Abstract
Multiple intergenic single-nucleotide polymorphisms (SNPs) near hedgehog interacting protein (HHIP) on chromosome 4q31 have been strongly associated with pulmonary function levels and moderate-to-severe chronic obstructive pulmonary disease (COPD). However, whether the effects of variants in this region are related to HHIP or another gene has not been proven. We confirmed genetic association of SNPs in the 4q31 COPD genome-wide association study (GWAS) region in a Polish cohort containing severe COPD cases and healthy smoking controls (P = 0.001 to 0.002). We found that HHIP expression at both mRNA and protein levels is reduced in COPD lung tissues. We identified a genomic region located ∼85 kb upstream of HHIP which contains a subset of associated SNPs, interacts with the HHIP promoter through a chromatin loop and functions as an HHIP enhancer. The COPD risk haplotype of two SNPs within this enhancer region (rs6537296A and rs1542725C) was associated with statistically significant reductions in HHIP promoter activity. Moreover, rs1542725 demonstrates differential binding to the transcription factor Sp3; the COPD-associated allele exhibits increased Sp3 binding, which is consistent with Sp3's usual function as a transcriptional repressor. Thus, increased Sp3 binding at a functional SNP within the chromosome 4q31 COPD GWAS locus leads to reduced HHIP expression and increased susceptibility to COPD through distal transcriptional regulation. Together, our findings reveal one mechanism through which SNPs upstream of the HHIP gene modulate the expression of HHIP and functionally implicate reduced HHIP gene expression in the pathogenesis of COPD.
INTRODUCTION
Chronic obstructive pulmonary disease (COPD), the third leading cause of death in the USA (1), is a complex disease strongly influenced by cigarette smoking and genetic predisposition (2,3). Alpha-1 antitrypsin deficiency is an uncommon major genetic risk factor for COPD (4), but candidate gene association studies have had limited success in the identification of additional COPD genetic risk determinants (5). Genome-wide association studies (GWAS) of COPD have provided compelling evidence for disease susceptibility loci on chromosomes 4q24 (6), 4q31 (7,8) and 15q25 (7). The COPD-susceptibility locus at chromosome 4q31 has also been significantly associated with lung function in general population samples (9,10) and with COPD in subsequent replication studies (11–13). Of note, most of these previous COPD association studies have included COPD subjects with a broad range of COPD severity. Only one set of severe to very severe COPD cases (from the National Emphysema Treatment Trial) has been reported in previous case–control association analysis of the chromosome 4q31 COPD-susceptibility locus (7); rs13118928 was associated with severe COPD [odds ratio (OR) 0.7; P = 0.002] in this analysis.
The region of the strongest association on chromosome 4q31 includes a block of genetic variants in linkage disequilibrium, which is ∼79 kb in length (from rs12504628 to rs1542726). This intergenic COPD-susceptibility locus is located ∼51 kb away (Fig. 1A) from hedgehog interacting protein (HHIP), which encodes an inhibitory protein for sonic hedgehog (SHH); hedgehog is a crucial signaling pathway for the development of the lungs and other organs (14–16). Of note, previous GWAS have associated other HHIP single-nucleotide polymorphisms (SNPs) with height (8,17).
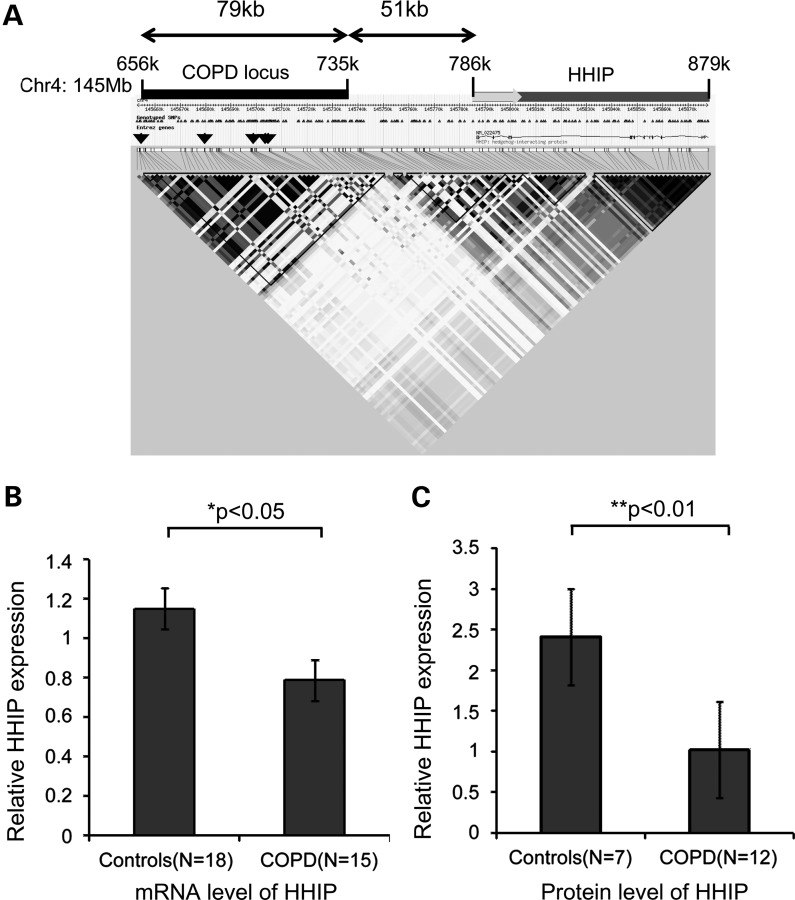
Figure 1.
COPD GWAS locus on chromosome 4q31 and HHIP expression levels in lung tissues. (A) Regional diagram for the COPD association signal on chromosome 4q31 around the HHIP gene based on genotypes from HapMap. Linkage disequilibrium (r2) is represented by gray scale and calculated using the CEU reference panel in Haploview. Black bar indicates the LD block containing the COPD-susceptibility locus; grey bar indicates the HHIP gene and grey arrow indicates the transcription direction of the HHIP gene. Arrowheads mark the locations of published COPD GWAS associations: rs12504628, rs13147758, rs1828591, rs1980057 and rs13118928. (B and C) Expression of HHIP is decreased in COPD compared with control lung tissues as determined by real-time PCR TaqMan assay for mRNA levels (B) and by western blot for protein levels (C). Two probes used for the HHIP gene in the gene expression assay showed similar trends and results from one representative probe are shown in the left panel. Means ± standard errors are shown.
Since the 4q31 locus has been consistently identified in multiple genetic association studies of lung function and COPD-related phenotypes, we investigated whether significant associations could be replicated for severe COPD. Subsequently, comprehensive evaluation to identify the causal variants and to investigate the functional impact of this region on COPD pathogenesis was undertaken. Given the pivotal role of HHIP during lung development, we hypothesized that cis-regulatory elements for HHIP are contained in this COPD-susceptibility locus. To address this hypothesis, we applied multiple approaches to localize the functional variants in this genomic region.
RESULTS
rs13118928 at 4q31 is associated with severe COPD
There were 315 subjects with severe COPD and 330 smoking control subjects enrolled in Poland (Table 1). There were more male than female case and control subjects, but gender was evenly distributed between both groups (P = 0.5). The severe COPD cases had greater pack-years smoking history (44.5 versus 33.4 pack-years, P < 0.0001) and were less likely to be active smokers (25.4 versus 49.4% current smokers, P < 0.0001). As expected, cases had significantly worse lung function, with an average forced expiratory volume in one second (FEV1) of 30.4% predicted in cases compared with 102.5% predicted in control subjects (P < 0.0001). As shown in Table 2, rs13118928 was significantly associated with severe COPD [OR 0.68, 95% confidence interval (CI) (0.53, 0.87), P = 0.002] by logistic regression analysis after adjustment for age, gender and pack-years of smoking.
Table 1.
Clinical characterization of severe COPD cases and healthy smoking controls in Poland
| Cases | Controls | P-value | |
|---|---|---|---|
| n | 315 | 330 | |
| Age (years) | 62.0 (7.3) | 58.3 (7.3) | <0.0001 |
| Male, n (%) | 220 (69.8) | 222 (67.3) | 0.50 |
| Pack-years | 44.5 (22.3) | 33.37 (15.0) | <0.0001 |
| Current smoker, n (%) | 80 (25.4) | 163 (49.4) | <0.0001 |
| FEV1 (% predicted) | 30.4% (9.6) | 102.5% (12.1) | <0.0001 |
| FEV1/FVC | 0.36 (0.10) | 0.76 (0.04) | <0.0001 |
Several COPD cases had borderline values for FEV1 (% predicted) but were included. Means (standard deviations) are presented except where specified.
FEV1, forced expiratory volume in one second; FVC, forced vital capacity.
Table 2.
Genetic association analysis of SNPs upstream from HHIP with severe COPD
| SNP | Chr | Gene | Reference allele | OR [CI] | P-valuea |
|---|---|---|---|---|---|
| rs13118928 | 4 | HHIP | G | 0.68 [0.53,0.87] | 0.002 |
| rs6537296 | 4 | HHIP | G | 0.67 [0.53, 0.86] | 0.001 |
| rs1542725 | 4 | HHIP | T | 0.68 [0.53, 0.86] | 0.0017 |
aTwo-sided P-values are reported.
Expression of HHIP is decreased in COPD lungs
We assessed whether HHIP gene expression is perturbed in lung tissues of COPD subjects. We measured the HHIP expression in lung tissues of COPD subjects and in smokers who have normal lung function. By real-time polymerase chain reaction (PCR), we found that mRNA levels of HHIP were significantly reduced in COPD subjects compared with control subjects with normal lung function (P < 0.05) using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference gene (Fig. 1B). A similar trend was observed using TBP (TATA-binding protein) as the reference gene (P = 0.057, data not shown). Significant decreases at the HHIP protein level were observed in COPD lungs with western blot analysis (Fig. 1C). In contrast, mRNA levels of the other gene flanking the COPD-susceptibility locus, GYPA (glycophorin A, situated ∼450 kb away), were barely detectable in these COPD lung tissues (data not shown). Together, our results demonstrate that decreased HHIP expression is associated with COPD and strongly suggest that HHIP is the most plausible candidate gene within this COPD-susceptibility locus.
DNA re-sequencing identified common variants in the COPD-susceptibility locus, intervening region and the HHIP gene
To develop a comprehensive list of common genetic variants in this genomic region, we began by using MaCH v1.06 in each of the three cohorts described in a recent COPD GWAS (18) to impute genotypes using CEU samples from both the HapMap Phase II and 1000 Genomes reference populations (6). In addition, to identify genetic variants that may be commonly found only in COPD cases, we sequenced the upstream COPD-susceptibility locus on chromosome 4q31, the intervening region leading to HHIP, and all of the exons and introns of HHIP, using long-range PCR followed by next-generation sequencing in DNA isolated from 29 severe, early-onset COPD subjects (2). A total of 493 SNPs, including 176 novel variants (not present in dbSNP130), were found (Supplementary Material, Table S1). Twenty-one SNPs were found in the genomic region ∼5 kb upstream of the transcription start site for HHIP; however, no common non-synonymous SNPs were found within the coding region of HHIP, suggesting that regulatory elements likely confer COPD susceptibility in this region.
Identification of a long-range enhancer for HHIP in the COPD 4q31 GWAS region
Since HHIP lies in a considerable linear distance from the COPD GWAS SNPs, and multiple lines of evidence have shown that critical developmental genes often have distal regulation (19,20), we hypothesized that the functional SNPs may exert long-distance regulation on HHIP expression, similar to functional variants on 8q24 affecting c-Myc expression (21–24). We explored this possibility by applying chromosome conformation capture (3C) assays (25) in an immortalized human bronchial epithelial cell line (Beas-2B) and in fetal lung fibroblasts (MRC-5) (Fig. 2A). This technique assesses long-range interactions of a constant genomic fragment with a series of DNA fragments spanning a potentially large genomic region. In our assays, we interrogated a constant fragment containing the promoter of HHIP against four fragments spanning this COPD-associated GWAS locus (145655k–145735k) and five fragments in between them (primer sequences are listed in Supplementary Material, Table S2). After PCR analysis, the interaction frequency of any given two fragments was calculated by semi-quantitative PCR, and ligation of distal chromatin segments was verified by sequencing 3C-PCR products. As expected, we detected strong interactions between the HHIP promoter and directly adjacent fragments in both cell types (peak near the anchoring fragment in Fig. 2A). The interactions became less prominent with increasing distance from the HHIP promoter; however, an additional strong interaction peak was detected between the HHIP promoter and a 7 kb fragment (145701k–145708k) contained within this COPD-susceptibility locus (Fig. 2A and Supplementary Material, Fig. S1A). Hence, we localized a potential HHIP gene regulatory element within the 79 kb GWAS region to a 7 kb fragment that harbors multiple associated SNPs from previous GWAS in COPD (7,10,26).
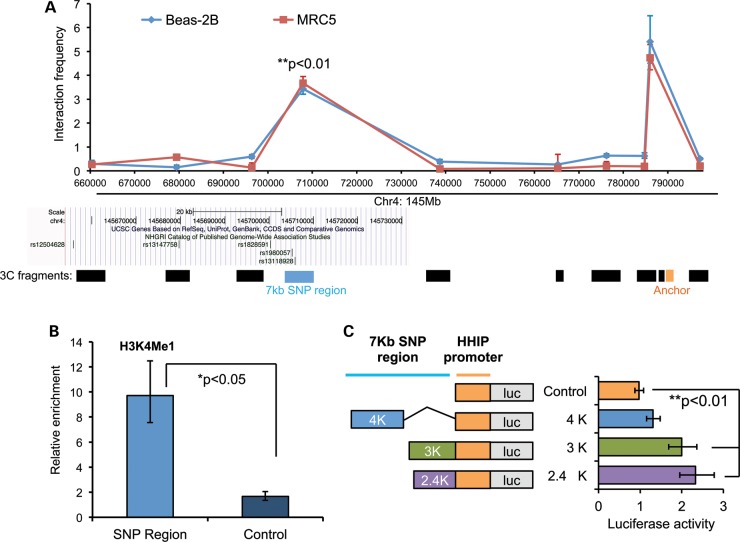
Figure 2.
Identification of a long-range enhancer for HHIP in the COPD GWAS locus on chromosome 4q31. (A). Long-range interaction between the COPD-susceptibility locus and the HHIP promoter in Beas-2B (bronchial epithelial) and MRC5 (lung fibroblast) cells detected by 3C-PCR. The graph demonstrates 3C interaction frequency of the constant fragment containing the HHIP promoter (orange bar) with other target fragments (black bars). The y-axis refers to 3C-PCR products normalized to the interaction frequency of fragments from the BAC clone. Geometric means and standard errors were from duplicate PCR reactions. Middle section: snapshot of LD block from UCSC genome browser with COPD GWAS variants. Lower section: 3C fragments at various lengths after BglII digestion. (B) ChIP-qPCR revealed relative enrichment of H3K4Me1 in the 3C interaction fragment (light blue column) compared with the control region (dark blue column). Geometric means and standard errors are shown from three independent repeats. Control region: 145668717–145668900; COPD GWAS SNP region: 145705250–145705431; (C) reporter assays revealed enhancer activity of COPD GWAS SNP region. Left panel: three pieces of the HHIP COPD GWAS SNP region were cloned to the 5′-end of the native HHIP promoter (control, orange bars) in the pGL3 basic reporter vector. Right panel: relative promoter activity in Beas-2B cells transfected with indicated constructs. Geometric means and standard errors were calculated from three independent repeats done in triplicate wells. GWAS regions—4K: 145701K–145705K; 3K: 145705K–145708K; 2.4K: 145705843–145708234; HHIP promoter: 145,786,102–145,787,277 based on hg18.
Given the strong physical interaction between this GWAS locus and the HHIP promoter detected by 3C, we hypothesized that by forming chromatin loops, this 7 kb region may function as an enhancer for the HHIP promoter. To test this, we first observed enrichment of an enhancer marker, histone 3 lysine 4 mono-methylation (H3K4Me1) (27–29), in this 7 kb region (Fig. 2B) by chromatin immunoprecipitation (ChIP) assays in Beas-2B cells. These results indicate that a functional cis-regulatory element is likely contained in this region. Moreover, comparative genomics data from the UCSC genome browser indicated that this DNA region contains highly conserved sequence among mammals (Supplementary Material, Fig. S1B), which suggested that important regulatory elements may be located in this region. To further test this hypothesis, we cloned deletion constructs of this 7 kb interaction fragment at various lengths (designated 2.4K, 3K and 4K) into reporter constructs that contained the HHIP promoter (designated as control)—the anchoring fragment in the 3C assays (Fig. 2C) followed by the firefly luciferase reporter gene. Luciferase assays revealed strongest enhancer activity within the 2.4K fragment (145705843–145708234, hg18). Thus, we demonstrated that similar to other developmental genes (27), HHIP is regulated by a cis-regulatory enhancer from a great distance, ∼85 kb away. Of interest, this enhancer region is directly adjacent to two published GWAS SNPs associated with COPD (7,26).
Risk haplotype for COPD is associated with decreased HHIP promoter activity
Based on our resequencing data of this genomic region described earlier, we identified two common SNPs located inside the 2.4K region: rs6537296 and rs1542725 (Supplementary Material, Table S3). These two SNPS were in strong LD (r2 > 0.99, D′ 1.0) with rs13118928 (Supplementary Material, Table S4). As expected based on this LD structure, both rs1542725 [OR 0.67, 95% CI (0.53, 0.87), P = 0.002] and rs6537296 [OR 0.67, 95% CI (0.53, 0.86), P = 0.001] were also significantly associated with severe COPD in our Polish study population (Table 2).
We set out to determine whether the region including these two SNPs (rs1542725 and rs6537296) was critical for the enhancer activity of the 2.4K region. We cloned ∼500 bp around these two SNPs into reporter constructs containing the HHIP promoter. Approximately 2.5-fold increased promoter activity was consistently observed in the minimal risk locus that contains these two SNPs both in Beas-2B cells (Fig. 3A) and MRC5 cells (Supplementary Material, Fig. S2). This enhancer activity was independent of orientation (Fig. 3A).
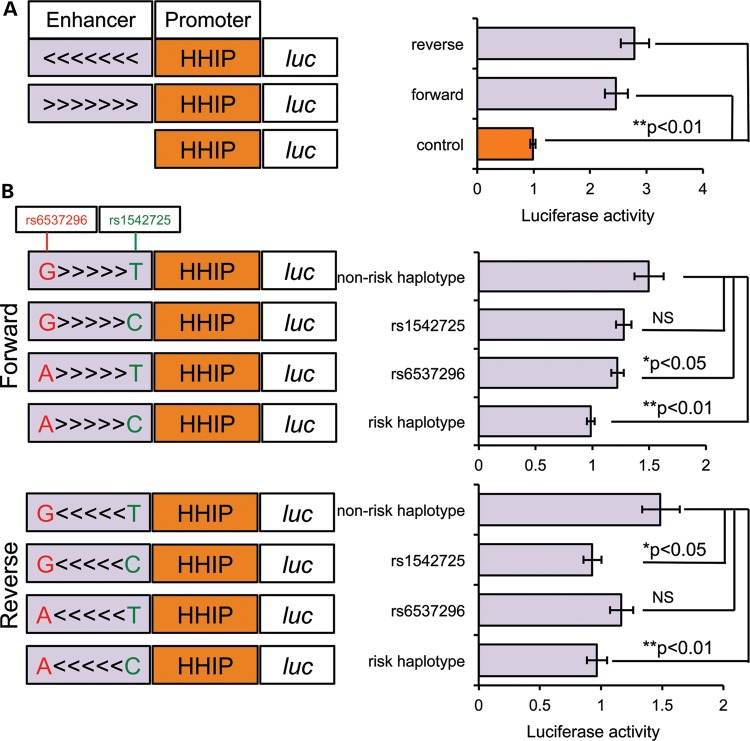
Figure 3.
COPD risk allele is associated with lower HHIP promoter activity. (A) The minimal COPD GWAS SNP region (∼500 bp around two key SNPs, light purple column) cloned at forward orientation (>>>) and reverse orientation (<<<) showed enhancer activity for the HHIP promoter (‘control’, orange) measured by dual-luciferase in Beas-2B cells. Shown are ratios of firefly luciferase expression to Renilla luciferase expression normalized to the mean ratio from the control construct. Geometric means and standard errors are shown for five independent repeats done in triplicate or quadruplicate. (B) The effects of rs1542725 and rs6537296 were evaluated in the reporter assays. Single-nucleotide alterations were introduced individually or combined into minimal enhancer constructs at forward (upper panel) and reverse (lower panel) orientations. Shown are ratios of firefly luciferase expression to Renilla luciferase expression normalized to the mean ratio from the risk haplotype (rs6537296A and rs1542725C). Geometric means and standard errors were calculated from eight (forward) or three (reverse) independent repeats.
In the association analyses, rs6537296A and rs1542725C were risk alleles associated with severe COPD (Table 2). To evaluate whether enhancer activity is allele-specific, we measured luciferase activities in Beas-2B cells transfected with single- or double-mutation constructs at rs6537296 and rs1542725. Compared with the non-risk haplotype (rs6537296G and rs1542725T), single-mutation constructs for rs6537296 and rs1542725 showed quantitatively modest but statistically significant decreases in enhancer activity at forward and reverse orientations, respectively (Fig. 3B). Moreover, double mutations at both SNPs consistently showed approximately a one-third decrease in enhancer activity compared with the non-risk GT haplotype in both forward and reverse orientations (Fig. 3B, P < 0.01). Hence, the AC haplotype associated with increased risk of COPD exhibited lower enhancer activity for the promoter of HHIP and might therefore correlate with lower HHIP expression.
rs1542725 affects binding to Sp1 family member Sp3
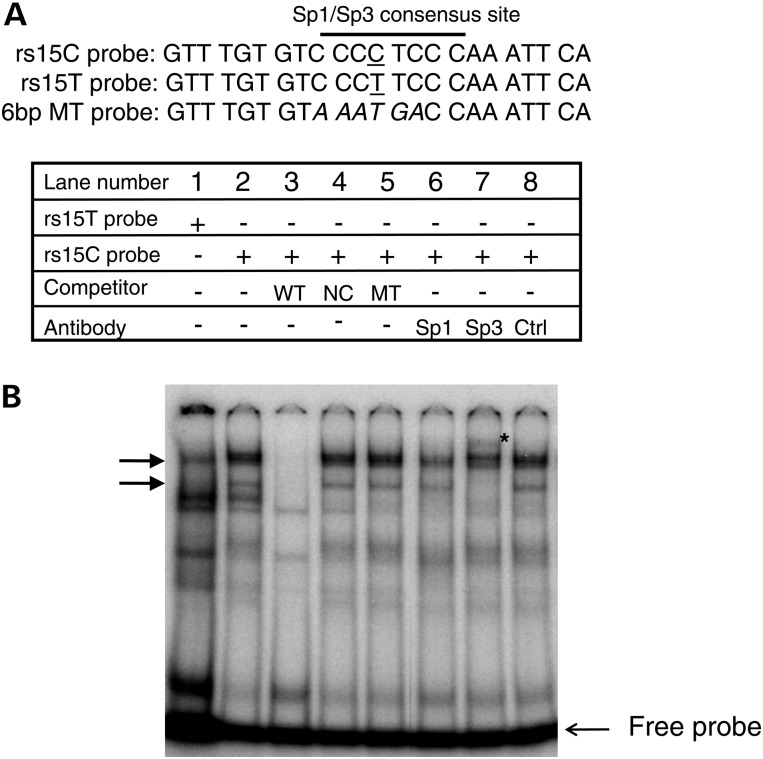
One of our identified COPD-associated SNPs (rs1542725) is located within a conserved GC-box like motif that may function as a binding site for the transcription factors Sp1 and Sp3 based on TRANSFAC analysis (Fig. 4A, Supplementary Material, Fig. S3A). We hypothesized that this SNP affects the binding of Sp1 and/or Sp3 to this region. We performed an electrophoretic mobility shift assay (EMSA) using radio-labeled probes spanning rs1542725 (Fig. 4A). When we mixed annealed probes with nuclear extract from Beas-2B cells, we observed a nucleoprotein complex in which the top two bands exhibited increased intensity with probe spanning the rs1542725C allele (arrows, Fig. 4B, lane 2) compared with a radio-labeled probe spanning the rs1542725T allele (non-risk allele) (Fig. 4B, lane 1). These top two bands were specific, as shown by complete elimination with an identical competitor (Fig. 4B, lane 3) and retention of the binding complex with addition of a non-identical competitor (NIC) (Fig. 4B, lane 4). Competition with an oligonucleotide with a mutation of the central 6 bp spanning the rs1545725C sequence failed to disrupt binding of the top two bands (Fig. 4B, lane 5), thus confirming binding of the nuclear protein to the central region of the rs1542725 sequence. Interestingly, mild reduction in the intensity of the top two complexes was seen with addition of an Sp1 antibody (Fig. 4B, lane 6), and addition of an Sp3 antibody produced disruption of the top two bands and a supershifted band (Fig. 4B, lane 7), supporting the presence of Sp3 in the nucleoprotein complex. In contrast, no change in the complex was seen with the addition of an unrelated antibody (Fig. 4B, lane 8). These results were consistently observed in multiple repeats (Supplementary Material, Fig. S4B). Moreover, competition with a non-radiolabeled consensus sequence for Sp1/Sp3 eliminated the top two bands (Supplementary Material, Fig. S3B, lane 5). Furthermore, addition of radio-labeled probe in the absence of nuclear extract showed no nucleoprotein complexes (data not shown). Taken together, our EMSA data support that Sp1 family member Sp3 binds to a central 6 bp region spanning rs1542725 in vitro. The rs1542725 C variant, which is associated with COPD and reduced HHIP gene expression, exhibited stronger Sp3 binding than the rs1542725T allele, consistent with the role of Sp3 as a transcriptional repressor.
Figure 4.
rs1542725 alters a Sp3 binding site. (A) Three probes used in EMSA are shown with nucleotides with underlines for rs1542725 and in italic for 6 bp central mutations spanning rs1542725. (B) P32 labeled oligonucleotide containing the sequence spanning rs1542725 with either C allele (rs1542725C, lanes 2–8) or T allele (rs1542725T, lane 1) was incubated with nuclear extract from Beas-2B cells. rs1542725C probe was incubated with identical competitor probe (WT) in lane 3, the NIC probe (NC) in lane 4 and the 6 bp mutant probe (MT) in lane 5. Sp1, Sp3 and unrelated Oct-1 control (Ctrl) antibodies were added in lanes 6, 7 and 8, respectively. Star (*) indicates the supershifted Sp3 band in lane 7. Arrows indicate specific nuclear proteins bound by probe.
DISCUSSION
The existence of a susceptibility locus on chromosome 4q31 influencing pulmonary function in general population samples (9,10) and moderate-to-severe COPD (7) has been strongly supported by previous genetic association studies. In a Caucasian study population from Poland, we confirmed that SNPs in this same genomic region are associated with severe to very severe COPD, which is substantially less common and potentially more strongly influenced by genetic factors (2) than moderate COPD. Several lines of evidence in the current study clearly implicate HHIP as the COPD-susceptibility gene in this region. First, we detected reduced mRNA and protein levels of HHIP in COPD lung tissues. Secondly, a genomic region containing a subset of associated SNPs forms a chromosomal loop that interacts with the HHIP promoter from ∼85 kb away and functions as an enhancer. Thirdly, a COPD risk haplotype within this enhancer region is associated with reduced HHIP promoter activity. Both of the SNPs within this haplotype appear to have functional effects on HHIP expression, and we found a potential biological mechanism for one of the COPD risk alleles in this region (rs1542725 C allele), which demonstrates increased binding to the transcription factor Sp3. This ubiquitously expressed transcription factor Sp3 has repressive activity related to its post-translational modification (30). Differential Sp3 binding at a functional SNP (rs1542725) within the chromosome 4q31 COPD GWAS locus could lead to reduced HHIP levels and increased susceptibility to COPD.
GWAS have been very successful in discovering the general location of many common susceptibility variants for complex traits. However, the identification of the functional variants within GWAS regions remains challenging. In some cases, as with the Complement Factor H gene in age-related macular degeneration (31), a non-synonymous SNP with likely functional impact is identified within the associated region. However, most common variants identified by GWAS are located in non-coding regions (32). These non-coding regions may contain cis- or trans-acting regulatory elements for nearby or distant genes. Indeed, successful identification of functional GWAS variants in non-coding regions has recently been reported for colorectal cancer (21), lipoprotein levels (33) and coronary artery disease (34). Given the difficulty of identifying functional genetic variants in non-coding genomic regions, it is not unreasonable to ask whether such efforts are worth the intensive investigation required. We contend that the identification of functional variants in such regions is extremely important for at least two reasons. First, the identification of functional variants can conclusively prove which gene is actually involved in disease susceptibility. Secondly, study of functional variants can lead to insights into pathophysiological mechanisms for disease.
It is not entirely surprising to identify a distal enhancer for HHIP, a critical developmental gene that is involved in fetal lung branching morphogenesis. SHH, an initiator signaling molecule for the Hedgehog pathway, has an enhancer called limb located ∼1 Mb upstream from the coding region (35,36). Similarly, SOX9 has long-range regulatory elements located over 1 Mb away from the coding sequence (37). Here, we showed that a putative enhancer element, harboring COPD GWAS SNPs, regulates HHIP expression from ∼85 kb away. The underlying mechanism by which enhancers are ‘tethered’ with the proximal promoter of a target gene from a great linear distance remains poorly defined. A recent study showed that in addition to the transcriptional apparatus, other proteins essential for chromosome segregation, mediator and cohesin, can form rings to connect DNA segments across substantial genomic distances and facilitate long-range regulation between a distal enhancer and core promoter (38). Further investigation to define the protein complex associated with the HHIP promoter and distal enhancer element may provide additional mechanistic insights regarding the distal-regulation of HHIP.
HHIP, through its extracellular domain, binds to sonic hedgehog (SHH) and prevents SHH from activating the Hedgehog signaling pathway (39), a critical pathway for embryonic lung development. Although the hedgehog pathway has been related to acute airway epithelial injury and small cell lung cancer (40), a key role for this pathway in COPD had not been considered until the identification of the COPD-susceptibility locus on chromosome 4q31 using GWAS (41). Decreased HHIP expression leads to over-activation of the Hedgehog pathway in multiple types of cancer, which in turn contributes to uncontrolled cellular proliferation (42,43). In our study, we showed that HHIP expression is decreased in lung tissue from COPD cases compared with control subjects with normal lung function (Fig. 1B). The COPD risk haplotype confers decreased enhancer activity for the HHIP promoter (Fig. 3B), indicating that lower HHIP expression may exacerbate smoking-induced COPD pathogenesis. The discovery of alpha-1 antitrypsin deficiency more than 40 years ago led to the development of the protease–antiprotease imbalance hypothesis for COPD (44), which remains a central model for COPD pathogenesis. Further mechanistic studies on the Hedgehog signaling pathway in the context of smoking may provide novel insights into the pathogenesis of COPD.
Taken together, our study has provided novel mechanistic insights into the functional impact of the 4q31 locus on COPD susceptibility. However, our work does have some limitations. First, since our genetic association results replicated previous findings in a severe COPD population (7), we did not include another replication population. Secondly, although our study population in Poland was Caucasian and likely reasonably homogeneous, we did not formally test for population stratification. Thirdly, our 3C assessment of the GWAS locus was not completely comprehensive due to some extreme sizes of fragments (>10 or <1 kb) inside the locus after BglII digestion; thus, it is possible that additional gene regulatory elements are located in that region. Fourthly, the enhancer activity of the COPD risk and non-risk haplotypes showed a statistically significant difference, but the magnitude of the differences was quantitatively modest (Fig. 3B, ∼35% decrease). Of interest, a functional variant identified on chromosome 8q24 for colorectal cancer (21) had a similar magnitude of allele-specific effects, suggesting that functional variants that confer moderate differences in reporter assays can be biologically important in complex disease susceptibility. It is also possible that there are additional functional variants in this interaction segment. Fifthly, although we showed evidence for differential binding of Sp3 to rs1542725 in the presence or absence of the risk allele, it is likely that other transcription factors bind in this region and regulate HHIP expression. Finally, abnormal HHIP expression has been observed in multiple tumor types due to hypermethylation of its promoter region, such as hepatocellular carcinoma (42), gastrointestinal cancer (45), neuroblastoma (46) and pancreatic cancer (43). Studies of the impact of epigenetic alterations on HHIP expression and COPD susceptibility will be required.
In summary, we have demonstrated that the COPD GWAS locus on chromosome 4q31 shows physical and functional long-range interactions with the HHIP gene and contains an enhancer element. Furthermore, the decreased expression of HHIP associated with increased binding of Sp3 to the COPD-associated rs1542725C allele could contribute to the development of COPD. These findings implicate HHIP and the Hedgehog pathway in COPD pathogenesis.
MATERIALS AND METHODS
Study population from Poland
Subjects were participants in the Transcontinental COPD Genetics Study (TCGS) and were recruited at clinical centers throughout Poland. Eligible participants were Caucasians from Poland between 40 and 80 years of age with at least 10 pack-years of cigarette smoking history. Subjects were excluded for: ongoing respiratory disorders other than COPD; lung surgery involving one or more lobes; abdominal, chest or eye surgery in the 3 months prior to enrollment; myocardial infarction in the 3 months prior to enrollment; inability to use albuterol; pregnancy; or use of antibiotics or systemic steroids for COPD exacerbations in the month prior to enrollment. Subjects were excluded if they had a first or second degree relative already enrolled in the TCGS. Eligible cases generally had severe to very severe COPD based on GOLD staging (47) with post-bronchodilator FEV1/FVC (forced vital capacity) < 0.7 and FEV1 ≤ 50% predicted; however, several subjects with borderline spirometric values were included after investigator review. Eligible control subjects had post-bronchodilator FEV1/FVC ≥ 0.7 and FEV1≥ 80%. All subjects completed a modified American Thoracic Society Respiratory Questionnaire (48), performed standardized spirometry using an Easy-one spirometer (NDD, Inc.) and provided blood samples for genotyping. All subjects signed written informed consent prior to enrollment. This study was approved by the IRBs at both the Institute of Tuberculosis and Lung Diseases in Warsaw and Brigham and Women's Hospital in Boston.
Genotyping and genetic association analysis in the Polish study population
Three SNPs located in the COPD GWAS locus near HHIP on chromosome 4 were examined for their association with COPD. These SNPs included rs13118928, rs6537296 and rs1542725, which were chosen based on prior studies demonstrating an association between rs13118928 and COPD (7) as well as DNA resequencing performed in this study. These three SNPs were genotyped using TaqMan assays with an ABI 7900 (Applied Biosystems, Inc.). We performed logistic regression analysis for the association of these SNPs with the presence or absence of COPD, adjusting for age, gender and pack-years of smoking.
Cell lines and human tissues
Human Beas-2B bronchial epithelial cells (#CRL-9609) and human MRC-5 fetal lung fibroblasts (#CCL-171) were purchased from ATCC and cultured in complete Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, penicillin (50 units/ml), streptomycin (50 μg/ml) and gentamicin (10 µg/ml). Lymphoblastoid cell lines from severe, early-onset COPD subjects were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin (50 units/ml), streptomycin (50 μg/ml) and gentamicin (10 µg/ml). Human lung tissue samples from 18 COPD patients (FEV1< 80%) and 15 control subjects with normal lung function were obtained from the Lung Tissue Research Consortium, a national biorepository of lung tissues.
Real-time PCR
Total RNA from human lung tissues stored in RNAlater (Ambion) solution was extracted using the RNeasy Mini Kit (Qiagen). Genomic DNA was eliminated by on-column digestion during RNA extraction using RNase-Free DNase Set (Qiagen). cDNA was synthesized using Multiscribe Reverse Transcriptase Kit (Applied Biosystems: #4368814). HHIP expression was quantified by real-time PCR using TaqMan assays (Applied Biosystems, #4369510) in an ABI Prism 7900 instrument; TaqMan primers used in the assays were HHIP (ABI #Hs01011015_m1 and HS01011009_m1), GYPA (ABI# Hs00266777_m1), GAPDH (ABI#Hs00266705_g1) and TBP (ABI#Hs00427620_m1). All samples were tested in duplicate PCR under the following cycling conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 60°C. The average cycle number at threshold [Ct] was normalized against GAPDH and TBP. The expression level of HHIP was calculated based on the 2−ΔΔCt method.
Western blot in human lung tissues
Protein extractions from lung tissues were performed as previously described (49). Western blotting was performed as previously described (50). Antibodies used were α-actin (Chemicon, #MAB1501) and HHIP (Abcam, #39208). Secondary antibodies were horseradish peroxidase-linked anti-mouse or anti-rabbit IgG (GE Healthcare). Detection was done with enhanced chemiluminescence (Perkin Elmer Life Sciences, Inc.). Band densities were quantified by Adobe Photoshop software and HHIP expression was normalized to loading with α-actin.
HHIP region DNA sequencing
Long-range PCR amplicons were designed to cover ∼229 kb, from the 5′ end of the linkage disequilibrium block previously associated with COPD to ∼4 kb past the HHIP 3′-UTR. DNA was extracted from blood or lymphoblastoid cell lines cultured at low passage from 29 subjects with severe, early-onset COPD. Long-range PCR amplicons were prepared and sequenced using the Illumina GAIIx. Single-end reads were aligned using BWA 0.5.8 (51). Average coverage per base was >1000×, with >95% of bases covered at least 20×. Sequence data were subject to base quality recalibration and indel realignment using the Genome Analysis Toolkit version 1.04218 (GATK) (52). Variants were called with the Unified Genotyper using multi-sample calls and filtered using the parameters: ‘QUAL < 50.0 || AB > 0.75 || QD < 5.0 || HRun > 5 || SB > −0.10’. Detailed examination identified variants excluded from multisample calls due to false indels and errors resulting in triallelic sites; thus GATK was rerun using single sample mode and these variants were added back into the calls. Indels were called using the GATK IndelGenotyperV2, without filtering.
Chromosome conformation capture assay (3C)-PCR
Interactions of distal genomic DNA regions with the HHIP promoter were evaluated by 3C-PCR using approaches that were previously described (25). Briefly, ∼8 × 107 human Beas-2B and MRC5 cells were cross-linked and lysed, and chromatin was digested with BglII. Crosslinked and digested fragments were ligated with T4 ligase (Invitrogen, # 15224-025) for 6h at 16°C. 3C products were detected with 3C unidirectional PCR primers in duplicate or triplicate PCR reactions. 3C-PCR primers were designed targeting DNA fragments at sizes between 1 and 10 kb after BglII digestion. We also limited primer designs for highly repeated genomic regions due to possible low specificity. All distal PCR ligation products from both BAC and cell lines were confirmed by sequencing. DNA fragment interaction frequencies were calculated by normalizing PCR signals from cell lines to BAC control for each primer pair. At least two biological replicates for each cell line were independently prepared and amplified for 3C-PCR. Primers used for 3C are listed in Supplementary Material, Table S2.
ChIP-PCR assay
ChIP was performed by using EZ-CHIP kits (Millipore, #17-371) according to the manufacturer's protocol. Briefly, Beas-2B cells were grown to reach sub-confluency. Approximately 1 × 107 cells were cross-linked by 1% formaldehyde in growth media for 10min at room temperature. After quenching with glycine, cells were lysed in sodium dodecyl sulfate lysis buffer in the presence of protease inhibitors. Chromatin samples from Beas-2B cells were sonicated to a size range of 200–600 bp and analyzed on agarose gels. Aliquots of 2 million cells were used for each immunoprecipitation. After preclearing with protein G agarose, a 1% aliquot of supernatant was collected as input DNA. Solubilized chromatin was subjected to immunoprecipitation with antibody against H3K4Me1 (ab8895, Abcam), and rabbit IgG (sc-2027, Santa Cruz) was used as a negative control. Immunoprecipitation was performed for 16h at 4 °C. DNA from ChIP preparation was quantified by SYBR Green qPCR (Kapabiosystems, #kk4601) by series dilution of input DNA for each primer set. Relative quantification was performed as described in the manufacturer's protocol (Applied Biosystems). Each ChIP was performed in three biological replicates from Beas-2B cells cross-linked and immunoprecipitated on different dates. The primers used in ChIP are listed in Supplementary Material, Table S2.
Molecular cloning and reporter assays
HHIP promoter 1.2K (145,786,102–145,787,277, hg18) amplified from BAC clone (Invitrogen, CTD-3027L19) was cloned into pGL3 basic vector (Promega) in BglII and HindIII digestion sites. Various segments of the COPD GWAS region were cloned to the 5′ end of the HHIP promoter in pGL3 basic vector.
For reporter assays, 80% confluent Beas-2B cells were seeded into a 24-well plate at a density of 5 × 104 cells/well. Forty-eight hours post-transfection with various reporter constructs along with TK-renilla, cells were collected to measure promoter activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. Luminescence signals were captured in a Wallac VICTOR3 1420 plate reader (Perkin Elmer). Firefly signals driven by the HHIP promoter were normalized to TK-renilla signals after background subtractions. All transfections were performed either in triplicate or quadruplicate. Mutant constructs for SNPs rs653296 and rs1542725 were made using an in situ mutagenesis kit (Agilent, #200521). All plasmids used for reporter assays were confirmed by sequencing. Independent transfection and reporter assays were performed four to six times.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (53,54) with double-stranded oligonucleotide radio-labeled probes surrounding rs1542725 in the risk locus. Additional double-stranded oligonucleotides that were used as unlabeled competitors include a consensus Sp1 binding site, a NIC [an AT-rich sequence from the P-selectin promoter (54)], and an oligonucleotide in which the central 6 bp surrounding the rs1542725 risk locus were mutated. Unlabeled competitor probes were added into reaction at 100-fold concentration compared with labeled probes. Detailed sequences of probes and competitors are listed in Supplementary Material, Table S2. Nuclear extracts were harvested from Beas-2B cells as described previously (54), and nuclear protein was quantified by the Bradford dye-binding method (Bio-Rad). The 32P radio-labeled probes, with or without the addition of unlabeled competitors, were incubated with 10 μg of nuclear extract for 30 min at 4°C prior to electrophoresis. In separate experiments to test for the presence of specific proteins within the observed complexes, the nuclear protein mixture was incubated for 30 min at room temperature with antibodies against Sp1 (Santa Cruz, sc-59 X), Sp3 (Santa Cruz, sc-644X) or an unrelated antibody Oct-1 (Santa Cruz, sc-232X).
Statistics for gene expression and cell-based assays
For comparisons between cases and controls of mRNA and protein levels in lung tissues, Wilcoxon's rank sum test in SAS (Cary, NC, USA) was used. For reporter assays, luciferase activity levels were assessed using general linear models with PROC GLM in SAS; values were compared with a reference group within an experimental repeat, and results from multiple experiments were included. Interaction frequencies in 3C assays and ChIP assays with H3K4Me1 were analyzed using a similar approach. For 3C assays, in addition to main effects for the primer and cell type (BAC versus Beas-2B or MRC-5), an interaction term between the 7 kb fragment in the COPD GWAS region and cell type was included. For ChIP assays, relative enrichment was normalized to an IgG control. Reporter, 3C and ChIP assays were analyzed after natural logarithm transformation; geometric means and standard errors calculated on the logarithmic scale were transformed back to the linear scale in the figures.
AUTHOR CONTRIBUTIONS
Study design: E.K.S., X.Z.; ChIP assays: X.Z., D.T.; EMSA: R.M.B., M.D.L.; genetic association analysis: M.H., E.K.S.; statistical analysis: X.Z., E.K.S., M.H., B.R.; 3C and reporter assays: X.Z., A.M.G., K.L., M.A.P.; HHIP region DNA sequencing: M.H.C., E.K.S., A.L.D. and B.J.K.; real-time PCR and western blot in lung tissues: X.Z., J.D.M., A.M.K.C.; manuscript writing: X.Z., R.M.B., M.H., J.Z., I.H., P.S., C.P.H., M.H.C., B.R., B.A.R., Q.L., M.D.L., M.A.P., A.M.G., S.T.W., A.M.K.C. and E.K.S. All authors reviewed and approved the manuscript.
SUPPLEMENTARY MATERIAL
FUNDING
This study utilized biological specimens and data provided by the Lung Tissue Research Consortium (LTRC) supported by the NIH. This work was supported by US National Institutes of Health (NIH) grants R01 HL075478, R01 HL084323, P01 HL083069 and P01 HL105339.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Karl Münger (Brigham and Women's Hospital, Boston, USA) for his valuable comments on the manuscript.
Conflict of Interest statement. E.K.S. has received grant support and consulting and speaker's fees from GlaxoSmith Kline, consulting and speaker's fees from Astra-Zeneca and speaker's fees from Bayer and Wyeth. X.Z., R.M.B., M.H., J.Z., I.H., P.S., C.P.H., M.H.C., A.L.D., B.J.K., B.R., B.A.R., Q.L., M.A.P., A.M.G., M.D.L., S.T.W., A.M.K.C., J.D.M., K.L. and D.T. do not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
REFERENCES
- 1.Miniño A.M., Xu J., Kochanek K.D. Deaths: Preliminary Data for 2008. Natl Vital Stat. Rep. 2010;59:1–72. [PubMed] [Google Scholar]
- 2.Silverman E.K., Chapman H.A., Drazen J.M., Weiss S.T., Rosner B., Campbell E.J., O'Donnell W.J., Reilly J.J., Ginns L., Mentzer S., et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 3.McCloskey S.C., Patel B.D., Hinchliffe S.J., Reid E.D., Wareham N.J., Lomas D.A. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am. J. Respir. Crit. Care Med. 2001;164:1419–1424. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 4.Silverman E.K., Sandhaus R.A. Clinical practice. Alpha1-antitrypsin deficiency. N. Engl. J. Med. 2009;360:2749–2757. doi: 10.1056/NEJMcp0900449. [DOI] [PubMed] [Google Scholar]
- 5.Hersh C.P., Demeo D.L., Lange C., Litonjua A.A., Reilly J.J., Kwiatkowski D., Laird N., Sylvia J.S., Sparrow D., Speizer F.E., et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am. J. Respir. Cell Mol. Biol. 2005;33:71–78. doi: 10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho M.H., Boutaoui N., Klanderman B.J., Sylvia J.S., Ziniti J.P., Hersh C.P., DeMeo D.L., Hunninghake G.M., Litonjua A.A., Sparrow D., et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai S.G., Ge D., Zhu G., Kong X., Shianna K.V., Need A.C., Feng S., Hersh C.P., Bakke P., Gulsvik A., et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sovio U., Bennett A.J., Millwood I.Y., Molitor J., O'Reilly P.F., Timpson N.J., Kaakinen M., Laitinen J., Haukka J., Pillas D., et al. Genetic determinants of height growth assessed longitudinally from infancy to adulthood in the northern Finland birth cohort 1966. PLoS Genet. 2009;5:e1000409. doi: 10.1371/journal.pgen.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock D.B., Eijgelsheim M., Wilk J.B., Gharib S.A., Loehr L.R., Marciante K.D., Franceschini N., van Durme Y.M., Chen T.H., Barr R.G., et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Repapi E., Sayers I., Wain L.V., Burton P.R., Johnson T., Obeidat M., Zhao J.H., Ramasamy A., Zhai G., Vitart V., et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soler Artigas M., Wain L.V., Repapi E., Obeidat M., Sayers I., Burton P.R., Johnson T., Zhao J.H., Albrecht E., Dominiczak A.F., et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am. J. Respir. Crit. Care Med. 2011;184:786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Durme Y.M., Eijgelsheim M., Joos G.F., Hofman A., Uitterlinden A.G., Brusselle G.G., Stricker B.H. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam Study. Eur. Respir. J. 2010;36:89–95. doi: 10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 13.Young R.P., Whittington C.F., Hopkins R.J., Hay B.A., Epton M.J., Black P.N., Gamble G.D. Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur. Respir. J. 2010;36:1375–1382. doi: 10.1183/09031936.00033310. [DOI] [PubMed] [Google Scholar]
- 14.Chuang P.T., McMahon A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 15.Kawahira H., Ma N.H., Tzanakakis E.S., McMahon A.P., Chuang P.T., Hebrok M. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development. 2003;130:4871–4879. doi: 10.1242/dev.00653. [DOI] [PubMed] [Google Scholar]
- 16.Chuang P.T., Kawcak T., McMahon A.P. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Abecasis G.R.S.M. 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 19.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 20.Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 21.Pomerantz M.M., Ahmadiyeh N., Jia L., Herman P., Verzi M.P., Doddapaneni H., Beckwith C.A., Chan J.A., Hills A., Davis M., et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sotelo J., Esposito D., Duhagon M.A., Banfield K., Mehalko J., Liao H., Stephens R.M., Harris T.J., Munroe D.J., Wu X. Long-range enhancers on 8q24 regulate c-Myc. Proc. Natl Acad. Sci. USA. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasserman N.F., Aneas I., Nobrega M.A. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmadiyeh N., Pomerantz M.M., Grisanzio C., Herman P., Jia L., Almendro V., He H.H., Brown M., Liu X.S., Davis M., et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl Acad. Sci. USA. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miele A., Gheldof N., Tabuchi T.M., Dostie J., Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr. Protoc. Mol. Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21 11. [DOI] [PubMed] [Google Scholar]
- 26.Wilk J.B., Chen T.H., Gottlieb D.J., Walter R.E., Nagle M.W., Brandler B.J., Myers R.H., Borecki I.B., Silverman E.K., Weiss S.T., et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visel A., Rubin E.M., Pennacchio L.A. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 29.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Stielow B., Kruger I., Diezko R., Finkernagel F., Gillemans N., Kong-a-San J., Philipsen S., Suske G. Epigenetic silencing of spermatocyte-specific and neuronal genes by SUMO modification of the transcription factor Sp3. PLoS Genet. 2010;6:e1001203. doi: 10.1371/journal.pgen.1001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manolio T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 33.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M., et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagai T., Amano T., Tamura M., Mizushina Y., Sumiyama K., Shiroishi T. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development. 2009;136:1665–1674. doi: 10.1242/dev.032714. [DOI] [PubMed] [Google Scholar]
- 36.Ferrai C., Pombo A. 3D chromatin regulation of Sonic hedgehog in the limb buds. Dev. Cell. 2009;16:9–11. doi: 10.1016/j.devcel.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 37.Gordon C.T., Tan T.Y., Benko S., Fitzpatrick D., Lyonnet S., Farlie P.G. Long-range regulation at the SOX9 locus in development and disease. J. Med. Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- 38.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop B., Aricescu A.R., Harlos K., O'Callaghan C.A., Jones E.Y., Siebold C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat. Struct. Mol. Biol. 2009;16:698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watkins D.N., Berman D.M., Burkholder S.G., Wang B., Beachy P.A., Baylin S.B. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 41.Shi W., Chen F., Cardoso W.V. Mechanisms of lung development: contribution to adult lung disease and relevance to chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6:558–563. doi: 10.1513/pats.200905-031RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tada M., Kanai F., Tanaka Y., Tateishi K., Ohta M., Asaoka Y., Seto M., Muroyama R., Fukai K., Imazeki F., et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin. Cancer Res. 2008;14:3768–3776. doi: 10.1158/1078-0432.CCR-07-1181. [DOI] [PubMed] [Google Scholar]
- 43.Martin S.T., Sato N., Dhara S., Chang R., Hustinx S.R., Abe T., Maitra A., Goggins M. Aberrant methylation of the Human Hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer Biol. Ther. 2005;4:728–733. doi: 10.4161/cbt.4.7.1802. [DOI] [PubMed] [Google Scholar]
- 44.Janoff A. Elastases and emphysema. Current assessment of the protease-antiprotease hypothesis. Am. Rev. Respir. Dis. 1985;132:417–433. doi: 10.1164/arrd.1985.132.2.417. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi H., Yamamoto H., Akutsu N., Nosho K., Adachi Y., Imai K., Shinomura Y. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. J. Pathol. 2007;213:131–139. doi: 10.1002/path.2216. [DOI] [PubMed] [Google Scholar]
- 46.Shahi M.H., Schiapparelli P., Afzal M., Sinha S., Rey J.A., Castresana J.S. Expression and epigenetic modulation of sonic hedgehog-GLI1 pathway genes in neuroblastoma cell lines and tumors. Tumour Biol. 2011;32:113–127. doi: 10.1007/s13277-010-0105-x. [DOI] [PubMed] [Google Scholar]
- 47.Global Strategy for the Diagnosis, Management and Prevention of COPD, Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2010. Available from http://www.goldcopd.org/
- 48.Ferris B.G. Epidemiology Standardization Project (American Thoracic Society) Am. Rev. Respir. Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 49.Chen Z.H., Kim H.P., Sciurba F.C., Lee S.J., Feghali-Bostwick C., Stolz D.B., Dhir R., Landreneau R.J., Schuchert M.J., Yousem S.A., et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. 2008;3:e3316. doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X., Munger K. Clld7, a candidate tumor suppressor on chromosome 13q14, regulates pathways of DNA damage/repair and apoptosis. Cancer Res. 2010;70:9434–9443. doi: 10.1158/0008-5472.CAN-10-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baron R.M., Carvajal I.M., Liu X., Okabe R.O., Fredenburgh L.E., Macias A.A., Chen Y.H., Ejima K., Layne M.D., Perrella M.A. Reduction of nitric oxide synthase 2 expression by distamycin A improves survival from endotoxemia. J. Immunol. 2004;173:4147–4153. doi: 10.4049/jimmunol.173.6.4147. [DOI] [PubMed] [Google Scholar]
- 54.Baron R.M., Lopez-Guzman S., Riascos D.F., Macias A.A., Layne M.D., Cheng G., Harris C., Chung S.W., Reeves R., von Andrian U.H., et al. Distamycin A inhibits HMGA1-binding to the P-selectin promoter and attenuates lung and liver inflammation during murine endotoxemia. PLoS ONE. 2010;5:e10656. doi: 10.1371/journal.pone.0010656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.