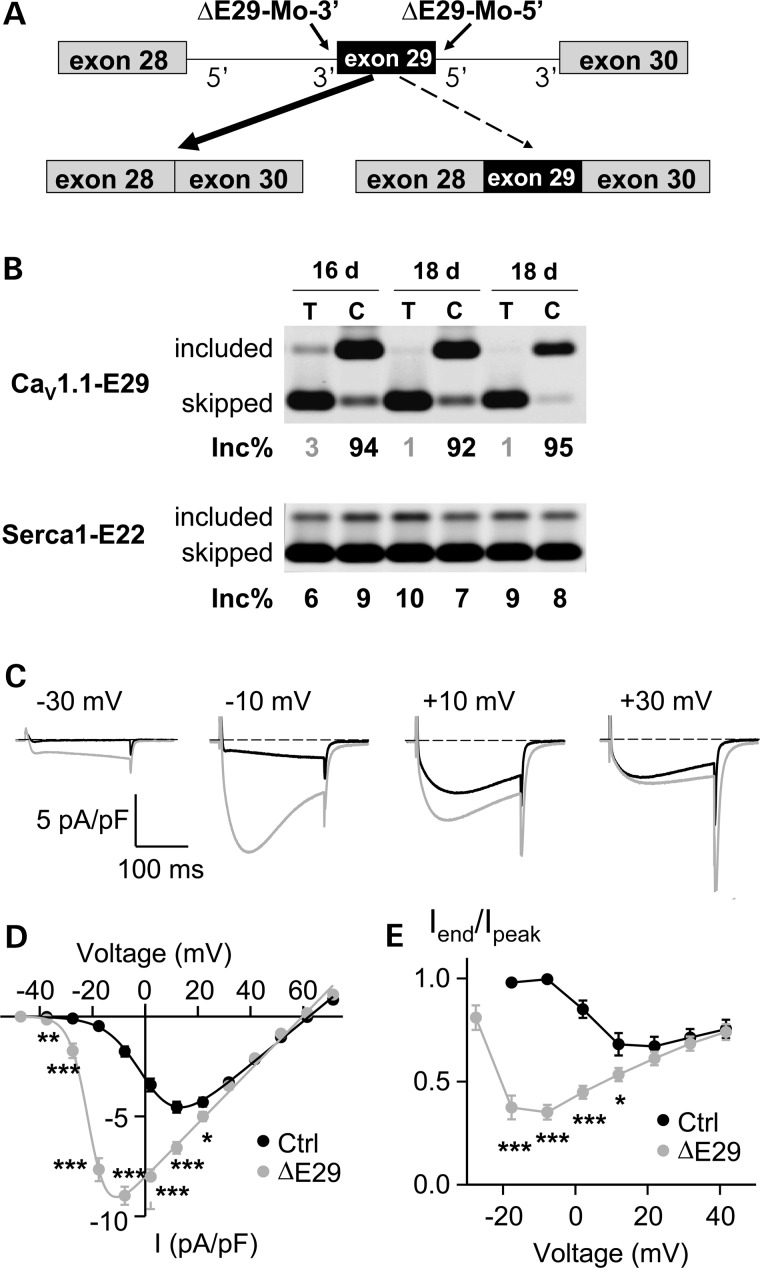
Skipping of CaV1.1 E29 increases L-type Ca2+ channel conductance and voltage sensitivity in mouse flexor digitorum brevis (FDB) muscle fibers
When GFP-tagged E29-deleted CaV1.1 (CaV1.1Δ29) was over-expressed in dysgenic (CaV1.1-null) myotubes, the channel was correctly targeted into triads and displayed increased L-type Ca2+ channel conductance, activation/inactivation kinetics and voltage sensitivity for activation (43). However, the degree to which these functional effects of the ΔE29 isoform are observed for physiological expression levels in fully differentiated muscle fibers is unclear because CaV1.1 channel properties are strongly influenced by auxiliary subunits and other proteins in the muscle membrane (52). To test this directly, we used splice-shifting antisense oligonucleotides (ASOs) to drive CaV1.1-E29 skipping in muscle fibers of WT mice. Flexor digitorum brevis (FDB) muscles of 3-week-old mice were injected and electroporated with morpholinos targeting the 3′ and 5′ splice sites of CaV1.1-E29 (Fig. 6A). Previously, we showed that this procedure is effective for loading ASOs into muscle fibers and modulating alternative splicing for up to 14 weeks (27,53). RT–PCR assays conducted 16–18 days after electroporation confirmed that ▵E29 morpholinos induced nearly complete E29 skipping without affecting Serca1-E22 inclusion (Fig. 6B, lower panel). In contrast, injection and electroporation of vehicle (saline, Fig. 6B, upper panel) or control morpholino (reverse sequence, Supplementary Material, Fig. S2) in the contralateral FDB muscle had no effect on E29 splicing. In ASO-injected FDB muscles, the inclusion of E29 was reduced from 92–95% down to 1–3%. Consistent with previous studies in myotubes (43), the patch clamp recordings of FDB fibers demonstrated that E29 deletion caused a dramatic hyperpolarizing shift in the voltage dependence of channel activation (Fig. 6C and D). Specifically, the voltage required for half-maximal channel activation (V0.5) was shifted from +1.4 ± 1.3 mV in control to −21.0 ± 0.7 mV following ASO-mediated E29 skipping. In addition, maximal L-type Ca2+ channel conductance (Gmax) was significantly increased (∼27%) following E29 deletion (Supplementary Material, Table S1), although E29 deletion produced a much larger increase in Gmax (>3-fold) following expression in CaV1.1-null myotubes (43). As a result of these two effects, peak L-type Ca2+ current density in E29-deleted fibers was significantly increased for all voltages from −40 to +20 mV (Fig. 6D). Modest effects on channel reversal potential (Vrev) and steepness of voltage activation (kg) were also observed (Supplementary Material, Table S1).
Figure 6.
CaV1.1-E29 skipping enhances L-type Ca2+ channel conductance and voltage sensitivity in mouse FDB muscle fibers. (A) Diagram of splice shifting antisense morpholino oligonucleotides used to induce CaV1.1 E29 skipping. The ▵E29 morpholinos target sequences near the 3′ and 5′ splice sites of CaV1.1 E29. (B) RT–PCR assay for CaV1.1-E29 (upper) and Serca1-E22 (lower) splicing 16 and 18 days following a single injection and electroporation of ▵E29 morpholinos into FDB muscle. ‘C’ denotes control FDB muscle that was injected and electroporated with vehicle (saline) alone. RNA for the assay was recovered from FDB fibers remaining after completion of patch-clamp recordings. (C) Representative calcium current (ICa) recordings in control (Ctrl, black) and CaV1.1-E29 skipping (ΔE29, grey) fibers obtained during 200 ms depolarizations to −30, −10, +10 and +30 mV. (D) Average (±SE) ICa–V relationships for control (black circles) and ΔE29-deleted fibers (grey circles) fitted by equation (1) with the following parameters for control: Gmax = 108 nS/nF, V0.5 = 1.1 mV, kg = 6.9 mV, Vrev = 62.2 mV and for ▵E29: Gmax = 135 nS/nF, V0.5 = −21.3 mV, kg = 3.5 mV, Vrev = 59.7 mV. Data were obtained from n = 14 fibers for each group. *P < 0.05, **P < 0.01 and ***P < 0.001. (E) The voltage dependence of fractional inactivation, represented as Iend/Ipeak, plotted against test depolarization voltage. All symbols have the same meaning as in (D). Data are presented as mean ± SEM, 14 fibers were recorded for both control and ▵E29 morpholino-treated fibers.
ASO-induced E29 skipping also resulted in an increase and hyperpolarizing shift in fractional channel inactivation. Specifically, average maximum fractional inactivation was only 0.32 at +10 mV in control and 0.65 at −10 mV following ΔE29 skipping (Fig. 6E). Thus, a similar marked shift to more hyperpolarized potentials was observed for the voltage dependencies of both channel activation and inactivation. Consistent with the findings of Tuluc et al. (43), the apparent rate of channel activation at the peak of the respective current–voltage relationships was significantly (P < 0.001) faster following E29 deletion (14.0 ± 0.9 ms at −10 mV) compared with control (23.8 ± 1.3 ms at +10 mV, P < 0.001). However, the rate of channel activation was not significantly different between control and E29-deleted fibers at stronger test potentials where current density and fractional inactivation were similar (e.g. from +30 to +50 mV). Overall, results following ASO-induced E29 skipping in adult muscle are consistent with the findings of Tuluc et al. (43) in myotubes with regard to increased CaV1.1 conductance, inactivation kinetics and voltage sensitivity for activation, although the effects of E29 exclusion on channel activation and conductance were somewhat less pronounced in the context of adult muscle fibers.
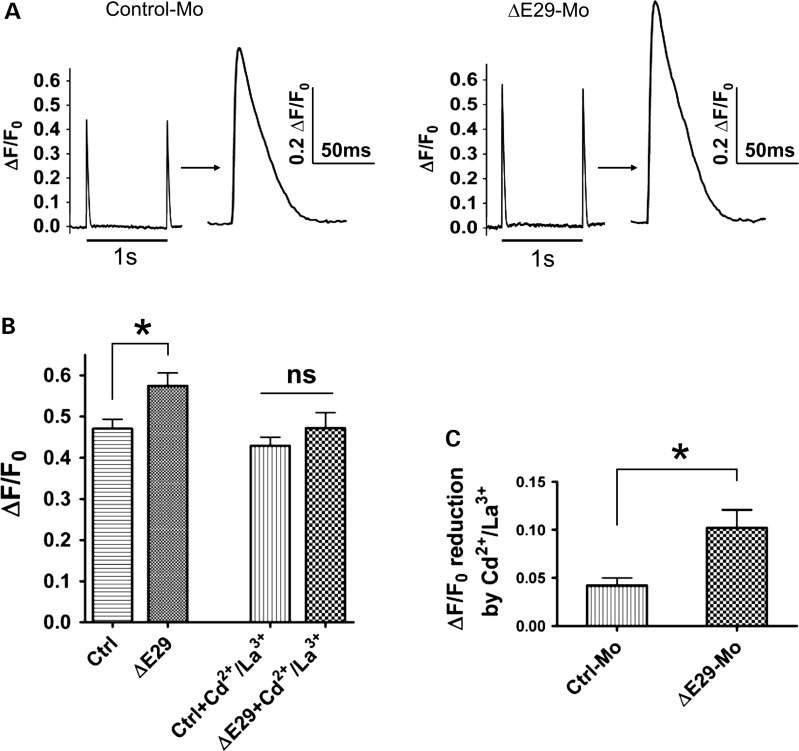
Exclusion of CaV1.1 E29 enhances electrically evoked Ca2+ release in mouse FDB muscle fibers
To test whether CaV1.1ΔE29 channels with increased voltage sensitivity and Ca2+ current density significantly impacts Ca2+ release during EC coupling in adult muscle fibers, we recorded electrically evoked Ca2+ transients in FDB fibers from mice treated with either control or ΔE29 morpholinos for 6–7 weeks. Near complete E29 skipping by electroporation of ▵E29 morpholinos in these experiments was confirmed by RT–PCR (Supplementary Material, Fig. S2, mice 7–10). Ten single-supramaximal electrical stimuli delivered at 1 Hz were applied to FDB fibers loaded with the low affinity Ca2+ dye, mag-fluo-4 and bathed in control extracellular Ringer's solution. Figure 7A shows representative traces of electrically evoked Ca2+ release for FDB fibers from mice treated with either control (left) or E29 (right) morpholinos. A statistically significant (P < 0.05, t-test) increase in peak electrically evoked relative mag-fluo-4 fluorescence (ΔF/F0) was observed in E29-skipped fibers (Fig. 7B). Specifically, average peak ▵F/F0 increased 22% from 0.47 ± 0.02 (n = 16) in the control-treated FDB fibers to 0.58 ± 0.03 (n = 18) in ▵E29-treated FDB fibers. This difference in evoked release was eliminated 2min after addition of Ringer' supplemented with 0.5 mm Cd2+ and 0.2 mm La3+ to block Ca2+ influx. Quantitative analysis revealed that the Cd/La-sensitive component of the electrically evoked Ca2+ transient was significantly larger (P < 0.05, t-test) in E29-skipped FDB fibers (Fig. 7C). These results indicate that CaV1.1▵E29 channels trigger a larger electrically evoked Ca2+ transient in adult muscle that is sensitive to block by extracellular Cd2+ and La3+. These results support a small, but significant, gain-of-function increase in depolarization-induced Ca2+ transients by E29-deleted CaV1.1 channels in adult muscle fibers, consistent with prior studies following over-expression of CaV1.1ΔE29 channels in dysgenic myotubes (43).
Figure 7.
CaV1.1-E29 skipping increases electrically evoked Ca2+ transients in FDB fibers. (A) Representative normalized mag-fluo-4 fluorescence transients (ΔF/F0) elicited during a 1 Hz train of electrical stimulation in mouse FDB fibers treated with either control morpholino (left, control-Mo) or ▵E29 morpholinos (right, ▵E29-Mo). Following measurement of Ca2+ transients, remaining fibers were collected for mRNA extraction to quantify the amount of CaV1.1-E29 skipping by RT–PCR (Supplementary Material, Fig. S2, mice 7–10, and data not shown). (B) Bar graph summarizing peak electrically evoked mag-fluo-4 transients (ΔF/F0) in mouse FDB fibers treated with either control (n = 16 fibers) or ▵E29 morpholinos (n = 18 fibers). Mag-fluo-4 transients were recorded both before (left) and after (right) addition of 0.5 mm Cd2+ and 0.2 mm La3+ for 2 min. *P < 0.05, t-test. No significant difference was found between the two groups in the presence of 0.5 mm Cd2+ and 0.2 mm La3+ (P > 0.3, t-test). FDB fibers were obtained from five mice each treated with either ▵E29 or control morpholinos. (C) Cd2+/La3+-sensitive component of electrically evoked mag-fluo-4 transients (ΔF/F0) in mouse FDB fibers treated with either control (n = 16 fibers) or ▵E29 morpholinos (n = 18 fibers). Addition of 0.5 mm Cd2+ and 0.2 mm La3+ produced a significantly greater reduction in the peak electrically evoked mag-fluo-4 transient in FDB fibers treated with ▵E29 morpholinos (*P< 0.05, t-test).
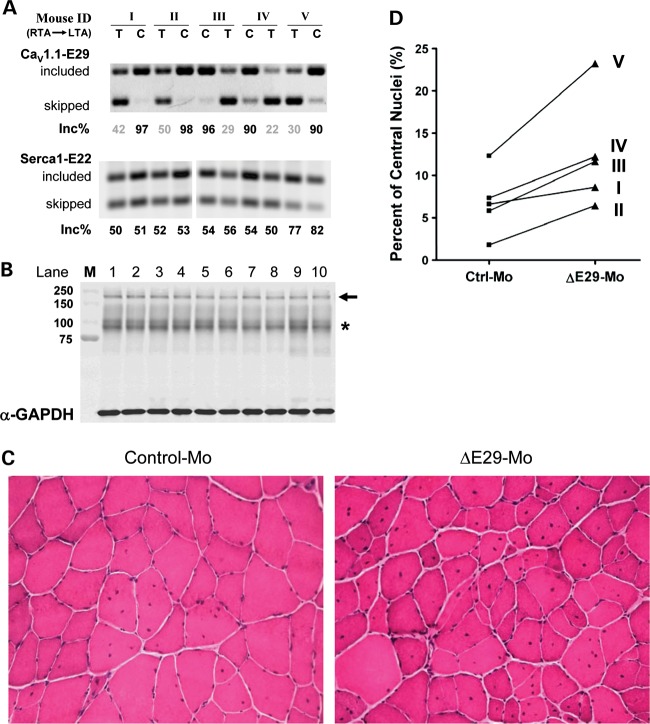
CaV1.1 E29 skipping aggravates myopathy in the HSALR mouse model of DM1
The degree of myopathy in HSALR mice is less severe than in human DM1 patients. As a first step to examine the potential role of CaV1.1 mis-splicing in myopathy, we used splice shifting ASOs to convert E29 splicing in HSALR mice to the pattern observed in human DM1. We postulated that E29 skipping, when combined with hyperexcitability (myotonia) and other splicing derangements that exist in this model, may exacerbate the myopathy. Morpholino ASOs were injected and electroporated into TA muscle of HSALR mice. RT–PCR analysis 3 months later confirmed that E29 splicing was repressed to levels comparable with moderately affected human DM1 muscle (E29 inclusion was 35 ± 5.0 and 94 ± 1.7% for ▵E29 and control morpholinos, respectively) (Fig. 8A). Immunoblots verified that total levels of CaV1.1 protein were not affected (Fig. 8B). Histological analysis of the entire cross-sectional area of TAs showed a significant increase in muscle fibers with central nuclei in muscles injected with ▵E29 ASOs (12.5 ± 2.9%), when compared with the contralateral TAs that received control ASO (6.8 ± 1.7%) (n = 5 mice, Fig. 8C and D, P < 0.05, paired t-test, Supplementary Material, Table S2).
Figure 8.
CaV1.1-E29 skipping in TA of HSALR mice increases the percentage of fibers with central nuclei. (A) RT–PCR analysis of CaV1.1-E29 (upper) and Serca1-E22 (lower) splicing in TA muscles of HSALR mice, 3 months following a single injection and electroporation of ΔE29 (T) or control (C) morpholinos. The left TA (LTA) or right TA (RTA) muscles of the same mouse were randomly selected for the treatment with either control or ΔE29 morpholinos. (B) Immunoblot of the CaV1.1 protein in TA muscles treated with ΔE29 or control morpholinos for the same 10 samples shown in (A). The CaV1.1-specific antibody detects a band around 200 kDa (arrow); * indicates non-specific cross-reactive bands. M, molecular weight marker. (C) Representative images of hematoxylin- and eosin-stained sections showing an increased fraction of fibers with central nuclei in TA muscles treated with ΔE29-morpholino (lower) compared with control morpholino (upper). (D) Quantification of the percentage of fibers with central nuclei in sections of contralateral TA muscles treated with control- (left) or ΔE29-skipping morpholinos (right) (P < 0.05, paired t-test).
DISCUSSION
MBNL1 and CUGBP1 promote the maturation of skeletal and cardiac muscle by controlling a key set of alternatively spliced exons (15,50,51). Many of these exons display a stereotypical pattern of developmental regulation in mice, switching from immature to mature splice products during the first 2 weeks of postnatal life. This period of extensive muscle remodeling is characterized by maturation of several structures, including the transverse tubule system (TTS), junctional triad and neuromuscular junction. A fundamental aspect of DM pathogenesis is that expression of CUGexp or CCUGexp RNA disrupts the function of splicing factors, causing reversion of splicing switches to their fetal/neonatal set points. One group of transcripts that are heavily impacted encode key proteins involved in calcium homeostasis and EC coupling (Table 1).
Table 1.
Genes related to Ca2+ regulation and EC coupling that display misregulated alternative splicing in DM1 patients and DM mouse models
| Gene and exon | Splice outcome in DM1 | Human DM1 | HSALR mice | Mbnl1 knockout mice | Functional consequence |
|---|---|---|---|---|---|
| CLCN1 exon7a (25,26) | Inclusion | +++ | +++ | +++ | Loss of Cl– conductance, repetitive action potentials |
| SERCA1 exon22 (15,44) | Skipping | +++ | +++ | +++ | Unknown |
| RyR1 AS1 (37,44) | Skipping | ++ | ++ | ? | ↑ depolarization-dependent Ca2+ release |
| CaV1.1 exon29 (this report) | Skipping | +++ | + | + | Altered gating, ↑ Ca2+ influx, ↑ Ca2+ transient |
| Junctin/Junctate (20) | skipping/3′-end formation | ? | ++ | ++ | Unknown |
‘+++’, ‘++’ or ‘+’ indicate large, moderate or minor changes of an alternative exon in DM1 patients or mouse models. ‘?’ indicates that alternative exon has not been determined.
In this respect, DM provides a window on muscle development by revealing the effects of expressing immature splice products in adult muscle cells. The splicing defect, or spliceopathy, does not lead to the production of ‘aberrant’ splice products, such as those resulting from point mutations in splice donor or acceptor sites. Instead, the DM-associated isoforms are natural splice products, although they are not optimal for adult muscle fibers. For example, in DM1, the inclusion of CLCN1 exon 7a (E7a), an exon normally repressed by MBNL1, causes a frameshift and loss of chloride channel function. In neonatal mice, chloride channels are dispensable because fibers are small and transverse tubules are rudimentary, but their absence in adult muscle causes a failure to counterbalance the activity-dependent potassium accumulation in the TTS, resulting in depolarization and myotonia. MBNL1 also regulates the splicing of BIN1 exon 11 (E11), whose inclusion promotes the correct formation of T tubules. Reduced BIN1-E11 splicing is proposed to result in TTS alterations that contribute to muscle weakness in DM1 (54). DM1 also causes spliceopathy of the RyR1 Ca2+ release channel thus enhancing depolarization-induced Ca2+ release (44), an alteration suggested to exacerbate myopathy through activation of Ca2+-dependent proteases (37).
Against this background of splicing changes affecting muscle excitability and EC coupling, we now report an effect of DM on alternative splicing of CaV1.1 E29. This exon is >93% included in TA muscles from healthy people, but in DM1 the fractional inclusion drops below 30% in TA muscles that show severe weakness. Considering that spliceopathy entails the re-emergence of immature splice products, it is possible that mis-splicing of E29 may represent a non-specific consequence of muscle degeneration/regeneration (55). However, the absence of E29 skipping in FSHD, a form of muscular dystrophy with chronicity similar to DM, and the occurrence of E29 skipping in TA muscle that is mildly affected and not discernably weak, would argue against this possibility. To our knowledge, this study is the first to show a splicing defect whose severity is correlated with muscle weakness across a broad spectrum of DM1-affected individuals, suggesting that E29 splicing may provide a useful biomarker of DM1 progression and therapeutic response. DM1 now becomes the third neuromuscular disorder, after malignant hyperthermia and hypokalemic periodic paralysis (56–58), associated with alterations of CaV1.1.
The mechanism of CaV1.1-E29 mis-splicing in DM1 is complex. Splicing of this exon is sensitive to levels of MBNL1, as shown by reduced E29 inclusion in C2C12 myoblasts and cardiac muscle following Mbnl1 knockdown or knockout. However, knockout of Mbnl1 resulted in only a small effect on E29 splicing in adult mouse skeletal muscle. Thus, there are other factors that participate in E29 regulation in mice, or compensatory changes that sustain E29 inclusion under conditions of constitutive MBNL1 deficiency. It is also possible that other MBNL isoforms promote E29 inclusion, although presently there is no evidence for upregulation of other family members, MBNL2 and MBNL3, in MBNL1 knockout mice. The proximity of several (U)GCAUG elements in the intron downstream of E29 suggests that this exon is also regulated by splicing factors in the RNA-binding Fox (Rbfox) family (59). Consistent with this proposal, a recent study showed that Rbfox1 knockout caused a reduction in E29 splicing in the brain (60). However, at present, there is no evidence for altered Rbfox activity in DM1.
In contrast to human DM1, expression of CUGexp RNA in transgenic mice resulted in only modest effects on E29 splicing. Although splicing changes in DM1 are generally conserved in HSALR mice (15), exceptions have been noted (54). While these discrepancies may result from species differences in splicing regulation, there are also indications that RNA toxicity and spliceopathy, as it occurs in human DM1, is not fully replicated in HSALR mice, either because the repeat expansion is much shorter (220 repeats in HSALR mice versus 2000–4000 repeats in adult DM1 skeletal muscle) (61) or due to differences in the RNA sequences flanking the repeat. For example, a difference from human DM1 is that levels of CUGBP1 protein and splicing outcomes for CUGBP1-dependent exons (e.g. Capzb exon 8) are not affected in the HSALR mice (15). It is noteworthy, therefore, that overexpression of CUGBP1 also caused partial repression of E29, and that antagonistic regulation by MBNL1 and CUGBP1 was previously observed for other exons (51). Thus, misregulated splicing of CaV1.1-E29 may require the combined effects of Mbnl protein sequestration and CUGBP1 upregulation.
Differences in splicing regulation may contribute to the overall milder phenotype of HSALR mice when compared with human DM1. To more fully reconstitute DM1 spliceopathy in this model, we used ASOs to further repress E29 splicing in TA muscle. In this mouse model, the level of CUGexp expression in TA is lower than in other hindlimb muscles (27), and consequently the histopathology of TA in young mice is relatively slight, consisting mainly of increased central nuclei. We found that induction of partial CaV1.1-E29 skipping for 12 weeks aggravated the pathology in HSALR TA muscle, as evidenced by a further increase in central nuclei. Notably, increased central nuclei in the absence of muscle necrosis or other signs of muscle regeneration are the earliest histologic feature of human DM1 (62). It will be interesting to determine if long-term, global induction of E29 skipping, using systemically administered ASOs, will further exacerbate the myopathy.
CaV1.1 functions as both an L-type Ca2+ channel and a voltage sensor for skeletal muscle EC coupling (41). The calcium ion conduction of CaV1.1 is evolutionarily conserved in mammals, but not required for its voltage sensing or EC coupling activities (38,63). We found that E29 skipping in FDB muscle fibers alters the gating properties of CaV1.1, resulting in increased channel conductance and hyperpolarized voltage dependence of channel activation and inactivation. Together, these functional changes in CaV1.1 channel gating resulted in a marked increase in L-type Ca2+ current density at moderate depolarization voltages (from −40 to +20 mV) and a modest increase in peak electrically evoked Ca2+ transient magnitude that was sensitive to block by extracellular Cd2+/La3+. Thus, the Cd2+/La3+-sensitive component of the evoked Ca2+ transient in E29-deleted fibers may reflect a gain-of-function increase in Ca2+ influx through CaV1.1 that is further amplified by local Ca2+-dependent activation of RyR1 within the triad junction. These data confirm previous observations obtained following transient expression of CaV1.1Δ29 in myotubes (43), and extend the conclusions to mature muscle fibers that exhibit a fully developed TTS, a normal complement of auxilliary proteins and a physiological expression level of untagged CaV1.1 channels. However, ASO-induced E29 skipping of endogenous CaV1.1 in adult muscle fibers resulted in a more limited effect on channel conductance and kinetics of activation compared with that observed following CaV1.1-Δ29 expression in myotubes.
While mis-splicing of CaV1.1 E29 has clear effects on Ca2+ channel function, the long-term implications for muscle maintenance and performance are unclear. Excitation-coupled calcium entry (ECCE) is an important pathway for calcium entry into skeletal muscle cells during high frequency stimulation (64,65), which involves Ca2+ influx through CaV1.1 channels (66). Since ECCE contributes a significant fraction of the total myoplasmic Ca2+ transient during repetitive activity (64,65), the contribution of this pathway would be expected to increase in the context of greater E29 skipping and myotonia. In the short term, ECCE may help sustain myoplasmic calcium levels during prolonged activity, as calcium stores begin to deplete. Thus, enhanced ECCE activity due to E29 mis-splicing may explain the paradoxical observation that DM1 muscles, although weak, are more fatigue resistant than healthy controls (67,68). Increased E29 skipping and enhanced CaV1.1 channel gating may also explain the earlier observation that myotubes from DM1 patients exhibit increased nifedipine-sensitive Ca2+ influx (69). However, the longer-term consequences may be harmful to muscle, especially when increased ECCE activity (CaV1.1 mis-splicing) is coupled with high-frequency repetitive discharges (myotonia, CLCN1 mis-splicing) and altered Ca2+ release and reuptake from the sarcoplasmic reticulum (RyR1 and SERCA1 mis-splicing). The combined effect may lead to chronic calcium overload, which has long been proposed to be a major contributor to progressive myopathy in Duchenne muscular dystrophy (30,31).
MATERIALS AND METHODS
Muscle testing and biopsy
The study was reviewed and approved by the Research Subjects Review Board of the University of Rochester. All participants gave their written informed consent. Muscle strength was determined by standardized manual muscle testing as previously described (46,47). Needle muscle biopsy of TA was performed as previously described (70). Muscle tissue was immediately flash frozen and stored at −80°C until use.
Cell culture, treatments and transfections
C2C12 myoblasts were grown in high-glucose Dulbecco's modified Eagle's medium (Invitrogen) with 1.5 g/l sodium bicarbonate and 1 mm sodium pyruvate, supplemented with 10% fetal bovine serum. Transfections were done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Briefly, si-RNAs (10 μl, 20 μm per well) were mixed with 2 μg empty pcDNA3 vector and co-transfected into C2C12 cells grown in six-well plates using Lipofectamine 2000 (Invitrogen). si-RNAs siMbnl1-No.5 (TARGETplus siRNA J-065216-05) and siMbnl1-No.8 (TARGETplus siRNA J-065216-08) were obtained from Dharmacon.
RT–PCR
For human muscle tissue, RNA was isolated using TRIzol (Invitrgen) and re-purified using RNeasy Mini Kits with on-column DNase treatment (QIAGEN), all according to the manufacturer's recommendations. cDNA was prepared using the WT-Ovation RNA Amplification kits (NuGEN) according to the manufacturer's instruction. For cells or mouse tissue, RNA was isolated with RNeasy Mini Kit (QIAGEN) and reverse transcribed with random hexamers and Superscript III (Invitrogen). For splicing assays, cDNA was PCR amplified (25–28 cycles) using primer pairs in which one primer was 5′-end labeled with 56-FAM. Primer sequences are provided in Supplementary Material, Table S3. PCR products were analyzed on agarose gels using a Typhoon fluorimager (Amersham Biosciences) and quantified using ImageQuant TL software (Amersham Biosciences).
Immunoblots
Tissue culture cell lysates for immunoblot were isolated in radioimmune precipitation assay buffer [150 mm NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mm Tris at pH 8.0] as previously described (71). Whole-cell lysates were resolved on 4–20% Tris–Glycine Gels (Invitrogen) in Tris–Glycine SDS running buffer. The proteins were transferred onto nitrocellulose membranes (Whatman) or Immobilon-FL polyvinylidene fluoride (PVDF) membranes (Millipore). Membranes were probed under standard conditions with α-Mbnl1 antibody (1:1000), α-CUGBP1 [3B1] (1:500, Abcam), α-CaV1.1 [MA3-920] (1:400, Affinity BioReagents) and α-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:10 000, Ambion). Blots were then probed with horseradish peroxidase-conjugated secondary antibodies (1:10 000, GE Healthcare) and developed using SuperSignal West Pico reagents (Pierce Biotechnology) and Kodak BioMax XAR film. For blotting with fluorophore-conjugated secondary antibodies, the PVDF membranes were probed with ECL Plex Cy3-conjugated goat α-mouse (1:5000; GE Healthcare) and Cy5-conjugated goat α-rabbit (1:2500; GE Healthcare) secondary antibodies, and scanned on a Typhoon fluorimager (Amersham Biosciences). For CaV1.1 expression in mouse TA muscle, a goat anti-mouse IRDye 800CW secondary antibody was used for detection and the blots were scanned with an ODYSSEY Infrared Imaging System (LI-COR Biosciences).
Splice shifting ASOs for CaV1.1-E29
Two antisense morpholino oligonucleotides, ▵E29-Mo-3′ and ▵E29-Mo-5′ (Gene Tools LLC), were designed to block the 3′ and 5′ mouse CaV1.1-E29 splice sites. The control morpholino was the inverted sequence of the morpholino targeting the 5′ splice site (rev-▵E29-Mo-5′), and did not match any known sequence in the mouse transcriptome. Morpholino sequences are included in Supplementary Material, Table S3.
cDNA and morpholino injections
All animal experiments were approved by the University Committee on Animal Resources at the University of Rochester. The protocol applied is similar to our previous studies with minor adjustment (27,72). Briefly, WT FVBn or HSALR mice (22) were anesthetized by intraperitoneal injection of 100 mg/kg ketamine, 10 mg/kg xylazine and 3 mg/kg acepromazine. TA muscles was pretreated by intramuscular injection of bovine hyaluronidase (15 μl, 0.4 U/μl) (Sigma-Aldrich) for 2 h (73). For CUGBP1 over-expression, 30 μg of CUGBP1 expression plasmid or control pcDNA3 vectors in a total volume of 20 μl phosphate buffered saline (PBS) were injected into TA muscles using a 30-gauge needle. To induce CaV1.1-E29 skipping, 20 μg ▵E29-Mo-3′ and 20 μg ▵E29-Mo-5′ morpholino in a total volume of 20 μl PBS was injected into TA muscle, or 10 μg ▵E29-Mo-3′ and 10 μg ▵E29-Mo-5′ morpholino in 10 μl PBS was injected into FDB muscle. An equal amount of rev-▵E29-Mo-5′ control morpholino was applied to the other leg of the same mouse as a control. Injected TA and FDB muscles were then electroporated. Electroporation parameters were 100 V/cm, 10 pulses at 1 Hz and 20 ms duration per pulse. For studies of CaV1.1-E29 skipping on muscle morphology, the determination of which TA muscle received ▵E29 morpholino was randomized and the frequency of muscle fibers showing central nuclei was determined across the entire transverse section of TA.
Whole-cell patch-clamp recording
The whole-cell patch-clamp technique was used to assess CaV1.1 channel currents (ICa) in FDB fibers isolated as described previously (28). FDB fibers were then bathed in an external recording solution containing (in mm): 157 TEA-methanesulfonate, 2 CaCl2, 10 HEPES, 1 MgCl2, 0.5 anthracene-9-carboxylic acid (9-AC), 0.1 N-benzyl-p-toluenesulfonamide (BTS), pH7.4 adjusted with TEA-OH. The patch pipette internal solution contained (in mm): 140 Cs-methanesulfonate, 10 HEPES, 20 Na-EGTA, 4 MgCl2, pH7.4 adjusted with CsOH. The patch pipette resistance when placed in the external solution was 0.6–1.0 Mohm. Fibers were voltage clamped at a holding potential of −80 mV. FDB fiber capacitance ranged from 1.33 to 2.21 nF with mean value 1.79 ± 0.05 nF (n = 28). There was no statistically significant difference in fiber capacitance between the control- and ▵E29 morpholino-treated groups. Series resistance was compensated up to 80%. Data were sampled every 120 µs and filtered using a low pass Bessel filter with 2 kHz cut-off frequency. ICa was activated by 200 ms depolarizing pulses ranging from −50 to +70 mV in 10 mV increments delivered every 10 s.
Ca2+ current (ICa) data analysis
All Ca2+ current data analysis was performed using Igor Pro 6 (Lake Oswego, OR, USA) and Clampfit 9 (Sunnyvale, CA, USA) software. Peak Ca2+ currents measured during each depolarization were normalized to cell capacitance and plotted against the corresponding test potential in order to obtain current–voltage relationship (ICa–V). ICa–V data that were then fitted by the following modified Boltzman equation:
| (1) |
Inactivation was estimated by calculating the ratio of ICa at the end of the 200 ms depolarizing pulse (IEnd) to the peak current (IPeak). Fractional inactivation (IEnd/IPeak with 1 representing no inactivation and 0 representing complete inactivation) was plotted against test potential. The activation phase of ICa was fitted using the following single exponential function:
where I(t) is the current at time t after the depolarization, A0 the steady-state current amplitude, τ0 the time constant of activation and C the steady state peak current. In all cases, the fitting procedure started 8 ms after the initiation of the voltage pulse (>10 × τm).
All data are presented as mean ± standard error of mean. Statistical significance was evaluated by Student's t-test and differences were considered statistically significant at P < 0.05.
Measurement of electrically evoked Ca2+ transients with mag-fluo-4
The magnitude of electrically evoked Ca2+ transients was determined in FDB fibers using the low affinity (in vitro Kd = 22 µm; Invitrogen) Ca2+ dye, mag-fluo-4, as described previously (74,75). Briefly, single FDB fibers were loaded with 5 µm mag-fluo-4 AM in Ringer's solution containing (in mm) (146 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, pH 7.4 with NaOH) for 30 min at room temperature and then incubated for >20 min at room temperature in a dye-free Ringer's solution supplemented with 25 µm BTS to inhibit movement due to contraction. Mag-fluo-4 was excited at 480 ± 30 nm, emission was monitored at 535 ± 40 nm using a ×40 (1.35 NA) oil-immersion objective and collected at 10 kHz using a photomultiplier detection system. Fibers were stimulated with pulse trains delivered at a frequency of 1 Hz using an adjacent stimulation electrode. Maximal ΔF/F0 values were calculated, where ΔF is the peak change in mag-fluo-4 emission from baseline and F0 is the baseline fluorescence recorded immediately before stimulation. Three different stimulation events were averaged and reported as a single value for each fiber. Identical measurements were also measured 2 min after local application of Ringer's solution supplemented with 0.5 mm Cd2+ + 0.2 mm La3+.
SUPPLEMENTARY MATERIAL
FUNDING
This work comes from the University of Rochester Wellstone Muscular Dystrophy Cooperative Research Center (U54NS48843) and Center for RNA Biology, with support from NIH AR049077 and Mid-Career Investigator Award (C.A.T.) AR/NS48143 and NIH AR44657 (R.T.D.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Thomas Cooper for the CUGBP1 expression plasmid and Dr Maurice Swanson for α-CUGBP1 antibody.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harper P.S. Myotonic Dystrophy. London: W.B. Saunders Company; 2001. [Google Scholar]
- 2.Norwood F.L., Harling C., Chinnery P.F., Eagle M., Bushby K., Straub V. Prevalence of genetic muscle disease in Northern England: in-depth analysis of a muscle clinic population. Brain. 2009;132:3175–3186. doi: 10.1093/brain/awp236. doi:10.1093/brain/awp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. doi:10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 4.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. doi:10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 5.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. doi:10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. doi:10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warf M.B., Berglund J.A. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. doi:10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rau F., Freyermuth F., Fugier C., Villemin J.P., Fischer M.C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D., et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. doi:10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 9.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. doi:10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 10.Timchenko N.A., Cai Z.J., Welm A.L., Reddy S., Ashizawa T., Timchenko L.T. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. doi:10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 11.Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. doi:10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 12.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. doi:10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 13.Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J., Cooper T.A. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. doi:10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. doi:10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. doi:10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 16.Pelletier R., Hamel F., Beaulieu D., Patry L., Haineault C., Tarnopolsky M., Schoser B., Puymirat J. Absence of a differentiation defect in muscle satellite cells from DM2 patients. Neurobiol. Dis. 2009;36:181–190. doi: 10.1016/j.nbd.2009.07.009. doi:10.1016/j.nbd.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Salisbury E., Schoser B., Schneider-Gold C., Wang G.L., Huichalaf C., Jin B., Sirito M., Sarkar P., Krahe R., Timchenko N.A., et al. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in myotonic dystrophy 2 patients. Am. J. Pathol. 2009;175:748–762. doi: 10.2353/ajpath.2009.090047. doi:10.2353/ajpath.2009.090047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.E., Cooper T.A. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. doi:10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper T.A., Wan L., Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. doi:10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne R.J., Lin X., Welle S., Sobczak K., O'Rourke J.R., Swanson M.S., Thornton C.A. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. doi:10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du H., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A., et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. doi:10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. doi:10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 23.Ward A.J., Rimer M., Killian J.M., Dowling J.J., Cooper T.A. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. doi:10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koshelev M., Sarma S., Price R.E., Wehrens X.H., Cooper T.A. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum. Mol. Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. doi:10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mankodi A., Takahashi M.P., Jiang H., Beck C.L., Bowers W.J., Moxley R.T., Cannon S.C., Thornton C.A. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. doi:10.1016/S1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 26.Charlet B.N., Savkur R.S., Singh G., Philips A.V., Grice E.A., Cooper T.A. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. doi:10.1016/S1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. doi:10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lueck J.D., Mankodi A., Swanson M.S., Thornton C.A., Dirksen R.T. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J. Gen. Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. doi:10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg J., Jiang H., Thornton C.A., Cannon S.C. Truncated ClC-1 mRNA in myotonic dystrophy exerts a dominant-negative effect on the Cl current. Neurology. 2004;63:2371–2375. doi: 10.1212/01.wnl.0000148482.40683.88. [DOI] [PubMed] [Google Scholar]
- 30.Turner P.R., Fong P.Y., Denetclaw W.F., Steinhardt R.A. Increased calcium influx in dystrophic muscle. J. Cell Biol. 1991;115:1701–1712. doi: 10.1083/jcb.115.6.1701. doi:10.1083/jcb.115.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner P.R., Westwood T., Regen C.M., Steinhardt R.A. Increased protein degradation results from elevated free calcium levels found in muscle from mdx mice. Nature. 1988;335:735–738. doi: 10.1038/335735a0. doi:10.1038/335735a0. [DOI] [PubMed] [Google Scholar]
- 32.Millay D.P., Goonasekera S.A., Sargent M.A., Maillet M., Aronow B.J., Molkentin J.D. Calcium influx is sufficient to induce muscular dystrophy through a TRPC-dependent mechanism. Proc. Natl Acad. Sci. USA. 2009;106:19023–19028. doi: 10.1073/pnas.0906591106. doi:10.1073/pnas.0906591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenzon N.M., Beam K.G. Disease causing mutations of calcium channels. Channels (Austin) 2008;2:163–179. doi: 10.4161/chan.2.3.5950. [DOI] [PubMed] [Google Scholar]
- 34.Lyfenko A.D., Goonasekera S.A., Dirksen R.T. Dynamic alterations in myoplasmic Ca2+ in malignant hyperthermia and central core disease. Biochem. Biophys. Res. Commun. 2004;322:1256–1266. doi: 10.1016/j.bbrc.2004.08.031. doi:10.1016/j.bbrc.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 35.Benders A.A., Wevers R.A., Veerkamp J.H. Ion transport in human skeletal muscle cells: disturbances in myotonic dystrophy and Brody's disease. Acta Physiol. Scand. 1996;156:355–367. doi: 10.1046/j.1365-201X.1996.202000.x. doi:10.1046/j.1365-201X.1996.202000.x. [DOI] [PubMed] [Google Scholar]
- 36.Benders A.A., Groenen P.J., Oerlemans F.T., Veerkamp J.H., Wieringa B. Myotonic dystrophy protein kinase is involved in the modulation of the Ca2+ homeostasis in skeletal muscle cells. J. Clin. Invest. 1997;100:1440–1447. doi: 10.1172/JCI119664. doi:10.1172/JCI119664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura T., Lueck J.D., Harvey P.J., Pace S.M., Ikemoto N., Casarotto M.G., Dirksen R.T., Dulhunty A.F. Alternative splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell Calcium. 2009;45:264–274. doi: 10.1016/j.ceca.2008.11.005. doi:10.1016/j.ceca.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanabe T., Beam K.G., Adams B.A., Niidome T., Numa S. Regions of the skeletal muscle dihydropyridine receptor critical for excitation-contraction coupling. Nature. 1990;346:567–569. doi: 10.1038/346567a0. doi:10.1038/346567a0. [DOI] [PubMed] [Google Scholar]
- 39.Catterall W.A., Perez-Reyes E., Snutch T.P., Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol. Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. doi:10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 40.Ertel E.A., Campbell K.P., Harpold M.M., Hofmann F., Mori Y., Perez-Reyes E., Schwartz A., Snutch T.P., Tanabe T., Birnbaumer L., et al. Nomenclature of voltage-gated calcium channels. Neuron. 2000;25:533–535. doi: 10.1016/s0896-6273(00)81057-0. doi:10.1016/S0896-6273(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 41.Tanabe T., Beam K.G., Powell J.A., Numa S. Restoration of excitation-contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature. 1988;336:134–139. doi: 10.1038/336134a0. doi:10.1038/336134a0. [DOI] [PubMed] [Google Scholar]
- 42.Flucher B.E., Tuluc P. A new L-type calcium channel isoform required for normal patterning of the developing neuromuscular junction. Channels (Austin) 2011;5:1–7. doi: 10.4161/chan.5.6.17951. [DOI] [PubMed] [Google Scholar]
- 43.Tuluc P., Molenda N., Schlick B., Obermair G.J., Flucher B.E., Jurkat-Rott K. A CaV1.1 Ca2+ channel splice variant with high conductance and voltage-sensitivity alters EC coupling in developing skeletal muscle. Biophys. J. 2009;96:35–44. doi: 10.1016/j.bpj.2008.09.027. doi:10.1016/j.bpj.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura T., Nakamori M., Lueck J.D., Pouliquin P., Aoike F., Fujimura H., Dirksen R.T., Takahashi M.P., Dulhunty A.F., Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. doi:10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 45.Hino S., Kondo S., Sekiya H., Saito A., Kanemoto S., Murakami T., Chihara K., Aoki Y., Nakamori M., Takahashi M.P., et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. doi:10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 46.Personius K., Pandya S., Tawil R., King W.M., McDermott M.P., Group F.-D. Facioscapulohumeral dystrophy natural history study: Standardization and reliability of testing procedures. Phys. Ther. 1994;74:253–263. doi: 10.1093/ptj/74.3.253. [DOI] [PubMed] [Google Scholar]
- 47.Mendell J.R., Moxley R.T., Griggs R.C., Brooke M.H., Fenichel G.M., Miller J.P., King W., Signore L., Pandya S., Florence J. Randomized, double-blind six-month trial of prednisone in Duchenne's muscular dystrophy [see comments] N. Engl. J. Med. 1989;320:1592–1597. doi: 10.1056/NEJM198906153202405. doi:10.1056/NEJM198906153202405. [DOI] [PubMed] [Google Scholar]
- 48.Lueck J.D., Lungu C., Mankodi A., Osborne R.J., Welle S.L., Dirksen R.T., Thornton C.A. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am. J. Physiol. Cell Physiol. 2007;292:C1291–C1297. doi: 10.1152/ajpcell.00336.2006. doi:10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 49.Kanadia R.N., Shin J., Yuan Y., Beattie S.G., Wheeler T.M., Thornton C.A., Swanson M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. doi:10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. doi:10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 51.Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. doi:10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flucher B.E., Obermair G.J., Tuluc P., Schredelseker J., Kern G., Grabner M. The role of auxiliary dihydropyridine receptor subunits in muscle. J. Muscle Res. Cell Motil. 2005;26:1–6. doi: 10.1007/s10974-005-9000-2. doi:10.1007/s10974-005-9000-2. [DOI] [PubMed] [Google Scholar]
- 53.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fugier C., Klein A.F., Hammer C., Vassilopoulos S., Ivarsson Y., Toussaint A., Tosch V., Vignaud A., Ferry A., Messaddeq N., et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat. Med. 2011;17:720–725. doi: 10.1038/nm.2374. doi:10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 55.Orengo J.P., Ward A.J., Cooper T.A. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann. Neurol. 2011;69:681–690. doi: 10.1002/ana.22278. doi:10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee E.H. Ca2+ channels and skeletal muscle diseases. Prog. Biophys. Mol. Biol. 2010;103:35–43. doi: 10.1016/j.pbiomolbio.2010.05.003. doi:10.1016/j.pbiomolbio.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Platt D., Griggs R. Skeletal muscle channelopathies: new insights into the periodic paralyses and nondystrophic myotonias. Curr. Opin. Neurol. 2009;22:524–531. doi: 10.1097/WCO.0b013e32832efa8f. doi:10.1097/WCO.0b013e32832efa8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Striessnig J., Bolz H.J., Koschak A. Channelopathies in Cav1.1, Cav1.3, and Cav1.4 voltage-gated L-type Ca2+ channels. Pflugers Arch. 2010;460:361–374. doi: 10.1007/s00424-010-0800-x. doi:10.1007/s00424-010-0800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Z.Z., Zheng S., Nikolic J., Black D.L. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol. Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. doi:10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.H., Shiue L., Ares M., Jr., Mody I., Black D.L. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 2011;43:706–711. doi: 10.1038/ng.841. doi:10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thornton C.A., Johnson K., Moxley R.T., 3rd Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 1994;35:104–107. doi: 10.1002/ana.410350116. doi:10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- 62.Vassilopoulos D., Lumb E.M. Muscle nuclear changes in myotonic dystrophy. Eur. Neurol. 1980;19:237–240. doi: 10.1159/000115152. doi:10.1159/000115152. [DOI] [PubMed] [Google Scholar]
- 63.Dirksen R.T., Beam K.G. Role of calcium permeation in dihydropyridine receptor function. Insights into channel gating and excitation-contraction coupling. J. Gen. Physiol. 1999;114:393–403. doi: 10.1085/jgp.114.3.393. doi:10.1085/jgp.114.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherednichenko G., Hurne A.M., Fessenden J.D., Lee E.H., Allen P.D., Beam K.G., Pessah I.N. Conformational activation of Ca2+ entry by depolarization of skeletal myotubes. Proc. Natl Acad. Sci. USA. 2004;101:15793–15798. doi: 10.1073/pnas.0403485101. doi:10.1073/pnas.0403485101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cherednichenko G., Ward C.W., Feng W., Cabrales E., Michaelson L., Samso M., Lopez J.R., Allen P.D., Pessah I.N. Enhanced excitation-coupled calcium entry in myotubes expressing malignant hyperthermia mutation R163C is attenuated by dantrolene. Mol. Pharmacol. 2008;73:1203–1212. doi: 10.1124/mol.107.043299. doi:10.1124/mol.107.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bannister R.A., Pessah I.N., Beam K.G. The skeletal L-type Ca(2+) current is a major contributor to excitation-coupled Ca(2+) entry. J. Gen. Physiol. 2009;133:79–91. doi: 10.1085/jgp.200810105. doi:10.1085/jgp.200810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres C., Moxley R.T., Griggs R.C. Quantitative testing of handgrip strength, myotonia, and fatigue in myotonic dystrophy. J. Neurol. Sci. 1983;60:157–168. doi: 10.1016/0022-510x(83)90135-1. doi:10.1016/0022-510X(83)90135-1. [DOI] [PubMed] [Google Scholar]
- 68.Zwarts M.J., van Weerden T.W. Transient paresis in myotonic syndromes. A surface EMG study. Brain. 1989;112:665–680. doi: 10.1093/brain/112.3.665. doi:10.1093/brain/112.3.665. [DOI] [PubMed] [Google Scholar]
- 69.Jacobs A.E., Benders A.A., Oosterhof A., Veerkamp J.H., van Mier P., Wevers R.A., Joosten E.M. The calcium homeostasis and the membrane potential of cultured muscle cells from patients with myotonic dystrophy. Biochim. Biophys. Acta. 1990;1096:14–19. doi: 10.1016/0925-4439(90)90006-b. [DOI] [PubMed] [Google Scholar]
- 70.Welle S., Thornton C., Jozefowicz R., Statt M. Myofibrillar protein synthesis in young and old men. Am. J. Physiol. 1993;264:E693–E698. doi: 10.1152/ajpendo.1993.264.5.E693. [DOI] [PubMed] [Google Scholar]
- 71.Boutz P.L., Chawla G., Stoilov P., Black D.L. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. doi:10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler T.M., Thornton C.A. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. doi:10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 73.McMahon J.M., Signori E., Wells K.E., Fazio V.M., Wells D.J. Optimisation of electrotransfer of plasmid into skeletal muscle by pretreatment with hyaluronidase—increased expression with reduced muscle damage. Gene Ther. 2001;8:1264–1270. doi: 10.1038/sj.gt.3301522. doi:10.1038/sj.gt.3301522. [DOI] [PubMed] [Google Scholar]
- 74.Capote J., Bolanos P., Schuhmeier R.P., Melzer W., Caputo C. Calcium transients in developing mouse skeletal muscle fibres. J. Physiol. 2005;564:451–464. doi: 10.1113/jphysiol.2004.081034. doi:10.1113/jphysiol.2004.081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loy R.E., Orynbayev M., Xu L., Andronache Z., Apostol S., Zvaritch E., MacLennan D.H., Meissner G., Melzer W., Dirksen R.T. Muscle weakness in Ryr1I4895T/WT knock-in mice as a result of reduced ryanodine receptor Ca2+ ion permeation and release from the sarcoplasmic reticulum. J. Gen. Physiol. 2011;137:43–57. doi: 10.1085/jgp.201010523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.