Abstract
This review integrates historical biochemical and modern genetic findings that underpin our understanding of the low-density lipoprotein (LDL) dyslipidemias that bear on human disease. These range from life-threatening conditions of infancy through severe coronary heart disease of young adulthood, to indolent disorders of middle- and old-age. We particularly focus on the biological aspects of those gene mutations and variants that impact on sterol absorption and hepatobiliary excretion via specific membrane transporter systems (NPC1L1, ABCG5/8); the incorporation of dietary sterols (MTP) and of de novo synthesized lipids (HMGCR, TRIB1) into apoB-containing lipoproteins (APOB) and their release into the circulation (ANGPTL3, SARA2, SORT1); and receptor-mediated uptake of LDL and of intestinal and hepatic-derived lipoprotein remnants (LDLR, APOB, APOE, LDLRAP1, PCSK9, IDOL). The insights gained from integrating the wealth of genetic data with biological processes have important implications for the classification of clinical and presymptomatic diagnoses of traditional LDL dyslipidemias, sitosterolemia, and newly emerging phenotypes, as well as their management through both nutritional and pharmaceutical means.
Keywords: intestinal sterol absorption and efflux, cellular cholesterol synthesis, lipoprotein assembly, gallstones, LDL uptake
INTRODUCTION
This review covers the dietary and biochemical origins and fates of key classes of sterol molecules in humans, namely, cholesterol and the relatively under-recognized and often unappreciated noncholesterol sterols and stanols; the intra- and intercellular systems that govern their transport; and the contribution of innate genetic programs to the biochemically observed levels of plasma LDL-cholesterol (LDL-C). The reasons for these foci are both biological and medical. The former is the burgeoning knowledge of the normal physiological roles that cholesterol performs within cell membranes in supporting receptor-mediated signaling activities (1–4), the movement of diverse molecules through different membrane-bound compartments (5–8), and multiple other cell functions (9–11), including myelination (12). The latter, medical rationale is itself duplex: to raise awareness of several newly recognized, genetically determined lipid metabolic phenotypes/disorders and how they are identified in the clinical setting; and, in the light of these and other better-known genetic disorders, to argue the case for replacing the traditional, but now 45-year-old, Fredrickson and Lees, essentially phenotypic, classification of hyperlipidemia (13) with one rising from biological and genetic foundations. Toward this end, we provide a commentary for the biological bases of the effects of gene variants on LDL, sterol disorders, and subclinical phenotypes, other than those of primary cholesterol biosynthesis (14) and the egress of cholesterol (and other lipids) from late endosomes and lysosomes to other cellular compartments (15–17).
On the basis of the results from recent genome-wide association studies (GWAS), we primarily focus on sterol absorption and hepatobiliary cholesterol efflux (Fig. 1, 2); the incorporation of cholesterol and other lipid moieties, including noncholesterol sterols (18–22) into apolipoprotein (apo)B-containing lipoproteins for their transport to distant sites; and receptor-mediated uptake of apoB-containing lipoproteins, especially LDL. Implicit in this approach is the view that the pathological consequences of deranged LDL levels can arise from the type, properties, and amounts of individual lipid classes conveyed by LDL particles (21–24).
Fig. 1.
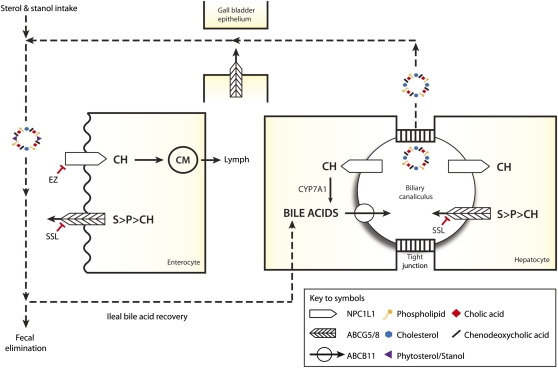
Simplified view of enterohepatic sterol metabolism intestinal uptake of sterols, including cholesterol (CH), phytosterols (P), stanols (S), and fat-soluble vitamins (not shown for clarity), into enterocytes is facilitated by the unidirectional NPC1L1 transporter, a process blocked by ezetimibe (EZ). Net sterol absorption is increased by recessive mutations of either ABCG5 or ABCG8, which encodes the ATP-binding cassette transporters G5 and 8, which preferentially efflux S and P back into the intestinal lumen. A proportion of enterocyte CH and esterified CH (not shown) is packaged into chylomicrons (CM) and released into lymph. In hepatocytes, ABC transporters pump sterols (ABCG5/8), bile acids (ABCB11), and phospholipids (ABCB4, not shown) into biliary canaliculi. Effluxed CH can be recaptured by NPC1L1 and reabsorbed. Mixed micelles formed in biliary canaliculi from phospholipids, cholesterol, and bile acids are stored in the gall bladder until their release is stimulated. Noncholesterol sterols reduce the incorporation of cholesterol into mixed micelles (350), thereby reducing cholesterol uptake by the NPC1L1-mediated process (351, 352). The majority of bile acids are recovered by the ileum. Emerging data (not shown for clarity) indicate that the ABCG5/8 transporter may also facilitate noncholesterol sterol stimulation of intestinal cholesterol excretion through a nonbiliary route (353, 354) Cholesterol 7 α -hydroxylase (CYP7A1) initiates the classic bile acid synthetic pathway to replace those eliminated in feces.
Fig. 2.
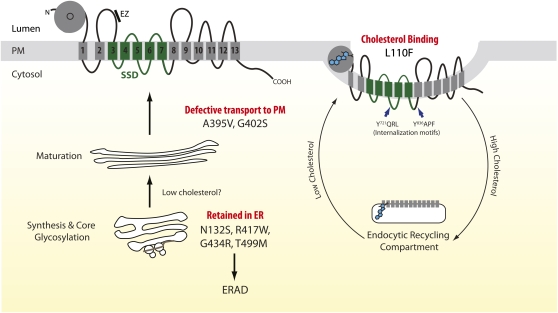
Proposed membrane topology of NPC1L1 and a model of cholesterol-regulated trafficking of NPC1L1 to and from the plasma membrane. The crystal structure of the N-terminal domain of NPC1L1 (amino acids 22-265, denoted by gray ball) reveals a fold almost identical to that formed by the homologous NPC1 region (355). However, in marked contrast to NPC1, its cholesterol-binding pocket (indicated with circle) is closed to solvent, suggesting a gating mechanism whereby multiple movements around the entrance to the NPC1L1 pocket are required to promote the expansion of its entrance and subsequent entry of a cholesterol molecule. The binding pocket is larger than that found in NPC1, allowing for broader substrate specificity, in particular for sterols with substitutions at C4 (e.g., lanosterol). The ethyl group at C24 on β -sitosterol may result in an unfavorable steric clash, consistent with in vitro assays: specifically, β -sitosterol competed poorly with cholesterol for binding to the isolated N-terminal NPC1L1 domain (355). Classification of the molecular mechanisms by which nonsynonymous coding variants may decrease NPC1L1-mediated cholesterol/nonsterol cholesterol uptake is based on the data of Wang et al. (51). N132S, R417W (extracellular loop 1), G434R (extracellular loop 1), and T499M (extracellular loop 1) variants, which reside in predicted N-linked glycosylation sites, are retained in the ER. The L110F variant resides close to the entrance of the NPC1L1 cholesterol (noncholesterol sterol?) binding pocket (355). Sixteen highly conserved amino acid residues line the binding pocket. No variants of these amino acids have been identified. Additionally, no splice site mutations and only one C-terminal truncating mutation (R1014X, transmembrane helix 9) have been identified. The membrane orientations of the two large luminal loops (amino acids 369-633; 861-1112) are deduced from protease protection and immunofluorescence studies performed on selectively permeabilized cells (356). Two putative YXXØ motifs (Y721QRL and Y836APF, blue arrows) may interact with the μ subunit of AP-2 and facilitate clathrin-mediated endocytosis (357). Ezetimibe (indicated by a bar) binds to a site (amino acids 510-571) distinct from that of cholesterol (358). For clarity, only the most mechanistically informative NS coding variants found in “low cholesterol absorbers” (47) are shown. ERAD, ER-associated degradation; PM, plasma membrane; SSD, sterol sensing domain.
NEW INSIGHTS INTO STEROL ABSORPTION AND HEPATOBILIARY SECRETION
Humans exhibits net selective absorption of dietary cholesterol over its biochemical look-alikes, the noncholesterol sterols (e.g., sitosterol) and stanols (e.g., 5 α -sitostanol), for example, ∼50% of dietary cholesterol versus <5% noncholesterol sterols and 5 α -stanols (20, 25, 26). In recent years, two fundamental discoveries have provided an unexpected explanation for this phenomenon, thereby considerably expanding our knowledge of how the membrane transport systems employed by enterocytes and, it turns out, the hepatobiliary system bring about net selective absorption of cholesterol (Figs. 1, 2). The new players on the block to be considered are Niemann-Pick C1 like-1 protein (NPC1L1), the major, and relatively unselective inward transporter of all three sterol lipid classes and vitamin E (27), and ABCG5 and ABCG8, the two halves of an ATP-binding cassette (ABC) transporter, which preferentially ejects 5 α -stanols and noncholesterol sterols from enterocytes (28–30) and the hepatobiliary system (30, 31) into the bowel and bile, respectively. Additionally, a synopsis on the newly described association between the CYP7A1 locus and LDL-C levels is provided (32), given the central role of cholesterol-7 α -hydroxylase in both cholesterol catabolism and bile acid production (Fig. 1).
Sterol absorption functions of NPC1L1
NPC1L1 was identified through studies probing the mechanism by which ezetimibe impairs intestinal cholesterol absorption (33). NPC1L1 was found to be highly expressed in the jejunum (but not in other tissues of the mouse) and to have the hallmarks of a plasma membrane transporter, including a signal peptide, predicted transmembrane and sterol sensing domains, and multiple N-linked glycosylation sites within its extracellular loops (Fig. 2). Moreover, NPC1L1-deficient mice displayed >70% reduction in net intestinal cholesterol adsorption, and the residual low level of cholesterol absorption could not be blocked by ezetimibe. In humans, NPC1LI is not only localized to the brush border of enterocytes (33, 34) where it acts as a unidirectional transporter of cholesterol and noncholesterol sterols into these absorptive cells (35), but it also resides on the apical (biliary canalicular) membranes of hepatocytes where it retrieves biliary cholesterol for its transport back into these cells (18, 35), presumably to inhibit excessive biliary cholesterol loss (Fig. 1).
In humans, the effects of ezetimibe on sterol metabolism and LDL-C levels are well documented (36–40). For example, when von Bergman and colleagues (36–39) examined its effects (10 mg/d, two-week treatment period, two-week washout period) on several metabolic indices in a placebo-controlled, cross-over study involving 18 men (mean age, 25.8 years; BMI, 25.5kg/m2) with mild to moderate hypercholesterolemia (LDL-C 130-180 mg/dl), they found that ezetimibe decreased fractional cholesterol absorption rates on average by >50% (ezetimibe 22.7 ± 25.8%; placebo 49.8 ± 13.8%); plasma total- and LDL-cholesterol levels by 15.1 and 20.4%, respectively; and campesterol and sitosterol by 48% and 41%, respectively. Accompanying these decreases were significant rises in de novo cholesterol synthesis (89%, primarily hepatic) and fecal excretion of neutral sterols (72%). Moreover, in a similarly designed study involving strict vegetarians whose dietary cholesterol intake was low (29.4 ± 16.8 and 31.4 ± 14.4 mg/day during the placebo and ezetimibe phases, respectively), ezetimibe decreased mean fractional cholesterol absorption by 58% (placebo 48.2 ± 8.2%; ezetimibe 20.2 ± 6.2%, P < 0.001). This was paired with increased fecal excretion of neutral (81%, P < 0.001) and acidic (35%, P = 0.052) sterols, plus de novo cholesterol synthesis (72%, P < 0.001) (41). By contrast, total plasma cholesterol and LDL-C levels were decreased (9.9 and 17.3%, both P < 0.001), suggesting the cholesterol-lowering effect of ezetimibe, at least in vegetarians, is mediated largely through inhibiting endogenous (biliary) cholesterol absorption (Fig. 1).
The most direct evidence that NPC1L1 is a relatively nonselective transporter of sterols derives from studies performed in mice (42, 43). Thus, in NPC1L1− / − mice [14C]cholesterol and [3H]sitosterol uptake into the proximal third of the small intestine were reduced to similar degrees: 64 and 52%, respectively, compared with control animals (42). This was accompanied by a ∼4-fold increase in intestinal cholesterol synthesis and elevated intestinal HMG-CoA reductase (HMGCR) mRNA levels but no change in ABCG5 and ABCG8 mRNA (Fig. 1). In ABCG5/8-deficient mice, NPC1L1 deficiency largely prevented the accumulation of phytosterols that normally occurs in these animals (43), as well as in ABCG5 − / − (29) and ABCG8− / − (30) only mice.
NPC1L1 genetic determinants affect circulating sterol and LDL-C levels
Consistent with the high heritability figures for the otherwise very variable population values of cholesterol (20, 44), campesterol, and β -sitosterol absorption rates (45, 46), genetic studies have unearthed robust associations between NPC1L1 variants, surrogate markers of sterol absorption efficiency and LDL-C levels (47, 48) (Table 1) .
TABLE 1.
Sterol absorption: genetic determinants affecting LDL-C and phytosterol levels and the risk of coronary heart disease and gallstone disease
| Gene | Sample (Reference) | Typed Variant (Proxy)a | Positionb(Effect) | Allelec(MAF) | Effect on LDL-C | Other Associated Traits/Comments |
| NPC1L1 | 5,804 members of PROSPER cohort (mean age 75 ± 3 year) with or at risk of vascular disease (48) | 1) rs41279633 | c.-18A>Cd | Cd(0.15) | C/C +4.2/1.6% (men/women) versus A/A, (combined P = 0.02). | Increased risk of CHD death/nonfatal MI for C/C. HR 1.67 [95% CI 1.10-2.54], (P = 0.04). |
| 2) rs17655652 | c.-133A>G | G(0.33) | G/G 2 4.4% versus A/A, (P = 0.023). Women only; men, NS. | |||
| 1) rs2072183 (rs41279633) | c.816C>G(L272L) | G(0.20) | G/G +3.8/2.9% (men/women) versus C/C, (combined P = 0.02). | Increased risk of CHD death/nonfatal MI for G/G. HR 1.50 [95% CI 1.09-2.06], (P = 0.04). | ||
| 1,003 statin-treated patients of Caucasian origin (EASE cohort) (53) | 1) rs41279633 | c.-18C>A | A(0.16) | Greater reduction in LDL-C with ezetimibe treatment: C/C, 2 24.36%; C/A, 2 28.22%; A/A, 2 26.28% (P = 0.002). | ||
| 2) rs17655652 | c.-133A>G | G(0.30) | Smaller reduction in LDL-C with ezetimibe treatment: A/A, 2 26.28%; A/G, − 24.89%; G/G, 2 21.99%, (P = 0.02). | |||
| 65 heterozygous ADH-1 patients/50 statin-resistant primary hypercholesterolemia patients (54) | 1) rs2072183 (rs41279633) | c.816C>G(L272L) | G(0.25/0.26) | Greater reduction in LDL-C with ezetimibe treatment: C/C 2 18.9/29.2%; C/G+G/G 2 23.3/33.7% (P < 0.06/0.07). | ||
| GLGC (32) | 1) rs2072183 (rs41279633) | c.816C>G(L272L) | G(0.25) | Z = 2 6.527, (P = 7 × 10 ) | Total cholesterol, 2 2.01 mg/dl per copy (Z = 2 6.636, P = 3 × 10 ) | |
| 2) rs17655652 | c.-133A>G | G(NA) | Z = 2 5.083, (P = 4 × 10 ) | Total cholesterol, Z = 2 4.778, (P = 2 × 10 ) | ||
| ABCG8 | 142 healthy Americans (94) | 1) rs11887534 | c.55G>C (D19H) | C | Not determined | Carriers (n = 14) lower plasma phytosterol (Ca 233 versus 338 μ g/dl, Si 177 versus 257 μ g/dl, P < 0.01). |
| 262 Finnish with mild to moderate hypercholesterolemia (95) | 1) rs11887534 | c.55G>C (D19H) | C(0.15) | 2 13% in carriers (P < 0.05) | Increased frequency in low (0.26) versus intermediate (0.13)/high cholesterol ‘absorbers’ (0.06) (P<0.001) | |
| Meta-analysis of four studies: healthy individuals, primarily Caucasian origin (96) | 1) rs11887534 | c.55G>C (D19H) | C(0.12) | Not determined | Lower Ca:cholesterol ratio (∼cholesterol absorption) in carriers (n = 83) versus noncarriers (n = 591), (WMD 2 0.50 µg/mg, [95% CI 2 0.80 to 2 0.20 µg/mg], P = 0.001). Higher La:cholesterol ratio (∼cholesterol synthesis) in carriers (n = 79) versus noncarriers (n = 541), (WMD +0.26 µg/mg, [95% CI 0.10 to 0.41 µg/mg], P = 0.001). | |
| Initial population study (n = 1,495): replication cohorts (n = 1,157/n = 1,760), European descent (97) | 1) rs41360247 (rs11887534) | c.322+206T>C | C(0.07/ 0.07/0.06) | 2 ∼10% per copy, measured in third cohort only (P = 0.039). | Ca 2 10% (P = 4x10 ), Si 2 17% (P = 3 × 10 ) and Br 2 13% (P = 1 × 10 ) per C allele. | |
| 2) rs4245791 | c.322+981C>T | C(0.33/ 0.32/0.33) | Not determined | Ca +11% (P = 2x10 ), Si +19% (P = 2 × 10 ), Br +13% (P = 4 × 10 ) per C allele. | ||
| 96 German gallstone disease (GD) cases and 205 controls/ Replication in 1105 cases and 873 controls (98) | 1) rs11887534 | c.55G>C (D19H) | C(0.18/ 0.05/0.10/0.05) | Not determined | Associated with GD (P = 2x10 / P = 4x10 ). Carrier OR combined samples, 2.2 [95% CI 1.8-2.6], P = 1.14 × 10 . Cholesterol gallstones OR, 3.3 [95% CI, 2.5-4.3]. | |
| 226 Indian GD patients plus 222 controls (102) | 1) rs11887534 | c.55G>C (D19H) | C(0.08/0.04) | Not determined | Associated with GD (P = 0.017). OR = 2.274 [95% 1.171-4.41]. Increased biliary cholesterol in patients (P < 0.001). | |
| Danish population study (n = 62,279) with mean follow-up period of 31 years for development of gallstone disease (103) | 1) rs11887534 | c.55G>C (D19H) | C(0.06) | G/C 2 1.6% and C/C 2 2.4% versus noncarriers (P < 0.001) | Total cholesterol 2 3.5% (G/C) and 2 4.5% (C/C) versus noncarriers (P < 0.001). 11% of all gallstones attributable to D19H. HR, 1.9 [95% CI 1.7-2.1] for G/C, 3.3 [95% CI 2.3-4.6] for C/C. | |
| GLGC (32) | 1) rs11887534 | c.55G>C (D19H) | C(NA) | Z = 2 11.715 (P = 1x10 ) +2.75 mg/dl per copy (Z = 14.476, P = 2 × 10 ) | Total cholesterol, Z = 2 11.13 (P = 9 × 10 ) Total cholesterol +3.01 mg/dl per copy (Z = 14.076, P = 4 × 10 ) | |
| 2) rs4299376 (rs4245791) | c.166-718G>T | G(0.30) | ||||
| CYP7A1 | GLGC (32) | 1) rs2081687 | ∼14 kb downstream | T(0.35) | Z = 5.64, (P = 2 × 10 ) | Total cholesterol +1.23 mg/dl per allele (Z = 7.037, P = 2 × 10 ) |
Independent variants/associations are numbered. “Proxy” variant is in strong linkage disequilibrium with genotyped variant.
Relative to the initiating ATG.
Minor allele is listed with frequency (MAF) in study population.
Minor and major alleles in reference publication reversed compared with 1000 Genomes reference panel.
Br, brassicasterol; Ca, campesterol; 95% CI, 95% confidence interval; CHD, coronary heart disease; HR, hazard ratio; La, lathosterol; MI, myocardial infarction; NA, not available; NS, not significant; OR, odds ratio; Si, sitosterol; WMD, weighted mean difference.
Rare NPC1L1 variants.
Cohen et al. (47), sequenced the coding regions of NPC1L1 in 256 individuals from the Dallas Heart Study (population-based sample of Dallas County residents, comprising 1,043 whites, 1,832 African Americans, and 601 Hispanics) who had the highest (n = 128) and lowest (n = 128) campesterol:lathosterol (Ca:L) ratios. The rationale was that this ratio serves as a good marker for rates of intestinal cholesterol absorption (20, 49). Thirteen nonsynonymous (NS) coding sequence variants were identified in both the “high” and “low” cholesterol-absorber groups compared with five and 19 such variants present only in the “high” and “low” cholesterol-absorbing groups, respectively. The cumulative frequency of the 19 NS alleles (plus one nonsense mutation) in the “low” cholesterol-absorbing group was higher than in the “high” cholesterol-absorbing group (26/256 alleles compared with 5/256 alleles, P < 0.001). In the African-American contingent, the 20 variants reached an appreciable frequency: 6.2% compared with 1.8% and 1.7% in the whites and Hispanics, respectively. Moreover, in this subpopulation, this group of alleles was associated with a lower mean plasma Ca:L ratio (1.7 ± 1.2 versus 2.2 ± 2.4, P = 0.005) and LDL-C values (96 ± 36 mg/dl versus 105 ± 37 mg/dl, P = 0.005). Consistent with estimated rates of cholesterol absorption, stable isotope methodology confirmed that two NS variants (no others tested) were indeed associated with lower net cholesterol absorption in two pedigrees, one with the I647N variant (n = 37 family members) and the other with the R693C allele (n = 16) (50).
Crucially, functional data now corroborate the genetic evidence that the 20 rare NPC1L1 alleles found solely in the “low” cholesterol absorption group impair NPC1L1-mediated cholesterol uptake (50, 51). Implicated mechanisms include defective glycosylation (e.g., N132S), aberrant folding (e.g., S620C), enhanced degradation through the endoplasmic reticulum (ER)-associated protein degradation pathway (e.g., S881L), impaired transport to the plasma membrane (e.g., A395V), decreased cholesterol-binding (e.g., L110F), as well as perturbed recycling of NPC1L1 to the plasma membrane in response to low cellular/ER-cholesterol concentrations (Fig. 2). As such, the experimental approach of resequencing genes in individuals at the extreme of the population distribution to identify putative causal variants has amply borne fruit. However, whether NPC1L1 null alleles exist in “low” cholesterol absorbers or, conversely, gain-of-function variants increase cholesterol/non-cholesterol sterol uptake remains to be established. Additionally, further investigations are required to explain the rather surprising in vitro finding, given the in vivo data (42, 43), that only cholesterol (ergosterol, β -sitosterol, campesterol, stigmasterol, and brassicasterol tested) had a significant capacity to internalize NPC1L1 via clathrin/AP2-mediated endocytosis (52).
Common NPC1L1 variants.
Table 1 summarizes the associations between relatively common NPC1L1 variants and LDL-C levels (48, 53, 54). In the PROSPER cohort, homozygotes with the rare allele at the c.-18C>A locus had significantly higher (albeit modest) LDL-C levels than cohort members who had two copies of the common allele, while the heterozygote individuals had intermediate levels (48). Supporting this association, CHD death and nonfatal myocardial infarction (MI) during the 3.2 year follow-up period were more common in the rare allele carriers. However, whether this association is attributable to the causative NPC1L1 allele(s) merely enhancing cholesterol uptake remains an open question. LDL particles are major carriers of phytosterols (21), lathosterol (21), sphingomyelin, and ceramide (22), none of which was analyzed in the patient cohort.
Table 1 also shows the ezetimibe (10 mg/day for 6 weeks) add-on to statin (any dose, any brand) for effectiveness trial (EASE) found association between the rare allele at the c.-18C>A locus and LDL-C lowering response (53), complementing the PROSPER study. However, neither study performed functional analyses to address whether the observed effects were attributable to the c.-18A allele, to an allele(s) in strong linkage disequilibrium (LD) (e.g., c.861C>G, L272L [rs2072183]; V1296V [rs217434]), or to both. So far, three studies have reported on rs2072183 (Table 1). Most recently, the meta-analysis of 46 GWAS comprising ∼100,000 individuals of European descent ascertained in multiple countries (hereafter referred to as GLGC sample/cohort) estimates the effect of the rs2072183 minor allele on LDL-C levels to be a modest 2.01 mg/dl/copy.
Table 1 highlights another noteworthy result to emerge from the PROSPER and EASE studies: the “promoter” variant c.-133A>G. In PROSPER, women, but not men, with the A allele had higher LDL-C levels than those who did not. Additionally, following randomization to the pravastatin arm of the study (40 mg/day), women homozygous for the A allele displayed greater reduction in LDL-C than those with the G allele. Similarly, in EASE, the mean percentage drop in LDL-C levels following addition of ezetimibe to the patients’ statin medications was highest in Caucasians with the AA genotype. Because the LD between the alleles at this locus and the g.-18C>A is relatively low, two independent haplotypic arrangements at the NPC1L1 locus appear to contribute to LDL-C levels.
The associations between common NPC1L1 variants and LDL-C levels prompt three important questions, all of which have yet to be addressed. First, are the alleles themselves neutral and simply in LD with functional variants that alter the primary sequence of NPC1L1, thereby promoting, for example, its recycling to the plasma membrane for further rounds of cholesterol/sterol uptake (Fig. 2)? Second, does decreased allelic expression of NPC1L1 translate into decreased functional protein and a consequent decrease in cellular and plasma sterol levels? Third, could the rare alleles at the c.-133A>G or c.-18C>A loci or the alleles in LD represent gain-of-function variants that increase plasma LDL-C levels by increasing NPC1L1 transcript numbers and the amount of NPC1L1 available for transport to the plasma membrane? Regarding NPC1L1 expression, Davis et al. (42) showed that the proximal small intestine of the NPC1L1− /+ mice fed standard chow diet contained ∼80% of wild-type levels of NPC1L1 mRNA, suggesting that haploinsufficiency in enterocytes induces a compensatory rise in transcription from the intact allele. Nonetheless, the mice had lower plasma sitosterol (37% reduction, P = <0.05) and campesterol (50%, P = 0.05) concentrations, presumably attributable to reduced NPC1L1 abundance and, in turn, NPC1L1-mediated sterol uptake, although neither of these parameters was measured. Moreover, in marked contrast to wild-type mice, NPC1L1− /+ animals were resistant to a high-cholesterol diet (1% cholesterol, 0.5% sodium cholate for seven days); thus, their plasma cholesterol levels remained virtually the same as when they consumed the standard chow diet. Moreover, they accumulated less cholesterol in both their bile (7.36 ± 0.58 μ mol/ml versus 11.1 ± 1.01 μ mol/ml, P < 0.05) and liver (79% lower, P < 0.01) than the control mice receiving the same cholesterol-enriched diet. Although these results support the premise that human NPC1L1 variants could modulate LDL-C levels by directly affecting NPC1L1 transcript numbers, their translatability, or both, other data reveal a disconnect between determinants of NPC1L1 gene expression (55, 56) and NPC1L1 protein levels (57). Thus, in a crossover study involving 22 men with modestly raised LDL-C (i.e., >50th percentile value for their age (38.1 ± 9.8 years), it was found that atorvastatin (12 weeks, 40 mg/day or placebo) increased intestinal NPC1L1 expression by 18.7% (P = 0.03), as judged by RNA concentrations in duodenum biopsy samples. However, the atorvastatin-induced increases in NPC1L1 transcript numbers were not correlated with changes in NPC1L1 protein values (average increase 33.5% [not significant]). Notwithstanding, plasma levels of sitosterol and campesterol, two surrogate markers of intestinal cholesterol absorption, were markedly increased (69.7% and 64.7%, respectively; both P < 0.0001), as were intestinal RNA levels of HMGCR (59%), LDL receptor (LDLR) (52%, Fig. 4) and proprotein convertase subtilisin-like/kexin type 9 (PCSK9) (187%, Fig. 4). As such, it would be interesting to establish the nature of the molecular events mediating the atorvastatin-induced increases in intestinal NPC1L1 and HMGCR expression, as well as how these relate to cholesterol/sterol concentrations in human enterocytes and the cellular distribution of the NPC1L1 protein (Fig. 2).
Fig. 4.
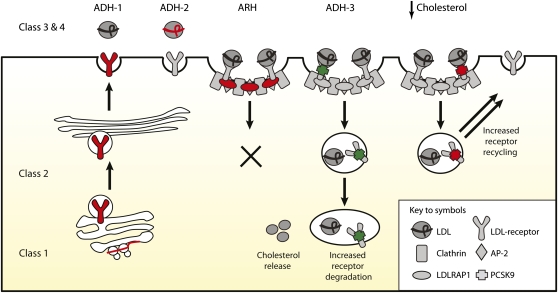
Schematic overview of the cellular processes mediating LDL-C uptake and genetic disorders. Five major classes of LDLR mutations cause ADH. The mutations prevent (i) production of immunologically detectable protein; (ii) ER exit of encoded gene product: complete (a) or partial (b); (iii) binding of apoB100 (a) and apoE (b) ligands; (iv) constitutive endocytosis, including of LDLR-apoB100 (a) and of VLDL-apoE (b); and (v) release of internalized LDLR ligand (not shown for clarity). ADH-2 is caused by APOB mutations that block the binding of apoB100 to the LDLR. The precise residues of the LDLR-mediating apoB100 binding are not known, but at the neutral pH of the cell surface, it seems to involve four of the cysteine-rich, complement-like repeats (R3-7, each ∼40 amino acids) and the β -propeller (YWTD, Fig. 5B) within its extracellular domain (118, 217). The ARH gene product LDLDRAP1 interacts with both the C-terminal 802FDNPVY807 internalization motif of the LDLR plus the clathrin-coated pit machinery to mediate hepatic uptake of LDLR/LDL-apoB100 complexes but not of LDLR-VLDL-remnant complexes, which are internalized via an 802FDNPVY807-independent mechanism. ADH-3 is caused by gain-of-function PCSK9 mutations. Loss-of-function mutations/variants increase LDLR recycling to the plasma membrane and, hence, LDL-C/VLDL clearance from the circulation. The ADH-3 gene product PCSK9 undergoes autocleavage in the ER (not shown), an event that occurs between amino acids Q152 and S153, and it requires the canonical triad of catalytic residues (D186, H226, and S386). The cleaved prodomain forms a tight complex with the catalytic (amino acids 153-451) and C-terminal (452-692) domains of newly cleaved PCSK9, and this complex is secreted into plasma. At the cell surface, PCSK9 binds in a calcium-dependent manner the EGF-like repeat A in the EGF domain of the LDLR (361). The mutant PCSK9 D374Y protein binds cell surface LDLR ∼30-fold more tightly than wild-type PCSK9 (362). Moreover, in the acidic pH of endosomes (and in marked contrast to wild-type PCSK9) it remains exclusively in a high-affinity LDLR-PCSK9 complex, preventing LDLR recycling back to the plasma membrane. PCSK9 may serve to target LDLR to the lysosome for degradation. Other mechanisms of PSCK9-mediated degradation of LDLR are outlined in Ref. 327. Red, loss-of-function mutation; green, gain-of-function mutation.
Efflux functions of ABCG5/8
The ABCG5/8 locus became the focus of attention when two groups (58, 59) showed that mutations in either ABCG5 or ABCG8 cause the rare recessive disorder sitosterolemia (SSL) [OMIM #210250]. Subsequently, it was shown that ABCG5 and ABCG8 were required to form a heterodimer in the endoplasmic reticulum (ER) to reach the surfaces of cultured cells and mouse hepatocytes (60). Moreover, ABCG5 and its obligate partner ABCG8 require bile salt micelles to mediate cholesterol efflux from dog gall-bladder epithelial cells (61). The study of ABCG5 − / − (29) and ABCG8 − / − (30) only mice further confirmed that null alleles of either gene are sufficient to cause sitosterolemia.
Understanding the efflux functions of the ABCG5/8 heterodimer requires consideration of its cellular expression pattern and the biochemical milieu to which such cells are exposed (Fig. 1). Northern blotting showed that gene expression is largely confined to human small intestine and liver (58) and that immunocytochemistry resolves the specific cellular and subcellular distribution of the corresponding protein within these organs. ABCG5 and ABCG8 are restricted to small intestinal enterocytes, hepatocytes, and biliary epithelium (two organs but three distinct cell-types) in both mouse (60) and human (62). More specifically, mouse small intestinal enterocytes express ABCG5 (ABCG8 not studied) on their absorptive brush border, as well as in a cytoplasmic punctuate fashion (60). Human enterocytes exhibit the same apical staining for ABCG5 and ABCG8, although the former has an additional diffuse cytoplasmic component (62).
In mouse hepatobiliary tissues, data on cellular expression patterns of ABCG5 and ABCG8 protein remain incomplete. By immunofluorescence microscopy, native ABCG5 protein epitopes are certainly expressed on the hepatocyte apical (i.e., biliary canalicular) membranes. However, the argument for ABCG8 hepatocytic colocalization is based upon visualization of specific epitopes in a transgenic strain expressing 14 copies of human ABCG5 and ABCG8. Issues of biliary ductal expression were not addressed (60). In man, immunocytochemical data on hepatobiliary cellular expression patterns of the two transporters are more comprehensive and comprehensible, largely tallying with mouse findings. Thus, both proteins could be identified in hepatocytes framing biliary canaliculi cells, with ABCG5 dominating and displaying a more restricted apical distribution, whereas ABCG8 was expressed on the plasma membrane generally and probably intracellular membrane systems as well. Bile ductular and gall bladder epithelia both immunocytochemically expressed ABCG5 and ABCG8 (62). Cell culture and derivative cDNA studies provide further evidence of distinct biliary epithelial and hepatocytic expression (60, 63).
Kinetic studies performed on sitosterolemia patients and their parents (25, 26, 64, 65) and, more recently, mice (31, 66) have helped adduce the sterol efflux functions of ABCG5/8. For instance, in an early study, Salen and colleagues (26) measured weekly, over a 10-week period, sterol absorption and turnover rates in a 28-year-old female sitosterolemic patient and her obligate heterozygote parents, all of whom received control diets containing ∼500 mg/day cholesterol and 100 mg/day sitosterol. Both the patient and her parents absorbed more cholesterol (patient, mother, father, and controls, respectively, 62 ± 7.6%, 59 ± 6.7%, 84 ± 9%, and 48 ± 4%) and sitosterol (34 ± 6%, 15 ± 10%, 17 ± 8%, and 5 ± 4%) than control participants, but despite these increases, only the proband displayed notably increased plasma cholesterol (233 ± 12, 210 ± 26, 194 ± 14, and 185 ± 6 mg/dl), sitosterol (21.0 ± 2.0, 0.95 ± 0.17, 0.36 ± 0.09, and 0.22 ± 0.20 mg/dl) and apoB (131, 77, 72, and 69 mg/dl) levels. Thus, while both parents displayed near-normal capacities to eliminate sterols, especially noncholesterol sterols, from their bodies, their sitosterolemic daughter did not.
Investigations on wild-type (66), ABCG5− / − (29), ABCG8− / − (30), and ABCG5/8-deficient mice (31) have produced data largely concordant with human findings. Thus, Igel et al. (66) examined sterol metabolism in wild-type mice that had received an intragastric dose of plant oil, containing rough equal quantities of three deuterated sterols and their corresponding 5 α -stanols. They found the mice rapidly ( ∼15 min) internalized all of the lipids with similar efficiencies, as judged by the sterol/stanol:cholesterol ratios in their upper intestinal enterocytes (i.e., campesterol:cholesterol 0.91 ± 0.05, sitosterol:cholesterol 0.75 ± 0.11, cholestanol:cholesterol 0.70 ± 0.08, campestanol:cholesterol 0.52 ± 0.08, and sitostanol:cholesterol 0.41 ± 0.08). However, thereafter, marked differences occurred. First, in the elimination of sterols and stanols from enterocytes (sitostanol > campestanol > sitosterol > campesterol = cholestanol > cholesterol); next in plasma accumulation (cholesterol > campsterol > cholestanol> sitosterol; [sitostanol and campestanol, not detected]); and then in their elimination in bile (campesterol > cholestanol > sitosterol > cholesterol).
In marked contrast to wild-type mice, ABCG5/8-deficient animals retain noncholesterol sterols in both their plasma and liver (i.e., sitosterol, 85 and 116×; campesterol, 14 and 16×; and cholestanol, 2 and 1.5× relative to wild-types animals). Cholesterol levels, in comparison to the human situation, were decreased by ∼50% (31) and associated with decreased hepatic mRNA levels for 13 enzymes on the cholesterol biosynthetic pathway. Thus in ABCG5/8− / − mice, high noncholesterol sterol(s) levels decrease cholesterol biosynthesis, as occurs in man. Moreover, their bile contains ∼90% less sterol than wild-type animals, with the bile:liver sterol ratios, indicating that the preferential efflux of noncholesterol sterols into bile (30) was also lost in these animals, consistent with data from studies performed in ABCG8-deficient mice, which measured the amounts of [14C]cholesterol and [3H]sitostanol that entered bile via HDL (30).
ABCG5/8 genetics
Below, we summarize the clinical presentation of sitosterolemia, the spectrum of reported ABCG5/8 mutations, and the phenotype of the carrier state (Table 2), as well as the associations among relatively common ABCG5/8 variants, plasma sterol profiles, and risk of gallstone disease and coronary heart disease (CHD; Table 1).
TABLE 2.
ABCG8 haploinsufficiency and ABCG5 missense mutation increase net sterol absorption in a Micronesian population
| Noncarriers | ABCG8p.Q24HfsX8 Carriers | ABCG5p.D450H Carriers | P (ANOVA) | |
| Frequency (%) | – | 11.1 | 1.8 | |
| Campesterol:cholesterol (∼sterol absorption) | 1.33 ± 0.52 | 2.00 ± 0.87 | 2.78 ± 1.01 | <0.0001a |
| Lathosterol:cholesterol (∼cholesterol synthesis) | 1.52 ± 0.67 | 1.33 ± 0.55 | 0.84 ± 0.38 | <0.0001a |
| Total cholesterol (mg/dl) | 165.48 ± 34.35 | 170.20 ± 34.48 | 167.55 ± 34.92 | 0.022b |
Sitosterolemia mutations.
Clinically, sitosterolemia (also known as phytosterolemia) manifests either in children as tendon and tuberous xanthomas (67–71) or in young adults with severe CHD attributable to massive accumulation of sterols and stanols in monocyte-derived macrophages (25, 68, 72–75). Other clinical manifestations include arthralgia and intermittent arthritis ascribed to sitosterol deposits (45, 70); liver disease (45); and hematological abnormalities (45, 69, 76, 77), including abnormally shaped, fragile erythrocytes and large platelets. In one patient, adrenal insufficiency was present (76).
Biochemically, sitosterolemia is characterized by raised ( ∼30×) plasma levels of plant (e.g., sitosterol, the main dietary sterol, campesterol, and stigmasterol), their 5 α -saturated derivatives (e.g., 5 α -cholestanol and 5 α -sitostanol) and shell-fish sterols (22-dehydrocholesterol, brassicasterol, C-26 sterol, and 24-methylene cholesterol) (25, 72, 78). Plasma cholesterol levels may also be severely elevated, especially in children (58, 59, 71, 79). At the tissue level (except brain), sterols and stanols accumulate in a range of cell types in approximately the same ratio as present in blood (72, 80).
So far, ∼40 different ABCG5 or G8 mutations have been described in 65+ sitosterolemia families (59, 67–71, 73, 76, 79, 81–84). In most kindred, the mutations definitively reside on both copies of either one gene or the other, but not on one chromosome of each. A few of the mutations are over-represented in certain ethnic groups (e.g., ABCG5, R389H in Japanese, Chinese (71, 85), ABCG8, W361X in Europeans (45, 59, 81), implying founder effects, but otherwise, each mutation is confined to one or two kindred. Many of the mutations are predicted to encode truncated ABCG5 or 8 polypeptides, resulting in the loss of their single nucleotide binding domain, putative transmembrane domain (six α -helices), glycosylated extracellular loop ( ∼70 amino acids), or short, highly conserved cytosolic C-terminal tail. Some (mutations) are missense, including five affecting the ATP binding cassette motif of either ABCG5 or ABCG8 and, thus, the presumed coupling of ATP hydrolysis to sterol transport (61, 86, 87). Others appear to impair the formation of stable G5G8 heterodimers and their subsequent trafficking out of the ER (88). A few may affect the ABCG5/8 sterol binding sites, which have yet to be mapped.
Modest effect of sitosterolemia carrier status on LDL-C levels.
The high carrier rate (11.1%) of a particular truncating mutant ABCG8 allele (Table 2) among individuals of the Micronesian Island of Kosrae illuminates the outcome of ABCG8 haploinsufficiency on sterol metabolism (83, 89). Compared with noncarriers, such individuals have sterol profiles indicative of increased net sterol absorption plus decreased endogenous cholesterol synthesis (Table 2). Hence, plasma cholesterol and LDL-C levels are only moderately increased.
Amazingly, nearly 2% of Kosrae (i.e., three large extended families and one apparently unrelated individual) carry a single missense mutation (D450H) (89), affecting the ABCG5 putative transmembrane domain. Intriguingly, this variant has greater impact on plasma noncholesterol sterol levels than the more common and truncating (i.e., null) mutant ABCG8 allele (Table 2). Furthermore, inferred endogenous cholesterol synthesis rates are more markedly decreased in ABCG5 missense mutation carriers than in ABCG8 null carriers, which may explain why mean plasma cholesterol values in islanders with the ABCG5 missense allele are midway between those islanders with no ABCG5/8 mutation and the ABCG8 null allele carriers (Table 2).
Historical but more direct evidence of reduced cholesterol synthesis in sitosterolemia patients (as well as heterozygote carriers) derives from the era preceding genetic diagnosis (64, 90–93). Nguyen and colleagues (93), for example, showed that hepatocytes from two affected sisters (ages 27 and 29 years) contained 24% less cholesterol than control subjects, as well as barely detectable levels of HMGCR mRNA. HMGCR protein (0.18 ± 0.04 versus 1.43 ± 0.41 units/mg cell protein) and activity (15.0 ± 2.0 versus 98.1 ± 28.8 pmol/mg protein/min) were also markedly reduced. It seems, however, that certain cell types may compensate for any potential shortfall in hepatic-derived cholesterol by increasing uptake of their exogenous supplies, as both peripheral blood mononuclear cells and hepatocytes themselves exhibited substantial increases in LDL-C high-affinity (receptor-mediated) binding (253 versus 95.1 ± 8.2 ng/mg). Moreover in the follow-up study (91) involving an additional sitosterolemic sister, neither colestipol (bile acid sequestrant) nor ileal bypass surgery (aimed at lowering sterol absorption and stimulating bile acid synthesis) increased de novo cholesterol synthesis, as measured in mononuclear leukocytes. Rather, receptor-mediated uptake of LDL was increased.
Opposing effects of ABCG8 variants on CHD and gallstone disease risk.
Prior to GLGC (32), three small studies had indicated that the D19H ABCG8 variant (or allele in strong LD (e.g., rs41360247), represents a gain-of-function (94–96) as 19H carriers had plasma sterol profiles suggestive of increased sterol efflux plus a compensatory rise in de novo cholesterol synthesis (Table 1). Accordingly, in the GLGC cohort, 19H allele ownership translates into only a modest reduction in LDL-C levels (Table 1). In another GWAS (97), an allele in strong LD (r2 = 0.93) with the 19H ABCG8 allele (not genotyped) displayed association with markedly lower plasma noncholesterol sterol levels and, more importantly, decreased CHD risk (Table 1).
The strong association of ABCG8 19H variant (or allele in LD) with gallstone disease was originally detected in German patients suffering from all types of stones and even more convincingly for stones in which cholesterol was the major component (Table 1) (98). Since then, associations between 19H ownership and gallstone disease have been reported in Chileans (98), Romanians (99), Swedes (100), Taiwanese (101), Indians (102), and the Danish general population (103). Tantalizingly, in Indian patients, 19H ownership was associated with higher cholesterol levels in bile (102). Sadly, in this study, noncholesterol sterols were not measured.
Genetic data for two further ABCG8 alleles (rs4299376, rs4245791) also point to the clinical importance of the ABCG5/8 transporter in regulating long-term, whole-body sterol homeostasis (Table 1). In the GLGC cohort, the rare allele at rs4299376 (in strong LD with the rare allele at rs4245791) was associated with increased LDL-C and 40% lower levels of hepatic ABCG8 mRNA (P = 0.009). Similarly, in the case-control study of Teupser et al. (97), the proxy for this allele displayed association with increased plasma phytosterol and cholesterol, and increased CHD risk.
In summary, the convergence of biological and genetic data indicate that the rare allele at ABCG5/8 single-nucleotide polymorphism (SNP) site rs41360247 (or allele[s] in LD; e.g., ABCG8 19H), increases sterol efflux, thereby lowering plasma phytosterols and LDL-C levels, and CHD risk (but increased risk of gallstone disease). Conversely, the rare allele at SNP site rs4245791 (or allele in LD) is associated with increased plasma phytosterol concentrations, modestly raised LDL-C, and increased CHD risk.
Downstream CYP7A1 variant associated with increased LDL-C levels
Variant rs2081687, located ∼14 kb 3′ of CYP7A1, is the first to be robustly associated with LDL-C levels in the population at large (Table 1). Even if this phenotype arises from a so-far undiscovered variant(s) in strong LD with rs2081687, rather than the variant itself, the genetic architecture of this genomic region suggests the causative lesion will reside outside the structural gene, its complex 5 ′ promoter elements, and 3′ untranslated region (UTR) (104, 105). Hence, an unsuspected, distant 3′ sequence may regulate CYP7A1 expression and, in turn, cholesterol conversion to bile acids (Fig. 1).
HMGCR variant associated with LDL-C levels
It seems to have been known for a considerable time, albeit anecdotally, that plasma cholesterol levels are abnormally low (i.e., <5th percentile for age and sex) in patients with cholesterol biosynthetic disorders such as Smith-Lemli-Opitz syndrome (106). However, it has only recently become apparent that a HMGCR variant (rs384662) within intron 13 is associated with modest reductions in plasma LDC-C levels (comparable to ABCG8 LDL-C-lowering variant) in the population at large (Table 3). This discovery originated with a GWAS, which identified association among three HMCGR variants in strong LD with the intron 13 variant and LDL-C levels in Kosraen Polynesians (107). Subsequent analyses replicated the association in three white (107), two Japanese (108) and the GLGC cohort populations In the Japanese, the intron 13 variant is also associated with increased risk of myocardial infarction (Table 3).
TABLE 3.
“Functional” HMGCR variant associated with increased LDL-C levels
| Gene | Sample (Reference) | Typed Variant (Proxy)a | Positionb(Effect) | Allelec(MAF) | Effect on LDL-C | Other Associated Traits |
| HMGCR | 2,346 inhabitants of Micronesian island of Kosrae, plus1464 type 2 diabetes cases plus 1,467 controls (Diabetes Genetics Initiative data) (107) | 1) rs3846662 | c.1722+45A>G | G(0.40) | G/G Kosraens +11.6 mg/dl versus A/A (P < 2 × 10 ). For combined dataset, P < 1 × 10 ). | Total cholesterol (combined dataset, P < 2 × 10 ) |
| 2 Japanese population studies (n = 2,686 and n = 2,110) (108) | 1) rs3846662 | c.1722+45A>G | G(0.51/0.50) | β = 3.56, P = 4.91 × 10 | MI, OR 1.15 [95% CI 1.04-1.28], P = 0.0075) | |
| GLGC (32) | 1) rs12916 (rs384662) | c.*372T>C | C(0.39) | +2.45 mg/dl per copy (Z = 14.079, P = 5 × 10 ) | Total cholesterol +2.84 mg/dl per copy (Z = 14.363, P = 9 × 10 ) |
Independent variants/associations are numbered. “Proxy” variant is in strong linkage disequilibrium with genotyped variant.
Relative to the initiating ATG or stop codon (*).
Minor allele is listed with frequency (MAF) in study population.
95% CI, 95% confidence interval; OR, odds ratio.
It turns out that the intron 13 variant rs384662 modulates splicing efficiency of HMGCR pre-mRNA, culminating in the production of fewer naturally occurring nonfunctional transcripts (i.e., minus exon 13 and catalytic residues). Hence, in vivo this variant is envisaged to increase HMGCR activity and cholesterol synthesis, producing a compensatory reduction in LDL-C uptake to maintain cellular cholesterol homeostasis (107).
NEW INSIGHTS INTO APOB-CONTAINING LIPOPROTEIN ASSEMBLY AND SECRETION
In the sections that follow, we provide an update on apoB and the microsomal triglyceride transfer protein (MTP), while acknowledging that the assembly of apoB-containing lipoprotein serves two complementary functions: (i) the delivery of lipids to distant cells and (ii) the removal of excess lipids that might otherwise lead to disease (e.g., liver cirrhosis). Another section highlights a potentially new function for angiopoetin-like protein 3 (ANGPLT3) and reports on the association of a DOCK7/ANGPLT3 variant with both LDL-C and triglyceride (TG) levels. Other sections focus on the intracellular trafficking of nascent chylomicron and VLDL. The last section summarizes the emerging evidence that the tribbles homolog 1 (Trib1) increases VLDL production through enhanced de novo lipogenesis. Although, helpful for understanding the hypocholesterolemia that develops in apoB-specific familial hypobetalipoproteinemia (FHBL) and ANGPLT3-specific combined hypolipidemia, it is beyond the scope of this review to cover the array of molecular processes regulating presecretory degradation of nascent apoB-containing lipoproteins (109–111).
ApoB structures and lipoprotein production
The specific structures required to initiate the cotranslational assembly of apoB-containing lipoproteins (Fig. 3) are formed by the N-terminal region ( β α 1) of apoB (amino acids 1-782; apoB17) (112–114), whereas sequences beyond apoB19.5/22 (i.e., amino acids 884/1000) supply the lipid binding structures for forming a stable, secretion-competent, lipoprotein particle (115, 116). These sequences include amphipathic β -strand and α -helix structures in the B1 (apoB20-apoB41; amino acids ∼908 to ∼1860) and α 2 (apoB42-48; amino acids ∼1900-2152) domains of apoB, respectively, and in particular sequences between apoB32 and apoB41 ( ∼1450-1860), which have a marked ability to recruit triglyceride into assembling lipoproteins (117). The amphipathic β -strand structures formed by apoB37-41 (amino acids 1694-1880) and apoB56.7-88.2 (amino acids 2571-4000) also appear to irreversibly anchor apoB to the lipid cores of VLDL and LDL (114, 118).
Fig. 3.
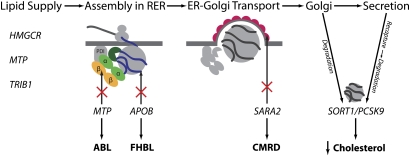
Multiple gene products participate in the assembly, intracellular trafficking, and secretion of chylomicrons and VLDL. Microsomal triglyceride protein (MTP), the abetalipoproteinemia gene product, forms a heterodimer with protein disulphide isomerase (PDI), an ER-chaperone-like protein. The PDI is envisaged to retain MTP at the site of apoB translocation, as it alone contains the “KDEL” ER-retention sequence. In one model, MTP acquires the lipids (for transfer to apoB) from the membranes of the ER via two helical peptides situated at the entrance to its lipid-binding cavity (dark green). The first helix interacts with a membrane in a manner similar to viral fusion peptides and is necessary for the acquisition of neutral lipid. The second helix, which contains an abetalipoproteinemia missense mutation, is required for the transfer of these lipids to the lipid-binding cavity of MTP (359). The schematic also illustrates that the highly conserved Interpro1747 domains (yellow, β -barrel; light green, α -helical domain) (360) of MTP and apoB physically interact, presumably to ensure MTP can cotranslationally load the lipid-binding structures of apoB with lipid as they enter the ER. APOB mutations that affect the folding of the N-terminus of apoB or its interaction with MTP or that remove the apoB's C-terminal lipid binding structures cause FHBL. Nascent VLDL and chylomicrons leave the ER in coat protein (COP) II vesicular structures that are assembled on the cytosolic surface of the ER. In CMRD, chylomicron-like particles are made, but these are retained intracellularly due to recessive mutations of SARA2, which encodes the Sar1b isoform of Sar1. In post-ER compartments, nascent VLDL has two distinct fates: it may interact with sortilin (encoded by SORT1) and, potentially, PCSK9 and be targeted for degradation in lysosomes; or alternatively, it may be secreted. Some newly secreted VLDL is recaptured by the LDLR-PCSK9 axis (not shown for clarity).
ApoB-specific FHBL.
Clinically, FHBL ([OMIM #107730], Table 4) is an autosomal dominant monogenic disorder reserved to describe those individuals who have (i) low levels of total cholesterol (TC; <5th age-sex-percentile values of the reference population), LDL-C (<75 mg/dl), and apoB (<50 mg/dl), and no underlying lifestyle (e.g., vegan, malnutrition) or medical cause (e.g., hyperthyroidism (119), chronic liver disease); and (ii) a parent (or parents) with the same biochemical trait. No reliable frequency estimates of the prevalence of FHBL (as defined by biochemical criteria and dominant transmission of the lipid trait) exist because no systematic population/family studies have been performed. We therefore eagerly await data from large-scale sequencing of people from the general population with a lipid phenotype indicative of FHBL and descriptions of the ensuing clinical associations.
TABLE 4.
Monogenic disorders of lipoprotein assembly and secretion: representative lipid and lipoprotein levels
| Lipid/Lipoprotein Levelsc (mg/dl) | |||||||||
| Disorder/Gene/CS (Reference) | Subject (Agea) | Mutation State | Mutation (Effectb) | TC | LDL-C | TG | HDL-C | ApoB | TC:HDL-Cd |
| FHBL/APOB/CS1 (120) | Proband (15) | Compound heterozygous | c.1315C>T/c.537+1G>T (ApoB9/apoB3.4-4.5) | 36 | 0 | 12 | 36 | NA | 1 |
| Mother (40) | Heterozygous | c.1315C>T (ApoB9) | 90 | NA | 40 | NA | NA | NA | |
| Father (43) | Heterozygous | c.537+1G>T (ApoB3.4-4.5) | 135 | 47 | 445 | 38 | NA | 3.55 | |
| Brother (14) | Heterozygous | c.537+1G>T (ApoB3.4-4.5) | 97 | 29 | 59 | 56 | NA | 1.73 | |
| FHBL/APOB/CS2 (121) | Proband (4) | Homozygous | c.2169delT (ApoB15.56) | 11 | 3 | 21 | 4 | ∼0 | 2.75 |
| Mother (27) | Heterozygous | 66 | 19 | 58 | 34 | 35 | 1.94 | ||
| Father (40) | Heterozygous | 62 | 15 | 22 | 42 | 35 | 1.48 | ||
| Brother (1) | Homozygous | 27 | 1 | 4 | 24 | ∼0 | 1.13 | ||
| FHBL/APOB/CS3 (130) | Proband (21) | Homozygous | c.4216+2T>C (ApoB27.6) | 19 | NA | 6 | NA | ∼0 | NA |
| Mother | Heterozygous | 180 | NA | 33 | NA | 67 | NA | ||
| Brother | Heterozygous | 80 | NA | 43 | NA | 31 | NA | ||
| Sister | Heterozygous | 99 | NA | 24 | NA | 27 | NA | ||
| FHBL/APOB/CS4 (131) | Proband (57) | Homozygous | c.5344C>T (ApoB38.7) | 84 | NA | 57 | 77 | 3 | 1.09 |
| Son (32) | Heterozygous | 124 | NA | 43 | 68 | 27 | 1.82 | ||
| Son (30) | Heterozygous | 127 | NA | 62 | 71 | 27 | 1.79 | ||
| FHBL/APOB/CS5 (132) | Proband (48) | Homozygous | c.6240T>A (ApoB45.2) | 58 | ∼0 | 60 | 51 | 7 | 1.14 |
| FHBL/APOB/CS6 (133) | Proband (10) | Heterozygous | c.3741T>A (ApoB26.87) | 89 | 18 | 20 | 67 | 10 | 1.33 |
| Mother (35) | Heterozygous | 110 | 60 | 59 | 38 | 31 | 2.89 | ||
| Father (37) | Wild-type | 175 | 92 | 258 | 31 | 70 | 5.65 | ||
| Brother (6) | Wild-type | 155 | 90 | 67 | 51 | 78 | 3.04 | ||
| FHBL/APOB/CS7 (139) | Proband (77) | Heterozygous | c.9754_9755insA (ApoB70.5) and ApoE ϵ2/ϵ2 | 128 | 41 | 89 | 61 | NA | 2.09 |
| Son (29) | Heterozygous | 116 | 29 | 124 | 50 | NA | 2.31 | ||
| Wife (67) | Wild-type | 193 | 94 | 186 | 53 | NA | 3.65 | ||
| Daughter (38) | Wild-type | 166 | 99 | 89 | 56 | NA | 2.97 | ||
| FHBL/APOB (155) | Proband (47) | Homozygous | c.1468C>T (p.R463W) | 36 | 3 | 59 | 23 | 7 | 1.56 |
| Brother (37) | Heterozygous | 98 | 31 | 44 | 50 | 28 | 1.96 | ||
| Daughter (26) | Heterozygous | 97 | 32 | 53 | 50 | 30 | 1.92 | ||
| ABL/MTP (158) | Proband (<1) | Homozygous | c.1867+5G>A (aberrant splicing) | 23.4 | NA | <8.8 | 17.6 | 0.1 | 1.33 |
| Mother | Heterozygous | 187.5 | NA | 52.6 | 39.1 | 74 | 4.80 | ||
| Father | Heterozygous | 257.8 | NA | 122.6 | 45.3 | 112 | 5.69 | ||
| Sister | Heterozygous | 136.7 | NA | 96.4 | NA | 83 | NA | ||
| ABL/MTP (121) | Proband (13) | Homozygous | c.923G>A (p.W308X) | 27 | ∼0 | ∼0 | 24 | ∼0 | 1.13 |
| Mother (36) | Heterozygous | 104 | 51 | 44 | 45 | 35 | 2.31 | ||
| Father (37) | Heterozygous | 170 | 108 | 114 | 40 | 61 | 4.25 | ||
| Brother (2) | Wild-type | 124 | 73 | 71 | 36 | 45 | 3.44 | ||
| ABL/MTP (121) | Proband (10) | Homozygous | c.618-3T>G (p.Q206fsE234X) | 20 | ∼0 | 6 | 20 | ∼0 | 1.00 |
| Mother (39) | Heterozygous | 175 | 92 | 55 | 83 | 112 | 2.11 | ||
| Father (43) | Heterozygous | 190 | 110 | 70 | 66 | 91 | 2.88 | ||
| Brother (1) | Wild-types | 149 | 92 | 196 | 57 | 121 | 2.61 | ||
| CMRD/SARA2 (178) | Proband (10) | Homozygous | c.537T>A (p.S179R) | 109 | NA | 120 | 23 | NA | 4.74 |
| Mother | Heterozygous | 175 | NA | 65 | 61 | NA | 2.87 | ||
| Father | Heterozygous | 211 | NA | 183 | 49 | NA | 4.31 | ||
| Sister | Wild-type | 164 | NA | 77 | 55 | NA | 2.98 | ||
| CMRD/SARA2 (182) | Proband (2) | Compound heterozygous | c.32G>A (p.G11D)/ c.224A>G (p.D75G) | 70 | 50 | 114 | 0 | 54 | High |
| Mother (37) | Heterozygous | c.32G>A (p.G11D) | 152 | 96 | 50 | 46 | 82 | 3.30 | |
| Father (35) | Heterozygous | c.224A>G (p.D75G) | 198 | 129 | 57 | 57 | 88 | 3.47 | |
| Brother (5) | Wild-type | 166 | 109 | 51 | 46 | 108 | 3.25 | ||
| Familial Combined Hypolipidemia/ANGPTL3 (170) | Proband (23) | Compound Heterozygous | p.S17X/p.E129X | 50.5 | 30.8 | 22.3 | 15.8 | NA | 3.20 |
| Father (67) | Heterozygous | p.E129X | 145.4 | 98.8 | 97.8 | 35.0 | NA | 4.15 | |
| Mother (64) | Heterozygous | p.S17X | 126.0 | 73.2 | 51.0 | 44.2 | NA | 2.85 | |
| Brother (43) | Compound Heterozygous | p.S17X/ p.E129X | 56.0 | 34.7 | 24.3 | 16.3 | NA | 3.43 | |
| Sister (44) | Compound Heterozygous | p.S17X/p.E129X | 57.4 | 36.8 | 19.1 | 17.8 | NA | 3.22 | |
| Niece (18) | Heterozygous | p.S17X | 129.8 | 71.5 | 55.5 | 49.0 | NA | 2.65 | |
| Niece (16) | Heterozygous | p.E129X | 154.8 | 102.7 | 106.8 | 34.7 | NA | 4.46 | |
| Brother (40) | Wild-type | None | 227.3 | 147.6 | 224.2 | 37.6 | NA | 6.05 | |
Age in years.
Predicted effect of mutations is specified in parentheses.
Levels ∼0 were undetectable; “NA” indicates that measurements were not available.
Differential diagnosis: TC:HDL-C ratio in CMRD and ANGPTL3-specific familial combined hypolipidemia > TC:HDL-C ratio in APOB-specific FHBL.
CS, case study. Probands are highlighted in bold.
Below we report seven eloquent case studies of apoB-specific FHBL (i.e., FHBL phenotype caused by pathogenic APOB mutations) to illustrate that compound heterozygote/homozygous patients have a hugely variable phenotype (Table 4) and that the most common reported causes of this condition involve mutations that prematurely block the translation of apoB mRNA. Although the clinical data show an overall association between increasing apoB protein length and chronologically later and milder clinical phenotype, it must be acknowledged that there are still too few case studies to discern the individual effects of fat-soluble vitamin deficiencies and other confounding factors, such as advanced age and lifestyle choices (e.g., excess energy intake), to accurately predict the severity of clinical manifestations that may develop for any given apoB mutation. We also note that more apoB missense mutations within the apoB-lipoprotein-initiating domain (after case studies) may emerge as patients with fatty liver and/or hypocholesterolemia are screened for such variants.
Case study 1.
A Caucasian male infant was hospitalized at 4 months of age for investigation of failure to thrive with severe fat malabsorption (120). His diagnosis of homozygous/compound heterozygote apoB-specific FHBL was originally suspected from his lipid profile and that of his parents, both of whom were severely hypocholesterolemic (Table 4). Molecular analyses confirmed his diagnosis (Table 4), identifying two different mutations, both of which almost certainly blocked the initiation of apoB-containing lipoprotein assembly in all cell types. At 10 months, he was treated with medium-chain triglycerides, resulting in weight gain and decreased steatorrhea. However, he developed micronodular cirrhosis, portal hypertension, and esophageal varices by age 11, plus severe neurological deficiencies that included ataxia, dysarthria, severe impairment of position and vibratory sensation, and absent deep tendon reflexes. His neurological symptoms were stabilized by intramuscular and intravenous vitamin E therapy. However, in his last three years of life he suffered seizures, a cerebrovascular accident, recurrent massive gastrointestinal hemorrhages, and finally cardiac arrest at age 18.
Case study 2.
A 4-year-old Tunisian girl was born from consanguineous parents (121). She had a long history of diarrhea, intolerance to fat-rich meals, and marked hypolipidemia (Table 4). Histological and biochemical analyses revealed fat-laden enterocytes, resulting from a homozygous mutation that removed >95% of apoB's lipid binding structures. Her obligate heterozygote parents had greatly reduced total cholesterol and LDL-C levels (Table 4), potentially attributable to either decreased production or increased catabolism of VLDL-apoB100, LDL-apoB100, or both (122–124). The parents did not clinically manifest fat malabsorption. However, when Hooper et al. (125) formally tested the postprandial fat responses of other similarly asymptomatic, heterozygote, FHBL subjects (apoB100/apoB6.9, n = 3; apoB100/apoB25.8, n = 1; apoB100/apoB40.3, n = 2), their plasma triglyceride and apoB-48 levels peaked earlier than in the controls (n = 10), and these peaks were markedly reduced in magnitude. Thus, these findings support the anecdotal reports of intestinal manifestations suggestive of mild fat malabsorption in apoB100/apoB6.7 (126) and apoB100/B8.2 (127) individuals, when they refrained from their fat-restricted diet. By contrast, normal postprandial fat-absorption responses were found in heterozygous apoB48.4 and apoB76 FHBL subjects (128, 129), implying that only those FHBL heterozygotes with apoBs shorter than apoB-48 have a reduced capacity to assemble dietary lipids into chylomicrons.
Case study 3.
A 21 year-old Caucasian woman from consanguineous parents was investigated because of chronic fatty diarrhea (130). Inspection of jejunal and ileal biopsies revealed accumulation of fat vacuoles. Acanthocytes were present, but the patient displayed neither neuropathy nor retinopathy. Laboratory analyses revealed trace amounts of plasma apoB27.6 associated with lipoprotein particles sized between normal LDL and HDL, plus a homozygous splice site mutation compatible with the observed truncated apoB species (Table 4). As seen in other heterozygote apoB-specific FHBL individuals, her mother, brother, and sister were clinically well and had less than half the expected plasma concentration of cholesterol, triglycerides, and apoB (Table 4). The patient received vitamin A and E supplementation, which may have contributed to her developing liver fibrosis two year later (130).
Case study 4.
The proband, a 57-year-old Japanese mother homozygous for an apoB mutation predicted to truncate apoB after amino acid 1755 (apoB38.7), was referred for evaluation of a liver mass (131). She had no history of diarrhea or steatorrhea, and plasma vitamin E levels were within the normal range. However, she was severely hypocholesterolemic (Table 4). Blood studies showed she was anemic, with blood film acanthocytosis. She also had proteinuria, extensive calcification of the major arteries, and cholelithiasis (see case study 6). Her medical history was significant for asthma, night blindness, and type II diabetes, all diagnosed at age 36 and managed by diet and insulin. At age 48, she had a retinal hemorrhage attributed to hemorrhagic glaucoma, resulting in right eye blindness, and at age 54, she developed hemopytsis secondary to bronchiectasis. Retinal pigmentation, hard exudates, and extensive photocoagulation scars were noted in both optic fundi. Neurological examination revealed no abnormal pyramidal, cerebellar, or posterior column abnormalities but she did have paraesthesia in both hands (“stocking-glove” type hypoaesthesia, absent deep tendon reflexes in the lower extremities, and positive Romberg's sign).
Case study 5.
Homozygous FHBL was diagnosed in a 48-year-old Caucasian mother (three normal pregnancies) following routine cholesterol screening (132). She denied any unusual dietary habits, symptoms of fat malabsorption, night blindness, or neurological or visual impairment. She was found to have a homozygous nonsense mutation that retained the apoB sequences required to form triglyceride-rich lipoproteins (Table 4) and the anchoring of apoB to its lipid core (114, 118). Accordingly, in marked contrast to her severe hypocholesterolemia, her plasma triglyceride, apoA1, HDL-C and vitamin E levels were within the normal range.
Case study 6.
A 9-year-old patient presented with epigastric pain, cholestastic jaundice, and acute cholecystitis (133). Liver ultrasound revealed grossly dilated common and intrahepatic bile ducts due to obstruction by a gallstone, with many more within her gallbladder. Laboratory analyses led to a diagnosis of heterozygous apoB-specific FHBL (Table 4) in her and her clinically well mother. This case study reminds us that gallstone disease is not uncommon in heterozygous apoB-specific FHBL patients (apoB90/40 (134); apoB100/apoB46 (135); apoB100/83 (136)) and that it may relate, at least in part, to their near-normal levels of intestinal cholesterol absorption (i.e., 47.5 ± 6.3 versus 53.8 ± 11.7% in controls (137, 138)).
Case study 7.
A 77-year-old proband was investigated for marked hypocholesterolemia following routine cholesterol screening (139). Laboratory analysis revealed that he and his son were heterozygous for apoB70.5 and homozygous for apoϵ2 (Table 4). Moreover, they had the classical features of dysbetalipoproteinemia superimposed onto heterozygous apoB-specific FHBL. Thus, their fasting plasma samples contained apoB48 in the triglyceride-rich fraction plus β -migrating VLDL, indicative of impaired clearance of apoE-containing lipoproteins (Fig. 5).
Fig. 5.
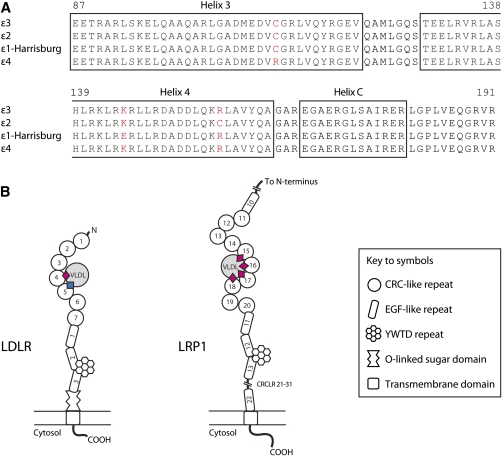
Alignment of representative human ApoE sequences in the region critical for binding to the LDLR and LRP1 and proposed model of binding. (A) The amino acid residue (C158) that distinguishes apoE ϵ2 and ϵ3 resides in the fourth helix (amino acids 130-164) of the N-terminal four-helix bundle (amino acids 24-164). In the lipid-free state, R158 (ϵ3 and ϵ4) forms a pair of salt bridges with amino acid residues E96 and D154 (256). Its removal in apoE ϵ2 induces a compensatory reorganization; e.g., D154 forms a salt bridge with R150 changing the charge of one local surface (amino acids 140-150) presented to the LDLR (363). Structural and site-directed mutagenesis indicate that the side chains of amino acid residues K143 and K146 make important contribution to LDLR binding (363). The naturally occurring apoE ϵ3:K146E mutation [(commonly referred to as apoE ϵ1-Harrisburg (K146E)] causes a dominantly inherited form of dyslipidemia characterized by markedly raised plasma levels of total- and VLDL- (but not LDL-) cholesterol, triglycerides, and apoE (364, 365). The mutant protein has markedly reduced ability in vitro to bind to the LDLR and to displace LDL (7.7% activity of apoE ϵ3) from the surface of human fibroblasts (266, 366). Binding to proteoglycan in vitro is also reduced (i.e., to ∼10% and 20% of apoE ϵ3 and ϵ2, respectively) (266). The C112R substitution in apoE ϵ4 occurs in the third helix (87–122) of the four-helix bundle, where it modifies the packing between the N-terminal (amino acids 1-191) and C-terminal (amino acids 216-299) domains (367, 368). In apoE ϵ4, changes to the local structure around amino acid 264 enhance lipid binding, and this isoform binds more avidly to VLDL than apoE ϵ3 but less strongly than apoE ϵ3 to HDL3 (369, 370). (B) Two distinct binding events are envisaged to cement the interaction between apoE and the LDLR (371). In the Guttman model (371), the cysteine-rich, complement-like repeat (CRCLR) 4 of the LDLR interacts with the charged surface presented by amino acids 140-150 of apoE (pink diamond), whereas the CRCLR5 binds to a distinct epitope (blue square) that is only exposed when bound to a lipoprotein. Amino acids E3 (372) and R172 (372, 373) may form part of this second binding site. In comparison, the high-affinity binding between lipid-bound apoE (130–149) and CRCLR of LRP1 (374, 375) is explained in terms of an avidity model: in essence, the strong interaction is envisaged to derive from the culmination of multiple weak interactions between apoE and the CRCLR of LRP1. ApoE residues E131, E132, V135, A138, and K143 (fourth helix) form the interface between apoE (130–149) and CRCLR 17 of LRP1 (375). LRP1, in contrast to the LDLR, in vitro binds lipid-bound apoE ϵ2 with as equal affinity as lipid-bound apoE ϵ3 (376).
A full understanding of the effects of apoB truncations on organ cholesterol homeostasis is hampered by incomplete (by contemporary standards) phenotypic and genetic characterization of apoB-specific FHBL patients. This is unfortunate because it is clear that such mutations (i) will affect lipid homeostasis in multiple cell types (e.g., cardiomyocytes (140), granulose cells (141), T cells (142), renal tubular epithelial cells (143)) and could contribute to multiple cell phenotypes (case study 5); (ii) may coexist with another inherited disorder (especially within consanguineous pedigrees); and (iii) may be influenced by the ever-present individual variation in compensatory mechanisms, as well as different apoE genotypes (case study 7 (144), Fig. 5). In this respect, three reports have shown that VLDL production is reduced in apoE deficiency (145, 146) and apo ϵ2 homozygosity (147). However, in vitro data have indicated that the expansion of the neutral lipid core of VLDL in the Golgi apparatus promotes apoE binding (148–150) and that this apolipoprotein is not required for VLDL maturation (149).
Arguably, insights into the molecular consequences of apoB-specific FHBL are best gleaned from model systems. Of these, the human apoB38.9 mouse model provides important data (151). In these mice, hepatic cholesterol levels were normal, despite impaired hepatic lipid export into the circulation via VLDL. However, hepatic ABCG5/8 mRNA levels were increased, suggesting that cholesterol efflux into the bile was enhanced (Fig. 1). Additionally, mRNA levels of cholesterol synthetic genes were decreased, as was hepatic cholesterol synthesis. LDLR mRNA was also decreased, indicative of reduced hepatic cholesterol uptake. Hence, it seems that the apoB38.9 mice called upon multiple mechanisms to maintain hepatic cholesterol homeostasis. However, and in keeping with the human situation, where fatty liver is a common complication of apoB-specific FHBL (152, 153), these mice were less successful in dealing with their reduced capacity to export triglycerides via VLDL (154): both the heterozygous and homozygous animals developed fatty liver.
Indeed, a diagnosis of apoB-specific FHBL should be considered whenever a patient is found to have fatty liver and LDL-C levels <75 mg/dl. In our series of 110 such patients, 51 had apoB100-truncating mutations (152, 153) and 6 had heterozygous missense variants (A31P, G275S, L324M, R463W [n = 3]) within the apoB lipoprotein-initiating domain (Fig. 3). In fact, the R463W mutation was originally identified in the homozygous state in a 47-year-old Christian-Lebanese woman (155) following detection of marked hypocholesterolemia during a routine lipid screen (Table 4), and it was shown to block the assembly/secretion of both apoB48- and apoB100-containing lipoproteins in vitro (155). In vivo, R463W patients accumulate fat in their intestinal enterocytes and exhibit a blunted postprandial rise in blood lipid levels (156), despite their apparent absence of clinically significant intestinal fat malabsorption (153, 155, 156).
Multiple common APOB variants modulate LDL-C levels
The GLGC study (Table 5) (32) lends support to the two propositions put forward by Benn et al. (157). First, relatively common NS coding sequence variants predicted to alter the structure of the apoB-lipoprotein-initiating domain (e.g., T71I in mature protein) contribute to the population variance in apoB, total, and LDL-cholesterol levels. Second, an additional set of independent apoB alleles modulate LDL-C levels, potentially by increasing apoB transcription. The GLGC results also indicate that causative apoB variants have a larger impact on LDL-C levels than common NPC1L1 and ABCG8 lesions.
TABLE 5.
Multiple common genetic determinants influencing LDL-C levels through the ApoB-lipoprotein assembly and secretion axis
| Gene | Sample(Reference) | Typed Variant (Proxy) a | Positionb(Effect) | Allelec(MAF) | Effect on LDL-C | Other Associated Traits |
| APOB | Prospective study of 9,185 Danish individuals (157) | 1) rs1367117 | c.293C>T (T71I)d | T(0.33) | T/T +11.60 mg/dl versus C/C, (P < 0.001) | Total cholesterol, apoB |
| 2) rs10199768 | c.2817 2 1223C>A | C(0.30) | C/A 2 3.09 mg/dl versus A/A, (P < 0.01) | Total cholesterol, apoB | ||
| 3) rs693 | c.7545C>T (T2488T) d | T(0.48) | T/T +9.67 mg/dl versus C/C, (P < 0.001) | Total cholesterol, apoB | ||
| 4) rs1801701 | c.10913G>A (R3611Q) d | A(0.09) | G/A +4.25 mg/dl versus G/G, (P < 0.01) | Total cholesterol, apoB | ||
| 5) rs1042031 | c.12541G>A(E4154K) d | A(0.17) | A/A 2 6.96 mg/dl versus G/G, (P < 0.05) | ApoB | ||
| GLGC (32) | 1) rs1367117 | c.293C>T (T71I) | T (0.30) | +4.05 mg/dl per copy (Z = 22.7, P = 4 × 10 ). | Total cholesterol, +4.16 mg/dl per copy (Z = 20.803, P = 4 × 10 ),Triglyceride (Z = 6.25, P = 4 × 10 ) | |
| 2) rs10199768 | c.2817 2 1223C>A | A (NA) | Z = 21.388 (P = 2 × 10 ) | Total cholesterol, Z = 19.518, (P = 8 × 10 )Triglyceride (Z = 8.382, P = 5 × 10 ) | ||
| 3) rs693 | c.7545C>T (T2488T) | T (NA) | Z = 20.022 (P = 4 × 10 ). | Total cholesterol, Z = 18.18, (P = 7 × 10 )Triglyceride (Z = 9.829, P = 8 × 10 ) | ||
| 4) rs1801701 | c.10913G>A (R3611Q) | A (NA) | Z = 6.838 (P = 8 × 10 ) | Total cholesterol, Z = 5.948, (P = 3 × 10 ). (Triglyceride P = 0.13) | ||
| 5) rs1042031 | c.12541G>A (E4154K) | A (NA) | Z = 2 9.982 (P = 2 × 10 ) | Total cholesterol, Z = 2 9.399, (P = 6 × 10 ). (Triglyceride P = 0.03) | ||
| ANGPTL3 | GLGC (32) | 1) rs3850634 (rs2131925) | ∼13 kb upstream (intronic DOCK7) | G (0.32) | 2 1.59 mg/dl per copy(Z = 2 8.726, P = 3 × 10 ) | Total cholesterol, 2 2.60 mg/ dl per copy(Z = 2 13.416, P = 5 × 10 )Triglyceride, Z = 2 13.278, (P = 3 × 10 ) |
| SORT1 | 1,132 Jamaicans of African ancestry (210) | 1) rs12740374 (rs629301) | ∼35 kb downstream | G (0.26) | β = 2 0.25, P = 4.6 × 10 | |
| Japanese population study (n = 21,010) (211) | 1) rs599839 (rs629301) | ∼30 kb downstream | G (0.08) | 2 4.7 mg/dl per copy (P ≈ 3.1 × 10 ) | ||
| GLGC (32) | 1) rs629301 | ∼34 kb downstream | G (0.22) | 2 5.65 mg/dl per copy (Z = 2 27.854, P = 1 × 10 ) | Total cholesterol, 2 5.41 mg/ dl per copy (Z = 2 24.350, P = 6 × 10 ). Reduced CAD risk (P = 4 × 10 ) | |
| TRIB1 | Danish population study (n = 50,309) (215) | 1) rs2954029 | ∼40 kb downstream | T (0.48) | T/T 2 4.25 mg/dl versus A/A (P < 0.001) | Triglyceride, T/T 2 14.17 mg/dl versus A/A (P < 0.001) |
| Japanese population study (n = 21,010) (211) | 1) rs17321515 (rs2954029) | ∼36 kb downstream | A (0.48) | 2 1.7 mg/dl per copy ( β = 2 0.04, P = 1×10 ) | Triglyceride, 2 3.5 mg/dl per copy ( β = 2 0.04, P = 5×10 ) | |
| GLGC (32) | 1) rs2954029 | ∼40 kb downstream | T (0.47) | Z = 2 11.176, (P = 5 × 10 ) | Triglyceride 2 5.64 mg/dl per copy (Z = 2 15.651, P = 3 × 10 ) and total cholesterol, Z = 2 12.464, (P = 1 × 10 ) |
Independent variants/associations are numbered. “Proxy” variant is in strong linkage disequilibrium with genotyped variant.
Relative to the initiating ATG.
Minor allele is listed with frequency (MAF) in study population.
APOB mutations are numbered according to Benn et al. (157) rather than current Human Genome Variation Society nomenclature.
NA, not available.
Update on the role of MTP function in cholesterol homeostasis
MTP mutations are the major, if not sole, cause of the rare recessive condition abetalipoproteinemia (ABL; [OMIM #200100], Fig. 3) (158). This condition is biochemically characterized by exceedingly low plasma levels of cholesterol plus triglyceride, as well as a total absence of all apoB-containing lipoproteins (Table 4). Its differential diagnosis from homozygous apoB-specific FHBL includes “normal” lipid levels in the parents, and exceedingly low plasma triglyceride levels. For information regarding the full spectrum of MTP mutations in ABL, the clinical manifestations, and management of this debilitating disorder, see Refs. 159–161. Here, we review the role of MTP in governing cholesterol homeostasis, and we note that the GLGC study detected no genome-wide significant associations between MTP variants and LDL-C levels (32).
The role of apoE in delivering cholesterol to specific body sites in ABL is largely unknown. An early study involving 9 patients with clinical features of this condition and 50 controls (162) revealed that the patients had increased concentrations of both plasma apoE (44.8 ± 8.2 μ g/ml versus 36.3 ± 11.1 μ g/ml, P < 0.025) and an apoE cholesterol-enriched HDL subfraction ( ∼1.5-fold) that had the capacity to compete with 125I-LDL for specific binding sites on the surface of cultured human skin fibroblasts. However, the apoE genotype status of these patients was not determined (Fig. 5). Subsequently, an analysis of the plasma decay curves of injected radiolabeled autologous lipoproteins (131I-apoAI and 125I-apoϵ3) in the HDL density range of 2 unrelated ABL patients and 22 control individuals revealed that the overall catabolism of apoϵ3 was increased by ∼25% in the patients, as was apoE production. Moreover, the catabolism of apoAI in apoE-containing HDL occurred at a faster rate than that in HDL without apoE, potentially explaining the lower plasma apoAI levels in ABL patients (163). Thus, quantitatively, neither the catabolic nor the production rate of apoE in ABL is impaired, despite the lack of apoB-containing lipoprotein assembly.
Two studies suggest that cholesterol biosynthesis may be increased in ABL, although note that these analyses were performed on individuals with no confirmatory molecular diagnosis. In the first study, cholesterol synthesis was increased by ∼2-fold compared with aged-matched controls (164), with the kinetic data conspicuously indicating that the increased synthesis was largely “driven” by enhanced fecal loss of intestinal cholesterol (i.e., both dietary and biliary). In the second study, Kuriyama et al. (165) showed that their patient had nearly 2-fold higher blood levels of dolichols (synthesized through the cholesterol biosynthetic pathway) than control individuals. Additionally, the dolichols were largely ( ∼80%) associated with HDL in both the patient and control subjects.
Recent studies on two different mouse models of MTP deficiency also contribute to our understanding of the different ways that the MTP:PDI heterodimer may influence organ cholesterol levels (166, 167). Xie et al. (167) produced mice with (conditional) intestine-specific deletion of the major, but not minor, Mttp species (168). They found that mean cholesterol absorption was reduced by 70% in the knockout animals and that enterocyte Mttp mRNA levels were positively correlated with cholesterol absorption (R2 = 0.69). They inferred from their studies that, in mice, as much as 30% of intestinal cholesterol reached the circulation via an apoB/MTP-independent mechanism. Of potential significance, they also found that enterocytes from their Mttp-deficient mice contained control levels of cholesterol and, not surprisingly, increased HMGCR and Acat2 mRNA levels. Liver HMGCR mRNA was also increased (by ∼4-fold) and ex vivo hepatic cholesterol synthesis was similarly elevated. Despite these increases, their mean liver cholesterol content was marginally decreased ( ∼10%), most likely as a result of increased VLDL secretion and cholesterol efflux into bile, given that hepatic ABCG5/8 mRNA were increased. In the second study, Iqbal et al. (166) first showed that the livers of mice deficient for both intestinal and hepatic Mttp contained 132% more unesterified cholesterol than did control animals and, unexpectedly, a 90% reduction (rather than increase) in cholesteryl esters. Next, they established that the MTP:PDI heterodimer very likely enhances cellular cholesterol esterification through its transfer of ACAT reaction products (i.e., cholesteryl esters) from their site of synthesis onto assembling apoB lipoproteinsm, leading them to speculate that ACAT-product inhibition may serve to prevent excessive accumulation of neutral lipids in ER membranes and thus to maintain cellular integrity. On the other hand, they noted that the observed increase in free cholesterol in their MTP-deficient animals could explain the toxicity associated with pharmacological inhibition of MTP activity.
In summary, both humans (164, 165, 169) and mice (166, 167), data indicate that the levels of blood/tissue cholesterol/cholesteryl esters in ABL reflect both the classically taught defective production of apoB-containing lipoproteins, which restricts cholesterol absorption (dietary and biliary-derived), as well as enhanced biliary cholesterol secretion and de novo cholesterol biosynthesis in multiple cell types. It should be emphasized, however, that point-for-point comparison of mouse and human is unlikely ever to be achieved, and some important differences may exist.
ANGPTL3-specific familial combined hypolipidemia
In both man (170) (Table 4) and mouse (171, 172), compound heterozygous/homozygous angiopoietin-like 3 (ANGPTL3) mutations have been associated with a low LDL-C phenotype. The human study involved exome sequencing of two siblings from a large European family exhibiting a combined hypolipidemia phenotype of extremely low plasma cholesterol, LDL-C, HDL-C, and triglyceride levels (Table 4). Adopting an analysis strategy that considered the combined hypolipidemia phenotype in this pedigree to be autosomal recessive in inheritance led to identification of a single bi-allelically mutated gene, ANGPTL3 (Table 4). However, close inspection of the lipid profiles in this family suggests an additive (rather than recessive) genetic model for both the low LDL-C and triglyceride traits, prompting Kathiresan and colleagues (170) to remind us that hepatic (171, 173) and secreted (174) ANGPTL3 perform distinct functions. In the liver, it acts to promote the production/secretion of apoB-containing lipoproteins whilst inhibiting lipoprotein clearance; the ANGPTL3 mutation carriers in the Kathiresan family displayed decreased rates of VLDL-apoB production (compound heterozygotes: 8.6 ± 0.1 mg/kg/day; heterozygote:16.4 ± 4.9 mg/kg/day; no mutations: 26.9 ± 4.6 mg/kg/day, P = 0.001, additive model) and elevated fractional catabolism of LDL-apoB (compound heterozygotes: 0.52 ± 0.013 pools/h; heterozygote 0.045 ± 0.01 pools/h; no mutations: 0.027 ± 0.004 pools/h, P = 0.005, additive model). In comparison, in the circulation ANGPTL3 inhibits the activities of lipoprotein lipase (175, 176) and endothelial lipase (177) and modulates triglyceride and HDL-C levels.
Importantly, two population-based studies support the ANGPTL3/LDL-C connection. Thus, sequencing of ANGPTL3 in Dallas Heart Study participants (n = 3,551) showed that carriers of frame shift mutations (n = 12) had lower LDL-C levels than did noncarriers (median 77.5 mg/dl versus 104 mg/dl in noncarriers, P = 0.03). Additionally, plasma triglyceride levels were lower (median 72 mg/dl versus 96 mg/dl, P = 0.08). In the GLGC sample, the DOCK7/ANGPTL3 variant rs2131925 displayed robust association with plasma cholesterol, LDL-C, and triglyceride (Table 5).
Chylomicron retention disease
Chylomicron retention disease (CMRD; also known as Anderson disease) ([OMIM #246700], Table 4) is a rare autosomal recessive disorder caused by mutations of SARA2. It typically presents in infancy with failure to thrive in association with severe fat malabsorption (178–183). Biochemically, CMRD is characterized by a selective absence of apoB48 in plasma, very low levels of total cholesterol, LDL-C, and HDL-C, plus substantially reduced apoB100 and A1 (Table 4). Plasma triglyceride levels, in comparison, tend to fall within the normal range, which differs from ABL and the genetically severe forms of homozygous apoB-specific FHBL (Table 4) (184). For an up-to-date description of the subclinical phenotypes found in CMRD patients, see Refs. 185 and 186.
The cause of the exceptionally low plasma cholesterol levels in CMRD is still not understood. An early study (187), performed on six CMRD children (mean age 8 ± 3 years), two to three years after their placement on a diet restricted in long-chain triglycerides and supplemented with medium-chain triglycerides, revealed that they manifested low levels of both LDL-C (38.0 ± 19.4 versus 110.8 ± 20.5 mg/dl) and HDL-C (12.8 ± 2.3 versus 59.0 ± 11.3 mg/dl). By contrast, VLDL-C (27.3 ± 11.0 versus 13.0 ± 5.7 mg/dl) and plasma triglyceride (135 ± 38 versus 65 ± 17 mg/dl) levels were increased. This increase was ascribed to their substantially decreased postheparin LPL and hepatic lipase activities, as they had larger VLDL than the control children. In comparison, the CMRD children had smaller LDL, and these contained more (2.3-fold) triglyceride and less (2.5-fold) cholesteryl esters. Their HDL particles were also smaller (i.e., HDL3 size) and contained decreased (3.4-fold) amounts of cholesteryl esters. Importantly, the molecular diagnosis of many of these patients was confirmed in a subsequent study (188).
Another challenge in understanding the pathogenesis of CMRD is why functional compensation for Sar1b deficiency is not provided by Sar1a, a protein with which it shares tissue and subcellular distribution, as well as ∼90% amino acid homology (188–190). It is apparent from structural studies that most of the amino acid differences between the two Sar1 isoforms reside on one surface of these two small GTPases at some distance from their GDP/GTP binding sites (188, 190). Moreover, Sar1 exchanges GDP for GTP on the cytosolic side of the ER membrane, and this exchange initiates COPII-coated vesicle assembly (191). Sar1-GTP hydrolysis also regulates assembling COPII-vesicle fission, along with its captured cargo (192, 193).
On the basis of structural (194, 195), organ bath (187), histological (179), and of course, clinical data, the current thinking is that Sar1b is required to promote the transport of nascent chylomicrons out of the ER in specialized COPII-transport carriers. Moreover, CMRD patients homozygous for Sara2 null mutations (described in two families) may produce few, if any, of these putative specialized COPII-transport carriers, whereas patients with mutations affecting the GDP/GTP binding site of Sar1b (majority of affected families) may retain some ability to initiate the assembly of such carriers at the site of chylomicron production but not their fission. As such, they also lead to retention of precursor chylomicrons in the ER.
Sortilin: post-ER trafficking of ApoB-containing lipoproteins
Sortilin, the archetypal member of a mammalian family of receptors defined by a unique vacuolar protein-sorting 10 domain (196), contains a single transmembrane and a short cytoplasmic domain composing the typical motifs that mediate intracellular trafficking events (197–200) and endocytosis (201–203). Its RNA is expressed at high levels in several tissues and cell types, including skeletal muscle, heart, and adipose tissue, but it is expressed at relatively low levels in the liver (204).
Two groups have investigated how human genetic variation at the chromosome 1p13 locus (205, 206), specifically, Sortilin 1 (Sort1), might influence plasma LDL-C levels (Table 4). Rader and colleagues (206) showed that in mice Sort1 overexpression and knockdown decreased and increased, respectively, total plasma cholesterol and LDL-C levels, which, at least in part, were attributable to changes in VLDL-apoB100 secretion. Independently, the Nykjaer group (205) presented evidence that sortilin interacts with apoB100 in the medial- to trans-Golgi compartment, presumably to determine the fate of nascent apoB100-containing lipoproteins: presecretory degradation or secretion (Fig. 3) (109–111).
Paradoxically - and important to resolve - is why the Nykjaer (205) and Rader (206) groups observed opposite effects of Sort1 expression on total cholesterol and LDL-C levels. There is reason to think that part of the answer may reside in the different targeting strategies used by the two groups to “knock out” Sort1, especially as, contrary to expectation, both Sort1-deficient mice were born viable and showed no gross abnormalities (207, 208). In the “Rader” mice, two 5 ′ sites were targeted (i.e., intron 1/exon 2, which encodes the N-terminus of the Vps 10 domain; plus intron 3) (208), and knockout was deemed successful based on the absence of detectable sortilin in cell lysates from the brain, liver, lung, and testis of the knockout animals. In the “Nykjaer” mice, inactivation was “achieved” by targeting exon 14, thereby disrupting the reading frame of Sort1 after amino acid 564, leaving open the possibility that their mice may produce a C-terminally truncated form of Sort1.
Consistent with their knockout animals, the Rader group found that liver-specific Sort1 knockdown in chow-fed “humanized” mice (i.e., Apob48-null/high-apoB100 expressers) increased total plasma cholesterol (46%) and LDL-C ( ∼220%) levels (206). Similar, but less dramatic, results were seen in other mouse models (e.g., Apobec1/Ldr− / − ), but sadly, no data were presented for apoB48-only mice, which could have served as a useful control given the data from Nykjaer and colleagues (205) that sortilin may only interact with apoB100. On the basis of ex vivo measurements, the observed dyslipidemia in the “Rader” Sort1 − / − mice was ascribed to increased VLDL secretion.
Important differences in study design could also explain the reported diametrically opposed effects of high-level Sort1 expression on plasma LDL-C levels. Rader and colleagues (206) used “humanized” apoB100-only transgenic mice models (i.e., comparable plasma cholesterol to man through increased apoB-lipoprotein assembly), while the Nykjaer group (205) used wild-type mice that express both apo48 and 100 in liver and have ∼4× lower plasma cholesterol levels than man. The two groups also used different adenoviruses to deliver Sort1. Focusing on the Rader experiments, Sort1 was delivered to chow-fed mice via an adenovirus vector known to appropriately target genes for specific expression in the liver. Compared with the “humanized” mice receiving the null adenovirus construct, Sort1 overexpression led to dramatic drops in total plasma cholesterol: 70% and 46% at weeks 2 and 6, respectively. These reductions were paired with a 73% reduction in LDL-C (at the 2-week time point) and markedly fewer medium-small and very small LDL particles, with the peak LDL-particle size increasing from 20.9 nm to 22.0 nm (P = 0.05). The Sort1 construct also induced reductions in total plasma cholesterol and LDL-C in other mice models (e.g., Apobec1/APOB/ldlr+/ − , 2 44%, 2 70%; Apobec1/ldlr− / − , − 26%, − 29%) at the 2-week time point. Moreover, in all mouse models examined, high-level Sort1 expression decreased VLDL secretion (range 30-70%). Potentially pertinent, LDL-C uptake was increased in HEK 293 cells transiently transfected with Sort1 (209).
Sort1 genetics.
Sort1 resides in a 98 kbp genomic interval containing multiple SNPs that associate with both reduced plasma LDL-C levels (Table 5) and a reduced risk of myocardial infarction in multiple populations, including Europeans (32), Africans (210), and Japanese (211). However, SNPs displaying the strongest association reside at some considerable distance from the 3′ end of Sort1 (Table 5) and actually closer to MYBPHL, PSRC1, and CELSR2, which, respectively, encode myosin binding protein H-like protein, a microtubule destabilizing protein and a nonclassic-type cadherin. Consequently, >1 causal variant(s) may underlie the LDL-C and myocardial infarction association signals, with the causative allele(s) impacting differently, both quantitatively and qualitatively, and in a cell-specific fashion to influence these two phenotypes. In two independent series of liver samples, the Rader group (206) found that the rare allele at tagging SNP rs646776 (Table 5) was associated with ∼10-fold higher levels of PSRC1 and SORT1 mRNA (P values for PSCR1: 2 × 10 − 271, 9 × 10 − 7; SORT1: 2 × 10 − 300, 1 × 10 − 11), a finding that both prompted and is consistent with their rodent Sort1 study results. In comparison, no SNP and transcript associations were detected in adipose tissue.
Trib1: primary role in hepatic lipogenesis modulates VLDL production
Trib1, a homolog of Drosophila tribbles, is evolutionarily conserved in mouse and humans (212). It expression is ubiquitous and certainly includes the liver (213) and coronary arteries (214). Indeed, preliminary data indicate that Trib1 expression is elevated in the coronary arteries of patients with advanced CHD (214).
Drosophila tribbles regulates String activity and, hence, mitosis during ventral furrow formation (212). Much more interesting, however, for the present discussion, is the Trib1 modulation of mouse liver levels of mRNA encoding three key enzymes of lipogenesis (acetyl-CoA carboxylase, fatty acid synthase, and stearoyl-CoA desaturase 1) and of plasma cholesterol and triglyceride concentrations (213). Specifically, when Burkhardt et al. (213) induced high-level Trib1 expression in mice livers (i.e., using the appropriate AAV8 adenovirus vector), plasma cholesterol and triglyceride levels fell by ∼45% and ∼57%, respectively. Similar decreases were also observed in Ldlr− / − animals (27% and 34%), as well as in humanized, transgenic apoB100 mice on a background of LDLR-haploinsufficiency (25% and 26%). Furthermore, in these mice, the Trib1-induced reductions in cholesterol and triglyceride were paired with reduced apoB (23%), indicative of an overall reduction in the number of plasma apoB-containing lipoproteins. In comparison, Trib1-deficient mice had elevated plasma cholesterol (54%) and triglyceride (33%) levels and increased numbers of circulating VLDL and LDL particles. Importantly, the triglyceride phenotype was largely reversed by high-level hepatic Trib1 l expression (via use of adenovirus construct), suggesting that the elevated triglyceride levels in the Trib1-deficient animals was specifically attributable to the lack of hepatocyte Trib1 mRNA. Moreover, restoration of Trib1 expression decreased VLDL-triglyceride production to control levels.
The association of mouse Trib1 overexpression or underexpression with plasma cholesterol and triglyceride levels is clear-cut, but the underlying mechanisms are much less so. The data imply modulation of lipogenic gene expression influences the amounts of lipid available for VLDL secretion (Fig. 3), but understanding the relative importance of each enzyme affected, and how, must await further research.
Downstream TRIB1 variants associated with decreased LDL-C.
Table 5 shows that the variants assigned to TRIB1 reside at some distance from its 3′ end (32, 211, 215). However, results in mice make this assignment plausible, especially as the variant (or allele[s] in LD) is associated with both plasma cholesterol and triglyceride levels. Indeed, in a Japanese study, the estimated effect on LDL-C and triglyceride levels returned exactly the same standardized coefficient (0.04) (211). However, the demonstration that TRIB1 mutations cause either combined hyperlipidemia or FHBL (gain-of-function) in non-apoB/PCSK9/ANGPTL3 patients would help reinforce the importance of TRIB1 in the lipid arena.
RECEPTOR-MEDIATED CATABOLISM OF LDL-PARTICLES
All known causes of monogenic hypercholesterolemia, whether it be autosomal dominant (ADH) or recessive hypercholesterolemia (ARH) involve mutations that block cellular cholesterol uptake via the LDLR pathway (Fig. 4, Tables 6, 7). Starting with familial hypercholesterolemia (FH or ADH-1, [OMIM #606945]), five major classes of LDLR mutations can be discerned through functional assays that probe the integrity of production and the individual cellular properties of the encoded gene product (216–219). These are (i): production of the 893 amino acid protein; (ii) its transport to the cell surface; (iii) ligand binding at neutral pH; (iv) delivery of LDLR (constitutive endocytosis) and LDLR/ligand complexes to endosomes, where acidic conditions and low free calcium concentration promote ligand release (220); and (v) recycling of LDLR to the cell surface for further rounds of lipid uptake. The all-important mechanisms regulating the egress of lipoprotein-released cholesterol from late endosomes and lysosomes to other cell compartments (221–223) are beyond the scope of this review.
TABLE 6.
Monogenic disorders of LDL-cholesterol clearance: representative phenotypes
| Reference | Disorder (Gene) | Mutationa | N | Age (years) | TC (mg/dl) | LDL-C (mg/dl) | HDL-C (mg/dl) | TG (mg/dl) | CHD (%) | Comment/Differential Diagnosis |
| (245) | ADH-1(LDLR) | Hm-null | 14 | 13 ± 12 | 826 ± 125 | 768 ± 125 | 31 ± 7 | 129 ± 37 | 71 | ADH-1 Hm-null most severe. ARH similar to ADH-1 Hm defective. |
| Hm-defective | 26 | 28 ± 16 | 660 ± 108 | 604 ± 106 | 31 ± 9 | 114 ± 46 | 62 | |||
| ARH (LDLRAP1) | Hm p.W22X and/ or p.H144QfsX26 | 42 | 31 ± 13 | 629 ± 95 | 566 ± 94 | 44 ± 8 | 97 ± 34 | 41 | ||
| (54) | ADH-1(LDLR) | Ht-null | 43 | 56 ± 13 | 432 ± 64 | 351 ± 61 | 54 ± 15 | 138 ± 61128 ± 62 | 42 | Ht-defective is less severe than Ht-null (lipids and CHD incidence). |
| Ht-defective | 13 | 54 ± 15 | 354 ± 51 | 277 ± 45 | 52 ± 14 | 15 | ||||
| (246) | Ht-null | 48 | 42 | 404 ± 86 | 326 ± 84 | 51 ± 16 | 116 | 27 | ||
| Ht-defective | 62 | 43 | 379 ± 78 | 296 ± 79 | 55 ± 14 | 118 | 13 | |||
| (378) | Ht-null | 172 | 11 | 311 | 247 | 49 | 73 | b | ||
| Ht-defective | 202 | 11 | 287 | 224 | 49 | 71 | b | |||
| (380) | Ht-null | 123 | 9 ± 5 | 305 | 237 | 51 53 | 75 | 62c | ||
| Ht-defective | 77 | 10 ± 5 | 258 | 193 | 72 | 32c | ||||
| (381) | ADH-2(APOB) | Hm p.R3500Q | 1 | 34 | 394 | 329 | 45 | 95 | 100 | Hm ADH-2 less severe than both forms of Hm ADH-1 but similar to Ht receptor-defective ADH-1. Additive effect of mutant alleles |
| (295) | Ht p.R3500Q | 23 | 46 | 344 ± 46 | 259 ± 46 | 54 ± 12 | 124 ± 44 | 9 | ||
| (246) | ||||||||||
| Hm p.R3500Q | 2 | 59 ± 6 | 391 ± 18 | 327 ± 5 | 56 ± 15 | NA | 50 | 4.41-fold higher odds [95% CI 2.69-7.21] of having detectable CAC in carriers versus noncarriers. | ||
| Ht p.R3500Q | 106 | 42 ± 15 | 260 ± 49 | 193 ± 43 | 54 ± 14 | NA | 3 | |||
| Ht p.R3500Q | 11 | 43 | 349 ± 67 | 263 ± 61 | 59 ± 14 | 95 | 9 | |||
| (339) | ADH-3(PCSK9) | Ht p.S127R | 18 | 34 ± 14 | 365 ± 58 | 277 ± 58 | NA | NA | NA | 2 families |
| (382) | Ht p.D374Y | 6 | 24 ± 14 | 416 ± 97 | 350 ± 98 | 43 ± 11 | 99 ± 47 | 0 | 2 families | |
| (383) | Ht p.D374Y | 13 | 21 ± 15 | 526 ± 112 | 425 ± 159 | 46 ± 15 | 151 ± 44 | 46 | 4 families | |
| (299) | ADH1/ADH-2 (LDLR/APOB)f | p.W66G/p.R3500Qp. | 11 | 10 | 649 | 583 | NA NA | NA | 100 | Mutations have an additive effect. |
| E207K/p.R3500Q | 55 | 479 | 411 | NA | 100 | |||||
| (300) | p.L479P/p.R3500Q | 1 | 12 | 340 | 162d | 54d | 15d | 0e | ||
| (301) | p.D206E/p.R3500Q | 6 | 27 ± 16 | 509 ± 111 | 446 ± 110 | 38 ± 13 | 111 ± 58 | 0 | ||
| (343) | ADH-1/ADH-3 (LDLR/PCSK9f | p.E228K/p.R496W | 1 | 35 | 580 | 518 | 50 | 60 | 100 | Mutations have an additive effect. |
| p.Y419X/p.N425S | 1 | 62 | 487 | 405 | 70 | 61 | 100 | |||
| p.Y419X/p.N425S | 1 | 48 | 522 | 445 | 48 | 148 | 100 |
Age and lipid levels are means ± standard deviation where applicable. Hm, homozygous; Ht, heterozygous.
Effect of mutation on protein is given.
A subset of individuals with receptor-null alleles had greater mean carotid intima-media thickness, a marker for atherosclerosis and future cardiovascular outcome, than those with receptor-defective alleles (difference, 0.020 ± 0.01 mm; P = 0.01) (379).
Incidence of CHD relates to family rather than to individual.
Levels measured while patient treated with lipid-lowering medication.
Father died of early myocardial infarction.
Individuals heterozygous for both mutations.
TABLE 7.
Influence of APOE and APOB polymorphisms upon LDL-C levels in heterozygous ADH-1 patients
| Reference | Age Groupa | Modifier | N | LDL-C (mg/dl) |
| (253) | Children | ϵ2ϵ3 | 13 | 205 ± 30 |
| ϵ3ϵ3 | 36 | 254 ± 48 | ||
| (254) | Adults | ϵ2ϵ2/ϵ2ϵ3 | 47 | 258 ± 9 |
| ϵ3ϵ3 | 425 | 282 ± 4 | ||
| ϵ2ϵ4/ϵ3ϵ4/ϵ4ϵ4 | 98 | 309 ± 7 | ||
| (252) | Adults | ϵ2ϵ2/ϵ2ϵ3 | 288b | 299 ± 11 |
| ϵ3ϵ3 | 311 ± 6 | |||
| ϵ3ϵ4/ϵ4ϵ4 | 313 ± 8 | |||
| (254) | Adults | APOB c.-516 C/C | 327 | 280 ± 4 |
| APOB c.-516 C/T | 131 | 291 ± 6 | ||
| APOB c.-516 T/T | 16 | 351 ± 16 |
Children < 15 years old.
Total number of study individuals=288.
One of the following sections provide a critique of what is and isn't known about the binding of LDL-apoB to the LDLR; the identities of mutations known to cause familial defective apoB (FDB or ADH-2, [OMIM #144010]); and the common NS coding sequence variants that might regulate LDL-C levels through effecting an LDL-apoB100:LDLR interaction(s). Another section covers the ARH gene product LDLR-associated protein 1 (LDLRAP1), a clathrin-associated sorting protein that ushers, in a cell-specific fashion, LDLR-LDL complexes into clathrin-coated structures (Fig. 4), describing the clinical and phenotypic (Table 6) presentation of this rare recessive disorder. The fourth section focuses on determinants of PCSK9-regulated recycling of LDLR to the plasma membrane (Tables 6, 8), a process that is defective in ADH-3 ([OMIM #607787], Table 6) and enhanced in PCSK9-defective FHBL (Table 9). We also point out differences in lipid phenotypes that aid the differential diagnosis of this seemingly rare condition, apoB-specific FHBL, and ANGPTL3-combined hypolipidemia (Tables 4, 9). The final section touches upon the inducible degrader of LDLR (IDOL), an E3 ubiquitin ligase that stimulates LDLR degradation through ubiquitination of its C-terminal domain, thereby limiting the cellular uptake of LDL and VLDL.
TABLE 8.
Lipoprotein clearance: genetic determinants affecting LDL- and total-cholesterol levels
| Gene | Sample (Reference) | Typed Variant (Proxy)a | Positionb(Effect) | Allelec (MAF) | Effect on LDL-C | Other Associated Traits/Comments |
| APOE | 1,943 white participants in longitudinal CARDIA study (258) | 1) rs35136575 | ∼27 kb downstream | G (0.26) | − 8.66 mg/dl G/G versus C/C (P ≤ 0.001) | |
| Prospective study of 2767 white-British men (384) | 2) rs4420638 (rs429358)d | ∼10 kb downstream | G (0.19) | +20.5 mg/dl G/G versus A/A (P < 0.01) (versus ϵ2 and ϵ3 carriers) | ApoB, +11 mg/dl G/G versus A/A (P < 0.01); Triglyceride, + 10.6 mg/dl G/G versus A/A (NS) | |
| Meta-Analysis of 8 individual European GWAS (385) | 2) rs4420638 (rs429358)d | ∼10 kb downstream | G (0.18) | β = 0.059fP = 2 × 10 -40(versus ϵ2 and ϵ3 carriers) | Triglyceride, β = 0.042e P = 6 × 10 -7 | |
| GLGC (32, 377)f | 2) rs4420638 (rs429358)d | ∼10 kb downstream | G (0.17) | +7.14 mg/dl per copy (Z = 25.801, P = 9 × 10 -147) (versus ε2 and ε3 carriers) | Total cholesterol, +6.83 mg/dl per copy (Z = 22.388, P = 5.2 × 10 -111); Triglyceride, Z = 9.640 P = 5 × 10 -22) | |
| Prospective study of 2767 white-British men (384) | 3) APOE ϵ2/ ϵ3/ϵ4 genotype | NA | NA | +17 mg/dl ϵ3/ϵ3 versus ϵ2 carriers; +5 mg/dl ϵ4 carriers versus ϵ3/ϵ3 (P < 0.01) | ApoB, +10 mg/dl ϵ3/ϵ3 versus ϵ2 carriers; +3 mg/dl ϵ4 carriers versus ϵ3/ϵ3 (P < 0.01); CHD, HR for ϵ2 carriers relative to ϵ3/ϵ3, 0.65 [95% CI 0.43-0.98], (P < 0.05) | |
| 10,035 men and 12,134 women in EPIC study(386) | 3) APOE ϵ2/ ϵ3/ϵ4 genotype | NA | NA | Men: ϵ4/ϵ4, +7.8 mg/dl versus ϵ3/ϵ3 and +30.9 mg/dl versus ϵ2/ϵ2 (P < 0.001). Women: ϵ4/ϵ4, +23.2 mg/dl versus ϵ3/ϵ3 and +57.9/dl versus ϵ2/ϵ2 (P < 0.001). | CHD, HR for ϵ2 carriers relative to ϵ3/ϵ3, 0.88 [95% CI 0.77-0.99], (P < 0.05) | |
| 968 Canadians of South Asian, Chinese or European descent (387) | 3) APOE ϵ2/ ϵ3/ϵ4 genotype | NA | NA | ϵ4 carriers, +33 mg/dl (South Asians), +17 mg/dl (Chinese),+22 mg/dl (Europeans) versus ϵ2 carriers (P < 0.01) | ApoB, ϵ4 carriers +32 mg/dl (South Asians), +13 mg/dl (Chinese), +21 mg/dl (Europeans) versus ϵ2 carriers (P < 0.02) | |
| 1,507 postacute coronary syndrome patients randomized to pravastatin or atorvastatin treatment (388) | 3) APOEϵ2/ ϵ3/ϵ4 genotype | NA | NA | Pravastatin: ϵ2 carriers, 53.8% reduction in LDL-C versus 48.1% ϵ3/ϵ3 and 46.4% ϵ4 carriers (P = 0.0004). Atorvastatin: ϵ2 carriers, 22.1% reduction in LDL-C versus 21.8% ϵ3/ϵ3 and 16.6% ϵ4 carriers (P = 0.0004). | Mean baseline LDL-C levels: ϵ2 carriers, 100.0 ± 2.1 mg/dl; ϵ3/ϵ3, 115.9 ± 1.1 mg/dl; ϵ4 carriers, 118.7 ± 1.7 mg/dl (P = 1.5 × 10 -6). | |
| LDLR | German population-based cohorts (total n = 8819) (273) | 1) rs2228671 | c.81C>T(C27C) | T (0.11) | -7.34 mg/dl per copy (P = 1.5 × 10 -10) | |
| GLGC (32) | 1) rs6511720 (rs2228671) | c.67+2015G>T | T (0.11) | - 6.99 mg/dl per copy (Z = - 23.004, P = 4 × 10 -117). | Total cholesterol, Z = 2 20.89, (P = 7 × 10 -97) | |
| 2) rs688 | c.1773C>T (N591N) | T | Z = 11.342 (P = 8 × 10 -30). | Total cholesterol, Z = 9.046, (P = 1 × 10 -19) | ||
| LDLRAP1 | GLGC (32) | 1) rs12027135 | ∼94 kb upstream | A (0.45) | Z = 2 6.435, (P = 1 × 10 ) | Total cholesterol - 1.22 mg/dl per copy (P = 4 × 10 -11) |
| PCSK9 | 1,750 African American participants in longitudinal CARDIA study (389) | 1) rs28362263 2) rs28362286 | c.1327G>A (A443T) c.2037C>A (C679X) | A (0.10) A (<0.01) | − 9.2 mg/dl in carriers versus noncarriers (baseline levels) (P < 0.01) − 33.7 mg/dl in carriers versus noncarriers (baseline levels) (P < 0.01) | Total cholesterol − 10.3 mg/dl in carriers versus noncarriers (P < 0.01)Total cholesterol − 38.3 mg/dl in carriers versus noncarriers (P < 0.01) |
| 3,363 Black participants in ARIC study (390) | 2) rs28362286 3) rs67608943 | c.2037C>A (C679X) Y142X | A (0.02) G (0.008) | − 38 mg/dl in carriers versus noncarriers (P < 0.001) − 35 mg/dl in carriers versus noncarriers (P < 0.001) G/G +19% versus A/A (P = 0.001) | Total cholesterol − 43 mg/dl in carriers versus noncarriers (P < 0.001) Total cholesterol − 38 mg/dl in carriers versus noncarriers (P < 0.001) | |
| 372 subjects ( ∼90% white) with LDL-C 115-190 mg/dl (age = 35-75 years) (391) | 4) rs505151 | c.2009A>G (E670G) | G (0.08) | − 21.2 mg/dl in carriers versus noncarriers (P = 2 × 10 ) | Total cholesterol, G/G +15% versus A/A (P = 0.002) | |
| 1,872 Italian patients with premature MI and 1,865 controls (392) | 5) rs11591147 | c.137G>T (R46L) | T (0.01) | − 12.8 mg/dl per copy, (P = 1.6 × 10 -7) | Total cholesterol, − 20 mg/dl in carriers versus noncarriers (P = 2 × 10 -1). Early-onset MI OR = 0.67 [95% CI 0.46-0.97], (P = 0.036) | |
| 6,382 healthy American women (age ≥ 45 years)(393) | 5) rs11591147 | c.137G>T (R46L) | T (0.02) | − 21 mg/dl in carriers versus noncarriers (P < 0.001) | ApoB, − 11.2 mg/dl per copy (P = 2 × 10 -10) | |
| 9,524 white participants in ARIC study (390) | 5) rs11591147 | c.137G>T (R46L) | T (0.02) | − 13% in carriers versus noncarriers (P < 0.0001) | Total cholesterol, − 20 mg/dl in carriers versus noncarriers (P < 0.001). CHD, HR for carriers relative to noncarriers, 0.5 [95% CI 0.32-0.79], (P = 0.003) | |
| Meta-analysis of three Danish cohorts (n = 45,699) (394) | 5) rs11591147 | c.137G>T (R46L) | T (0.03) | − 17 mg/dl in carriers versus noncarriers (baseline levels) (P < 0.01). | Total cholesterol up to 9% lower in carriers versus noncarriers. CHD HR for carriers relative to noncarriers 0.70 [95% CI 0.58-0.86], (P = 0.001) | |
| 1,828 white participants in longitudinal CARDIA study (389) | 5) rs11591147 | c.137G>T (R46L) | T (0.02) | − 40 mg/dl in A/A versus G/G family members (P = 0.0014) and − 11 mg/dl in A/A versus G/G in replication cohort (P = 0.0002) | Total cholesterol − 15.9 mg/dl in carriers versus noncarriers (P < 0.01) | |
| 401 African Caribbeans belonging to seven multigenerational pedigrees plus replication in 1,750 African- Caribbean men (395) | 6) rs7517090g | c.523+2637 G>A | A (0.20) | Z = 7.008, (P = 4 × 10 -12) | ||
| GLGCf (32) | 4) rs505151 | c.2009G>A (E670G) | G | Z = − 9.159 (P = 5 × 10 -20) | Total cholesterol, Z = 6.182, (P = 6 × 10 -10) | |
| 7) rs11206510 (R46L)h | ∼9 kb upstream | C | +2.01 mg/dl per copy | Total cholesterol, Z = − 9.201 (P = 4 × 10 -20) | ||
| 8) rs2479409 | 571 bp upstream | G (0.30) | (Z = 11.061, P = 2 × 10 -28) | Total cholesterol, +1.96 mg/dl per copy, (Z = 10.136, P = 4 × 10 -24) | ||
| 9) rs516499 | c.1682 − 363A>G | G | Z = − 7.807, (P = 6 × 10 -15) | Total cholesterol, Z = − 7.046 (P = 2 × 10 -12) | ||
| IDOL | GLGC (32) | 1) rs3757354 | ∼2 kb upstream | T (0.22) | − 1.43 mg/dl per copy (Z = − 6.785, P = 1 × 10 -11) | Total cholesterol, − 1.46 mg/dl per copy (Z = − 5.944, P = 3 × 10 -9) |
Independent variants/associations are numbered. “Proxy” variant is in strong linkage disequilibrium with genotyped variant (r2 ≥ 0.8, unless indicated otherwise).
Relative to the initiating ATG.
Minor allele is listed with frequency (MAF) in study population.
rs429358 C allele = APOE ϵ4 allele. rs4420638 in linkage disequilibrium with rs429358 (r2 = 0.967) (377). APOE ϵ2 equivalent variant (rs7412) not genotyped in GLGC.
β represents the change in lipid level (natural log) per effect allele.
rs35136575, rs11591147, rs28362263, rs28362286, and rs7417090 not surveyed in GLGC.
Variant not found in European populations.
Signal attributed to R46L (377). Linkage disequilibrium between alleles, r2 = 0.10.
95% CI, 95% confidence interval; HR, hazard ratio; NS, not significant; OR, odds ratio.
TABLE 9.
PCSK9 deficiency: representative lipid levels in an African American Family and an Italian family
| Lipid/Lipoprotein Levels (mg/dl)
(Percentileb) |
||||||||
| Population (Reference) | Subject (Agea) | Mutation State | Mutation | TC | LDL-C | TG | HDL-C | TC:HDL-C |
| African American (345) | Proband (53) | Heterozygous | p.Y142X | 96 (<1st) | 39 (<1st) | 88 (25th) | 44 (55th) | 2.18 |
| Partner (51) | Heterozygous | p.R97del | 144 (<1 ) | 49 (<1 ) | 51 (<1 ) | 88 (90 ) | 2.82 | |
| Daughter (32) | Compound Heterozygous | p.R97del/ p.Y142X | 96 (<1 ) | 14 (<1 ) | 119 (70 ) | 65 (80 ) | 1.47 | |
| Daughter (28) | Wild-type | None | 152 (20 ) | 80 (15 ) | 92 (55 ) | 59 (65 ) | 2.57 | |
| Grandson (6) | Heterozygous | p.Y142X | 106 (<1 ) | 30 (<1 ) | 77 (75 ) | 65 (75 ) | 1.63 | |
| Granddaughter (13) | Wild-type | None | 174 (60 ) | 104 (60 ) | 110 (75 ) | 55 (65 ) | 3.16 | |
| Italian (346) | Proband (34) | Heterozygous | p.A68PfsX15 | 114 | 53 | NA | NA | NA |
| Father (67) | Heterozygous | p.A68PfsX15 | 182 | 82 | NA | NA | NA | |
| Mother (59) | Wild-type | None | 116 | 58 | NA | NA | NA | |
| Son (3) | Heterozygous | p.A68PfsX15 | 147 | 87 | NA | NA | NA | |
| Paternal Aunt (62) | Heterozygous | p.A68PfsX15 | 196 | 95 | NA | NA | NA | |
Age in years.
Percentile values are derived from comparisons with age- and sex-matched African American controls.
NA, not available. Probands are highlighted in bold.
Cataloguing LDLR mutations in ADH-1: cautionary tales and update
The large number (n = 1100+) of LDLR variants found in ADH-1 patients prompted the development of a database (http://www.ucl.ac.uk/ldlr) (224) that contains information regarding their segregation in families, geographic origins, and predicted effects on encoded gene products (224). Importantly, the database now holds the same information for the rather fewer PCSK9 and LDLRAP1 variants found in ADH-3 and ARH patients, respectively (225).
The ADH1:3/ARH database also provides information on mutational heterogeneity across specific populations and communities. In countries such as France (226) and Holland (227), >100 different LDLR mutations cause ADH-1, whereas in other countries, only one or two mutations are responsible (228–230). For example, in Ashkenazi Jews, the LDLR c.652-654 del GGT mutation (G197del mutation in mature protein) is by far the most common cause of ADH-1 (230). In fact, genetic analyses indicate that the most recent common ancestor bearing this mutation dates back to the start at the 14th century at the founding of the Jewish community in Lithuania (230). Similarly most (>80%) Lebanese ADH-1 patients have the c.2043 C>A (Cys660X; known as Lebanese allele) mutation (228).
It is important to emphasize that certain sequence variants entered into the ADH1:3/ARH database will have no pathogenic consequences, despite their cosegregation with hypercholesterolemia. For example, a sequence variant in the donor splice region of intron 14 (c.2140 +5 G>A) was originally deemed to be pathogenic, as it occurred in a number of ADH-1 patients from different backgrounds (e.g., Dutch, Austrian, Canadian) (231). However, subsequent studies revealed that it was present at comparable frequencies in ADH patients from the Simon Broome Register and healthy white men from the Northwick Park Heart Study II (0.022 versus 0.013, P = 0.23) (232). Moreover, RNA analysis on mononuclear cells from an ADH patient revealed that it had no effect on LDLR pre-mRNA splicing (233), whereas DNA sequencing showed that some ADH patients with this so-called mutation had, on the same chromosome, real pathogenic LDLR mutations (234). Thus, although the ADH1:3/ARH database is an extremely valuable tool for those interested in the molecular genetics of monogenic hypercholesterolemia, in vitro (e.g., heterologous cells systems), ex vivo (i.e., fibroblasts and/or lymphocytes from affected patients), or solid phase binding assays (218) should be performed on variants contained therein to demonstrate their pathogenic effects.
The relatively new multiplex ligation-dependent probe amplification assay technique has greatly facilitated the identification of LDLR deletions/insertions and rearrangements in ADH-1 patients (226, 235–240). In a Polish sample, this technique revealed that duplication of exons 4-8 was the second most common cause of ADH-1, affecting 4.2% of such patients (235). Additionally, in French ADH-1 patients (n = 1003, recruited from 11 regions of France), major rearrangements (n = 19) were found in 9.7% of the sample (226). As such, screening for large rearrangements of the LDLR is now recommended for the molecular diagnosis of ADH-1.
One of the most interesting observations on the molecular pathology of LDLR mutations concerns the pathogenic effects of apparently silent coding sequence variants, which can be easily overlooked. For instance, screening a series of patients with a definite clinical diagnosis of monogenic hypercholesterolemia led to the identification of two single-nucleotide substitutions, c.621 C>T (G186G in mature protein) and c.1216 C>A (R385R in mature protein) (241, 242), which were correctly predicted in silico to be pathogenic. Thus, the c.621 C>T transition introduces a new donor splice site, resulting in an in-frame 75 bp deletion (Gly 186 to Cys 210) from the mature mRNA (242), whereas the c.1216 C>A transversion creates a new acceptor site, resulting in formation of a truncated product (241). Hence, it is possible that in silico analysis is also correct in predicting that other silent LDLR coding sequencing variants [i.e., c.1813 C>T (L695L) and c.2140 G>C (E714E)] also cause ADH-1 through aberrant pre-mRNA splicing (226). Furthermore, we now appreciate that even mutations predicted to lead to an amino acid substitution may actually cause ADH-1 through aberrant splicing. One such example involves the c.2389 G>T transversion (last nucleotide of exon 16; V776L). Examining patients’ blood mononuclear cells revealed they produced an abnormal mRNA devoid of exon 16 (233), demonstrating the importance of functional analyses to establish the molecular mechanism by which pathogenic mutations exert their effects.
Finally, Civeira et al. (243) have highlighted the importance of screening for LDLR mutations in people who have personal and family lipid profiles indicative of familial combined hyperlipidemia (FCHL). In their series of 143 unrelated patients, which they acknowledged had an inherent selection bias toward those with more severe lipid abnormalities and therefore might not be representative of FCHL patients in the general population, the frequency of LDLR mutations was 19.6%. “FCHL” patients with a LDLR mutation (n = 28) had higher mean total cholesterol, LDL-C, and apoB and nonsignificantly lower triglyceride levels (e.g., cholesterol: 357 ± 54 versus 318 ± 56 mg/dl, P = 0.001; triglyceride 248 (217-361) versus 301 (226-464) mg/dl, P = 0.059) than those who did not. Thus, 41.7% of the 48 patients (i.e., 20) in their series with a total cholesterol or apoB >335 and 185 mg/dl, respectively, had a LDLR mutation, compared with 8.8% (i.e., 8/91) who had values below these levels. Although there is no formal proof that all of LDLR mutations found by Civeria et al. (243) were pathogenic, their study clearly demonstrates that screening for LDLR mutations should be performed in patients with a clinical diagnosis of FCHL who have very high total cholesterol and apoB levels.
Genotype-phenotype correlations in ADH-1.
One of the most challenging issues in ADH-1 pathology is defining the relationship among causative mutations, the associated biochemical disturbance (e.g., elevated LDL-C), and clinical/preclinical phenotypes, such as premature CHD, tendon xanthomas, and asymptomatic atherosclerosis. The problem arises in large part from the low frequency of each individual causative mutation encountered in genetically heterogeneous populations combined with patient ascertainment bias. One simple approach for overcoming this problem is to subdivide mutations into two broad categories: null alleles (also designated “LDLR-negative” mutations that are expected to completely abolish LDLR functions) and defective alleles (designated LDLR-defective mutations; i.e., variable, residual LDLR functional activities). Using these criteria, we compared LDL-C levels in both homozygous and heterozygous Italian patients (244). For homozygous patients, the different impact of mutation type was clearly discernible (Table 6) (245), with the LDLR-null patients having markedly higher LDL-C levels. Similarly, the heterozygous LDLR-negative subjects had higher LDL-C levels and a 2-fold higher prevalence of tendon xanthoma and CHD than did the LDLR-defective patients. Likewise, heterozygous Spaniards with a null allele (n = 269; mean age 43 years) had ∼30 mg/dl higher LDL-C and ∼2-fold increased prevalence of tendon xanthomas and CHD than did LDLR-defective patients (n = 162, mean age 46 years). Additionally, in this population, null allele ownership was documented to be associated with more advanced femoral atherosclerosis (246).
Intriguingly, data suggest that the differential effects of LDLR-negative and defective alleles on LDL-C levels are diminished by the β -thalassemia trait (247). Sardinians with this trait and a null LDLR allele (i.e., Fs572 mutation) had lower LDL-C than those with just the Fs572 mutation (222.7 ± 41.8 mg/dl versus 319.0 ± 64.2 mg/dl, P < 0.001). Additionally, the one ADH-1 Sardinian with this null mutation and full-blown β -thalassemia had the lowest plasma LDL-C of all (100.9 mg/dl). Similar findings were also evident when the analyses were extended to include LDLR-defective patients. Allelic heterogeneity, however, precluded an accurate estimate of the effect of the β -thalassemia trait on LDL-C levels in these patients.
Considering the biological basis of the thalassemia-LDLR interaction, two reports implicate high-erythropoietic activity, with the authors concluding that substantial amounts of cholesterol are utilized during active erythropoiesis (248, 249). Shalev et al. (249) observed that patients with chronic anemia associated with high-erythropoeitic activity (e.g., thalassemia intermedia) had significantly lower plasma total cholesterol, LDL-cholesterol, and HDL-cholesterol levels (80 ± 19, 35 ± 14, 31 ± 10 mg/dl) than did patients with low-erythropoietic activity anemias (139.75 ± 15.86, 82.33 ± 17.85, 49.38 ± 12.86 mg/dl) and healthy controls (156.80 ± 35.69, 83.10 ± 32.52, 51.95 ± 10.88 mg/dl). Moreover, whereas all patients with chronic anemia and increased erythropoietic activity had hypocholesterolemia, none with low erythropoietic activity did. Hence, it is rather disappointing, particularly given the relatively high incidence of hemoglobinopathies in Tunisia (250), that the unexpectedly mild lipid phenotype in their heterozygous Tunisian ADH-1 patients (total and LDL-cholesterol: 272.2 ± 54.1 and 216.6 ± 42.5 mg/dl, Table 6) was not investigated in terms of their thalassemia status, especially as homozygous Tunisian ADH-1 patients have the expected severe phenotype (total and LDL-cholesterol levels of 677.5 ± 120.7 and 623.0 ± 119.5 mg/dl, respectively) (251).
As highlighted in Table 7, apoE isoforms ϵ2 (rs7412) and ϵ4 (rs429358) may influence the effect of LDLR mutations on LDL-C levels in both children and adults (252–254), consistent with structural (255, 256), functional (257) and nonADH-1 genetic (Table 8) data. Thus, in (apparently unrelated) heterozygous ADH-1 French-Canadian children with the same 15 kb+ deletion null allele, those with an apoϵ2/3 genotype had markedly lower total plasma cholesterol, LDL-C, and apoB levels than apoϵ3 homozygotes, whereas LDL-C levels in children from the general population who had an apoϵ2/3 genotype were only decreased by 17.4 mg/dl (253). The ADH-1 adult studies also point in the same direction as young children studies, albeit with a more modest LDL-C lowering effect (252, 254). Conversely, ϵ4 allele (rs429358) ownership in the adult ADH-1 cohorts was associated with elevated LDL-C: 26.7 mg/dl in the Italians and 2 mg/dl in the Finnish (Table 7), consistent with population data (Table 8). By analogy, variants at the apoE/CI/C4/C2 gene cluster, other than the ϵ2 and ϵ4 alleles, are also likely to influence LDL-C levels in ADH-1 patients (Table 8); including one (i.e., rs35136575) modulating apo gene expression, given its location in an hepatic apoE/CI/C4/C2 control region (258).
Intriguingly, not only the magnitude but also the very direction of the effect of specific apoE isoforms on plasma cholesterol levels in response to a high-fat, Western type diet is likely to depend on a number of factors, including their interactions with prevailing levels of proteoglycans (259–261), LDLR-related protein 1 (262), and LDLR at the hepatic cell surface. Supporting evidence for this proposition derives from the study of transgenic apoE and Ldlr mice (257, 263, 264) in which native apoE was replaced by human APOE isoforms and an endogenous murine Ldlr allele was replaced by a human LDLR minigene to produce mRNA transcripts with a longer half-life. When exposed to a Western-style diet, apoϵ4/4 mice expressing ∼2-3-fold higher levels of LDLR developed higher plasma cholesterol concentrations than those with baseline LDLR expression (187 ± 19 versus 124 ± 8 mg/dl), whereas apoϵ3/3 littermates displayed the expected decrease (72 ± 8 versus 114 ± 12 mg/dl; P < 0.0001 for LDLR × ApoE isoform interaction). Similarly, in an independent experiment (263), the dyslipidemia of chow-fed apoϵ2/2 mice (plasma cholesterol 268 ± 12 mg/dl; triglyceride 157 ± 22 mg/dl) was normalized (plasma cholesterol 83 ± 4 mg/dl; triglyceride 40 ± 5 mg/dl) by constitutively high LDLR expression. Additionally, the cholesterol-elevating effect of the Western-style diet (cholesterol 546 ± 30 mg/dl) was ameliorated (172 ± 43 mg/dl). Through follow-up analyses, Maeda and colleagues (257) were able to conceptualize the counter-intituitive apoϵ4/4 results in terms of hepatic sequestration of apoE, chlyomicron- and VLDL-remnants on the surface of hepatocytes, and their subsequent internalization. In essence, although apoϵ4 has a greater affinity for the LDLR and heparan sulfate proteoglycan than either apoϵ3 or apoϵ2 (Fig. 5A) (261), it is poorly internalized and recycled (265). In the face of constitutively elevated LDLR expression, the effect is to reduce apoE availability for transfer to nascent chylomicrons and VLDL (either directly or indirectly via HDL) that ultimately enables the cellular uptake of cholesterol-rich lipoproteins (i.e., LDL and remnant particles), for example, by the LDLR and LRP1 (Fig. 5B). Apoϵ2 and ϵ3, on the other hand, with their reduced affinity for the LDLR (Fig. 5A) are compatible with elevated LDLR expression reducing plasma cholesterol levels. The big question, of course, is how this rodent model translates in man and, in particular, whether the relatively low plasma cholesterol levels seen in most ( ∼95%) apo ϵ2/2 individuals (Table 8) is mechanistically linked to the increased availability of this isoform for transfer to circulating lipoproteins in spite of its lower affinity (i.e., on human fibroblasts: 8.5 ± 1.0% of apoϵ3 (266) for the LDLR). Clearly, detailed in vivo kinetics will be crucial.
The GLGC study also substantiates our observation that a particular APOB allele (or allele in LD) has a LDL-C elevating effect in people with LDLR mutations (254). We found that heterozygote ADH-1 patients with the rare allele at SNP site rs934197 (-516 C>T), which is in strong LD with the T71I variant (Table 5), had higher LDL-C levels than noncarriers; specifically, 11 mg/dl in the heterozygous apoB ADH-1 patients and a staggering 71 mg/dl hike in the apoB homozygotes (Table 8), a result that warrants further clinical and laboratory investigations.
Finally, it is important to appreciate the data from Tybjaerg-Hansen et al. (267) that suggest the effect of LDLR mutations on LDL-C levels in patients seen within a clinical environment may be overestimated. On genotyping 9,255 adults from the general Danish population for the three LDLR mutations [W23X, W66G, and W556S (Class 2A, Fig. 3) that explain >40% of all clinical cases of ADH-1 in Denmark], the heterozygous carriers (n = 6) had, respectively, 112 mg/dl and 147 mg/dl higher total plasma cholesterol and LDL-C levels than had the general population noncarriers, whereas the same mutations in ADH-1/CHD patients (3/948) increased values by 158 mg/dl and 162 mg/dl, respectively. Furthermore, ADH-1 patients (28/63) with a clinical diagnosis of monogenic hypercholesterolemia had increases of 189 mg/dl and 205 mg/dl.
Mechanistic insights.
Conventional wisdom is that the LDLR primarily serves to mediate the uptake of extracellular lipoproteins, primarily LDL, and that disruption of this uptake explains the higher plasma LDL-C levels in LDLR-negative patients compared with those with a LDLR-defective allele. However, it now appears that this difference is not solely ascribed to failure of apoB100-mediated uptake of lipoprotein particles; it also includes significant contributions from both increased net secretion of VLDL and decreased clearance of VLDL catabolic by-products (268–271).
In outline, metabolic studies have established that VLDL-apoB100 production/secretion is increased in LDLR-negative patients (270, 271) and that in mice, the LDLR preferentially directs small, poorly lipidated apoB particles to a post-ER compartment for degradation (268, 269). Thus, Tremblay et al. (271) showed that the production/secretion of VLDL-apoB100 was increased by 50% and 109%, respectively, in six heterozygote patients and one homozygote individual that had the same LDLR-null allele. Likewise, Millar et al. (270) found a 68.8% increase in heterozygous LDLR-negative patients with the Lebanese C660X mutation (Class 2A, Fig. 4) but not in LDLR-defective patients (increase 21.2%; P = 0.48, for difference from controls). Likewise, compared with control livers, the livers from homozygous LDLR− / − and C678Y (LDLR retained in ER) mice have increased net secretion of apoB48- and apoB100-containing lipoproteins (268), whereas hepatocytes expressing LDLR mutant Y807C, which does reach the cell surface, do not. In fact, through a series of cleverly designed experiments that employed a LDLR mutant (i.e., I140D) defective in apoB, but not apoE, binding and internalization (Class 3A, Fig. 3), Blaisole et al. (268) reached the conclusion that the LDLR regulates VLDL-apoB secretion through selective (re)uptake of (VLDL-bound) apoE (Fig. 5). Moreover, this uptake and that of β -VLDL (272), in contrast to the LDL-apoB100 ligand, is mediated via a VLDL-induced 802FDNPVY807-independent internalization mechanism(s).
Common LDLR variants associated with LDL-C levels.
From the perspective of increased understanding of how LDLR transcript functionality is regulated, two independently segregating SNPs are noteworthy (Table 8). In multiple German and British population samples, the rare allele at SNP site rs2228671 displays association with decreased LDL-C (273) and CHD risk. Moreover, because adjustment for LDL-C levels by logistic regression and Mendelian randomization models abolished the rs2228671 and CHD signal, the data point to a link between the rare allele at this SNP site [or allele(s) in LD] and a lifelong reduction in LDL-C levels and CHD risk. In the GLGC cohort, the rs6511720 rare allele (which tags rs2228671) also displayed strong association with decreased LDL-C levels, with the estimated effect size being larger than those observed for APOB, SORT1, and TRIB1 variants (Tables 5, 8).
The story of the second LDLR variant (rs688) started with an in silico prediction that the rare allele at this SNP site neutralizes a putative exonic splicing enhancer within exon 12. Estus and colleagues (274, 275) then went on to show that the splicing efficiencies of minigene-derived transcripts containing the rare allele were, relative to common allele bearing transcripts, decreased by ∼9 and 15% in both HepG2 and SH-SY5Y (neuroblastoma) cells. Additionally, in human tissue samples, the rare variant was associated with less efficient exon 12 splicing: in female, postmortem liver and male anterior cingulate region of brain, the decreases were 8.6% (P = 0.024) and 8% (P = 0.041), respectively. Consistent with a potential reduction in LDLR protein production, the GLGC study associates the rs688 rare allele with increased LDL-C (32) (Table 8), wheres Estus and colleagues (275) present evidence of association with Alzheimer disease in elderly men [recessive model, OR 1.49, (95% CI 1.13-1.97), P = 0.005] but not in women, leading them to speculate that the rare rs688 allele decreases exon 12 splicing efficiency in vivo, leading to reduced functional LDLR and apoE clearance, increased amyloid pathology, and risk of Alzheimer disease.
Binding of LDL-apoB100 to LDLR
General acceptance that an apoB100-LDLR interaction(s) initiates the uptake of LDL-C stands in marked contrast to our level of ignorance over the precise molecular contacts that apoB100 makes with this receptor. Therefore, critical review of exactly what is known seems timely.
Early antibody studies mapped potential binding sequences to amino acids 2835-4189 (276–278), whereas comparisons with the apoE-LDLR-ligand binding sequence (Fig. 5) suggested two prime candidate regions: peptides A, amino acids 3147-3157 and B, 3359-3367 (279). However, despite extensive sequencing of the exonic region encoding these two apoB regions, no LDL-C raising mutations/variants have been identified in either region. Additionally, data from mice regarding the functional importance of peptide B in vivo are equivocal. Promisingly, Boren et al. (280) found that mutant human recombinant LDL containing amino acid changes in peptide B of the putative LDLR-apoB100 binding region (i.e., R3359S, R3362S, K3363A, R3364S, and R3367S) competed far less efficiently with labeled human plasma LDL for binding to human fibroblasts in vitro than similarly derived control recombinant human LDL. However, data regarding the effects of the defective mutant LDL binding on plasma LDL-C levels in their mice were conspicuous by their absence. Moreover, even the in vitro data could conceivably be explained by the introduced mutations perturbing LDLR binding residues/motifs outside the hypothetical peptide B region. The more recent mice studies of Johnson et al. (281) serve only to underscore the current ignorance of the apoB100-LDLR binding mechanism. They showed that Ldlr/Apobec1− / − mice expressing recombinant apoB100 with two foreign peptide sequences in place of peptides A and B had lower, rather than the expected higher, LDL-C than mice expressing control, recombinant apoB100. Investigations revealed that the mutant apoB mice were secreting abnormally large VLDL particles overendowed with apoE, which were cleared more rapidly from the circulation than control VLDL- apoB100 through an apoE-LRP-mediated mechanism, a salutary reminder to always consider the secretory as well as uptake pathways when studying LDL-C levels.
Immunoelectron microscopic analyses of human LDL (282) indicate that peptide A and B (amino acids 3147-3157 and 3359-3367) reside on one side of the LDL hemisphere close to where apoB100 amino acid residues ∼4275-4440 cross the ∼620A long, wedge-shaped, kinked ribbon (apoB1.5-89; amino acid residue 71-4050) encircling LDL. Subsequent competitive binding assays involving labeled human LDL-apoB100 and LDL containing recombinant apoB substantiate these data and indicate apoB amino acids R3500 and W4369 (apoB96.3) interact to facilitate normal apoB100-LDLR binding (280, 283). Moreover, LDLs containing C-terminal truncated forms of apoB retaining the hypothetical LDLR ligand binding peptides A and B, such as apoB77 and apoB95 (i.e., truncated at residues 3497 and 4329), compete more avidly for LDLR binding than full-length apoB100. Hence, the extremely low LDL-C levels seen in apoB-specific FHBL patients with C-terminal truncated apoBs in the range of ∼apoB77-95 (284, 285) may be attributable, at least in part, to enhanced LDL clearance.
Very recently, electron cryomicroscopy analyses (118) have suggested that the apoB100 structures that interact with the LDLR are stabilized by the cholesteryl-ester rich core of LDL and that they include a region formed by amino acids ∼2050-2600 (apoB45.2-57.3), which bind to the LDLR β -propeller (Fig. 5B), as well as apoB structure(s) within one of its β -sheet domains (i.e., residues 1000-2074 or 2600-4000). In summary, the LDL-apoB100:LDLR interaction(s) seems considerably more complex and extensive than originally thought.
APOB mutations, defective LDLR binding and ADH-2.
Grundy and colleagues were the first to report the association between moderate hypercholesterolemia, defective LDLR binding, and the apoB100 R3500Q mutation (286). Specifically, they identified a heterozygous index patient (LDL-C, 215 mg/dl) who cleared his own LDL from the circulation ∼2-fold (P < 0.001) less efficiently than control LDL and then showed that the patient's LDL in vitro was ∼90% less effective in competing with normal LDL for cellular uptake and degradation. Moreover, the patient's LDL had reduced capacity to stimulate intracellular cholesteryl ester synthesis. Similar, but numerically unspecified, in vitro results were obtained from four other family members, all of whom possessed the apoB100 R3500Q allele. In comparison, blood relatives with “normal” LDL binding (range not specified) did not. Subsequent studies have confirmed that the R3500Q allele(s) is a major cause of moderate to severe hypercholesterolemia in people of European descent (Table 6) and that it confers increased risk of premature atherosclerotic disease (226, 227, 235, 238, 287–292). In Asian populations, the recurrent R3500W mutation (293, 294) is reported to be the principle cause of ADH-2.
In the last year, it has emerged that ∼12% of the Old Order Amish residents of Lancaster County, Pennsylvania, have the R3500Q mutation (295) and that it is likely to have been introduced into this community at the end of the 1700s by a single Swiss ancestor. Today, Switzerland has one of the highest frequencies (0.41%) of the R3500Q allele in Europe (296). Returning to the Amish community, Shen et al. (295) performed a GWAS to identify variants associated with high LDL-C levels and coronary artery calcification in 841 asymptomatic individuals (age 43.7 ± 13.9 years) and found a cluster of 65 SNPs spanning a 12-megabase region near APOB displaying strong association with LDL-C levels. Subsequent genotyping for the R3500Q (rs5742904) mutation disclosed that it was in near-complete LD with the SNP alleles that had displayed the strongest association with LDL-C. Moreover, combining this GWAS sample with another 663 Amish revealed that it accounted for 26% of the variation in age- and sex-adjusted LDL-C levels in the population (Table 6). Moreover, R3500Q carriers had 4.41-higher odds of having radiologically detectable coronary artery calcification. In a control analysis, all other SNP associations (n = 65) with LDL-C concentrations disappeared when APOB R3500Q was considered as a covariate. Nonetheless, because many of the associating SNPs are in high LD (i.e., many carriers of the R3500Q mutation share a common extended haplotype potentially originating from a Celtic ancestor some 6,000-7,000 years ago (296)), it would have been nearly impossible without independent genetic (227, 293, 294) and supporting functional (280, 283) data to conclude that the R3500Q mutation (rather than a mutation(s) in LD) is the cause of moderate hypercholesterolemia in the Old Order Amish and other populations.
Boren et al. (280) showed that LDL containing recombinant apoB100 with a glutamine, lysine, or glycine at amino acid 3500 bound the LDLR ∼80% less efficiently than recombinant apoB100-R3500 in vitro. R3500, however, is unlikely to be directly involved in LDLR binding (or be the major determinant of LDLR binding) because LDL containing recombinant apoB80 R3500Q and apoB77 (truncated at amino acid 3497) displayed enhanced LDLR binding (280). Instead, it is envisaged that amino acid residue R3500 is required to attain the conformation of the yet undefined and potentially multiple apoB100 motifs that bind to the LDLR.
It is also now known that 2-10% of patients with a clinical diagnosis of FH have the apoB R3500Q mutation (289, 297, 298), including some who also have a LDLR mutation (299–301). Compound heterozygote ADH-1/2 patients tend to have higher LDL-C, more extensive xanthomatosis, and more severe premature CHD than heterozygote ADH-1 and homozygote ADH-2 patients (Table 6). Their phenotype, however, appears to be less severe than that seen in homozygous ADH-1 (300).
It has emerged that some people with familial ligand-defective apoB100 due to R3500 and R3531 mutations have plasma cholesterol levels within the normal range (302–306), despite defective LDL binding and accumulation of the mutant allotype in blood. In one study, Pullinger et al. (304) showed by using a monoclonal antibody and dynamic light scattering that the mass ratio of Gln3500 to Arg 3500 in the LDL of heterozygote apoB100R3500Q patients was ∼73:27 and that in vitro the mutant LDL had <10% of the normal affinity for the LDLR. In comparison, the mass ratio of Cys3531 to Arg3531 was 58:42 for heterozygote apoB100R3531C individuals, and the mutant LDL displayed 27% of normal affinity for the LDLR. Analyzing the lipid profiles of R3531C family members revealed a wide range of total cholesterol and LDL-cholesterol values (age- and sex-adjusted cholesterol and LDL-C: 152-326 and 68-238 mg/dl, respectively), with the average LDL-C increase (i.e., 32 mg/dl or 16%) being more modest than that observed for the R3500Q mutation (Table 6). In a second study, five heterozygote individuals [i.e., R3480P (n = 4); P3480W (n = 1)] from the Danish general population (n = 9,255) with ligand-defective apoB100 (binding affinity intermediate between that of LDL R3500Q and R3531C) had lower, not higher, plasma LDL-C and apoB (mean decreases 42.5 and 24 mg/dl, P = 0.03 and 0.02 for difference from noncarriers), with the potential explanation at least in part attributable to reduced conversion of VLDL to LDL (302). The mutation carriers also had reduced plasma VLDL-C levels, which may relate to impaired VLDL assembly given that both PANTHER and PolyPhen predict that a proline substitution at position 3480 would be deleterious (157).
It is important to emphasize that most, if not all, investigators in their search for ADH-2 mutations have restricted their analyses to specific apoB100 regions (e.g., encoding amino acids 2488-3901). Moreover, such analyses have typically gone ahead in the absence of a definitive biochemical diagnosis of ligand-defective apoB100, which may underestimate the spectrum of apoB mutations that cause/confer susceptibility to ADH-2. Finally, the finding of apoB NS coding sequence mutations (e.g., S3476L, S3488G, Y3533C, T3540M, I4350T, G4369D (298, 307), R4358, and R4367 (227)) in hypercholesterolemic patients is in itself insufficient for a definitive diagnosis of ADH-2: in vitro and in vivo functional analyses are generally required.
Nonsynonymous ApoB100 coding variants modulate plasma LDL-C.
As shown in Table 5, data from the GLGC (32) replicate the prior evidence of association between three independent apoB100 variants (R3611Q, E4154K, and N4311S) and LDL-C levels (157). Thus, for example, Benn et al. (157) showed that in the prospective Copenhagen City Heart Study, individuals with the R3611Q variant had higher LDL-C levels than noncarriers and that apoB100 levels were similarly increased, consistent with a potential defect in LDLR binding. In comparison, loss of the glutamic acid residue (or gain of a lysine residue) at amino acid 4154 (apoB91.6) was associated with decreased LDL-C and apoB. Heterozygote carriers of the N4311S variant also had lower LDL-C and apoB levels. The homozygote carriers, however, had values intermediate between the noncarriers and heterozygote carriers, which could relate to the complexities of LDLR-mediated uptake of LDL and VLDL, as well as the conversion of VLDL to LDL and the VLDL-apoB assembly process itself. Regarding this, the N4311S variant is in strong LD with a proline (i.e., P2712L) residue that is conserved from humans to zebra fish and sea urchin. Moreover, its replacement with leucine is predicted to be deleterious (157).
Recruiting LDL:LDLR into clathrin-coated structures for internalization
Meticulous clinical evaluation and painstaking laboratory characterization of the physiological/cellular defect(s) in patients with an atypically inherited form of familial hypercholesterolemia (i.e., ARH) led to the discovery that clathrin-mediated uptake of LDLR-apoB complexes (and of LDLR) in certain cell types (e.g., hepatocytes) require the adaptor protein LDLRAP1. Harada-Shiba et al. (308) began this story with a full clinical and biochemical description of two Japanese siblings who had lipid profiles and clinical symptoms (e.g., multiple xanthomas since childhood) similar to those in homozygous ADH-1 patients, despite evidence of normal functioning LDLR. Specifically, their cultured skin fibroblasts synthesized normal quantities of fully active LDLR protein, and these were normally downregulated with LDL or cholesterol loading. Moreover, genetic markers at the LDLR locus did not segregate with the hypercholesterolemia. Whole-body in vivo turnover studies, however, returned convincing evidence that the elevated LDL-C in these two siblings was not attributable to increased de novo cholesterol synthesis but, rather, to impaired LDL catabolism. This conclusion was subsequently substantiated by 125I-LDL turnover studies and a biodistribution and uptake analysis of 99mtechnetium-labeled LDL in three Sardinian ARH patients (309). Thus, these patients had a marked reduction in the fractional catabolic rate of LDL (patients versus five controls: 0.19 ± 0.1 and 0.36 ± 0.03 pools/day, P < 0.001), plus a significant increase in LDL production (20.7 ± 4.4 versus 14.0 ± 2.4 mg/kg/day, P < 0.01), although this increase is lower than that typically seen in homozygous ADH-1 patients. These changes were paired with a severe reduction in hepatic LDL uptake similar to that observed in ADH-1 homozygotes and decreased LDL uptake by the kidney and spleen (but not heart), findings which prompted Zuliani et al. (309) to speculate that the ARH lipid phenotype was caused by selective reduction in hepatic LDL uptake. Subsequently, Norman et al. (310) showed the defect resided in a component of the endocytic machinery required for LDLR and LDLR-LDL complex uptake through clathrin-coated pits; specifically, they found that LDLR was present on the cell surface of EBV-immortalized lymphocytes prepared from ARH patients and that the receptor bound LDL normally but was unable to internalize it.
It turns out that LDLRAP1 is required for the hepatic uptake of LDLR/LDL-apoB100 complexes but not of LDLR-VLDL-remnant complexes, which are internalized by an 802FDNPVY807-independent mechanism (Fig. 4). Thus, when Jones et al. (311) fed ldlr− / − and ldlrap1− / − mice a high sucrose diet (to stimulate hepatic VLDL production), the ldlrap1− / − animals developed less pronounced hyperlipidemia due to preservation of the LDLR-dependent clearance of VLDL remnants. This phenomenon in humans could explain the development of a relatively mild LDL-C phenotype in ARH patients compared with their compatriots with homozygous/compound heterozygous LDLR-null alleles (Table 6), especially as the LDL phenotypes of ARH and homozygous ADH-1 patients with LDLR-defective mutations are superimposable (Table 6).
As to the role of LDLRAP1 in mediating LDL uptake in only certain cell types, it seems noteworthy that ARH typically manifests in children as large tendon xanthomata, either planar or tuberous (310, 312–318), suggesting that LDLRAP1 is dispensable for LDLR activity in macrophages.
Genetics of autosomal recessive hypercholesterolemia.
Current studies indicate that in most parts of world autosomal recessive hypercholesterolemia (ARH) is likely to be an exceedingly rare condition (314–322). One exception is Sardinia, where the disease has an estimated frequency of 1:40.000, accounted for by two mutations (W22X and c.432 ins A) (313). In middle-aged people from this population, ARH carriers (1:143 individuals) had similar LDL-C levels to noncarriers (141.0 ± 0.41 versus 137 ± 0.41 mg/dl, P = 0.19) and a comparable rate of myocardial infarction (323). However, it should be noted that the study of only 3,410 islanders limited the power to detect small effect size and that in the GLGC sample a common LDLRAP1/Tmem57 variant displayed association with LDL-C levels (Table 8). However, whether the actual allele(s) causing this association signal is mediating its LDL-lowering effect through LDLRAP1 or Tmem57 is not known.
Returning internalized LDLR to the plasma membrane
As depicted in Fig. 4, PCSK9 binds hepatic LDLR, promoting its degradation and thereby impairing LDL/VLDL uptake by this receptor. This discovery has culminated in the development of a PCSK9-binding antibody that mimics the epidermal growth factor (EGF)A domain of the LDLR, which in vivo serves to effectively remove PCSK9’s ability to bind and promote LDLR degradation and significantly reduce LDL-C by 20-50% in monkeys and 40% in “humanized” mice (324). The mechanics of PCSK9/LDLR binding and intracellular degradation have been reviewed quite recently (325–327) and are not covered in depth here. Instead, we focus on correlations between PCSK9, de novo cholesterol biosynthesis, and LDL-C levels in humans (328, 329), as well as the growing consensus that PCSK9 participates in VLDL production/net secretion (330).
In humans, plasma PCSK9 levels normally display a diurnal rhythm that closely parallels that of cholesterol synthesis, whereas total plasma cholesterol levels remain relatively stable during these oscillations (328, 329). However, depleting hepatic cholesterol and prolonged fasting obliterates this natural rhythm. Thus, in healthy individuals consuming three standardized meals per day, plasma PCSK9 and lathosterol (measure of cholesterol synthesis) levels are strongly correlated, reaching a nadir between 3 and 9 PM and a peak at 4.30 AM (329). By contrast, short-term (12 h) depletion of hepatic cholesterol by cholestyramine induces parallel increases in plasma PCSK9 and cholesterol synthesis, which remain elevated for 2-3 days after cessation of these bile acid sequestrants (329), suggesting that changes in a hepatic regulatory pool(s) of cholesterol contribute to the regulation of plasma PCSK9 levels, a result substantiated by the finding that statins also cause a sustained increase in plasma PCSK9 levels (331). Conversely, during prolonged fasting, cholesterol synthesis and PCSK9 levels fall, reaching nadirs at 32 and 36 h, respectively (328), and somewhat unexpectedly, LDL-C levels steadily rise ( ∼20 mg/dl from fed state), reaching a peak at 32 h, when de novo cholesterol synthesis is at its lowest. This paradoxical rise in LDL-C levels, given the observed reduction in both cholesterol synthesis and LDLR degradation (via reduced PCSK9), suggests that prolonged fasting also reduces LDLR activity and thus LDL-C uptake and that this is mediated at least in part by the SREBP-2 transcriptional route, which also decreases PCSK9 and cholesterol biosynthetic gene expression (328, 332).
A recent study by Herbert et al. (330) substantiates the in vivo evidence that PCSK9 gain-of-function mutations increase the net secretion of VLDL (333). In brief, they produced and analyzed transgenic mice lines expressing wild-type human PCSK9 and PCSK9-D374Y (Fig. 4) at levels comparable to endogenous Pcsk9 and with the same tissue specificity. On a chow diet, the wild-type Pcsk9 mice had, compared with their BAC-negative littermates, reduced hepatic levels of LDLR, increased plasma cholesterol ( ∼1.8-fold, mostly in LDL fraction), and a comparable rate of triglyceride secretion. The livers of PCSK9-D374Y mice contained even less LDLR, and they developed more pronounced hypercholesterolemia (cholesterol increase, 2.7-fold). Additionally, they secreted ∼2-fold more triglyceride-rich lipoproteins into the circulation than the control animals. This result complements two earlier studies: the first study showed that hepatocytes from Pcsk9-deficient mice ex vivo secreted fewer apoB48-containing ( ∼33%) and apo100-containing (25%) lipoproteins than their wild-type litter mates (334), and the second study showed that these same mice secreted fewer chylomicrons and had attenuated postprandial hypertriglyceridemia (335).
Genetics of PCSK9.
Ten gain-of-function mutations have been described in ADH-3: two (S127R, D129G) residing in its prodomain (amino acids 31-152), seven (R215H, F216L, R218S, R357H, D374Y, D374H, N425S) within the catalytic domain (amino acids 153-451), and one to three (N425S, R469W, R496W) in the cysteine-rich C-terminal (452-692) residues (336). For the most commonly reported LDL-elevating mutations, S127R and D374Y (Table 6), functional assays have provided plausible genotype-phenotype correlations (337, 338). Thus, the S127R mutation in vitro significantly reduces autocleavage of the PCSK9 prodomain in the ER, a prerequisite for its subsequent trafficking and secretion, whereas PCSK-D374Y undergoes normal rates of autocleavage and secretion (338). Moreover, S127R is less potent than D374Y in reducing cellular LDL uptake (LDL-uptake EC50: wild-type, 56 ± 10, S127R, 13 ± 5.8; D374Y, 2.2 ± 0.4 nM) (338). Thus, the combination of reduced PCSK9-S127R secretion and of potency in blocking LDL-C uptake could well explain the reduced penetrance (0.94) of the S127R mutation (339), Table 6), whereas the normally secreted PCSK9-D374Y with its higher potency accords with the unusually severe hypercholesterolemia observed in ADH-3 patients with this particular mutation (Table 6). Thus, while some heterozygote ADH-3 patients have plasma cholesterol and LDL-C levels comparable to homozygous LDLR-null allele patients, others, consistent with in vitro assay results (337), have levels more similar to heterozygote ADH-1 patients with LDLR-defective alleles (Table 6). Finally, it should be noted that even though large-scale mutation screening projects have indicated that gain-of-function PSCK9 mutations are a rare (<1%) cause of ADH (245, 307, 340–342), such mutations can occur in heterozygote ADH-1 patients with a particular severe phenotype (Table 6), and these patients may respond better to aggressive LDL-C lowering treatment than homozygous LDLR-null allele patients (343).
It turns out that 11% of the African American subjects in the Dallas Heart Study with low plasma LDL-C levels harbor one of two PCSK9 nonsense mutations (i.e., Y142X, Tables 8, 9; C679X, Table 8) compared with ∼1.8% of medically unselected but ethnically and geographically matched individuals (344). The Y142X allele may contribute to FHBL in this group, insofar as it segregates with the “the LDL-C <75 mg/dl trait” (344, 345); however, internationally, the picture is less clear. Thus, in an Italian “FHBL” family with a similar truncating mutation (Ala68fsLeu82X) three of the four mutation-carrying family members had “raised” ( ∼85 mg/dl, ∼10th percentile) LDL-C levels, albeit possibly confounded by coexisting obesity (346) (Table 9). On the other hand, the sister and daughter of a 49-year-old Frenchman (LDL-C 16 mg/dl) with the dominant-negative double-mutant PCSK9 R104C/V114A had LDL-C levels of 57 mg/dl and 58 mg/dl, respectively (347). Thus, it seems that a preliminary diagnosis of PCSK9 deficiency (rather than apoB-specific FHBL) should be considered when a proband with very low LDL-C levels has family members with moderate rather than very low LDL-C levels and relatively high cholesterol:HDL-C ratios (Tables 4, 8). Plasma PCSK9 measurements may also be helpful. Indeed, despite the fact that plasma PCSK9 levels explain <10% of the population variance in LDL-C levels, recent data have shown that certain loss-of-function mutations are associated with lower PCSK9 concentrations (348). For example, Dallas Heart Study participants with an Y142X/C679X or R46L mutation had, respectively, 60% and 34% lower plasma PCSK9 levels than those had participants with no mutation (P < 0.0001).
At the population level, the two African mutations/polymorphisms (i.e., Y142X and C679X) have remarkably similar effects on LDL-C levels (Table 8), the average drop being ∼35-38 mg/dl compared with ∼20 mg/dl in Europeans/Americans with the R46L allele (Table 8). Encouragingly, the GLGC has now replicated the evidence for association between the rare alleles at SNP sites rs505151 (E670G) and rs11206510 (upstream) and LDL-C levels (Table 8), and perhaps more importantly, it has provided a platform for the first time on which to compare their potential effects on LDL-C levels relative to all other associated common variants across the entire genome in a single population, even for those genes with no prior connection to sterol and LDL metabolism (Table 10).
TABLE 10.
Additional genetic variants associated with LDL-cholesterol levels in the GLGC study
| Nearby Gene | SNP | Chr | Position | Minor allele (MAF) | Effect Size (SE)a | P | Identified in Previous Study? | Other Associated Traits |
| MOSC1 | rs2807834 | 1 | 219,037,216 | T (0.32) | - 1.09 (0.20) | 6 × 10 -11 | N | TC |
| IRF2BP2 | rs514230 | 1 | 232,925,220 | A (0.48) | - 1.13 (0.18) | 9 × 10 -12 | N | TC |
| TIMD4 | rs68882076 | 5 | 156,322,875 | T (0.35) | - 1.67 (0.19) | 2 × 10 -22 | Y | TC, TG |
| HFE | rs1800562 | 6 | 26,201,120 | A (0.06) | - 2.22 (0.39) | 6 × 10 -10 | N | TC |
| HLA | rs3277928 | 6 | 32,520,413 | A (0.16) | 1.83 (0.24) | 2 × 10 -15 | N | TC, TG |
| LPA | rs1564248 | 6 | 160,498,850 | C (0.17) | 1.95 (0.24) | 2 × 10 -17 | N | TC, HDL |
| DNAH11 | rs12670798 | 7 | 21,573,877 | C (0.23) | 1.26 (0.20) | 7 × 10 -10 | N | TC |
| PLEC1 | rs11136341 | 8 | 145,115,531 | G (0.40) | 1.40 (0.21) | 4 × 10 -13 | N | TC |
| ABO | rs649129 | 9 | 135,144,125 | T (0.22) | 2.05 (0.21) | 8 × 10 -22 | Y | TC |
| GPAM | rs1129555 | 10 | 113,900,711 | A (0.29) | 1.08 (0.20) | 2 × 10 -9 | N | TC |
| ST3GAL4 | rs11220462 | 11 | 125,749,162 | A (0.14) | 1.95 (0.26) | 1 × 10 -15 | N | TC |
| BRAP | rs11065987 | 12 | 110,556,807 | G (0.42) | - 0.97 (0.18) | 2 × 10 -9 | N | TC |
| HNF1A | rs1169288 | 12 | 119,901,033 | C (0.33) | 1.42 (0.19) | 1 × 10 -15 | Y | TC |
| NYNRIN | rs8017377 | 14 | 23,952,898 | T (0.48) | 1.17 (0.19) | 4 × 10 -11 | N | None |
| HPR | rs2000999 | 16 | 70,665,594 | A (0.20) | 3.00 (0.22) | 2 × 10 -22 | N | TC |
| OSBPL7 | rs7225700 | 17 | 42,746,803 | T (0.35) | - 0.87 (0.18) | 4 × 10 -9 | N | TC |
| CILP2 | rs10401969 | 19 | 19,268,718 | C (0.07) | - 3.11 (0.38) | 7 × 10 -22 | Y | TC, TG |
| MAFB | rs2902941 | 20 | 38,524,928 | G (0.33) | - 0.98 (0.19) | 1 × 10 -8 | Y | TC |
| TOP1 | rs6029526 | 20 | 39,370,229 | T (0.47) | 1.41 (0.17) | 3 × 10 -19 | N | TC |
Table does not include associated variants where triglyceride or HDL-cholesterol was the lead trait. Chr, chromosome; HDL, high-density lipoprotein cholesterol; MAF, minor allele frequency.
Effect size is expressed in mg/dl per copy of the minor allele.
Moderate impact of IDOL-mediated degradation of LDLR on LDL-C levels
In 2009, Zelcer et al. (349) showed that the sterol-responsive nuclear receptor liver X receptor (LXR) helps maintain cellular cholesterol homeostasis not only by promoting cholesterol efflux but by also suppressing cellular LDL uptake through transcriptional induction of IDOL expression. In brief, they (349) demonstrated that activated LXR increased IDOL mRNA levels in specific cell types and that in wild-type but not Ldlr− / − mice, (adenovirus-mediated) high-level hepatic expression of IDOL markedly reduced LDLR protein levels, and increased plasma concentrations of total and unesterified cholesterol ( ∼2-fold), LDL-C ( ∼6-fold), and apoB ( ∼3-fold). In comparison, PCSK9 levels were not altered. Notably, however, the totality of the data suggests that the LXR-IDOL pathway is more active in peripheral cells, such as macrophages and adrenal cells, than in hepatocytes, which may explain why in humans the estimated size effect of the rare allele at SNP site rs3757354 on LDL-C levels is considerably smaller than those observed for PCSK9, apoB, and LDLR (Table 8).
CONCLUDING COMMENT
In the past five years, our understanding of the biological and genetic bases of low and high LDL levels has been advanced by the acquisition of new insights into noncholesterol sterol and cholesterol absorption and elimination, apoB-containing lipoprotein production and intracellular trafficking, and the fine regulation of cellular cholesterol uptake via the LDLR. Genetic (e.g., in-depth sequence) analyses of patients with monogenic dyslipidemias and of individuals with plasma LDL-C levels at the extreme of the population distribution, as well as the identification of pathogenic mutations in diverse populations, have established important genotype-phenotype correlations. Independently, large GWAS studies have not only confirmed the role of genes previously known to affect plasma LDL-C levels but, more importantly, also identified a large set of previously unsuspected genes. Their impact on cholesterol metabolism and LDL-C levels is under active investigation. Indeed, recently developed mouse models have begun to provide some fascinating new knowledge about the cellular mechanisms contributing to the pathophysiology of cholesterol and apoB-containing lipoprotein diseases, thereby suggesting new molecular targets for hypolipidemic drugs. In the immediate future, new DNA sequencing technologies, such as exome sequencing, will provide a very powerful tool for identifying the causative mutations in those familial conditions of low and high LDL levels that are not explained by the usual suspects. The overall picture to emerge is that circulating LDL levels result from the output of a complex array of cellular activities and that even in people with conventional, straightforward, Mendelian clinical conditions of LDL (e.g., ADH and FHBL) metabolism, these conditions have a complex genetic basis. A major challenge will be to integrate the effects of lifestyle choices (e.g., diet, obesity, smoking) and epigenetic variation, often neglected in the study of genotype-phenotype correlations, with knowledge of DNA sequence determinants to arrive at a deeper understanding of the cause, course, treatment, and prevention of LDL dyslipidemias.
Acknowledgments
The authors thank Emma Duncan and Sophie Dean for their help in preparing this article.
Footnotes
Abbreviations:
- ABCG5
- ATP-binding cassette, subfamily G, member 5
- ABCG8
- ATP-binding cassette, subfamily G, member 8
- ABL
- abetalipoproteinemia
- ADH
- autosomal dominant hypercholesterolemia
- ANGPTL3
- angiopoietin-like 3
- ARH
- autosomal recessive hypercholesterolemia
- BMI
- body mass index
- CAC
- coronary artery calcification
- CAD
- coronary artery disease
- CHD
- coronary heart disease
- CMRD
- chylomicron retention disease
- CYP7A1
- cholesterol 7 α -hydroxylase
- EGF
- epidermal growth factor
- ER
- endoplasmic reticulum
- FCHL
- familial combined hyperlipidemia
- FDB
- familial defective apoB
- FH
- familial hypercholesterolemia
- FHBL
- familial hypobetalipoproteinemia
- GWAS
- genome-wide association study
- HMGCR
- HMG-CoA reductase
- IDOL
- inducible degrader of LDLR
- LD
- linkage disequilibrium
- LDL-C
- LDL-cholesterol
- LDLR
- LDL receptor
- LRP1, LDLRAP1
- LDLR-associated protein 1; LDLR-related protein 1
- LXR
- liver X receptor
- MI
- myocardial infarction
- MTP
- microsomal triglyceride transfer protein
- NPC1L1
- Niemann-Pick C1-like 1
- NS
- nonsynonymous
- PCSK9
- proprotein convertase subtilisin-like/kexin type 9
- SORT1
- Sortilin 1
- SNP
- single-nucleotide polymorphism
- SSL
- sitosterolemia
- TC
- total cholesterol
- TG
- triglyceride
- TRIB1
- tribbles homolog 1
- UTR
- untranslated region
This work was supported by grants from the University of Modena and Reggio Emilia and Fondazione Cassa di Risparmio di Modena (S.C. and P.T.); the British Heart Foundation (H.E.S. and C.C.S.); the Medical Research Council (C.C.S.); and the William Harvey Research Institute (C.C.S. and H.E.S.).
REFERENCES
- 1.Brezski R. J., Monroe J. G. 2007. B cell antigen receptor-induced Rac1 activation and Rac1-dependent spreading are impaired in transitional immature B cells due to levels of membrane cholesterol. J. Immunol. 179: 4464–4472 [DOI] [PubMed] [Google Scholar]
- 2.Pucadyil T. J., Chattopadhyay A. 2006. Role of cholesterol in the function and organization of G-protein coupled receptors. Prog. Lipid Res. 45: 295–333 [DOI] [PubMed] [Google Scholar]
- 3.Mistafa O., Stenius U. 2009. Statins inhibit Akt/PKB signaling via P2X7 receptor in pancreatic cancer cells. Biochem. Pharmacol. 78: 1115–1126 [DOI] [PubMed] [Google Scholar]
- 4.Sun Y., Ishibashi M., Seimon T., Lee M., Sharma S. M., Fitzgerald K. A., Samokhin A. O., Wang Y., Sayers S., Aikawa M., et al. 2009. Free cholesterol accumulation in macrophage membranes activates Toll-like receptors and p38 mitogen-activated protein kinase and induces cathepsin K. Circ. Res. 104: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridsdale A., Denis M., Gougeon P. Y., Ngsee J. K., Presley J. F., Zha X. 2006. Cholesterol is required for efficient endoplasmic reticulum-to-Golgi transport of secretory membrane proteins. Mol. Biol. Cell. 17: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glodowski D. R., Chen C. C., Schaefer H., Grant B. D., Rongo C. 2007. RAB-10 regulates glutamate receptor recycling in a cholesterol-dependent endocytosis pathway. Mol. Biol. Cell. 18: 4387–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippincott-Schwartz J., Phair R. D. 2010. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 39: 559–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vacca F., Giustizieri M., Ciotti M. T., Mercuri N. B., Volonte C. 2009. Rapid constitutive and ligand-activated endocytic trafficking of P2X receptor. J. Neurochem. 109: 1031–1041 [DOI] [PubMed] [Google Scholar]
- 9.Vrljic M., Nishimura S. Y., Moerner W. E., McConnell H. M. 2005. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys. J. 88: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feingold K. R. 2007. Thematic review series: skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J. Lipid Res. 48: 2531–2546 [DOI] [PubMed] [Google Scholar]
- 11.Ponce J., Brea D., Carrascal M., Guirao V., Degregorio-Rocasolano N., Sobrino T., Castillo J., Davalos A., Gasull T. 2010. The effect of simvastatin on the proteome of detergent-resistant membrane domains: decreases of specific proteins previously related to cytoskeleton regulation, calcium homeostasis and cell fate. Proteomics. 10: 1954–1965 [DOI] [PubMed] [Google Scholar]
- 12.Chrast R., Saher G., Nave K. A., Verheijen M. H. 2011. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J. Lipid Res. 52: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredrickson D. S., Lees R. S. 1965. A system for phenotyping hyperlipoproteinemia. Circulation. 31: 321–327 [DOI] [PubMed] [Google Scholar]
- 14.Porter F. D., Herman G. E. 2011. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52: 6–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abi-Mosleh L., Infante R. E., Radhakrishnan A., Goldstein J. L., Brown M. S. 2009. Cyclodextrin overcomes deficient lysosome-to-endoplasmic reticulum transport of cholesterol in Niemann-Pick type C cells. Proc. Natl. Acad. Sci. USA. 106: 19316–19321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum A. I., Maxfield F. R. 2011. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J. Neurochem. 116: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garver W. S., Jelinek D., Meaney F. J., Flynn J., Pettit K. M., Shepherd G., Heidenreich R. A., Vockley C. M., Castro G., Francis G. A. 2010. The National Niemann-Pick Type C1 Disease Database: correlation of lipid profiles, mutations, and biochemical phenotypes. J. Lipid Res. 51: 406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. 2007. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J. Clin. Invest. 117: 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Chang C. C., Westover E. J., Covey D. F., Chang T. Y. 2005. Investigating the allosterism of acyl-CoA:cholesterol acyltransferase (ACAT) by using various sterols: in vitro and intact cell studies. Biochem. J. 391: 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gylling H., Miettinen T. A. 2002. Inheritance of cholesterol metabolism of probands with high or low cholesterol absorption. J. Lipid Res. 43: 1472–1476 [DOI] [PubMed] [Google Scholar]
- 21.Simonen P. P., Gylling H., Miettinen T. A. 2007. The distribution of squalene and non-cholesterol sterols in lipoproteins in type 2 diabetes. Atherosclerosis. 194: 222–229 [DOI] [PubMed] [Google Scholar]
- 22.Wiesner P., Leidl K., Boettcher A., Schmitz G., Liebisch G. 2009. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J. Lipid Res. 50: 574–585 [DOI] [PubMed] [Google Scholar]
- 23.März W., Siekmeier R., Scharnagl H., Seiffert U. B., Gross W. 1993. Fast lipoprotein chromatography: new method of analysis for plasma lipoproteins. Clin. Chem. 39: 2276–2281 [PubMed] [Google Scholar]
- 24.Ståhlman M., Davidsson P., Kanmert I., Rosengren B., Borén J., Fagerberg B., Camejo G. 2008. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 49: 481–490 [DOI] [PubMed] [Google Scholar]
- 25.Gregg R. E., Connor W. E., Lin D. S., Brewer H. B., Jr 1986. Abnormal metabolism of shellfish sterols in a patient with sitosterolemia and xanthomatosis. J. Clin. Invest. 77: 1864–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salen G., Tint G. S., Shefer S., Shore V., Nguyen L. 1992. Increased sitosterol absorption is offset by rapid elimination to prevent accumulation in heterozygotes with sitosterolemia. Arterioscler. Thromb. 12: 563–568 [DOI] [PubMed] [Google Scholar]
- 27.Narushima K., Takada T., Yamanashi Y., Suzuki H. 2008. Niemann-pick C1-like 1 mediates alpha-tocopherol transport. Mol. Pharmacol. 74: 42–49 [DOI] [PubMed] [Google Scholar]
- 28.Duan L. P., Wang H. H., Wang D. Q. 2004. Cholesterol absorption is mainly regulated by the jejunal and ileal ATP-binding cassette sterol efflux transporters Abcg5 and Abcg8 in mice. J. Lipid Res. 45: 1312–1323 [DOI] [PubMed] [Google Scholar]
- 29.Plösch T., Bloks V. W., Terasawa Y., Berdy S., Siegler K., Van Der Sluijs F., Kema I. P., Groen A. K., Shan B., Kuipers F., et al. 2004. Sitosterolemia in ABC-transporter G5-deficient mice is aggravated on activation of the liver-X receptor. Gastroenterology. 126: 290–300 [DOI] [PubMed] [Google Scholar]
- 30.Wang H. H., Patel S. B., Carey M. C., Wang D. Q. 2007. Quantifying anomalous intestinal sterol uptake, lymphatic transport, and biliary secretion in Abcg8( 2 / 2 ) mice. Hepatology. 45: 998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L., von Bergmann K., Lutjohann D., Hobbs H. H., Cohen J. C. 2004. Selective sterol accumulation in ABCG5/ABCG8-deficient mice. J. Lipid Res. 45: 301–307 [DOI] [PubMed] [Google Scholar]
- 32.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altmann S. W., Davis H. R., Jr, Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204 [DOI] [PubMed] [Google Scholar]
- 34.Sané A. T., Sinnett D., Delvin E., Bendayan M., Marcil V., Menard D., Beaulieu J. F., Levy E. 2006. Localization and role of NPC1L1 in cholesterol absorption in human intestine. J. Lipid Res. 47: 2112–2120 [DOI] [PubMed] [Google Scholar]
- 35.Brown J. M., Rudel L. L., Yu L. 2007. NPC1L1 (Niemann-Pick C1-like 1) mediates sterol-specific unidirectional transport of non-esterified cholesterol in McArdle-RH7777 hepatoma cells. Biochem. J. 406: 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ezzet F., Wexler D., Statkevich P., Kosoglou T., Patrick J., Lipka L., Mellars L., Veltri E., Batra V. 2001. The plasma concentration and LDL-C relationship in patients receiving ezetimibe. J. Clin. Pharmacol. 41: 943–949 [DOI] [PubMed] [Google Scholar]
- 37.Sudhop T., Lutjohann D., Kodal A., Igel M., Tribble D. L., Shah S., Perevozskaya I., von Bergmann K. 2002. Inhibition of intestinal cholesterol absorption by ezetimibe in humans. Circulation. 106: 1943–1948 [DOI] [PubMed] [Google Scholar]
- 38.Foody J. M., Brown W. V., Zieve F., Adewale A. J., Flaim D., Lowe R. S., Jones-Burton C., Tershakovec A. M. 2010. Safety and efficacy of ezetimibe/simvastatin combination versus atorvastatin alone in adults ≥ 65 years of age with hypercholesterolemia and with or at moderately high/high risk for coronary heart disease (the VYTELD study). Am. J. Cardiol. 106: 1255–1263 [DOI] [PubMed] [Google Scholar]
- 39.Howard W. J. 2010. The role of ezetimibe in the prevention of cardiovascular disease: where do we stand after ARBITER 6-HALTS. Nutr. Metab. Cardiovasc. Dis. 20: 295–300 [DOI] [PubMed] [Google Scholar]
- 40.Lioudaki E., Ganotakis E. S., Mikhailidis D. P. 2011. Ezetimibe; more than a low density lipoprotein cholesterol lowering drug? An update after 4 years. Curr. Vasc. Pharmacol. 9: 62–86 [DOI] [PubMed] [Google Scholar]
- 41.Clarenbach J. J., Reber M., Lutjohann D., von Bergmann K., Sudhop T. 2006. The lipid-lowering effect of ezetimibe in pure vegetarians. J. Lipid Res. 47: 2820–2824 [DOI] [PubMed] [Google Scholar]
- 42.Davis H. R., Jr, Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. 2004. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J. Biol. Chem. 279: 33586–33592 [DOI] [PubMed] [Google Scholar]
- 43.Tang W., Ma Y., Jia L., Ioannou Y. A., Davies J. P., Yu L. 2009. Genetic inactivation of NPC1L1 protects against sitosterolemia in mice lacking ABCG5/ABCG8. J. Lipid Res. 50: 293–300 [DOI] [PubMed] [Google Scholar]
- 44.Bosner M. S., Lange L. G., Stenson W. F., Ostlund R. E., Jr 1999. Percent cholesterol absorption in normal women and men quantified with dual stable isotopic tracers and negative ion mass spectrometry. J. Lipid Res. 40: 302–308 [PubMed] [Google Scholar]
- 45.Miettinen T. A., Klett E. L., Gylling H., Isoniemi H., Patel S. B. 2006. Liver transplantation in a patient with sitosterolemia and cirrhosis. Gastroenterology. 130: 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boomsma D. I., Princen H. M., Frants R. R., Gevers Leuven J. A., Kempen H. J. 2003. Genetic analysis of indicators of cholesterol synthesis and absorption: lathosterol and phytosterols in Dutch twins and their parents. Twin Res. 6: 307–314 [DOI] [PubMed] [Google Scholar]
- 47.Cohen J. C., Pertsemlidis A., Fahmi S., Esmail S., Vega G. L., Grundy S. M., Hobbs H. H. 2006. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc. Natl. Acad. Sci. USA. 103: 1810–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polisecki E., Peter I., Simon J. S., Hegele R. A., Robertson M., Ford I., Shepherd J., Packard C., Jukema J. W., de Craen A. J., et al. 2010. Genetic variation at the NPC1L1 gene locus, plasma lipoproteins, and heart disease risk in the elderly. J. Lipid Res. 51: 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miettinen T. A., Tilvis R. S., Kesaniemi Y. A. 1990. Serum plant sterols and cholesterol precursors reflect cholesterol absorption and synthesis in volunteers of a randomly selected male population. Am. J. Epidemiol. 131: 20–31 [DOI] [PubMed] [Google Scholar]
- 50.Fahmi S., Yang C., Esmail S., Hobbs H. H., Cohen J. C. 2008. Functional characterization of genetic variants in NPC1L1 supports the sequencing extremes strategy to identify complex trait genes. Hum. Mol. Genet. 17: 2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L. J., Wang J., Li N., Ge L., Li B. L., Song B. L. 2011. Molecular characterization of the NPC1L1 variants identified from cholesterol low absorbers. J. Biol. Chem. 286: 7397–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. 2008. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 7: 508–519 [DOI] [PubMed] [Google Scholar]
- 53.Simon J. S., Karnoub M. C., Devlin D. J., Arreaza M. G., Qiu P., Monks S. A., Severino M. E., Deutsch P., Palmisano J., Sachs A. B., et al. 2005. Sequence variation in NPC1L1 and association with improved LDL-cholesterol lowering in response to ezetimibe treatment. Genomics. 86: 648–656 [DOI] [PubMed] [Google Scholar]
- 54.Pisciotta L., Fasano T., Bellocchio A., Bocchi L., Sallo R., Fresa R., Colangeli I., Cantafora A., Calandra S., Bertolini S. 2007. Effect of ezetimibe coadministered with statins in genotype-confirmed heterozygous FH patients. Atherosclerosis. 194: e116–e122 [DOI] [PubMed] [Google Scholar]
- 55.Iwayanagi Y., Takada T., Suzuki H. 2008. HNF4alpha is a crucial modulator of the cholesterol-dependent regulation of NPC1L1. Pharm. Res. 25: 1134–1141 [DOI] [PubMed] [Google Scholar]
- 56.Pramfalk C., Jiang Z. Y., Cai Q., Hu H., Zhang S. D., Han T. Q., Eriksson M., Parini P. 2010. HNF1alpha and SREBP2 are important regulators of NPC1L1 in human liver. J. Lipid Res. 51: 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tremblay A. J., Lamarche B., Lemelin V., Hoos L., Benjannet S., Seidah N. G., Davis H. R., Jr, Couture P. 2011. Atorvastatin increases intestinal expression of NPC1L1 in hyperlipidemic men. J. Lipid Res. 52: 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. 2000. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 290: 1771–1775 [DOI] [PubMed] [Google Scholar]
- 59.Lee M. H., Lu K., Hazard S., Yu H., Shulenin S., Hidaka H., Kojima H., Allikmets R., Sakuma N., Pegoraro R., et al. 2001. Identification of a gene, ABCG5, important in the regulation of dietary cholesterol absorption. Nat. Genet. 27: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Graf G. A., Yu L., Li W. P., Gerard R., Tuma P. L., Cohen J. C., Hobbs H. H. 2003. ABCG5 and ABCG8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J. Biol. Chem. 278: 48275–48282 [DOI] [PubMed] [Google Scholar]
- 61.Vrins C., Vink E., Vandenberghe K. E., Frijters R., Seppen J., Groen A. K. 2007. The sterol transporting heterodimer ABCG5/ABCG8 requires bile salts to mediate cholesterol efflux. FEBS Lett. 581: 4616–4620 [DOI] [PubMed] [Google Scholar]
- 62.Klett E. L., Lee M. H., Adams D. B., Chavin K. D., Patel S. B. 2004. Localization of ABCG5 and ABCG8 proteins in human liver, gall bladder and intestine. BMC Gastroenterol. 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tauscher A., Kuver R. 2003. ABCG5 and ABCG8 are expressed in gallbladder epithelial cells. Biochem. Biophys. Res. Commun. 307: 1021–1028 [DOI] [PubMed] [Google Scholar]
- 64.Lütjohann D., Björkhem I., Beil U. F., von Bergmann K. 1995. Sterol absorption and sterol balance in phytosterolemia evaluated by deuterium-labeled sterols: effect of sitostanol treatment. J. Lipid Res. 36: 1763–1773 [PubMed] [Google Scholar]
- 65.Salen G., Batta A. K., Tint G. S., Shefer S., Ness G. C. 1994. Inverse relationship between plasma cholestanol concentrations and bile acid synthesis in sitosterolemia. J. Lipid Res. 35: 1878–1887 [PubMed] [Google Scholar]
- 66.Igel M., Giesa U., Lutjohann D., von Bergmann K. 2003. Comparison of the intestinal uptake of cholesterol, plant sterols, and stanols in mice. J. Lipid Res. 44: 533–538 [DOI] [PubMed] [Google Scholar]
- 67.Lam C. W., Cheng A. W., Tong S. F., Chan Y. W. 2002. Novel donor splice site mutation of ABCG5 gene in sitosterolemia. Mol. Genet. Metab. 75: 178–180 [DOI] [PubMed] [Google Scholar]
- 68.Wang J., Joy T., Mymin D., Frohlich J., Hegele R. A. 2004. Phenotypic heterogeneity of sitosterolemia. J. Lipid Res. 45: 2361–2367 [DOI] [PubMed] [Google Scholar]
- 69.Su Y., Wang Z., Yang H., Cao L., Liu F., Bai X., Ruan C. 2006. Clinical and molecular genetic analysis of a family with sitosterolemia and co-existing erythrocyte and platelet abnormalities. Haematologica. 91: 1392–1395 [PubMed] [Google Scholar]
- 70.Togo M., Hashimoto Y., Iso O. N., Kurano M., Hara M., Kadowaki T., Koike K., Tsukamoto K. 2009. Identification of a novel mutation for phytosterolemia. Genetic analyses of 2 cases. Clin. Chim. Acta. 401: 165–169 [DOI] [PubMed] [Google Scholar]
- 71.Niu D. M., Chong K. W., Hsu J. H., Wu T. J., Yu H. C., Huang C. H., Lo M. Y., Kwok C. F., Kratz L. E., Ho L. T. 2010. Clinical observations, molecular genetic analysis, and treatment of sitosterolemia in infants and children. J. Inherit. Metab. Dis. 33: 437–443 [DOI] [PubMed] [Google Scholar]
- 72.Salen G., Horak I., Rothkopf M., Cohen J. L., Speck J., Tint G. S., Shore V., Dayal B., Chen T., Shefer S. 1985. Lethal atherosclerosis associated with abnormal plasma and tissue sterol composition in sitosterolemia with xanthomatosis. J. Lipid Res. 26: 1126–1133 [PubMed] [Google Scholar]
- 73.Hubacek J. A., Berge K. E., Cohen J. C., Hobbs H. H. 2001. Mutations in ATP-cassette binding proteins G5 (ABCG5) and G8 (ABCG8) causing sitosterolemia. Hum. Mutat. 18: 359–360 [DOI] [PubMed] [Google Scholar]
- 74.Heimer S., Tezak D. 2002. Structure of polydispersed colloids characterised by light scattering and electron microscopy. Adv. Colloid Interface Sci. 98: 1–23 [DOI] [PubMed] [Google Scholar]
- 75.Katayama T., Satoh T., Yagi T., Hirose N., Kurita Y., Anzai T., Asakura Y., Yoshikawa T., Mitamura H., Ogawa S. 2003. A 19-year-old man with myocardial infarction and sitosterolemia. Intern. Med. 42: 591–594 [DOI] [PubMed] [Google Scholar]
- 76.Mushtaq T., Wales J. K., Wright N. P. 2007. Adrenal insufficiency in phytosterolaemia. Eur. J. Endocrinol. 157(Suppl. 1): S61–S65 [DOI] [PubMed] [Google Scholar]
- 77.Rees D. C., Iolascon A., Carella M., O'Marcaigh A. S., Kendra J. R., Jowitt S. N., Wales J. K., Vora A., Makris M., Manning N., et al. 2005. Stomatocytic haemolysis and macrothrombocytopenia (Mediterranean stomatocytosis/macrothrombocytopenia) is the haematological presentation of phytosterolaemia. Br. J. Haematol. 130: 297–309 [DOI] [PubMed] [Google Scholar]
- 78.Bhattacharyya A. K., Connor W. E. 1974. Beta-sitosterolemia and xanthomatosis. A newly described lipid storage disease in two sisters. J. Clin. Invest. 53: 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rios J., Stein E., Shendure J., Hobbs H. H., Cohen J. C. 2010. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum. Mol. Genet. 19: 4313–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen L. B., Shefer S., Salen G., Horak I., Tint G. S., McNamara D. J. 1988. The effect of abnormal plasma and cellular sterol content and composition on low density lipoprotein uptake and degradation by monocytes and lymphocytes in sitosterolemia with xanthomatosis. Metabolism. 37: 346–351 [DOI] [PubMed] [Google Scholar]
- 81.Heimerl S., Langmann T., Moehle C., Mauerer R., Dean M., Beil F. U., von Bergmann K., Schmitz G. 2002. Mutations in the human ATP-binding cassette transporters ABCG5 and ABCG8 in sitosterolemia. Hum. Mutat. 20: 151. [DOI] [PubMed] [Google Scholar]
- 82.Cheng W. F., Yuen Y. P., Chow C. B., Au K. M., Chan Y. W., Tam S. C. 2003. Sitosterolaemia and xanthomatosis in a child. Hong Kong Med. J. 9: 206–209 [PubMed] [Google Scholar]
- 83.Sehayek E., Yu H. J., von Bergmann K., Lutjohann D., Stoffel M., Duncan E. M., Garcia-Naveda L., Salit J., Blundell M. L., Friedman J. M., et al. 2004. Phytosterolemia on the island of Kosrae: founder effect for a novel ABCG8 mutation results in high carrier rate and increased plasma plant sterol levels. J. Lipid Res. 45: 1608–1613 [DOI] [PubMed] [Google Scholar]
- 84.Mannucci L., Guardamagna O., Bertucci P., Pisciotta L., Liberatoscioli L., Bertolini S., Irace C., Gnasso A., Federici G., Cortese C. 2007. Beta-sitosterolaemia: a new nonsense mutation in the ABCG5 gene. Eur. J. Clin. Invest. 37: 997–1000 [DOI] [PubMed] [Google Scholar]
- 85.Lu K., Lee M. H., Hazard S., Brooks-Wilson A., Hidaka H., Kojima H., Ose L., Stalenhoef A. F., Mietinnen T., Bjorkhem I., et al. 2001. Two genes that map to the STSL locus cause sitosterolemia: genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by ABCG5 and ABCG8, respectively. Am. J. Hum. Genet. 69: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. 2008. Structural requirements for PCSK9-mediated degradation of the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 105: 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson B. J., Lee J. Y., Pickert A., Urbatsch I. L. 2010. Bile acids stimulate ATP hydrolysis in the purified cholesterol transporter ABCG5/G8. Biochemistry. 49: 3403–3411 [DOI] [PubMed] [Google Scholar]
- 88.Graf G. A., Cohen J. C., Hobbs H. H. 2004. Missense mutations in ABCG5 and ABCG8 disrupt heterodimerization and trafficking. J. Biol. Chem. 279: 24881–24888 [DOI] [PubMed] [Google Scholar]
- 89.Kenny E. E., Gusev A., Riegel K., Lutjohann D., Lowe J. K., Salit J., Maller J. B., Stoffel M., Daly M. J., Altshuler D. M., et al. 2009. Systematic haplotype analysis resolves a complex plasma plant sterol locus on the Micronesian Island of Kosrae. Proc. Natl. Acad. Sci. USA. 106: 13886–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Honda A., Salen G., Nguyen L. B., Tint G. S., Batta A. K., Shefer S. 1998. Down-regulation of cholesterol biosynthesis in sitosterolemia: diminished activities of acetoacetyl-CoA thiolase, 3-hydroxy-3-methylglutaryl-CoA synthase, reductase, squalene synthase, and 7-dehydrocholesterol delta7-reductase in liver and mononuclear leukocytes. J. Lipid Res. 39: 44–50 [PubMed] [Google Scholar]
- 91.Nguyen L., Salen G., Shefer S., Shore V., Tint G. S., Ness G. 1990. Unexpected failure of bile acid malabsorption to stimulate cholesterol synthesis in sitosterolemia with xanthomatosis. Comparison with lovastatin. Arteriosclerosis. 10: 289–297 [DOI] [PubMed] [Google Scholar]
- 92.Nguyen L. B., Cobb M., Shefer S., Salen G., Ness G. C., Tint G. S. 1991. Regulation of cholesterol biosynthesis in sitosterolemia: effects of lovastatin, cholestyramine, and dietary sterol restriction. J. Lipid Res. 32: 1941–1948 [PubMed] [Google Scholar]
- 93.Nguyen L. B., Shefer S., Salen G., Ness G. C., Tint G. S., Zaki F. G., Rani I. 1990. A molecular defect in hepatic cholesterol biosynthesis in sitosterolemia with xanthomatosis. J. Clin. Invest. 86: 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berge K. E., von Bergmann K., Lutjohann D., Guerra R., Grundy S. M., Hobbs H. H., Cohen J. C. 2002. Heritability of plasma noncholesterol sterols and relationship to DNA sequence polymorphism in ABCG5 and ABCG8. J. Lipid Res. 43: 486–494 [PubMed] [Google Scholar]
- 95.Gylling H., Hallikainen M., Pihlajamaki J., Agren J., Laakso M., Rajaratnam R. A., Rauramaa R., Miettinen T. A. 2004. Polymorphisms in the ABCG5 and ABCG8 genes associate with cholesterol absorption and insulin sensitivity. J. Lipid Res. 45: 1660–1665 [DOI] [PubMed] [Google Scholar]
- 96.Jakulj L., Vissers M. N., Tanck M. W., Hutten B. A., Stellaard F., Kastelein J. J., Dallinga-Thie G. M. 2010. ABCG5/G8 polymorphisms and markers of cholesterol metabolism: systematic review and meta-analysis. J. Lipid Res. 51: 3016–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teupser D., Baber R., Ceglarek U., Scholz M., Illig T., Gieger C., Holdt L. M., Leichtle A., Greiser K. H., Huster D., et al. 2010. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circ. Cardiovasc. Genet. 3: 331–339 [DOI] [PubMed] [Google Scholar]
- 98.Buch S., Schafmayer C., Volzke H., Becker C., Franke A., von Eller-Eberstein H., Kluck C., Bassmann I., Brosch M., Lammert F., et al. 2007. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat. Genet. 39: 995–999 [DOI] [PubMed] [Google Scholar]
- 99.Grünhage F., Acalovschi M., Tirziu S., Walier M., Wienker T. F., Ciocan A., Mosteanu O., Sauerbruch T., Lammert F. 2007. Increased gallstone risk in humans conferred by common variant of hepatic ATP-binding cassette transporter for cholesterol. Hepatology. 46: 793–801 [DOI] [PubMed] [Google Scholar]
- 100.Katsika D., Magnusson P., Krawczyk M., Grunhage F., Lichtenstein P., Einarsson C., Lammert F., Marschall H. U. 2010. Gallstone disease in Swedish twins: risk is associated with ABCG8 D19H genotype. J. Intern. Med. 268: 279–285 [DOI] [PubMed] [Google Scholar]
- 101.Kuo K. K., Shin S. J., Chen Z. C., Yang Y. H., Yang J. F., Hsiao P. J. 2008. Significant association of ABCG5 604Q and ABCG8 D19H polymorphisms with gallstone disease. Br. J. Surg. 95: 1005–1011 [DOI] [PubMed] [Google Scholar]
- 102.Siddapuram S. P., Mahurkar S., Duvvuru N. R., Mitnala S., Guduru V. R., Rebala P., Mansard M. J. 2010. Hepatic cholesterol transporter ABCG8 polymorphisms in gallstone disease in an Indian population. J. Gastroenterol. Hepatol. 25: 1093–1098 [DOI] [PubMed] [Google Scholar]
- 103.Stender S., Frikke-Schmidt R., Nordestgaard B. G., Tybjaerg-Hansen A. 2011. Sterol transporter adenosine triphosphate-binding cassette transporter G8, gallstones, and biliary cancer in 62,000 individuals from the general population. Hepatology. 53: 640–648 [DOI] [PubMed] [Google Scholar]
- 104.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmidt D. R., Holmstrom S. R., Fon Tacer K., Bookout A. L., Kliewer S. A., Mangelsdorf D. J. 2010. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J. Biol. Chem. 285: 14486–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tint G. S., Irons M., Elias E. R., Batta A. K., Frieden R., Chen T. S., Salen G. 1994. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N. Engl. J. Med. 330: 107–113 [DOI] [PubMed] [Google Scholar]
- 107.Burkhardt R., Kenny E. E., Lowe J. K., Birkeland A., Josowitz R., Noel M., Salit J., Maller J. B., Pe'er I., Daly M. J., et al. 2008. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler. Thromb. Vasc. Biol. 28: 2078–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hiura Y., Tabara Y., Kokubo Y., Okamura T., Goto Y., Nonogi H., Miki T., Tomoike H., Iwai N. 2010. Association of the functional variant in the 3-hydroxy-3-methylglutaryl-coenzyme a reductase gene with low-density lipoprotein-cholesterol in Japanese. Circ. J. 74: 518–522 [DOI] [PubMed] [Google Scholar]
- 109.Fisher E. A., Khanna N. A., McLeod R. S. 2011. Ubiquitination regulates the assembly of VLDL in HepG2 cells and is the committing step of the apoB-100 ERAD pathway. J. Lipid Res. 52: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rutledge A. C., Su Q., Adeli K. 2010. Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem. Cell Biol. 88: 251–267 [DOI] [PubMed] [Google Scholar]
- 111.Qiu W., Zhang J., Dekker M. J., Wang H., Huang J., Brumell J. H., Adeli K. 2011. Hepatic autophagy mediates endoplasmic reticulum stress-induced degradation of misfolded apolipoprotein B. Hepatology. 53: 1515–1525 [DOI] [PubMed] [Google Scholar]
- 112.Jiang Z. G., Carraway M., McKnight C. J. 2005. Limited proteolysis and biophysical characterization of the lipovitellin homology region in apolipoprotein B. Biochemistry. 44: 1163–1173 [DOI] [PubMed] [Google Scholar]
- 113.Jiang Z. G., Gantz D., Bullitt E., McKnight C. J. 2006. Defining lipid-interacting domains in the N-terminal region of apolipoprotein B. Biochemistry. 45: 11799–11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang L., Jiang Z. G., McKnight C. J., Small D. M. 2010. Interfacial properties of apolipoprotein B292-593 (B6.4-13) and B611-782 (B13-17). Insights into the structure of the lipovitellin homology region in apolipoprotein B. Biochemistry. 49: 3898–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ledford A. S., Cook V. A., Shelness G. S., Weinberg R. B. 2009. Structural and dynamic interfacial properties of the lipoprotein initiating domain of apolipoprotein B. J. Lipid Res. 50: 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Manchekar M., Richardson P. E., Sun Z., Liu Y., Segrest J. P., Dashti N. 2008. Charged amino acid residues 997-1000 of human apolipoprotein B100 are critical for the initiation of lipoprotein assembly and the formation of a stable lipidated primordial particle in McA-RH7777 cells. J. Biol. Chem. 283: 29251–29265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carraway M., Herscovitz H., Zannis V., Small D. M. 2000. Specificity of lipid incorporation is determined by sequences in the N-terminal 37 of apoB. Biochemistry. 39: 9737–9745 [DOI] [PubMed] [Google Scholar]
- 118.Ren G., Rudenko G., Ludtke S. J., Deisenhofer J., Chiu W., Pownall H. J. 2010. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proc. Natl. Acad. Sci. USA. 107: 1059–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iglesias P., Devora O., Garcia J., Tajada P., Garcia-Arevalo C., Diez J. J. 2010. Severe hyperthyroidism: aetiology, clinical features and treatment outcome. Clin. Endocrinol. (Oxf.). 72: 551–557 [DOI] [PubMed] [Google Scholar]
- 120.Huang L. S., Kayden H., Sokol R. J., Breslow J. L. 1991. ApoB gene nonsense and splicing mutations in a compound heterozygote for familial hypobetalipoproteinemia. J. Lipid Res. 32: 1341–1348 [PubMed] [Google Scholar]
- 121.Najah M., Di Leo E., Awatef J., Magnolo L., Imene J., Pinotti E., Bahri M., Barsaoui S., Brini I., Fekih M., et al. 2009. Identification of patients with abetalipoproteinemia and homozygous familial hypobetalipoproteinemia in Tunisia. Clin. Chim. Acta. 401: 51–56 [DOI] [PubMed] [Google Scholar]
- 122.Aguilar-Salinas C. A., Barrett P. H., Parhofer K. G., Young S. G., Tessereau D., Bateman J., Quinn C., Schonfeld G. 1995. Apoprotein B-100 production is decreased in subjects heterozygous for truncations of apoprotein B. Arterioscler. Thromb. Vasc. Biol. 15: 71–80 [DOI] [PubMed] [Google Scholar]
- 123.Elias N., Patterson B. W., Schonfeld G. 1999. Decreased production rates of VLDL triglycerides and ApoB-100 in subjects heterozygous for familial hypobetalipoproteinemia. Arterioscler. Thromb. Vasc. Biol. 19: 2714–2721 [DOI] [PubMed] [Google Scholar]
- 124.Latour M. A., Patterson B. W., Pulai J., Chen Z., Schonfeld G. 1997. Metabolism of apolipoprotein B-100 in a kindred with familial hypobetalipoproteinemia without a truncated form of apoB. J. Lipid Res. 38: 592–599 [PubMed] [Google Scholar]
- 125.Hooper A. J., Robertson K., Barrett P. H., Parhofer K. G., van Bockxmeer F. M., Burnett J. R. 2007. Postprandial lipoprotein metabolism in familial hypobetalipoproteinemia. J. Clin. Endocrinol. Metab. 92: 1474–1478 [DOI] [PubMed] [Google Scholar]
- 126.Lancellotti S., Di Leo E., Penacchioni J. Y., Balli F., Viola L., Bertolini S., Calandra S., Tarugi P. 2004. Hypobetalipoproteinemia with an apparently recessive inheritance due to a “de novo” mutation of apolipoprotein B. Biochim. Biophys. Acta. 1688: 61–67 [DOI] [PubMed] [Google Scholar]
- 127.Tarugi P., Lonardo A., Gabelli C., Sala F., Ballarini G., Cortella I., Previato L., Bertolini S., Cordera R., Calandra S. 2001. Phenotypic expression of familial hypobetalipoproteinemia in three kindreds with mutations of apolipoprotein B gene. J. Lipid Res. 42: 1552–1561 [PubMed] [Google Scholar]
- 128.Ruotolo G., Zanelli T., Tettamanti C., Ragogna F., Parlavecchia M., Vigano F., Catapano A. L. 1998. Hypobetalipoproteinemia associated with apo B-48.4, a truncated protein only 14 amino acids longer than apo B-48. Atherosclerosis. 137: 125–131 [DOI] [PubMed] [Google Scholar]
- 129.Takahashi K., Hikita M., Taira K., Kobayashi J., Bujo H., Saito Y. 2001. Clinical characterization of a case with familial hypobetalipoproteinemia caused by apo B-76, a new truncation of apolipoprotein B, combined with apo E2/E2 phenotype. Intern. Med. 40: 1015–1019 [DOI] [PubMed] [Google Scholar]
- 130.Talmud P. J., Krul E. S., Pessah M., Gay G., Schonfeld G., Humphries S. E., Infante R. 1994. Donor splice mutation generates a lipid-associated apolipoprotein B-27.6 in a patient with homozygous hypobetalipoproteinemia. J. Lipid Res. 35: 468–477 [PubMed] [Google Scholar]
- 131.Ohashi K., Ishibashi S., Yamamoto M., Osuga J., Yazaki Y., Yukawa S., Yamada N. 1998. A truncated species of apolipoprotein B (B-38.7) in a patient with homozygous hypobetalipoproteinemia associated with diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 18: 1330–1334 [DOI] [PubMed] [Google Scholar]
- 132.Young S. G., Bihain B., Flynn L. M., Sanan D. A., Ayrault-Jarrier M., Jacotot B. 1994. Asymptomatic homozygous hypobetalipoproteinemia associated with apolipoprotein B45.2. Hum. Mol. Genet. 3: 741–744 [DOI] [PubMed] [Google Scholar]
- 133.Lancellotti S., Zaffanello M., Di Leo E., Costa L., Lonardo A., Tarugi P. 2005. Pediatric gallstone disease in familial hypobetalipoproteinemia. J. Hepatol. 43: 188–191 [DOI] [PubMed] [Google Scholar]
- 134.Krul E. S., Kinoshita M., Talmud P., Humphries S. E., Turner S., Goldberg A. C., Cook K., Boerwinkle E., Schonfeld G. 1989. Two distinct truncated apolipoprotein B species in a kindred with hypobetalipoproteinemia. Arteriosclerosis. 9: 856–868 [DOI] [PubMed] [Google Scholar]
- 135.Young S. G., Hubl S. T., Chappell D. A., Smith R. S., Claiborne F., Snyder S. M., Terdiman J. F. 1989. Familial hypobetalipoproteinemia associated with a mutant species of apolipoprotein B (B-46). N. Engl. J. Med. 320: 1604–1610 [DOI] [PubMed] [Google Scholar]
- 136.Farese R. V., Jr, Garg A., Pierotti V. R., Vega G. L., Young S. G. 1992. A truncated species of apolipoprotein B, B-83, associated with hypobetalipoproteinemia. J. Lipid Res. 33: 569–577 [PubMed] [Google Scholar]
- 137.Pulai J. I., Averna M., Srivastava R. A., Latour M. A., Clouse R. E., Ostlund R. E., Schonfeld G. 1997. Normal intestinal dietary fat and cholesterol absorption, intestinal apolipoprotein B (ApoB) mRNA levels, and ApoB-48 synthesis in a hypobetalipoproteinemic kindred without any ApoB truncation. Metabolism. 46: 1095–1100 [DOI] [PubMed] [Google Scholar]
- 138.Pulai J. I., Zakeri H., Kwok P. Y., Kim J. H., Wu J., Schonfeld G. 1998. Donor splice mutation (665 + 1 G_T) in familial hypobetalipoproteinemia with no detectable apoB truncation. Am. J. Med. Genet. 80: 218–220 [PubMed] [Google Scholar]
- 139.Groenewegen W. A., Krul E. S., Averna M. R., Pulai J., Schonfeld G. 1994. Dysbetalipoproteinemia in a kindred with hypobetalipoproteinemia due to mutations in the genes for ApoB (ApoB-70.5) and ApoE (ApoE2). Arterioscler. Thromb. 14: 1695–1704 [DOI] [PubMed] [Google Scholar]
- 140.Nielsen L. B., Veniant M., Boren J., Raabe M., Wong J. S., Tam C., Flynn L., Vanni-Reyes T., Gunn M. D., Goldberg I. J., et al. 1998. Genes for apolipoprotein B and microsomal triglyceride transfer protein are expressed in the heart: evidence that the heart has the capacity to synthesize and secrete lipoproteins. Circulation. 98: 13–16 [DOI] [PubMed] [Google Scholar]
- 141.Gautier T., Becker S., Drouineaud V., Menetrier F., Sagot P., Nofer J. R., von Otte S., Lagrost L., Masson D., Tietge U. J. 2010. Human luteinized granulosa cells secrete apoB100-containing lipoproteins. J. Lipid Res. 51: 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaser A., Hava D. L., Dougan S. K., Chen Z., Zeissig S., Brenner M. B., Blumberg R. S. 2008. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur. J. Immunol. 38: 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Krzystanek M., Pedersen T. X., Bartels E. D., Kjaehr J., Straarup E. M., Nielsen L. B. 2010. Expression of apolipoprotein B in the kidney attenuates renal lipid accumulation. J. Biol. Chem. 285: 10583–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yue P., Isley W. L., Harris W. S., Rosipal S., Akin C. D., Schonfeld G. 2005. Genetic variants of ApoE account for variability of plasma low-density lipoprotein and apolipoprotein B levels in FHBL. Atherosclerosis. 178: 107–113 [DOI] [PubMed] [Google Scholar]
- 145.Ikewaki K., Cain W., Thomas F., Shamburek R., Zech L. A., Usher D., Brewer H. B., Jr, Rader D. J. 2004. Abnormal in vivo metabolism of apoB-containing lipoproteins in human apoE deficiency. J. Lipid Res. 45: 1302–1311 [DOI] [PubMed] [Google Scholar]
- 146.Mensenkamp A. R., Jong M. C., van Goor H., van Luyn M. J., Bloks V., Havinga R., Voshol P. J., Hofker M. H., van Dijk K. W., Havekes L. M., et al. 1999. Apolipoprotein E participates in the regulation of very low density lipoprotein-triglyceride secretion by the liver. J. Biol. Chem. 274: 35711–35718 [DOI] [PubMed] [Google Scholar]
- 147.Demant T., Bedford D., Packard C. J., Shepherd J. 1991. Influence of apolipoprotein E polymorphism on apolipoprotein B-100 metabolism in normolipemic subjects. J. Clin. Invest. 88: 1490–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gillard B. K., Lin H. Y., Massey J. B., Pownall H. J. 2009. Apolipoproteins A-I, A-II and E are independently distributed among intracellular and newly secreted HDL of human hepatoma cells. Biochim. Biophys. Acta. 1791: 1125–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gusarova V., Seo J., Sullivan M. L., Watkins S. C., Brodsky J. L., Fisher E. A. 2007. Golgi-associated maturation of very low density lipoproteins involves conformational changes in apolipoprotein B, but is not dependent on apolipoprotein E. J. Biol. Chem. 282: 19453–19462 [DOI] [PubMed] [Google Scholar]
- 150.Wang H., Gilham D., Lehner R. 2007. Proteomic and lipid characterization of apolipoprotein B-free luminal lipid droplets from mouse liver microsomes: implications for very low density lipoprotein assembly. J. Biol. Chem. 282: 33218–33226 [DOI] [PubMed] [Google Scholar]
- 151.Lin X., Chen Z., Yue P., Averna M. R., Ostlund R. E., Jr, Watson M. A., Schonfeld G. 2006. A targeted apoB38.9 mutation in mice is associated with reduced hepatic cholesterol synthesis and enhanced lipid peroxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G1170–G1176 [DOI] [PubMed] [Google Scholar]
- 152.Tarugi P., Averna M., Di Leo E., Cefalu A. B., Noto D., Magnolo L., Cattin L., Bertolini S., Calandra S. 2007. Molecular diagnosis of hypobetalipoproteinemia: an ENID review. Atherosclerosis. 195: e19–e27 [DOI] [PubMed] [Google Scholar]
- 153.Zhong S., Magnolo A. L., Sundaram M., Zhou H., Yao E. F., Di Leo E., Loria P., Wang S., Bamji-Mirza M., Wang L., et al. 2010. Nonsynonymous mutations within APOB in human familial hypobetalipoproteinemia: evidence for feedback inhibition of lipogenesis and postendoplasmic reticulum degradation of apolipoprotein B. J. Biol. Chem. 285: 6453–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Z., Fitzgerald R. L., Averna M. R., Schonfeld G. 2000. A targeted apolipoprotein B-38.9-producing mutation causes fatty livers in mice due to the reduced ability of apolipoprotein B-38.9 to transport triglycerides. J. Biol. Chem. 275: 32807–32815 [DOI] [PubMed] [Google Scholar]
- 155.Burnett J. R., Shan J., Miskie B. A., Whitfield A. J., Yuan J., Tran K., McKnight C. J., Hegele R. A., Yao Z. 2003. A novel nontruncating APOB gene mutation, R463W, causes familial hypobetalipoproteinemia. J. Biol. Chem. 278: 13442–13452 [DOI] [PubMed] [Google Scholar]
- 156.Noto D., Cefalu A. B., Cannizzaro A., Mina M., Fayer F., Valenti V., Barbagallo C. M., Tuttolomondo A., Pinto A., Sciume C., et al. 2009. Familial hypobetalipoproteinemia due to apolipoprotein B R463W mutation causes intestinal fat accumulation and low postprandial lipemia. Atherosclerosis. 206: 193–198 [DOI] [PubMed] [Google Scholar]
- 157.Benn M., Stene M. C., Nordestgaard B. G., Jensen G. B., Steffensen R., Tybjaerg-Hansen A. 2008. Common and rare alleles in apolipoprotein B contribute to plasma levels of low-density lipoprotein cholesterol in the general population. J. Clin. Endocrinol. Metab. 93: 1038–1045 [DOI] [PubMed] [Google Scholar]
- 158.Narcisi T. M., Shoulders C. C., Chester S. A., Read J., Brett D. J., Harrison G. B., Grantham T. T., Fox M. F., Povey S., de Bruin T. W., et al. 1995. Mutations of the microsomal triglyceride-transfer-protein gene in abetalipoproteinemia. Am. J. Hum. Genet. 57: 1298–1310 [PMC free article] [PubMed] [Google Scholar]
- 159.Berriot-Varoqueaux N., Aggerbeck L. P., Samson-Bouma M., Wetterau J. R. 2000. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu. Rev. Nutr. 20: 663–697 [DOI] [PubMed] [Google Scholar]
- 160.Chardon L., Sassolas A., Dingeon B., Michel-Calemard L., Bovier-Lapierre M., Moulin P., Lachaux A. 2009. Identification of two novel mutations and long-term follow-up in abetalipoproteinemia: a report of four cases. Eur. J. Pediatr. 168: 983–989 [DOI] [PubMed] [Google Scholar]
- 161.Zamel R., Khan R., Pollex R. L., Hegele R. A. 2008. Abetalipoproteinemia: two case reports and literature review. Orphanet J. Rare Dis. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Blum C. B., Deckelbaum R. J., Witte L. D., Tall A. R., Cornicelli J. 1982. Role of apolipoprotein E-containing lipoproteins in abetalipoproteinemia. J. Clin. Invest. 70: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ikewaki K., Rader D. J., Zech L. A., Brewer H. B., Jr 1994. In vivo metabolism of apolipoproteins A-I and E in patients with abetalipoproteinemia: implications for the roles of apolipoproteins B and E in HDL metabolism. J. Lipid Res. 35: 1809–1819 [PubMed] [Google Scholar]
- 164.Illingworth D. R., Connor W. E., Lin D. S., Diliberti J. 1980. Lipid metabolism in abetalipoproteinemia: a study of cholesterol absorption and sterol balance in two patients. Gastroenterology. 78: 68–75 [PubMed] [Google Scholar]
- 165.Kuriyama M., Yoshidome H., Nakahara K., Nakagawa H., Fujiyama J., Take H., Osame M. 1999. Blood dolichols in a patient with abetalipoproteinaemia. Ann. Clin. Biochem. 36: 176–179 [DOI] [PubMed] [Google Scholar]
- 166.Iqbal J., Rudel L. L., Hussain M. M. 2008. Microsomal triglyceride transfer protein enhances cellular cholesteryl esterification by relieving product inhibition. J. Biol. Chem. 283: 19967–19980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xie Y., Newberry E. P., Young S. G., Robine S., Hamilton R. L., Wong J. S., Luo J., Kennedy S., Davidson N. O. 2006. Compensatory increase in hepatic lipogenesis in mice with conditional intestine-specific Mttp deficiency. J. Biol. Chem. 281: 4075–4086 [DOI] [PubMed] [Google Scholar]
- 168.Mohler P. J., Zhu M. Y., Blade A. M., Ham A. J., Shelness G. S., Swift L. L. 2007. Identification of a novel isoform of microsomal triglyceride transfer protein. J. Biol. Chem. 282: 26981–26988 [DOI] [PubMed] [Google Scholar]
- 169.Goodman D. S., Deckelbaum R. J., Palmer R. H., Dell R. B., Ramakrishnan R., Delpre G., Beigel Y., Cooper M. 1983. Cholesterol turnover and metabolism in two patients with abetalipoproteinemia. J. Lipid Res. 24: 1605–1611 [PubMed] [Google Scholar]
- 170.Musunuru K., Pirruccello J. P., Do R., Peloso G. M., Guiducci C., Sougnez C., Garimella K. V., Fisher S., Abreu J., Barry A. J., et al. 2010. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 363: 2220–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Koishi R., Ando Y., Ono M., Shimamura M., Yasumo H., Fujiwara T., Horikoshi H., Furukawa H. 2002. Angptl3 regulates lipid metabolism in mice. Nat. Genet. 30: 151–157 [DOI] [PubMed] [Google Scholar]
- 172.Köster A., Chao Y. B., Mosior M., Ford A., Gonzalez-DeWhitt P. A., Hale J. E., Li D., Qiu Y., Fraser C. C., Yang D. D., et al. 2005. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 146: 4943–4950 [DOI] [PubMed] [Google Scholar]
- 173.Romeo S., Yin W., Kozlitina J., Pennacchio L. A., Boerwinkle E., Hobbs H. H., Cohen J. C. 2009. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 119: 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Moon H. D., Nakajima K., Kamiyama K., Takanashi K., Sakurabayashi I., Nagamine T. 2008. Higher frequency of abnormal serum angiopoietin-like protein 3 than abnormal cholesteryl ester transfer protein in Japanese hyperalphalipoproteinemic subjects. Clin. Chim. Acta. 398: 99–104 [DOI] [PubMed] [Google Scholar]
- 175.Liu J., Afroza H., Rader D. J., Jin W. 2010. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. J. Biol. Chem. 285: 27561–27570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shan L., Yu X. C., Liu Z., Hu Y., Sturgis L. T., Miranda M. L., Liu Q. 2009. The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. J. Biol. Chem. 284: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shimamura M., Matsuda M., Yasumo H., Okazaki M., Fujimoto K., Kono K., Shimizugawa T., Ando Y., Koishi R., Kohama T., et al. 2007. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler. Thromb. Vasc. Biol. 27: 366–372 [DOI] [PubMed] [Google Scholar]
- 178.Charcosset M., Sassolas A., Peretti N., Roy C. C., Deslandres C., Sinnett D., Levy E., Lachaux A. 2008. Anderson or chylomicron retention disease: molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol. Genet. Metab. 93: 74–84 [DOI] [PubMed] [Google Scholar]
- 179.Dannoura A. H., Berriot-Varoqueaux N., Amati P., Abadie V., Verthier N., Schmitz J., Wetterau J. R., Samson-Bouma M. E., Aggerbeck L. P. 1999. Anderson's disease: exclusion of apolipoprotein and intracellular lipid transport genes. Arterioscler. Thromb. Vasc. Biol. 19: 2494–2508 [DOI] [PubMed] [Google Scholar]
- 180.Nemeth A., Myrdal U., Veress B., Rudling M., Berglund L., Angelin B. 1995. Studies on lipoprotein metabolism in a family with jejunal chylomicron retention. Eur. J. Clin. Invest. 25: 271–280 [DOI] [PubMed] [Google Scholar]
- 181.Roy C. C., Levy E., Green P. H., Sniderman A., Letarte J., Buts J. P., Orquin J., Brochu P., Weber A. M., Morin C. L., et al. 1987. Malabsorption, hypocholesterolemia, and fat-filled enterocytes with increased intestinal apoprotein B. Chylomicron retention disease. Gastroenterology. 92: 390–399 [DOI] [PubMed] [Google Scholar]
- 182.Treepongkaruna S., Chongviriyaphan N., Suthutvoravut U., Charoenpipop D., Choubtum L., Wattanasirichaigoon D. 2009. Novel missense mutations of SAR1B gene in an infant with chylomicron retention disease. J. Pediatr. Gastroenterol. Nutr. 48: 370–373 [DOI] [PubMed] [Google Scholar]
- 183.Cefalù A. B., Calvo P. L., Noto D., Baldi M., Valenti V., Lerro P., Tramuto F., Lezo A., Morra I., Cenacchi G., et al. 2010. Variable phenotypic expression of chylomicron retention disease in a kindred carrying a mutation of the Sara2 gene. Metabolism. 59: 463–467 [DOI] [PubMed] [Google Scholar]
- 184.Bouma M. E., Beucler I., Aggerbeck L. P., Infante R., Schmitz J. 1986. Hypobetalipoproteinemia with accumulation of an apoprotein B-like protein in intestinal cells. Immunoenzymatic and biochemical characterization of seven cases of Anderson's disease. J. Clin. Invest. 78: 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Peretti N., Roy C. C., Sassolas A., Deslandres C., Drouin E., Rasquin A., Seidman E., Brochu P., Vohl M. C., Labarge S., et al. 2009. Chylomicron retention disease: a long term study of two cohorts. Mol. Genet. Metab. 97: 136–142 [DOI] [PubMed] [Google Scholar]
- 186.Peretti N., Sassolas A., Roy C. C., Deslandres C., Charcosset M., Castagnetti J., Pugnet-Chardon L., Moulin P., Labarge S., Bouthillier L., et al. 2010. Guidelines for the diagnosis and management of chylomicron retention disease based on a review of the literature and the experience of two centers. Orphanet J. Rare Dis. 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Levy E., Marcel Y., Deckelbaum R. J., Milne R., Lepage G., Seidman E., Bendayan M., Roy C. C. 1987. Intestinal apoB synthesis, lipids, and lipoproteins in chylomicron retention disease. J. Lipid Res. 28: 1263–1274 [PubMed] [Google Scholar]
- 188.Jones B., Jones E. L., Bonney S. A., Patel H. N., Mensenkamp A. R., Eichenbaum-Voline S., Rudling M., Myrdal U., Annesi G., Naik S., et al. 2003. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat. Genet. 34: 29–31 [DOI] [PubMed] [Google Scholar]
- 189.Georges A., Bonneau J., Bonnefont-Rousselot D., Champigneulle J., Rabes J. P., Abifadel M., Aparicio T., Guenedet J. C., Bruckert E., Boileau C., et al. 2011. Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson's disease (Chylomicron retention disease). Orphanet J. Rare Dis. 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shoulders C. C., Stephens D. J., Jones B. 2004. The intracellular transport of chylomicrons requires the small GTPase, Sar1b. Curr. Opin. Lipidol. 15: 191–197 [DOI] [PubMed] [Google Scholar]
- 191.Lee M. C., Miller E. A. 2007. Molecular mechanisms of COPII vesicle formation. Semin. Cell Dev. Biol. 18: 424–434 [DOI] [PubMed] [Google Scholar]
- 192.Stagg S. M., Gurkan C., Fowler D. M., LaPointe P., Foss T. R., Potter C. S., Carragher B., Balch W. E. 2006. Structure of the Sec13/31 COPII coat cage. Nature. 439: 234–238 [DOI] [PubMed] [Google Scholar]
- 193.Stagg S. M., LaPointe P., Razvi A., Gurkan C., Potter C. S., Carragher B., Balch W. E. 2008. Structural basis for cargo regulation of COPII coat assembly. Cell. 134: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bi X., Corpina R. A., Goldberg J. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 419: 271–277 [DOI] [PubMed] [Google Scholar]
- 195.Rao Y., Bian C., Yuan C., Li Y., Chen L., Ye X., Huang Z., Huang M. 2006. An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg2+. Biochem. Biophys. Res. Commun. 348: 908–915 [DOI] [PubMed] [Google Scholar]
- 196.Hermey G. 2009. The Vps10p-domain receptor family. Cell. Mol. Life Sci. 66: 2677–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen Z. Y., Ieraci A., Teng H., Dall H., Meng C. X., Herrera D. G., Nykjaer A., Hempstead B. L., Lee F. S. 2005. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J. Neurosci. 25: 6156–6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nielsen M. S., Jacobsen C., Olivecrona G., Gliemann J., Petersen C. M. 1999. Sortilin/neurotensin receptor-3 binds and mediates degradation of lipoprotein lipase. J. Biol. Chem. 274: 8832–8836 [DOI] [PubMed] [Google Scholar]
- 199.Wähe A., Kasmapour B., Schmaderer C., Liebl D., Sandhoff K., Nykjaer A., Griffiths G., Gutierrez M. G. 2010. Golgi-to-phagosome transport of acid sphingomyelinase and prosaposin is mediated by sortilin. J. Cell Sci. 123: 2502–2511 [DOI] [PubMed] [Google Scholar]
- 200.Yuan L., Morales C. R. 2010. A stretch of 17 amino acids in the prosaposin C terminus is critical for its binding to sortilin and targeting to lysosomes. J. Histochem. Cytochem. 58: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Canuel M., Lefrancois S., Zeng J., Morales C. R. 2008. AP-1 and retromer play opposite roles in the trafficking of sortilin between the Golgi apparatus and the lysosomes. Biochem. Biophys. Res. Commun. 366: 724–730 [DOI] [PubMed] [Google Scholar]
- 202.Hu F., Padukkavidana T., Vaegter C. B., Brady O. A., Zheng Y., Mackenzie I. R., Feldman H. H., Nykjaer A., Strittmatter S. M. 2010. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 68: 654–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lefrancois S., Zeng J., Hassan A. J., Canuel M., Morales C. R. 2003. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 22: 6430–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Petersen C. M., Nielsen M. S., Nykjaer A., Jacobsen L., Tommerup N., Rasmussen H. H., Roigaard H., Gliemann J., Madsen P., Moestrup S. K. 1997. Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J. Biol. Chem. 272: 3599–3605 [DOI] [PubMed] [Google Scholar]
- 205.Kjolby M., Andersen O. M., Breiderhoff T., Fjorback A. W., Pedersen K. M., Madsen P., Jansen P., Heeren J., Willnow T. E., Nykjaer A. 2010. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 12: 213–223 [DOI] [PubMed] [Google Scholar]
- 206.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N. E., Ahfeldt T., Sachs K. V., Li X., Li H., Kuperwasser N., Ruda V. M., et al. 2010. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 466: 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jansen P., Giehl K., Nyengaard J. R., Teng K., Lioubinski O., Sjoegaard S. S., Breiderhoff T., Gotthardt M., Lin F., Eilers A., et al. 2007. Roles for the pro-neurotrophin receptor sortilin in neuronal development, aging and brain injury. Nat. Neurosci. 10: 1449–1457 [DOI] [PubMed] [Google Scholar]
- 208.Zeng J., Racicott J., Morales C. R. 2009. The inactivation of the sortilin gene leads to a partial disruption of prosaposin trafficking to the lysosomes. Exp. Cell Res. 315: 3112–3124 [DOI] [PubMed] [Google Scholar]
- 209.Linsel-Nitschke P., Heeren J., Aherrahrou Z., Bruse P., Gieger C., Illig T., Prokisch H., Heim K., Doering A., Peters A., et al. 2010. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 208: 183–189 [DOI] [PubMed] [Google Scholar]
- 210.Gupta R., Ejebe K., Butler J., Lettre G., Lyon H., Guiducci C., Wilks R., Bennett F., Forrester T., Tayo B., et al. 2010. Association of common DNA sequence variants at 33 genetic loci with blood lipids in individuals of African ancestry from Jamaica. Hum. Genet. 128: 557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nakayama K., Bayasgalan T., Yamanaka K., Kumada M., Gotoh T., Utsumi N., Yanagisawa Y., Okayama M., Kajii E., Ishibashi S., et al. 2009. Large scale replication analysis of loci associated with lipid concentrations in a Japanese population. J. Med. Genet. 46: 370–374 [DOI] [PubMed] [Google Scholar]
- 212.Hegedus Z., Czibula A., Kiss-Toth E. 2006. Tribbles: novel regulators of cell function; evolutionary aspects. Cell. Mol. Life Sci. 63: 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Burkhardt R., Toh S. A., Lagor W. R., Birkeland A., Levin M., Li X., Robblee M., Fedorov V. D., Yamamoto M., Satoh T., et al. 2010. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J. Clin. Invest. 120: 4410–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sung H. Y., Guan H., Czibula A., King A. R., Eder K., Heath E., Suvarna S. K., Dower S. K., Wilson A. G., Francis S. E., et al. 2007. Human tribbles-1 controls proliferation and chemotaxis of smooth muscle cells via MAPK signaling pathways. J. Biol. Chem. 282: 18379–18387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Varbo A., Benn M., Tybjaerg-Hansen A., Grande P., Nordestgaard B. G. 2011. TRIB1 and GCKR polymorphisms, lipid levels, and risk of ischemic heart disease in the general population. Arterioscler. Thromb. Vasc. Biol. 31: 451–457 [DOI] [PubMed] [Google Scholar]
- 216.Boswell E. J., Jeon H., Blacklow S. C., Downing A. K. 2004. Global defects in the expression and function of the low density lipoprotein receptor (LDLR) associated with two familial hypercholesterolemia mutations resulting in misfolding of the LDLR epidermal growth factor-AB pair. J. Biol. Chem. 279: 30611–30621 [DOI] [PubMed] [Google Scholar]
- 217.Huang S., Henry L., Ho Y. K., Pownall H. J., Rudenko G. 2010. Mechanism of LDL binding and release probed by structure-based mutagenesis of the LDL receptor. J. Lipid Res. 51: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ranheim T., Kulseth M. A., Berge K. E., Leren T. P. 2006. Model system for phenotypic characterization of sequence variations in the LDL receptor gene. Clin. Chem. 52: 1469–1479 [DOI] [PubMed] [Google Scholar]
- 219.Tveten K., Holla O. L., Ranheim T., Berge K. E., Leren T. P., Kulseth M. A. 2007. 4-Phenylbutyrate restores the functionality of a misfolded mutant low-density lipoprotein receptor. FEBS J. 274: 1881–1893 [DOI] [PubMed] [Google Scholar]
- 220.Zhao Z., Michaely P. 2009. The role of calcium in lipoprotein release by the low-density lipoprotein receptor. Biochemistry. 48: 7313–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., Infante R. E. 2009. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 137: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Urano Y., Watanabe H., Murphy S. R., Shibuya Y., Geng Y., Peden A. A., Chang C. C., Chang T. Y. 2008. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc. Natl. Acad. Sci. USA. 105: 16513–16518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang M. L., Motamed M., Infante R. E., Abi-Mosleh L., Kwon H. J., Brown M. S., Goldstein J. L. 2010. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 12: 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Leigh S. E., Foster A. H., Whittall R. A., Hubbart C. S., Humphries S. E. 2008. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann. Hum. Genet. 72: 485–498 [DOI] [PubMed] [Google Scholar]
- 225.Leigh S. E., Leren T. P., Humphries S. E. 2009. Commentary PCSK9 variants: a new database. Atherosclerosis. 203: 32–33 [DOI] [PubMed] [Google Scholar]
- 226.Marduel M., Carrie A., Sassolas A., Devillers M., Carreau V., Di Filippo M., Erlich D., Abifadel M., Marques-Pinheiro A., Munnich A., et al. 2010. Molecular spectrum of autosomal dominant hypercholesterolemia in France. Hum. Mutat. 31: E1811–E1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fouchier S. W., Kastelein J. J., Defesche J. C. 2005. Update of the molecular basis of familial hypercholesterolemia in The Netherlands. Hum. Mutat. 26: 550–556 [DOI] [PubMed] [Google Scholar]
- 228.Abifadel M., Rabes J. P., Jambart S., Halaby G., Gannage-Yared M. H., Sarkis A., Beaino G., Varret M., Salem N., Corbani S., et al. 2009. The molecular basis of familial hypercholesterolemia in Lebanon: spectrum of LDLR mutations and role of PCSK9 as a modifier gene. Hum. Mutat. 30: E682–E691 [DOI] [PubMed] [Google Scholar]
- 229.Dedoussis G. V., Schmidt H., Genschel J. 2004. LDL-receptor mutations in Europe. Hum. Mutat. 24: 443–459 [DOI] [PubMed] [Google Scholar]
- 230.Durst R., Colombo R., Shpitzen S., Avi L. B., Friedlander Y., Wexler R., Raal F. J., Marais D. A., Defesche J. C., Mandelshtam M. Y., et al. 2001. Recent origin and spread of a common Lithuanian mutation, G197del LDLR, causing familial hypercholesterolemia: positive selection is not always necessary to account for disease incidence among Ashkenazi Jews. Am. J. Hum. Genet. 68: 1172–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lombardi M. P., Redeker E. J., Defesche J. C., Kamerling S. W., Trip M. D., Mannens M. M., Havekes L. M., Kastelein J. J. 2000. Molecular genetic testing for familial hypercholesterolemia: spectrum of LDL receptor gene mutations in The Netherlands. Clin. Genet. 57: 116–124 [DOI] [PubMed] [Google Scholar]
- 232.Whittall R. A., Matheus S., Cranston T., Miller G. J., Humphries S. E. 2002. The intron 14 2140+5G>A variant in the low density lipoprotein receptor gene has no effect on plasma cholesterol levels. J. Med. Genet. 39: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bourbon M., Duarte M. A., Alves A. C., Medeiros A. M., Marques L., Soutar A. K. 2009. Genetic diagnosis of familial hypercholesterolaemia: the importance of functional analysis of potential splice-site mutations. J. Med. Genet. 46: 352–357 [DOI] [PubMed] [Google Scholar]
- 234.Rabacchi C., Wunsch A., Ghisellini M., Marino M., Pisciotta L., Bertolini S., Calandra S. 2009. An apparent inconsistency in parent to offspring transmission of point mutations of LDLR gene in familial hypercholesterolemia. Clin. Chim. Acta. 406: 75–80 [DOI] [PubMed] [Google Scholar]
- 235.Chmara M., Wasag B., Zuk M., Kubalska J., Wegrzyn A., Bednarska-Makaruk M., Pronicka E., Wehr H., Defesche J. C., Rynkiewicz A., et al. 2010. Molecular characterization of Polish patients with familial hypercholesterolemia: novel and recurrent LDLR mutations. J. Appl. Genet. 51: 95–106 [DOI] [PubMed] [Google Scholar]
- 236.Holla O. L., Teie C., Berge K. E., Leren T. P. 2005. Identification of deletions and duplications in the low density lipoprotein receptor gene by MLPA. Clin. Chim. Acta. 356: 164–171 [DOI] [PubMed] [Google Scholar]
- 237.Taylor A., Martin B., Wang D., Patel K., Humphries S. E., Norbury G. 2009. Multiplex ligation-dependent probe amplification analysis to screen for deletions and duplications of the LDLR gene in patients with familial hypercholesterolaemia. Clin. Genet. 76: 69–75 [DOI] [PubMed] [Google Scholar]
- 238.Tosi I., Toledo-Leiva P., Neuwirth C., Naoumova R. P., Soutar A. K. 2007. Genetic defects causing familial hypercholesterolaemia: identification of deletions and duplications in the LDL-receptor gene and summary of all mutations found in patients attending the Hammersmith Hospital Lipid Clinic. Atherosclerosis. 194: 102–111 [DOI] [PubMed] [Google Scholar]
- 239.Wang J., Ban M. R., Hegele R. A. 2005. Multiplex ligation-dependent probe amplification of LDLR enhances molecular diagnosis of familial hypercholesterolemia. J. Lipid Res. 46: 366–372 [DOI] [PubMed] [Google Scholar]
- 240.Chiou K. R., Charng M. J. 2010. Detection of mutations and large rearrangements of the low-density lipoprotein receptor gene in Taiwanese patients with familial hypercholesterolemia. Am. J. Cardiol. 105: 1752–1758 [DOI] [PubMed] [Google Scholar]
- 241.Bourbon M., Sun X. M., Soutar A. K. 2007. A rare polymorphism in the low density lipoprotein (LDL) gene that affects mRNA splicing. Atherosclerosis. 195: e17–e20 [DOI] [PubMed] [Google Scholar]
- 242.Defesche J. C., Schuurman E. J., Klaaijsen L. N., Khoo K. L., Wiegman A., Stalenhoef A. F. 2008. Silent exonic mutations in the low-density lipoprotein receptor gene that cause familial hypercholesterolemia by affecting mRNA splicing. Clin. Genet. 73: 573–578 [DOI] [PubMed] [Google Scholar]
- 243.Civeira F., Jarauta E., Cenarro A., Garcia-Otin A. L., Tejedor D., Zambon D., Mallen M., Ros E., Pocovi M. 2008. Frequency of low-density lipoprotein receptor gene mutations in patients with a clinical diagnosis of familial combined hyperlipidemia in a clinical setting. J. Am. Coll. Cardiol. 52: 1546–1553 [DOI] [PubMed] [Google Scholar]
- 244.Bertolini S., Cantafora A., Averna M., Cortese C., Motti C., Martini S., Pes G., Postiglione A., Stefanutti C., Blotta I., et al. 2000. Clinical expression of familial hypercholesterolemia in clusters of mutations of the LDL receptor gene that cause a receptor-defective or receptor-negative phenotype. Arterioscler. Thromb. Vasc. Biol. 20: E41–E52 [DOI] [PubMed] [Google Scholar]
- 245.Pisciotta L., Priore Oliva C., Pes G. M., Di Scala L., Bellocchio A., Fresa R., Cantafora A., Arca M., Calandra S., Bertolini S. 2006. Autosomal recessive hypercholesterolemia (ARH) and homozygous familial hypercholesterolemia (FH): a phenotypic comparison. Atherosclerosis. 188: 398–405 [DOI] [PubMed] [Google Scholar]
- 246.Junyent M., Gilabert R., Zambon D., Pocovi M., Mallen M., Cofan M., Nunez I., Civeira F., Tejedor D., Ros E. 2008. Femoral atherosclerosis in heterozygous familial hypercholesterolemia: influence of the genetic defect. Arterioscler. Thromb. Vasc. Biol. 28: 580–586 [DOI] [PubMed] [Google Scholar]
- 247.Deiana L., Garuti R., Pes G. M., Carru C., Errigo A., Rolleri M., Pisciotta L., Masturzo P., Cantafora A., Calandra S., et al. 2000. Influence of beta(0)-thalassemia on the phenotypic expression of heterozygous familial hypercholesterolemia: a study of patients with familial hypercholesterolemia from Sardinia. Arterioscler. Thromb. Vasc. Biol. 20: 236–243 [DOI] [PubMed] [Google Scholar]
- 248.Ricchi P., Ammirabile M., Spasiano A., Costantini S., Di Matola T., Cinque P., Pagano L., Prossomariti L. 2009. Hypocholesterolemia in adult patients with thalassemia: a link with the severity of genotype in thalassemia intermedia patients. Eur. J. Haematol. 82: 219–222 [DOI] [PubMed] [Google Scholar]
- 249.Shalev H., Kapelushnik J., Moser A., Knobler H., Tamary H. 2007. Hypocholesterolemia in chronic anemias with increased erythropoietic activity. Am. J. Hematol. 82: 199–202 [DOI] [PubMed] [Google Scholar]
- 250.Chebil-Laradi S., Pousse H., Khelif A., Ghanem N., Martin J., Kortas M., Ennabli S., Mehdouani K., Chir N. B', Kharrat H., et al. 1994. [Screening of hemoglobinopathies and molecular analysis of beta-thalassemia in Central Tunisia]. Arch. Pediatr. 1: 1100–1105 [PubMed] [Google Scholar]
- 251.Jelassi A., Slimani A., Jguirim I., Najah M., Abid A., Boughamoura L., Mzid J., Fkih M., Maatouk F., Rouis M., et al. 2010. Moderate phenotypic expression of familial hypercholesterolemia in Tunisia. Clin. Chim. Acta. 411: 735–738 [DOI] [PubMed] [Google Scholar]
- 252.Vuorio A. F., Turtola H., Piilahti K. M., Repo P., Kanninen T., Kontula K. 1997. Familial hypercholesterolemia in the Finnish north Karelia. A molecular, clinical, and genealogical study. Arterioscler. Thromb. Vasc. Biol. 17: 3127–3138 [DOI] [PubMed] [Google Scholar]
- 253.Lambert M., Assouline L., Feoli-Fonseca J. C., Brun N., Delvin E. E., Levy E. 2001. Determinants of lipid level variability in French-Canadian children with familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 21: 979–984 [DOI] [PubMed] [Google Scholar]
- 254.Bertolini S., Pisciotta L., Di Scala L., Langheim S., Bellocchio A., Masturzo P., Cantafora A., Martini S., Averna M., Pes G., et al. 2004. Genetic polymorphisms affecting the phenotypic expression of familial hypercholesterolemia. Atherosclerosis. 174: 57–65 [DOI] [PubMed] [Google Scholar]
- 255.Dong L. M., Wilson C., Wardell M. R., Simmons T., Mahley R. W., Weisgraber K. H., Agard D. A. 1994. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J. Biol. Chem. 269: 22358–22365 [PubMed] [Google Scholar]
- 256.Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 252: 1817–1822 [DOI] [PubMed] [Google Scholar]
- 257.Altenburg M., Arbones-Mainar J., Johnson L., Wilder J., Maeda N. 2008. Human LDL receptor enhances sequestration of ApoE4 and VLDL remnants on the surface of hepatocytes but not their internalization in mice. Arterioscler. Thromb. Vasc. Biol. 28: 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Klos K., Shimmin L., Ballantyne C., Boerwinkle E., Clark A., Coresh J., Hanis C., Liu K., Sayre S., Hixson J. 2008. APOE/C1/C4/C2 hepatic control region polymorphism influences plasma apoE and LDL cholesterol levels. Hum. Mol. Genet. 17: 2039–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.de Man F. H., de Beer F., van de Laarse A., Smelt A. H., Leuven J. A., Havekes L. M. 1998. Effect of apolipoprotein E variants on lipolysis of very low density lipoproteins by heparan sulphate proteoglycan-bound lipoprotein lipase. Atherosclerosis. 136: 255–262 [DOI] [PubMed] [Google Scholar]
- 260.Futamura M., Dhanasekaran P., Handa T., Phillips M. C., Lund-Katz S., Saito H. 2005. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J. Biol. Chem. 280: 5414–5422 [DOI] [PubMed] [Google Scholar]
- 261.Yamauchi Y., Deguchi N., Takagi C., Tanaka M., Dhanasekaran P., Nakano M., Handa T., Phillips M. C., Lund-Katz S., Saito H. 2008. Role of the N- and C-terminal domains in binding of apolipoprotein E isoforms to heparan sulfate and dermatan sulfate: a surface plasmon resonance study. Biochemistry. 47: 6702–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Yu K. C., Chen W., Cooper A. D. 2001. LDL receptor-related protein mediates cell-surface clustering and hepatic sequestration of chylomicron remnants in LDLR-deficient mice. J. Clin. Invest. 107: 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Knouff C., Malloy S., Wilder J., Altenburg M. K., Maeda N. 2001. Doubling expression of the low density lipoprotein receptor by truncation of the 3′ -untranslated region sequence ameliorates type iii hyperlipoproteinemia in mice expressing the human apoe2 isoform. J. Biol. Chem. 276: 3856–3862 [DOI] [PubMed] [Google Scholar]
- 264.Malloy S. I., Altenburg M. K., Knouff C., Lanningham-Foster L., Parks J. S., Maeda N. 2004. Harmful effects of increased LDLR expression in mice with human APOE*4 but not APOE*3. Arterioscler. Thromb. Vasc. Biol. 24: 91–97 [DOI] [PubMed] [Google Scholar]
- 265.Heeren J., Grewal T., Laatsch A., Becker N., Rinninger F., Rye K. A., Beisiegel U. 2004. Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. J. Biol. Chem. 279: 55483–55492 [DOI] [PubMed] [Google Scholar]
- 266.Mann W. A., Lohse P., Gregg R. E., Ronan R., Hoeg J. M., Zech L. A., Brewer H. B., Jr 1995. Dominant expression of type III hyperlipoproteinemia. Pathophysiological insights derived from the structural and kinetic characteristics of ApoE-1 (Lys146 → Glu). J. Clin. Invest. 96: 1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tybjaerg-Hansen A., Jensen H. K., Benn M., Steffensen R., Jensen G., Nordestgaard B. G. 2005. Phenotype of heterozygotes for low-density lipoprotein receptor mutations identified in different background populations. Arterioscler. Thromb. Vasc. Biol. 25: 211–215 [DOI] [PubMed] [Google Scholar]
- 268.Blasiole D. A., Oler A. T., Attie A. D. 2008. Regulation of ApoB secretion by the low density lipoprotein receptor requires exit from the endoplasmic reticulum and interaction with ApoE or ApoB. J. Biol. Chem. 283: 11374–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Larsson S. L., Skogsberg J., Bjorkegren J. 2004. The low density lipoprotein receptor prevents secretion of dense apoB100-containing lipoproteins from the liver. J. Biol. Chem. 279: 831–836 [DOI] [PubMed] [Google Scholar]
- 270.Millar J. S., Maugeais C., Ikewaki K., Kolansky D. M., Barrett P. H., Budreck E. C., Boston R. C., Tada N., Mochizuki S., Defesche J. C., et al. 2005. Complete deficiency of the low-density lipoprotein receptor is associated with increased apolipoprotein B-100 production. Arterioscler. Thromb. Vasc. Biol. 25: 560–565 [DOI] [PubMed] [Google Scholar]
- 271.Tremblay A. J., Lamarche B., Ruel I. L., Hogue J. C., Bergeron J., Gagne C., Couture P. 2004. Increased production of VLDL apoB-100 in subjects with familial hypercholesterolemia carrying the same null LDL receptor gene mutation. J. Lipid Res. 45: 866–872 [DOI] [PubMed] [Google Scholar]
- 272.Michaely P., Zhao Z., Li W. P., Garuti R., Huang L. J., Hobbs H. H., Cohen J. C. 2007. Identification of a VLDL-induced, FDNPVY-independent internalization mechanism for the LDLR. EMBO J. 26: 3273–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Linsel-Nitschke P., Gotz A., Erdmann J., Braenne I., Braund P., Hengstenberg C., Stark K., Fischer M., Schreiber S., El Mokhtari N. E., et al. 2008. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease–a Mendelian Randomisation study. PLoS ONE. 3: e2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhu H., Tucker H. M., Grear K. E., Simpson J. F., Manning A. K., Cupples L. A., Estus S. 2007. A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased cholesterol. Hum. Mol. Genet. 16: 1765–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zou F., Gopalraj R. K., Lok J., Zhu H., Ling I. F., Simpson J. F., Tucker H. M., Kelly J. F., Younkin S. G., Dickson D. W., et al. 2008. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer's disease. Hum. Mol. Genet. 17: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Marcel Y. L., Hogue M., Theolis R., Jr, Milne R. W. 1982. Mapping of antigenic determinants of human apolipoprotein B using monoclonal antibodies against low density lipoproteins. J. Biol. Chem. 257: 13165–13168 [PubMed] [Google Scholar]
- 277.Milne R., Theolis R., Jr, Maurice R., Pease R. J., Weech P. K., Rassart E., Fruchart J. C., Scott J., Marcel Y. L. 1989. The use of monoclonal antibodies to localize the low density lipoprotein receptor-binding domain of apolipoprotein B. J. Biol. Chem. 264: 19754–19760 [PubMed] [Google Scholar]
- 278.Pease R. J., Milne R. W., Jessup W. K., Law A., Provost P., Fruchart J. C., Dean R. T., Marcel Y. L., Scott J. 1990. Use of bacterial expression cloning to localize the epitopes for a series of monoclonal antibodies against apolipoprotein B100. J. Biol. Chem. 265: 553–568 [PubMed] [Google Scholar]
- 279.Law A., Scott J. 1990. A cross-species comparison of the apolipoprotein B domain that binds to the LDL receptor. J. Lipid Res. 31: 1109–1120 [PubMed] [Google Scholar]
- 280.Borén J., Olin K., Lee I., Chait A., Wight T. N., Innerarity T. L. 1998. Identification of the principal proteoglycan-binding site in LDL. A single-point mutation in apo-B100 severely affects proteoglycan interaction without affecting LDL receptor binding. J. Clin. Invest. 101: 2658–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Johnson L. A., Altenburg M. K., Walzem R. L., Scanga L. T., Maeda N. 2008. Absence of hyperlipidemia in LDL receptor-deficient mice having apolipoprotein B100 without the putative receptor-binding sequences. Arterioscler. Thromb. Vasc. Biol. 28: 1745–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chatterton J. E., Phillips M. L., Curtiss L. K., Milne R., Fruchart J. C., Schumaker V. N. 1995. Immunoelectron microscopy of low density lipoproteins yields a ribbon and bow model for the conformation of apolipoprotein B on the lipoprotein surface. J. Lipid Res. 36: 2027–2037 [PubMed] [Google Scholar]
- 283.Borén J., Ekstrom U., Agren B., Nilsson-Ehle P., Innerarity T. L. 2001. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J. Biol. Chem. 276: 9214–9218 [DOI] [PubMed] [Google Scholar]
- 284.Gabelli C., Bilato C., Martini S., Tennyson G. E., Zech L. A., Corsini A., Albanese M., Brewer H. B., Jr, Crepaldi G., Baggio G. 1996. Homozygous familial hypobetalipoproteinemia. Increased LDL catabolism in hypobetalipoproteinemia due to a truncated apolipoprotein B species, apo B-87Padova. Arterioscler. Thromb. Vasc. Biol. 16: 1189–1196 [DOI] [PubMed] [Google Scholar]
- 285.Parhofer K. G., Daugherty A., Kinoshita M., Schonfeld G. 1990. Enhanced clearance from plasma of low density lipoproteins containing a truncated apolipoprotein, apoB-89. J. Lipid Res. 31: 2001–2007 [PubMed] [Google Scholar]
- 286.Soria L. F., Ludwig E. H., Clarke H. R., Vega G. L., Grundy S. M., McCarthy B. J. 1989. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc. Natl. Acad. Sci. USA. 86: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Real J. T., Chaves F. J., Ejarque I., Garcia-Garcia A. B., Valldecabres C., Ascaso J. F., Armengod M. E., Carmena R. 2003. Influence of LDL receptor gene mutations and the R3500Q mutation of the apoB gene on lipoprotein phenotype of familial hypercholesterolemic patients from a South European population. Eur. J. Hum. Genet. 11: 959–965 [DOI] [PubMed] [Google Scholar]
- 288.Viola S., Benlian P., Morali A., Dobbelaere D., Lacaille F., Rieu D., Ginies J. L., Maurage C., Meyer M., Lachaux A., et al. 2001. Apolipoprotein B Arg3500Gln mutation prevalence in children with hypercholesterolemia: a French multicenter study. J. Pediatr. Gastroenterol. Nutr. 33: 122–126 [DOI] [PubMed] [Google Scholar]
- 289.Gasparovic J., Basistova Z., Fabryova L., Wsolova L., Vohnout B., Raslova K. 2007. Familial defective apolipoprotein B-100 in Slovakia: are differences in prevalence of familial defective apolipoprotein B-100 explained by ethnicity? Atherosclerosis. 194: e95–e107 [DOI] [PubMed] [Google Scholar]
- 290.Bourbon M., Alves A. C., Medeiros A. M., Silva S., Soutar A. K. 2008. Familial hypercholesterolaemia in Portugal. Atherosclerosis. 196: 633–642 [DOI] [PubMed] [Google Scholar]
- 291.Dedoussis G. V., Genschel J., Bochow B., Pitsavos C., Skoumas J., Prassa M., Lkhagvasuren S., Toutouzas P., Vogt A., Kassner U., et al. 2004. Molecular characterization of familial hypercholesterolemia in German and Greek patients. Hum. Mutat. 23: 285–286 [DOI] [PubMed] [Google Scholar]
- 292.Taylor A., Tabrah S., Wang D., Sozen M., Duxbury N., Whittall R., Humphries S. E., Norbury G. 2007. Multiplex ARMS analysis to detect 13 common mutations in familial hypercholesterolaemia. Clin. Genet. 71: 561–568 [DOI] [PubMed] [Google Scholar]
- 293.Tai E. S., Koay E. S., Chan E., Seng T. J., Loh L. M., Sethi S. K., Tan C. E. 2001. Compound heterozygous familial hypercholesterolemia and familial defective apolipoprotein B-100 produce exaggerated hypercholesterolemia. Clin. Chem. 47: 438–443 [PubMed] [Google Scholar]
- 294.Yang K. C., Su Y. N., Shew J. Y., Yang K. Y., Tseng W. K., Wu C. C., Lee Y. T. 2007. LDLR and ApoB are major genetic causes of autosomal dominant hypercholesterolemia in a Taiwanese population. J. Formos. Med. Assoc. 106: 799–807 [DOI] [PubMed] [Google Scholar]
- 295.Shen H., Damcott C. M., Rampersaud E., Pollin T. I., Horenstein R. B., McArdle P. F., Peyser P. A., Bielak L. F., Post W. S., Chang Y. P., et al. 2010. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 170: 1850–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Miserez A. R., Laager R., Chiodetti N., Keller U. 1994. High prevalence of familial defective apolipoprotein B-100 in Switzerland. J. Lipid Res. 35: 574–583 [PubMed] [Google Scholar]
- 297.Humphries S. E., Cranston T., Allen M., Middleton-Price H., Fernandez M. C., Senior V., Hawe E., Iversen A., Wray R., Crook M. A., et al. 2006. Mutational analysis in UK patients with a clinical diagnosis of familial hypercholesterolaemia: relationship with plasma lipid traits, heart disease risk and utility in relative tracing. J. Mol. Med. 84: 203–214 [DOI] [PubMed] [Google Scholar]
- 298.Liyanage K. E., Hooper A. J., Defesche J. C., Burnett J. R., van Bockxmeer F. M. 2008. High-resolution melting analysis for detection of familial ligand-defective apolipoprotein B-100 mutations. Ann. Clin. Biochem. 45: 170–176 [DOI] [PubMed] [Google Scholar]
- 299.Benlian P., de Gennes J. L., Dairou F., Hermelin B., Ginon I., Villain E., Lagarde J. P., Federspiel M. C., Bertrand V., Bernard C., et al. 1996. Phenotypic expression in double heterozygotes for familial hypercholesterolemia and familial defective apolipoprotein B-100. Hum. Mutat. 7: 340–345 [DOI] [PubMed] [Google Scholar]
- 300.Taylor A., Bayly G., Patel K., Yarram L., Williams M., Hamilton-Shield J., Humphries S. E., Norbury G. 2010. A double heterozygote for familial hypercholesterolaemia and familial defective apolipoprotein B-100. Ann. Clin. Biochem. 47: 487–490 [DOI] [PubMed] [Google Scholar]
- 301.Rubinsztein D. C., Raal F. J., Seftel H. C., Pilcher G., Coetzee G. A., van der Westhuyzen D. R. 1993. Characterization of six patients who are double heterozygotes for familial hypercholesterolemia and familial defective apo B-100. Arterioscler. Thromb. 13: 1076–1081 [DOI] [PubMed] [Google Scholar]
- 302.Benn M., Nordestgaard B. G., Jensen J. S., Nilausen K., Meinertz H., Tybjaerg-Hansen A. 2005. Mutation in apolipoprotein B associated with hypobetalipoproteinemia despite decreased binding to the low density lipoprotein receptor. J. Biol. Chem. 280: 21052–21060 [DOI] [PubMed] [Google Scholar]
- 303.Hansen P. S., Defesche J. C., Kastelein J. J., Gerdes L. U., Fraza L., Gerdes C., Tato F., Jensen H. K., Jensen L. G., Klausen I. C., et al. 1997. Phenotypic variation in patients heterozygous for familial defective apolipoprotein B (FDB) in three European countries. Arterioscler. Thromb. Vasc. Biol. 17: 741–747 [DOI] [PubMed] [Google Scholar]
- 304.Pullinger C. R., Gaffney D., Gutierrez M. M., Malloy M. J., Schumaker V. N., Packard C. J., Kane J. P. 1999. The apolipoprotein B R3531C mutation. Characteristics of 24 subjects from 9 kindreds. J. Lipid Res. 40: 318–327 [PubMed] [Google Scholar]
- 305.Rabès J. P., Varret M., Devillers M., Aegerter P., Villeger L., Krempf M., Junien C., Boileau C. 2000. R3531C mutation in the apolipoprotein B gene is not sufficient to cause hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 20: E76–E82 [DOI] [PubMed] [Google Scholar]
- 306.Wenham P. R., Henderson B. G., Penney M. D., Ashby J. P., Rae P. W., Walker S. W. 1997. Familial ligand-defective apolipoprotein B-100: detection, biochemical features and haplotype analysis of the R3531C mutation in the UK. Atherosclerosis. 129: 185–192 [DOI] [PubMed] [Google Scholar]
- 307.Medeiros A. M., Alves A. C., Francisco V., Bourbon M. 2010. Update of the Portuguese Familial Hypercholesterolaemia Study. Atherosclerosis. 212: 553–558 [DOI] [PubMed] [Google Scholar]
- 308.Harada-Shiba M., Tajima S., Yokoyama S., Miyake Y., Kojima S., Tsushima M., Kawakami M., Yamamoto A. 1992. Siblings with normal LDL receptor activity and severe hypercholesterolemia. Arterioscler. Thromb. 12: 1071–1078 [DOI] [PubMed] [Google Scholar]
- 309.Zuliani G., Arca M., Signore A., Bader G., Fazio S., Chianelli M., Bellosta S., Campagna F., Montali A., Maioli M., et al. 1999. Characterization of a new form of inherited hypercholesterolemia: familial recessive hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 19: 802–809 [DOI] [PubMed] [Google Scholar]
- 310.Norman D., Sun X. M., Bourbon M., Knight B. L., Naoumova R. P., Soutar A. K. 1999. Characterization of a novel cellular defect in patients with phenotypic homozygous familial hypercholesterolemia. J. Clin. Invest. 104: 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Jones C., Garuti R., Michaely P., Li W. P., Maeda N., Cohen J. C., Herz J., Hobbs H. H. 2007. Disruption of LDL but not VLDL clearance in autosomal recessive hypercholesterolemia. J. Clin. Invest. 117: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Abera A. B., Marais A. D., Raal F. J., Leisegang F., Jones S., George P., Henderson H. E. 2007. Autosomal recessive hypercholesterolaemia: discrimination of ARH protein and LDLR function in the homozygous FH phenotype. Clin. Chim. Acta. 378: 33–37 [DOI] [PubMed] [Google Scholar]
- 313.Arca M., Zuliani G., Wilund K., Campagna F., Fellin R., Bertolini S., Calandra S., Ricci G., Glorioso N., Maioli M., et al. 2002. Autosomal recessive hypercholesterolaemia in Sardinia, Italy, and mutations in ARH: a clinical and molecular genetic analysis. Lancet. 359: 841–847 [DOI] [PubMed] [Google Scholar]
- 314.Harada-Shiba M., Takagi A., Miyamoto Y., Tsushima M., Ikeda Y., Yokoyama S., Yamamoto A. 2003. Clinical features and genetic analysis of autosomal recessive hypercholesterolemia. J. Clin. Endocrinol. Metab. 88: 2541–2547 [DOI] [PubMed] [Google Scholar]
- 315.Lind S., Olsson A. G., Eriksson M., Rudling M., Eggertsen G., Angelin B. 2004. Autosomal recessive hypercholesterolaemia: normalization of plasma LDL cholesterol by ezetimibe in combination with statin treatment. J. Intern. Med. 256: 406–412 [DOI] [PubMed] [Google Scholar]
- 316.Quagliarini F., Vallve J. C., Campagna F., Alvaro A., Fuentes-Jimenez F. J., Sirinian M. I., Meloni F., Masana L., Arca M. 2007. Autosomal recessive hypercholesterolemia in Spanish kindred due to a large deletion in the ARH gene. Mol. Genet. Metab. 92: 243–248 [DOI] [PubMed] [Google Scholar]
- 317.Rodenburg J., Wiegman A., Vissers M. N., Kastelein J. J., Stalenhoef A. F. 2004. A boy with autosomal recessive hypercholesterolaemia. Neth. J. Med. 62: 89–93 [PubMed] [Google Scholar]
- 318.Thomas H. P., Vogt A., Wilund K. R., Schliesser C., Steinhagen-Thiessen E., Kassner U. 2004. Autosomal recessive hypercholesterolemia in three sisters with phenotypic homozygous familial hypercholesterolemia: diagnostic and therapeutic procedures. Ther. Apher. Dial. 8: 275–280 [DOI] [PubMed] [Google Scholar]
- 319.Al-Kateb H., Bautz E. K., Luft F. C., Bahring S. 2003. A splice mutation in a Syrian autosomal recessive hypercholesterolemia family causes a two-nucleotide deletion of mRNA. Circ. Res. 93: e49–e50 [DOI] [PubMed] [Google Scholar]
- 320.Eden E. R., Patel D. D., Sun X. M., Burden J. J., Themis M., Edwards M., Lee P., Neuwirth C., Naoumova R. P., Soutar A. K. 2002. Restoration of LDL receptor function in cells from patients with autosomal recessive hypercholesterolemia by retroviral expression of ARH1. J. Clin. Invest. 110: 1695–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Garcia C. K., Wilund K., Arca M., Zuliani G., Fellin R., Maioli M., Calandra S., Bertolini S., Cossu F., Grishin N., et al. 2001. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 292: 1394–1398 [DOI] [PubMed] [Google Scholar]
- 322.Wilund K. R., Yi M., Campagna F., Arca M., Zuliani G., Fellin R., Ho Y. K., Garcia J. V., Hobbs H. H., Cohen J. C. 2002. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum. Mol. Genet. 11: 3019–3030 [DOI] [PubMed] [Google Scholar]
- 323.Filigheddu F., Quagliarini F., Campagna F., Secci T., Degortes S., Zaninello R., Argiolas G., Verna R., Pitzoi S., Frau F., et al. 2009. Prevalence and clinical features of heterozygous carriers of autosomal recessive hypercholesterolemia in Sardinia. Atherosclerosis. 207: 162–167 [DOI] [PubMed] [Google Scholar]
- 324.Ni Y. G., Di Marco S., Condra J. H., Peterson L. B., Wang W., Wang F., Pandit S., Hammond H. A., Rosa R., Cummings R. T., et al. 2011. A PCSK9-binding antibody that structurally mimics the EGF(A) domain of LDL-receptor reduces LDL cholesterol in vivo. J. Lipid Res. 52: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Abifadel M., Pakradouni J., Collin M., Samson-Bouma M. E., Varret M., Rabès J. P., Boileau C. 2010. Strategies for proprotein convertase subtilisin kexin 9 modulation: a perspective on recent patents. Expert Opin. Ther. Pat. 20: 1547–1571 [DOI] [PubMed] [Google Scholar]
- 326.Duff C. J., Hooper N. M. 2011. PCSK9: an emerging target for treatment of hypercholesterolemia. Expert Opin. Ther. Targets. 15: 157–168 [DOI] [PubMed] [Google Scholar]
- 327.Horton J. D., Cohen J. C., Hobbs H. H. 2009. PCSK9: a convertase that coordinates LDL catabolism. J. Lipid Res. 50(Suppl.): S172–S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Browning J. D., Horton J. D. 2010. Fasting reduces plasma proprotein convertase, subtilisin/kexin type 9 and cholesterol biosynthesis in humans. J. Lipid Res. 51: 3359–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Persson L., Cao G., Stahle L., Sjoberg B. G., Troutt J. S., Konrad R. J., Galman C., Wallen H., Eriksson M., Hafstrom I., et al. 2010. Circulating proprotein convertase subtilisin kexin type 9 has a diurnal rhythm synchronous with cholesterol synthesis and is reduced by fasting in humans. Arterioscler. Thromb. Vasc. Biol. 30: 2666–2672 [DOI] [PubMed] [Google Scholar]
- 330.Herbert B., Patel D., Waddington S. N., Eden E. R., McAleenan A., Sun X. M., Soutar A. K. 2010. Increased secretion of lipoproteins in transgenic mice expressing human D374Y PCSK9 under physiological genetic control. Arterioscler. Thromb. Vasc. Biol. 30: 1333–1339 [DOI] [PubMed] [Google Scholar]
- 331.Welder G., Zineh I., Pacanowski M. A., Troutt J. S., Cao G., Konrad R. J. 2010. High-dose atorvastatin causes a rapid sustained increase in human serum PCSK9 and disrupts its correlation with LDL cholesterol. J. Lipid Res. 51: 2714–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Dong B., Wu M., Li H., Kraemer F. B., Adeli K., Seidah N. G., Park S. W., Liu J. 2010. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 51: 1486–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ouguerram K., Chetiveaux M., Zair Y., Costet P., Abifadel M., Varret M., Boileau C., Magot T., Krempf M. 2004. Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler. Thromb. Vasc. Biol. 24: 1448–1453 [DOI] [PubMed] [Google Scholar]
- 334.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 102: 5374–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Le May C., Kourimate S., Langhi C., Chetiveaux M., Jarry A., Comera C., Collet X., Kuipers F., Krempf M., Cariou B., et al. 2009. Proprotein convertase subtilisin kexin type 9 null mice are protected from postprandial triglyceridemia. Arterioscler. Thromb. Vasc. Biol. 29: 684–690 [DOI] [PubMed] [Google Scholar]
- 336.Abifadel M., Rabes J. P., Devillers M., Munnich A., Erlich D., Junien C., Varret M., Boileau C. 2009. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Hum. Mutat. 30: 520–529 [DOI] [PubMed] [Google Scholar]
- 337.Fasano T., Sun X. M., Patel D. D., Soutar A. K. 2009. Degradation of LDLR protein mediated by ‘gain of function’ PCSK9 mutants in normal and ARH cells. Atherosclerosis. 203: 166–171 [DOI] [PubMed] [Google Scholar]
- 338.Pandit S., Wisniewski D., Santoro J. C., Ha S., Ramakrishnan V., Cubbon R. M., Cummings R. T., Wright S. D., Sparrow C. P., Sitlani A., et al. 2008. Functional analysis of sites within PCSK9 responsible for hypercholesterolemia. J. Lipid Res. 49: 1333–1343 [DOI] [PubMed] [Google Scholar]
- 339.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156 [DOI] [PubMed] [Google Scholar]
- 340.Allard D., Amsellem S., Abifadel M., Trillard M., Devillers M., Luc G., Krempf M., Reznik Y., Girardet J. P., Fredenrich A., et al. 2005. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum. Mutat. 26: 497. [DOI] [PubMed] [Google Scholar]
- 341.Taylor A., Wang D., Patel K., Whittall R., Wood G., Farrer M., Neely R. D., Fairgrieve S., Nair D., Barbir M., et al. 2010. Mutation detection rate and spectrum in familial hypercholesterolaemia patients in the UK pilot cascade project. Clin. Genet. 77: 572–580 [DOI] [PubMed] [Google Scholar]
- 342.van der Graaf A., Avis H. J., Kusters D. M., Vissers M. N., Hutten B. A., Defesche J. C., Huijgen R., Fouchier S. W., Wijburg F. A., Kastelein J. J., et al. 2011. Molecular basis of autosomal dominant hypercholesterolemia: assessment in a large cohort of hypercholesterolemic children. Circulation. 123: 1167–1173 [DOI] [PubMed] [Google Scholar]
- 343.Pisciotta L., Priore Oliva C., Cefalu A. B., Noto D., Bellocchio A., Fresa R., Cantafora A., Patel D., Averna M., Tarugi P., et al. 2006. Additive effect of mutations in LDLR and PCSK9 genes on the phenotype of familial hypercholesterolemia. Atherosclerosis. 186: 433–440 [DOI] [PubMed] [Google Scholar]
- 344.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165 [DOI] [PubMed] [Google Scholar]
- 345.Zhao Z., Tuakli-Wosornu Y., Lagace T. A., Kinch L., Grishin N. V., Horton J. D., Cohen J. C., Hobbs H. H. 2006. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79: 514–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Fasano T., Cefalu A. B., Di Leo E., Noto D., Pollaccia D., Bocchi L., Valenti V., Bonardi R., Guardamagna O., Averna M., et al. 2007. A novel loss of function mutation of PCSK9 gene in white subjects with low-plasma low-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol. 27: 677–681 [DOI] [PubMed] [Google Scholar]
- 347.Cariou B., Ouguerram K., Zair Y., Guerois R., Langhi C., Kourimate S., Benoit I., Le May C., Gayet C., Belabbas K., et al. 2009. PCSK9 dominant negative mutant results in increased LDL catabolic rate and familial hypobetalipoproteinemia. Arterioscler. Thromb. Vasc. Biol. 29: 2191–2197 [DOI] [PubMed] [Google Scholar]
- 348.Lakoski S. G., Lagace T. A., Cohen J. C., Horton J. D., Hobbs H. H. 2009. Genetic and metabolic determinants of plasma PCSK9 levels. J. Clin. Endocrinol. Metab. 94: 2537–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zelcer N., Hong C., Boyadjian R., Tontonoz P. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 325: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Matsuoka K., Kajimoto E., Horiuchi M., Honda C., Endo K. 2010. Competitive solubilization of cholesterol and six species of sterol/stanol in bile salt micelles. Chem. Phys. Lipids. 163: 397–402 [DOI] [PubMed] [Google Scholar]
- 351.Amiot M. J., Knol D., Cardinault N., Nowicki M., Bott R., Antona C., Borel P., Bernard J. P., Duchateau G., Lairon D. 2011. Phytosterol ester processing in the small intestine: impact on cholesterol availability for absorption and chylomicron cholesterol incorporation in healthy humans. J. Lipid Res. 52: 1256–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Jones P. J., AbuMweis S. S. 2009. Phytosterols as functional food ingredients: linkages to cardiovascular disease and cancer. Curr. Opin. Clin. Nutr. Metab. Care. 12: 147–151 [DOI] [PubMed] [Google Scholar]
- 353.Brufau G., Kuipers F., Lin Y., Trautwein E. A., Groen A. K. 2011. A reappraisal of the mechanism by which plant sterols promote neutral sterol loss in mice. PLoS ONE. 6: e21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Lin X., Racette S. B., Lefevre M., Ma L., Spearie C. A., Steger-May K., Ostlund R. E., Jr 2011. Combined effects of ezetimibe and phytosterols on cholesterol metabolism: a randomized, controlled feeding study in humans. Circulation. 124: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kwon H. J., Palnitkar M., Deisenhofer J. 2011. The structure of the NPC1L1 N-terminal domain in a closed conformation. PLoS ONE. 6: e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Wang J., Chu B. B., Ge L., Li B. L., Yan Y., Song B. L. 2009. Membrane topology of human NPC1L1, a key protein in enterohepatic cholesterol absorption. J. Lipid Res. 50: 1653–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Betters J. L., Yu L. 2010. NPC1L1 and cholesterol transport. FEBS Lett. 584: 2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Weinglass A. B., Kohler M., Schulte U., Liu J., Nketiah E. O., Thomas A., Schmalhofer W., Williams B., Bildl W., McMasters D. R., et al. 2008. Extracellular loop C of NPC1L1 is important for binding to ezetimibe. Proc. Natl. Acad. Sci. USA. 105: 11140–11145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Read J., Anderson T. A., Ritchie P. J., Vanloo B., Amey J., Levitt D., Rosseneu M., Scott J., Shoulders C. C. 2000. A mechanism of membrane neutral lipid acquisition by the microsomal triglyceride transfer protein. J. Biol. Chem. 275: 30372–30377 [DOI] [PubMed] [Google Scholar]
- 360.Shoulders C. C., Shelness G. S. 2005. Current biology of MTP: implications for selective inhibition. Curr. Top. Med. Chem. 5: 283–300 [DOI] [PubMed] [Google Scholar]
- 361.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612 [DOI] [PubMed] [Google Scholar]
- 362.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., et al. 2007. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat. Struct. Mol. Biol. 14: 413–419 [DOI] [PubMed] [Google Scholar]
- 363.Fisher C., Beglova N., Blacklow S. C. 2006. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 22: 277–283 [DOI] [PubMed] [Google Scholar]
- 364.Mann W. A., Gregg R. E., Sprecher D. L., Brewer H. B., Jr 1989. Apolipoprotein E-1Harrisburg: a new variant of apolipoprotein E dominantly associated with type III hyperlipoproteinemia. Biochim. Biophys. Acta. 1005: 239–244 [DOI] [PubMed] [Google Scholar]
- 365.Moriyama K., Sasaki J., Matsunaga A., Arakawa F., Takada Y., Araki K., Kaneko S., Arakawa K. 1992. Apolipoprotein E1 Lys-146-Glu with type III hyperlipoproteinemia. Biochim. Biophys. Acta. 1128: 58–64 [DOI] [PubMed] [Google Scholar]
- 366.Hoffer M. J., Niththyananthan S., Naoumova R. P., Kibirige M. S., Frants R. R., Havekes L. M., Thompson G. R. 1996. Apolipoprotein E1-Hammersmith (Lys146 → Asn;Arg147 → Trp), due to a dinucleotide substitution, is associated with early manifestation of dominant type III hyperlipoproteinaemia. Atherosclerosis. 124: 183–189 [DOI] [PubMed] [Google Scholar]
- 367.Dong L. M., Weisgraber K. H. 1996. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 271: 19053–19057 [DOI] [PubMed] [Google Scholar]
- 368.Hatters D. M., Budamagunta M. S., Voss J. C., Weisgraber K. H. 2005. Modulation of apolipoprotein E structure by domain interaction: differences in lipid-bound and lipid-free forms. J. Biol. Chem. 280: 34288–34295 [DOI] [PubMed] [Google Scholar]
- 369.Nguyen D., Dhanasekaran P., Nickel M., Nakatani R., Saito H., Phillips M. C., Lund-Katz S. 2010. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 49: 10881–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Tetali S. D., Budamagunta M. S., Simion C., den Hartigh L. J., Kalai T., Hideg K., Hatters D. M., Weisgraber K. H., Voss J. C., Rutledge J. C. 2010. VLDL lipolysis products increase VLDL fluidity and convert apolipoprotein E4 into a more expanded conformation. J. Lipid Res. 51: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Guttman M., Prieto J. H., Croy J. E., Komives E. A. 2010. Decoding of lipoprotein-receptor interactions: properties of ligand binding modules governing interactions with apolipoprotein E. Biochemistry. 49: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Sivashanmugam A., Wang J. 2009. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J. Biol. Chem. 284: 14657–14666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Morrow J. A., Arnold K. S., Dong J., Balestra M. E., Innerarity T. L., Weisgraber K. H. 2000. Effect of arginine 172 on the binding of apolipoprotein E to the low density lipoprotein receptor. J. Biol. Chem. 275: 2576–2580 [DOI] [PubMed] [Google Scholar]
- 374.Croy J. E., Brandon T., Komives E. A. 2004. Two apolipoprotein E mimetic peptides, ApoE(130-149) and ApoE(141-155)2, bind to LRP1. Biochemistry. 43: 7328–7335 [DOI] [PubMed] [Google Scholar]
- 375.Guttman M., Prieto J. H., Handel T. M., Domaille P. J., Komives E. A. 2010. Structure of the minimal interface between ApoE and LRP. J. Mol. Biol. 398: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Ruiz J., Kouiavskaia D., Migliorini M., Robinson S., Saenko E. L., Gorlatova N., Li D., Lawrence D., Hyman B. T., Weisgraber K. H., et al. 2005. The apoE isoform binding properties of the VLDL receptor reveal marked differences from LRP and the LDL receptor. J. Lipid Res. 46: 1721–1731 [DOI] [PubMed] [Google Scholar]
- 377.Sanna S., Li B., Mulas A., Sidore C., Kang H. M., Jackson A. U., Piras M. G., Usala G., Maninchedda G., Sassu A., et al. 2011. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 7: e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Koeijvoets K. C., Wiegman A., Rodenburg J., Defesche J. C., Kastelein J. J., Sijbrands E. J. 2005. Effect of low-density lipoprotein receptor mutation on lipoproteins and cardiovascular disease risk: a parent-offspring study. Atherosclerosis. 180: 93–99 [DOI] [PubMed] [Google Scholar]
- 379.Koeijvoets K. C., Rodenburg J., Hutten B. A., Wiegman A., Kastelein J. J., Sijbrands E. J. 2005. Low-density lipoprotein receptor genotype and response to pravastatin in children with familial hypercholesterolemia: substudy of an intima-media thickness trial. Circulation. 112: 3168–3173 [DOI] [PubMed] [Google Scholar]
- 380.Guardamagna O., Restagno G., Rolfo E., Pederiva C., Martini S., Abello F., Baracco V., Pisciotta L., Pino E., Calandra S., et al. 2009. The type of LDLR gene mutation predicts cardiovascular risk in children with familial hypercholesterolemia. J. Pediatr. 155: 199–204e2 [DOI] [PubMed] [Google Scholar]
- 381.Ceska R., Vrablik M., Horinek A. 2000. Familial defective apolipoprotein B-100: a lesson from homozygous and heterozygous patients. Physiol. Res. 49(Suppl. 1): S125–S130 [PubMed] [Google Scholar]
- 382.Leren T. P. 2004. Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia. Clin. Genet. 65: 419–422 [DOI] [PubMed] [Google Scholar]
- 383.Naoumova R. P., Tosi I., Patel D., Neuwirth C., Horswell S. D., Marais A. D., van Heyningen C., Soutar A. K. 2005. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: long-term follow-up and treatment response. Arterioscler. Thromb. Vasc. Biol. 25: 2654–2660 [DOI] [PubMed] [Google Scholar]
- 384.Ken-Dror G., Talmud P. J., Humphries S. E., Drenos F. 2010. APOE/C1/C4/C2 gene cluster genotypes, haplotypes and lipid levels in prospective coronary heart disease risk among UK healthy men. Mol. Med. 16: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Waterworth D. M., Ricketts S. L., Song K., Chen L., Zhao J. H., Ripatti S., Aulchenko Y. S., Zhang W., Yuan X., Lim N., et al. 2010. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 30: 2264–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Ward H., Mitrou P. N., Bowman R., Luben R., Wareham N. J., Khaw K. T., Bingham S. 2009. APOE genotype, lipids, and coronary heart disease risk: a prospective population study. Arch. Intern. Med. 169: 1424–1429 [DOI] [PubMed] [Google Scholar]
- 387.Burman D., Mente A., Hegele R. A., Islam S., Yusuf S., Anand S. S. 2009. Relationship of the ApoE polymorphism to plasma lipid traits among South Asians, Chinese, and Europeans living in Canada. Atherosclerosis. 203: 192–200 [DOI] [PubMed] [Google Scholar]
- 388.Mega J. L., Morrow D. A., Brown A., Cannon C. P., Sabatine M. S. 2009. Identification of genetic variants associated with response to statin therapy. Arterioscler. Thromb. Vasc. Biol. 29: 1310–1315 [DOI] [PubMed] [Google Scholar]
- 389.Huang C. C., Fornage M., Lloyd-Jones D. M., Wei G. S., Boerwinkle E., Liu K. 2009. Longitudinal association of PCSK9 sequence variations with low-density lipoprotein cholesterol levels: the Coronary Artery Risk Development in Young Adults Study. Circ. Cardiovasc. Genet. 2: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Cohen J. C., Boerwinkle E., Mosley T. H., Jr, Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272 [DOI] [PubMed] [Google Scholar]
- 391.Chen S. N., Ballantyne C. M., Gotto A. M., Jr, Tan Y., Willerson J. T., Marian A. J. 2005. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J. Am. Coll. Cardiol. 45: 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Guella I., Asselta R., Ardissino D., Merlini P. A., Peyvandi F., Kathiresan S., Mannucci P. M., Tubaro M., Duga S. 2010. Effects of PCSK9 genetic variants on plasma LDL cholesterol levels and risk of premature myocardial infarction in the Italian population. J. Lipid Res. 51: 3342–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Chasman D. I., Pare G., Zee R. Y., Parker A. N., Cook N. R., Buring J. E., Kwiatkowski D. J., Rose L. M., Smith J. D., Williams P. T., et al. 2008. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and Apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ. Cardiovasc. Genet. 1: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Benn M., Nordestgaard B. G., Grande P., Schnohr P., Tybjaerg-Hansen A. 2010. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J. Am. Coll. Cardiol. 55: 2833–2842 [DOI] [PubMed] [Google Scholar]
- 395.Miljkovic I., Yerges-Armstrong L. M., Kuller L. H., Kuipers A. L., Wang X., Kammerer C. M., Nestlerode C. S., Bunker C. H., Patrick A. L., Wheeler V. W., et al. 2010. Association analysis of 33 lipoprotein candidate genes in multi-generational families of African ancestry. J. Lipid Res. 51: 1823–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]