Abstract
Resistance to oseltamivir, the most widely used influenza antiviral drug, spread to fixation in seasonal influenza A(H1N1) between 2006 and 2009. This sudden rise in resistance seemed puzzling given the low overall level of the oseltamivir usage and the lack of a correlation between local rates of resistance and oseltamivir usage. We used a stochastic simulation model and deterministic approximations to examine how such events can occur, and in particular to determine how the rate of fixation of the resistant strain depends both on its fitness in untreated hosts as well as the frequency of antiviral treatment. We found that, for the levels of antiviral usage in the population, the resistant strain will eventually spread to fixation, if it is not attenuated in transmissibility relative to the drug-sensitive strain, but not at the speed observed in seasonal H1N1. The extreme speed with which the resistance spread in seasonal H1N1 suggests that the resistant strain had a transmission advantage in untreated hosts, and this could have arisen from genetic hitchhiking, or from the mutations responsible for resistance and compensation. Importantly, our model also shows that resistant virus will fail to spread if it is even slightly less transmissible than its sensitive counterpart—a finding of relevance given that resistant pandemic influenza (H1N1) 2009 may currently suffer from a small, but nonetheless experimentally perceptible reduction in transmissibility.
Keywords: influenza, antiviral agents, mathematical model
1. Introduction
Antiviral drugs are a major component of strategies for mitigating pandemic strains of influenza for which vaccines are not immediately available [1,2]. In addition, antiviral drugs are used for treatment and prophylaxis of both highly pathogenic influenza (e.g. avian influenza A (H5N1)) and seasonal influenza, particularly for those in high-risk groups. The most widely used antiviral drug is the neuraminidase-inhibitor oseltamivir (Tamiflu) [3]. Amino acid mutations have been identified that confer oseltamivir resistance on neuraminidases of both the N1 and N2 subtypes [4,5]. It had been believed that none of these mutations were likely to be of clinical significance, since studies showed that they attenuated viral transmission or replication in H3N2 strains [6–8] and seasonal H1N1 strains from 1999 and earlier [9–11]. However, one of these mutations (H274Y) went to fixation in seasonal H1N1 in a span of just a few years between 2006 and 2009 [12,13]. This rapid spread of resistance was enabled by the fact that by 2007, the H274Y mutation no longer caused detectable attenuation [14–16], owing at least in part to the acquisition of secondary mutations that remedied the defect in neuraminidase surface expression caused by H274Y [17]. However, the speed with which oseltamivir resistance spread remains puzzling, because global oseltamivir usage was low, and there was no significant correlation between a country's oseltamivir usage and the prevalence of resistance [12,18].
To test various hypotheses about the rapid global spread of seasonal H1N1 influenza, we developed stochastic models of the global transmission of influenza (figure 1). In these models, oseltamivir-sensitive and resistant influenza can infect people in 321 cities connected by air travel (figure 1a). A fraction of simulated infected individuals were treated with oseltamivir. We used the models to test how oseltamivir-resistant strains would spread under a variety of assumptions about their transmissibility relative to their oseltamivir-sensitive counterparts.
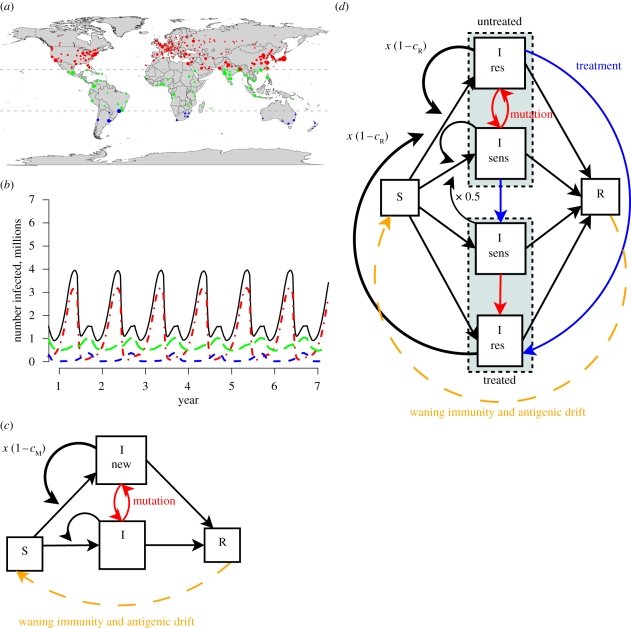
Figure 1.
A model of global influenza transmission. (a) Three hundred and twenty-one cities are represented in the model. Dot sizes are proportional to population. Cities in the temperate Northern Hemisphere (north of the Tropic of Cancer) are in red, cities in the temperate Southern Hemisphere (south of the Tropic of Capricorn) are in blue and cities in the tropics are in green and total are indicated in black. (b) The cities in the model have influenza seasons, as shown in this simulation plot. In general, influenza season occurs during the winter in the temperate Northern and Southern Hemispheres and during the rainy season in the tropics. (c) Within each city, susceptible individuals can be infected with influenza. After an infected individual recovers, that person becomes susceptible again as immunity wanes (orange dashed line). In addition, previously infected individuals become susceptible as flu strains antigenically drift. In a simple strain replacement model, there are two strains of influenza: an original strain and more transmissible mutant strain, which is 1 − cM times more transmissible. (d) In the antiviral resistance model, infected individuals may be treated with oseltamivir to reduce transmission of the sensitive strain (blue lines). A resistant strain can arise in treated or untreated individuals infected with the sensitive strain (red lines). The resistant strain is 1 − cR times as transmissible as the sensitive strain.
2. Stochastic model of influenza transmission
We developed a stochastic model of the global transmission of influenza, described in more detail in the electronic supplementary material, with parameters summarized in electronic supplementary material, table S1. The model includes the populations of 321 cities around the world. Individuals can be susceptible, infected or (temporarily) recovered. Infected individuals are infectious for six days with no latency period. Infectious individuals can transmit to susceptible people in the same city or travel to other cities where they can infect others. After six days of infection (electronic supplementary material, figure S1), individuals recover and are completely immune for three months, after which they become fully susceptible again. Although the duration of immunity is longer in reality, three months is sufficient to ensure that an individual is unlikely to become infected more than once in a single outbreak.
Transmissibility follows an annual cycle in each city, with periods of higher transmissibility (R0 = 1.1) representing the influenza season and periods of low transmissibility (R0 = 0.8) representing the rest of the year [19]. Generally, the temperate Northern and Southern Hemispheres have influenza seasons during their respective winters, and the tropical regions often have influenza seasons that coincide with their rainy seasons and have higher transmissibility out of season than the temperate regions (figure 1b). To account for the higher out-of-season transmissibility in the tropics, the transmissibility was set to values higher than R0 = 0.8 during the low season in tropical cities as described in Kenah et al. [19].
In order to isolate the effects of oseltamivir use on the spread of resistance in our model, we model two different kinds of mutations and run simulations in which one or both of these types can occur. One mutation (which we refer to with the subscript M) is an arbitrary mutation that alters the transmissibility of the virus, but not its sensitivity to oseltamivir. The other mutation (which we refer to with the subscript R) is a mutation that specifically confers oseltamivir resistance. In principle, there may be many M mutations that alter transmissibility, such as by causing antigenic changes that help the virus evade pre-existing immunity. However, there are insufficient data to pinpoint exactly which mutations altered viral transmissibility of seasonal H1N1 in the time frame we are studying. On the other hand, for seasonal H1N1, there is only one R mutation that alters drug resistance, the H274Y substitution. We allow our model to account for both types of mutations because, as we will discuss below, both may have played a role in the spread of oseltamivir resistance.
In a simple strain competition model that does not include antiviral drug use, there is an original strain and a mutant strain, which has a fitness cost of cM that makes it (1−cM) times as transmissible (figure 1c). When cM is negative, the mutant has a transmission advantage. Transitions from the original to mutant strain arise owing to mutations with probability μ = 10−6 per day in infected people, and reversion from the mutant strain to original occurs at the same rate. Note that μ corresponds to the combined probability that a new mutant strain arises within a host and becomes the strain transmitted by that host. In the model with antiviral drug use, infected individuals can take oseltamivir. As shown in figure 1d, oseltamivir usage reduces by 63 per cent the probability that an individual becomes infected with the drug-sensitive strain, and makes them 15 per cent less-infectious should they become infected with that strain, which reduces transmissibility (figure 1d). These numbers are based on estimates from clinical data [20]. The fraction of infected people treated in various countries is calculated from prescription data (electronic supplementary material, table S2). In untreated individuals, resistance appears spontaneously with probability μ = 10−6 per day and is lost at the same rate. In infected individuals undergoing oseltamivir treatment, resistance appears with probability μT = 0.04 per day (see the electronic supplementary material). We assume that the oseltamivir usage has no effect on transmission of the resistant strain. The resistant strain has a fitness cost of cR relative to sensitive, corresponding to a change in transmission by a factor of 1−cR. We also ran simulations in which both kinds of mutations (i.e. the M and R mutations) were allowed, so there were four strains of influenza corresponding to the possible combinations of these two mutations (electronic supplementary material).
We fit our models to the estimated global prevalence of the H274Y mutation, as described in §4 of the electronic supplementary material, figure S2.
3. Oseltamivir usage alone cannot explain the speed at which resistance went to fixation in seasonal H1N1
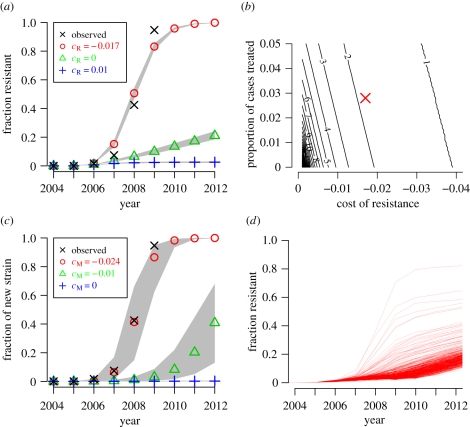
We used the models to examine how the usage of oseltamivir to treat influenza cases could increase the prevalence of drug-resistant influenza. We assumed that a drug-resistant strain would have a slight fitness cost in untreated hosts (lower transmission), but its transmissibility would not be further reduced in individuals taking the drug. When resistant strains are as little as 1 per cent less transmissible, they fail to spread appreciably (figure 2a). This finding concurs with the observed lack of resistant seasonal H1N1 strains prior to 2006, as experimental studies [10,11] have shown that resistant strains from 1991 and 1999 were severely attenuated.
Figure 2.
The resistant strain must be slightly more transmissible than the sensitive strain. (a) Simulated fraction of resistant isolates when the oseltamivir-resistant strain is 1% less transmissible (cR = 0.01), equally transmissible (cR = 0), or 1.7% more transmissible (cR = −0.017) than the oseltamivir-sensitive strain. The black crosses show the actual prevalence of resistance among sequenced seasonal H1N1 neuraminidases. The grey regions represent range covered by 95% of the 500 simulations. (b) The number of years for a resistant strain to rise from 1% to 50% prevalence for various values of cR and the proportion of cases treated in a simplified and deterministic model. Our estimate for the actual values is indicated by the red X. (c) Simulated fraction of mutant isolates in a simple strain-replacement model when no antivirals are used and the mutant strain is 0%, 1% or 2.4% more transmissible than the original strain (cM = 0.0, − 0.01, − 0.024). (d) Simulated fraction of resistant isolates when there are independent occurrences of an oseltamivir-resistance mutation that does not affect transmissibility (cR = 0) and a second mutation that confers at 2.4% increase in transmissibility (cM = −0.024). Each represents one of 500 runs of the stochastic simulation. There is wide variability in the spread of resistance depending on whether there is a stochastic co-occurrence of the two mutations early on. (a) Crosses, observed; circles, cR − 0.017; triangles, cR = 0; plus symbols, cR = 0.01. (b) Crosses, observed; circles, cM = −0.024; triangles, cM = −0.01; plus symbols, cM = 0.
Experimental studies have failed to detect any difference in fitness between oseltamivir-resistant strains from 2007 and later and their contemporaneous oseltamivir-sensitive counterparts [14,15]. When the resistant strain is exactly as transmissible as the drug-sensitive strain, oseltamivir usage is sufficient to drive a slow increase in the prevalence of resistant viruses (figure 2a). This result still holds when we assume that the efficacy of oseltamivir is extremely high (electronic supplementary material, figure S8). However, oseltamivir resistance was observed to increase extremely rapidly in seasonal H1N1 between 2006 and 2008, which is not compatible with the slow spread of resistance observed in the model.
Although some countries regularly used oseltamivir to treat influenza cases (electronic supplementary material, table S2), the worldwide average use of the drug was low. In a sensitivity analysis, we found that if the drug-resistant strain is not more transmissible than the sensitive strain, then about 30 per cent of all cases worldwide would need to take the drug in order to drive resistance to fixation as rapidly as observed (electronic supplementary material, figure S6). In our model, which over-represents the wealthier countries because of its focus on air travel, the fraction of cases that used oseltamivir was about 3 per cent. Therefore, we conclude that the oseltamivir use was at least an order of magnitude too low to select resistance as quickly as observed.
4. Oseltamivir-resistant seasonal H1N1 must have been more transmissible than the sensitive strains
One hypothesis that could explain the rapid spread of antiviral resistance is that the mutation that confers resistance also makes the virus slightly more transmissible in untreated hosts. Several authors have conjectured that H274Y might have slightly improved viral fitness, either by fine-tuning neuraminidase substrate affinity [16,21] or by improving haemagglutinin/neuraminidase balance [22,23]. In the model, enhanced transmissibility predictably speeds the fixation of the resistant strain, and a maximum-likelihood analysis indicates that the observed rise in resistance is best matched if the resistant strain is 1.7 per cent more transmissible (cR = −0.017, 95% CI = −0.019, − 0.016) than the sensitive strain (figure 2a). The prophylactic use of the drug had no significant effect on the results (electronic supplementary material, figure S5). We found that higher treatment-induced mutation rates (μT) did not significantly affect the results, while higher background mutation rates (μ) produced results inconsistent with observations (electronic supplementary material, figure S7).
To test the robustness of the estimate of cR with respect to the choice of modelling approach, we constructed a simpler and deterministic alternative model of strain replacement (described in the electronic supplementary material). Briefly, we assume a drug-sensitive and a drug-resistant strains are transmitted in a single, infinite population, a fraction of which is treated with the drug when infected. Consistent with the stochastic model, we found that for drug resistance to reach 50 per cent prevalence among circulating strains in a time span of about 1–2 years requires the resistant strain to be 1–2% more transmissible, and results are similar when up to 5 per cent of infected individuals are treated (figures 2b and electronic supplementary material, S4). Thus, the speed at which fixation occurs, once resistance is established, does not depend on stochastic effects, influenza seasonality or air travel. However, as described below, a stochastic and spatial model is needed to describe regional observations of antiviral resistance. Using the deterministic model, we found that the speed of fixation is more sensitive to the cost of resistance than to the fraction of cases treated (figure 2b).
An alternative hypothesis is that antiviral resistance does not make the virus inherently more transmissible, but rather that the resistance mutation ‘hitchhiked’ on another advantageous mutation. Such hitchhiking is thought to have explained the spread of adamantane resistance in seasonal H3N2 [24]. A number of authors have suggested that H274Y might have similarly hitchhiked on a haemagglutinin antigenic drift mutation [12,15,23,25]. Therefore, the use of oseltamivir may have had a negligible effect on the spread of resistance. To test this hypothesis, we used a model in which there is no antiviral agent use, and a new, more transmissible strain can arise through mutation (figure 1c). The new strain goes to fixation as quickly as did the oseltamivir resistance in seasonal H1N1, when it is 2.4 per cent more transmissible than the original strain (cM = −0.024, 95% CI = −0.028, − 0.014; figures 2c electronic supplementary material, figure S3b).
There are several possible hitchhiking scenarios involving H274Y and a second mutation conferring increased transmissibility. One of these two mutations could have stochastically occurred in the background of the other, leading to a virus that possessed both resistance and increased transmissibility (figure 2d). In most simulations with a model that includes both a resistance mutation that does not alter transmissibility in untreated hosts (cR = 0) and a mutation that increases transmissibility in all hosts (cM = −0.024), an antiviral sensitive strain with the transmissibility mutation quickly goes to fixation, then resistance slowly spreads in this background. But when the mutations stochastically co-occur early enough, resistance spreads at speeds comparable to that observed in seasonal H1N1. A second scenario is that reassortment may have combined independent occurrences of the two mutations. This could only have been the case if the two mutations occurred on different viral gene segments, since intra-segment reassortment in influenza is thought to be extremely rare [26]. A final scenario is that selection for improved haemagglutinin/neuraminidase balance [22,23] specifically favoured the combination of H274Y and a mutation near the haemagglutinin receptor-binding pocket that altered antigenicity, thereby improving transmissibility. This scenario is similar to the case shown in figure 2a in which the resistant strain is more transmissible, and would obviously lead to rapid fixation of resistance.
5. Little correlation between oseltamivir usage and resistance
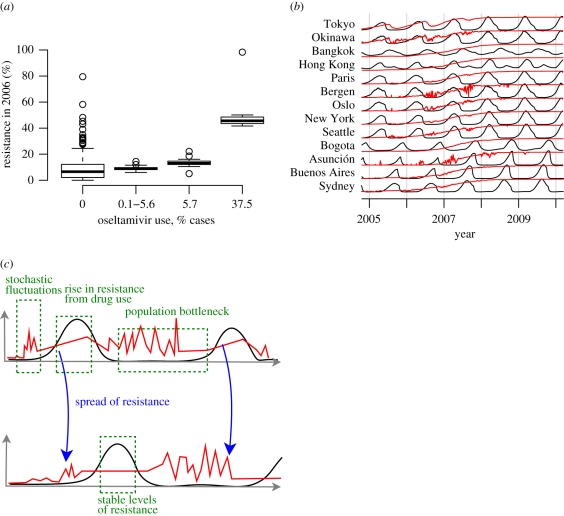
One of the most puzzling aspects of the appearance of oseltamivir resistance in seasonal H1N1 was the observed lack of correlation between the prevalence of resistance and oseltamivir usage [12,18]. We measured the correlation between the fraction of cases treated with oseltamivir in a city and the simulated fraction of residents infected with influenza who were infected with a resistant strain during a calendar year. In a simulation in which oseltamivir resistance hitchhikes on a fitness-enhancing mutation (cM = −0.024, cR = 0, corresponding to figure 2d), the prevalence of resistant strains in a city was not strongly correlated with the level of oseltamivir usage (R2 = 0.20 for the first year of the simulation and R2 = 0.14 for the second with p < 0.01 for both years, figure 3a).
Figure 3.
Simulated levels of oseltamivir resistance in individual cities. (a) The fraction of resistant strains from a single simulation in individual cities for the calendar year 2006 when resistance hitchhikes on a fitness-enhancing mutation (cM = −0.024, cR = 0). The thick horizontal lines indicate the median fractions of resistance, the boxes outline the first and third quartiles, and the whiskers show the range of 95% of the data. The first resistance mutations occur in 2005 in the simulations. (b) Simulated antiviral resistance in selected cities when resistance hitchhikes on a fitness-enhancing mutation (cM = −0.024, cR = 0), for the simulation shown in (a). Black lines plot infection prevalence in a city, and red lines plot the fraction of resistant strains. (c) Illustration of the transmission of resistant influenza strains between cities. The black lines plot infection prevalence, and the red lines plot the fraction of infected individuals who have the resistant strain.
During the influenza season, our model behaves deterministically, and the fraction of circulating resistant strains may increase when regional conditions favour transmission of the resistant strain (e.g. a high level of drug usage) (figure 3b, see curves for Tokyo and Okinawa). Outside the influenza season, when the number of cases is low in a region, the composition of circulating strains can be strongly influenced by both stochastic fluctuations and importation (figure 3b,c). Stochastic fluctuations may cause the prevalence of resistance to increase or decrease dramatically, while importation from passenger air travel allows a city to ‘sample’ the global proportion of resistant strains. Once the influenza season starts, the exponential transmission of all circulating strains of influenza can stabilize the proportion of resistant strains and ‘lock in’ the proportion of resistance set by stochastic fluctuations, particularly in small cities (figure 3b, see curves for Bergen and Asunción).
6. Interventions to slow the spread of antiviral resistance
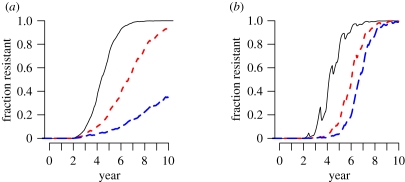
One possible strategy for slowing the spread of antiviral-resistant influenza is to target the source, i.e. infected individuals who are taking antivirals. We model this approach by further reducing the infectiousness of individuals taking antivirals by φ. This would represent the effects of control measures such as careful isolation of patients or increased adherence to proper hygiene. If the resistant strain is exactly as transmissible as the sensitive strain, then this strategy would slow the spread of resistance (figure 4a). On the other hand, if the drug resistance mutation confers a fitness advantage (i.e. cR < 0), then speed at which the resistant strain spreads would not change, but increased infection control for treated infected individuals might delay the beginning of spread (figure 4b).
Figure 4.
The effect of increased transmission control on the prevalence of resistance. (a) If 5% of all cases in the world are treated and the resistant and sensitive strains are equally transmissible in untreated individuals (cR = 0), then antiviral use drives selection of resistant strains. In these simulations, we assume that all cases in the world have the same probability of treatment. We alter the parameter φ, the relative transmissibility of treated infected individuals owing to increased measures to prevent transmission from treated individuals. Note that this parameter affects those infected with either the resistant or sensitive strain. We ran the model for φ = 1 (black solid line; no effort is made to reduce transmission from treated cases, black line), φ = 0.5 (red dashed line; treated cases are half as transmissible, red line) and φ = 0.25 (blue solid line; treated cases are 25% as transmissible, blue line). (b) If only a small fraction of individuals are treated (i.e. levels of oseltamivir use as described in the text) and the resistant strain is 1.7% more transmissible in untreated individuals (cR = −0.017), then reducing infections from treated cases does not appreciably slow the spread of resistant strains.
7. Discussion
Our simulations help clarify the forces that were necessary to allow the spread of oseltamivir resistance in seasonal H1N1. A drug-resistant virus would be unable to establish itself if it is even slightly less transmissible than the sensitive virus, and this explains the lack of observed resistance in seasonal H1N1 in the 1990s. However, by 2007, seasonal H1N1 had acquired mutations that eliminated the fitness difference between resistant and sensitive viruses [14–17] (electronic supplementary material, figure S2). Once this occurred, direct selection owing to oseltamivir usage could favour the spread of resistance. However, this selection would have been insufficient to fix resistance as rapidly as observed. Therefore, the selection owing to oseltamivir usage was likely assisted either by another mutation, or by some small additional transmissibility benefit conferred by the resistance mutation itself. In our analyses, we found that mutations that increased transmissibility among untreated individuals were associated with the most rapid increases in drug-resistant influenza, while factors that only accelerated the generation of resistant strains, such as elevated mutation and treatment rates, were associated with slower increases in resistance that was inconsistent with observations.
Our results suggest that once established (i.e. present at sufficient levels that stochastic fluctuations would not cause it to go extinct), a drug-resistant strain may spread rapidly during the influenza season, as was observed in Europe [27]. Conversely, the rapid decline in cases at the end of the season can also cause large fluctuations in the proportion of circulating strains. For example, the proportion of resistant strains may increase significantly during the influenza season in a city that treats a large number of cases but the proportion may decline at the end of the season (e.g. Tokyo in figure 3b). However, Japan, the largest consumer of oseltamivir, may not have had the required background of strains with secondary mutations to compensate for the resistance mutation until 2007 (electronic supplementary material, figure S2). Although deterministic models can describe the dynamics of strain replacement once new strains become established [28], the stochastic effects from the seasonality of influenza can drive local patterns of resistance. In fact, any local adaptation of influenza strains may disappear at the end of the season in temperate regions [29]. In addition, the rapid global mixing of influenza strains [30,31] could reduce the correlation between local rates of resistance and oseltamivir usage. Because regional trends in drug-resistance can be misleading, surveillance must be global.
Influenza must constantly evolve (drift antigenically) in response to intense immune pressure from human hosts, which probably drives the replacement of strains every 2–8 years [32,33]. With such frequent selective sweeps, it is perhaps not surprising that traits that have small impacts on viral fitness, such as oseltamivir resistance in seasonal H1N1 or adamantane resistance in seasonal H3N2 [24], spread so rapidly. Although the model in which a single mutation both conferred antiviral agent resistance, and increased transmissibility could produce results consistent with observations, we believe that it is more likely that hitchhiking on an antigenically drifted strain of flu occurred.
Our findings have implications for predicting the spread of resistance in pandemic influenza (H1N1) 2009. A small fraction of pandemic H1N1 viruses have been found to contain the H274Y mutation [34]. Most of the oseltamivir-resistant isolates were from immunocompromised patients or individuals taking oseltamivir, with only a few reported cases of resistant virus being transmitted in healthy untreated adults [34]. In early seasonal H1N1 isolates (i.e. 1991 and 1999), H274Y led to profound attenuation in experimental models [10,11]. In contrast, experimental studies of pandemic H1N1 have found that H274Y is associated with only a slight but nonetheless perceptible reduction in viral growth in tissue culture [35–39] or airborne transmission in animal models [36,39,40]. A question has been what such a slight effect on viral fitness might imply for the spread of resistance. Our simulations show that a resistant virus will fail to spread if it is even slightly less transmissible than the sensitive strain (figure 2a). This finding suggests that if H274Y causes even extremely minor attenuation of transmissibility in pandemic H1N1, then it will fail to spread widely without either secondary mutations that remedy the slight defect or adventitious hitchhiking.
To slow the spread of drug-resistant strains of influenza, we suggest increased infection control for treated individuals. This strategy could delay the establishment of resistant versions of currently circulating strains in the general population. However, our model assumes that a random selection of infected individuals are treated with the drug. If those taking antiviral agents are likely to be in contact with others also taking the same drug, such as in a hospital ward, our model's random mixing assumption is violated and the model may under- or overestimate the efficacy of this intervention depending on the clustering of those taking antiviral agents. Once established in a small fraction of the population, the speed at which resistance spreads might depend entirely on its transmissibility relative to other circulating strains, regardless of the level of drug use or infection control. Fortunately, resistance-granting mutations appear to often be detrimental, but the experience with seasonal H1N1 shows that such mutations can still quickly go to fixation given the correct combination of background secondary mutations and adventitious hitchhiking.
Acknowledgements
This work was partially supported by the National Institute of General Medical Sciences MIDAS grant U01-GM070749. B.F.K. was supported by the Fannie and John Hertz Foundation.
References
- 1.Ferguson N. M., Cummings D. A. T., Cauchemez S., Fraser C., Riley S., Meeyai A., Iamsirithaworn S., Burke D. S. 2005. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437, 209–214 10.1038/nature04017 (doi:10.1038/nature04017) [DOI] [PubMed] [Google Scholar]
- 2.Longini I. M., Jr, Nizam A., Xu S., Ungchusak K., Hanshaoworakul W., Cummings D. A. T., Halloran M. E. 2005. Containing pandemic influenza at the source. Science 309, 1083–1087 10.1126/science.1115717 (doi:10.1126/science.1115717) [DOI] [PubMed] [Google Scholar]
- 3.Ortiz J. R., Kamimoto L., Aubert R. E., Yao J., Shay D. K., Bresee J. S., Epstein R. S. 2008. Oseltamivir prescribing in pharmacy-benefits database, United States, 2004–2005 Emerg. Infect. Dis. 14, 1280–1283 10.3201/eid1408.080074 (doi:10.3201/eid1408.080074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubareva L. V., Kaiser L., Matrosvich M. N., Soo-Hoo Y., Hayden F. G. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183, 523–531 10.1086/318537 (doi:10.1086/318537) [DOI] [PubMed] [Google Scholar]
- 5.Whitley R. J., Hayden F. G., Reisinger K. S., Young N., Dutkowski R., Ipe D., Mills R. G., Ward P. 2001. Oral oseltamivir treatment of influenza in children. Pediatric Infect. Dis. J. 20, 127–133 10.1097/00006454-200102000-00002 (doi:10.1097/00006454-200102000-00002) [DOI] [PubMed] [Google Scholar]
- 6.Bouvier N. M., Lowen A. C., Palese P. 2008. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J. Virol. 82, 10 052–10 058 10.1128/JVI.01226-08 (doi:10.1128/JVI.01226-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr J., Ives J., Kelly L., Lambkin R., Oxford J., Mendel D., Tai L., Roberts N. 2002. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antivir. Res. 54, 79–88 10.1016/S0166-3542(01)00215-7 (doi:10.1016/S0166-3542(01)00215-7) [DOI] [PubMed] [Google Scholar]
- 8.Herlocher M. L., Carr J., Ives J., Elias S., Truscon R., Roberts N., Monto A. S. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antivir. Res. 54, 99–111 10.1016/S0166-3542(01)00214-5 (doi:10.1016/S0166-3542(01)00214-5) [DOI] [PubMed] [Google Scholar]
- 9.Abed Y., Goyette N., Bovin G. 2004. A reverse genetics study of resistance to neuraminidase inhibitors in an influenza A/H1N1 virus. Antivir. Therapy 9, 577–581 [PubMed] [Google Scholar]
- 10.Herlocher M. L., Truscon R., Elias S., Yen H. L., Roberts N. A., Ohmit S. E., Monto A. S. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190, 1627–1630 10.1086/424572 (doi:10.1086/424572) [DOI] [PubMed] [Google Scholar]
- 11.Ives J. A. L., Carr J. A., Mendel D. B., Tai C. Y., Lambkin R., Kelly L., Oxford J. S., Hayden F. G., Roberts N. A. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 55, 307–317 10.1016/S0166-3542(02)00053-0 (doi:10.1016/S0166-3542(02)00053-0) [DOI] [PubMed] [Google Scholar]
- 12.Baranovich T., et al. 2010. Emergence of H274Y oseltamivir-resistant A(H1N1) influenza viruses in Japan during the 2008–2009 season. J. Clin. Virol. 47, 23–28 10.1016/j.jcv.2009.11.003 (doi:10.1016/j.jcv.2009.11.003) [DOI] [PubMed] [Google Scholar]
- 13.Moscona A. 2009. Global transmission of oseltamivir-resistant influenza. N Engl. J. Med. 360, 953–956 10.1056/NEJMp0900648 (doi:10.1056/NEJMp0900648) [DOI] [PubMed] [Google Scholar]
- 14.Baz M., Abed Y., Simon P., Hamelin M. E., Boivin G. 2010. Effect of neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J. Infect. Dis. 201, 740–745 10.1086/650464 (doi:10.1086/650464) [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki Y., et al. 2010. A two-year survey of the oseltamivir-resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamavir. Virol. J. 7, 53. 10.1186/1743-422X-7-53 (doi:10.1186/1743-422X-7-53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rameix-Welti M.-A., Enouf V., Cuvelier F., Jeannin P., van der Werf S. 2008. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 4, e1000103 10.1371/journal.ppat.1000103 (doi:10.1371/journal.ppat.1000103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom J. D., Gong L. I., Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 10.1126/science.1187816 (doi:10.1126/science.1187816) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramarz P., Monnet D., Nicoll A., Yilmaz C., Ciancio B. 2009. Use of oseltamivir in 12 European countries between 2002 and 2007—lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses. Euro. Surveill. 14, 19112. [DOI] [PubMed] [Google Scholar]
- 19.Kenah E., Chao D. L., Matrajt L., Halloran M. E., Longini , I. M., Jr 2011. The global transmission and control of influenza. PLoS ONE 6, e19515. 10.1371/journal.pone.0019515 (doi:10.1371/journal.pone.0019515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halloran M. E., Hayden F. G., Yang Y., Longini I. M., Jr, Monto A. S. 2007. Antiviral effects on influenza viral transmission and pathogenicity: observations from household-based trials. Am. J. Epidemiol. 165, 212–21 10.1093/aje/kwj362 (doi:10.1093/aje/kwj362) [DOI] [PubMed] [Google Scholar]
- 21.Collins P. J., et al. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27, 6317–6323 10.1016/j.vaccine.2009.07.017 (doi:10.1016/j.vaccine.2009.07.017) [DOI] [PubMed] [Google Scholar]
- 22.Wagner R., Matrosovich M., Klenk H. D. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12, 159–166 10.1002/rmv.352 (doi:10.1002/rmv.352) [DOI] [PubMed] [Google Scholar]
- 23.Zaraket H., Saito R., Suzuki Y., Baranovich T., Dapta C., Caperig-Dapat I., Suzuki H. 2010. Genetic makeup of amantadine-resistant and oseltamivir-resistant human influenza A/H1N1 viruses. J. Clin. Microbiol. 48, 1085–1092 10.1128/JCM.01532-09 (doi:10.1128/JCM.01532-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen L., et al. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol. Biol. Evol. 24, 1811–1820 10.1093/molbev/msm103 (doi:10.1093/molbev/msm103) [DOI] [PubMed] [Google Scholar]
- 25.Niman H. L. 2009. Emergence and fixing of antiviral resistance in influenza A via recombination and hitch hiking. Nat. Precedings. See http://hdl.handle.net/10101/npre.2009.2832.1 [Google Scholar]
- 26.Boni M. F., Zhou Y., Taubenberger J. K., Holmes E. C. 2008. Homologous recombination is very rare or absent in human influenza A virus. J. Virol. 82, 4807–4811 10.1128/JVI.02683-07 (doi:10.1128/JVI.02683-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer A., et al. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg. Infect. Dis. 15, 552–560 10.3201/eid1504.081280 (doi:10.3201/eid1504.081280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.zur Wiesch P. A., Kouyos R., Engelstädter J., Regoes R. R., Bonhoeffer S. 2011. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect. Dis. 11, 236–47 10.1016/S1473-3099(10)70264-4 (doi:10.1016/S1473-3099(10)70264-4) [DOI] [PubMed] [Google Scholar]
- 29.Nelson M. I., Simonsen L., Viboud C., Miller M. A., Holmes E. C. 2007. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 3, e0030131. 10.1371/journal.ppat.0030131 (doi:10.1371/journal.ppat.0030131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedford T., Cobey S., Beerli P., Pascual M. 2010. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog. 6, e1000918. 10.1371/journal.ppat.1000918 (doi:10.1371/journal.ppat.1000918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A., Pybus O. G., Nelson M. I., Viboud C., Taubenberger J. K., Holmes E. C. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 10.1038/nature06945 (doi:10.1038/nature06945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plotkin J. B., Dushoff J., Levin S. A. 2002. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl Acad. Sci. USA 99, 6263–6268 10.1073/pnas.082110799 (doi:10.1073/pnas.082110799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith D. J., Lapedes A. S., de Jong J. C., Bestebroer T. M., Rimmelzwaan G. F., Osterhaus A. D. M. E., Fouchier R. A. M. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 10.1126/science.1097211 (doi:10.1126/science.1097211) [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. 2011 Update on oseltamivir resistance in influenza A(H1N1) 2009 viruses: 15 June 2011. See http://www.who.int/entity/csr/disease/influenza/2011_06_17_weekly_web_update_oseltamivir_resistance.pdf. (accessed on June 19 2011) [Google Scholar]
- 35.Brookes D. W., Miah S., Lackenby A., Hartgroves L., Barclay W. S. 2011. Pandemic H1N1 2009 influenza virus with the H275Y oseltamivir resistance neuraminidase mutation shows a small compromise in enzyme activity and viral fitness. J. Antimicrob. Chemother. 66, 466–70 10.1093/jac/dkq486 (doi:10.1093/jac/dkq486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan S., Boltz D. A., Seiler P., Li J., Bragstad K., Nielsen L. P., Webby R. J., Webster R. G., Govorkova E. A. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6, e1001022. 10.1371/journal.ppat.1001022 (doi:10.1371/journal.ppat.1001022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamelin M.-È., et al. 2010. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 6, e1001015. (doi:10.1371/journal.ppat.1001015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizzorno A., Bouhy Z., Abed Y., Boivin G. 2011. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J. Infect. Dis. 203, 25–31 10.1093/infdis/jiq010 (doi:10.1093/infdis/jiq010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seibert C. W., et al. 2010. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 84, 11 219–11 226 10.1128/JVI.01424-10 (doi:10.1128/JVI.01424-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiso M., et al. 2010. Characterization of oseltamivir-resistant 2009 H1N1 pandemic influenza A viruses. PLoS Pathog. 6, e1001079. 10.1371/journal.ppat.1001079 (doi:10.1371/journal.ppat.1001079) [DOI] [PMC free article] [PubMed] [Google Scholar]