Abstract
The total contribution of the acyl CoA:diacylglycerol acyltransferase (DGAT) enzymes, DGAT1 and DGAT2, to mammalian triacylglycerol (TG) synthesis has not been determined. Similarly, whether DGAT enzymes are required for lipid droplet (LD) formation is unknown. In this study, we examined the requirement for DGAT enzymes in TG synthesis and LDs in differentiated adipocytes with genetic deletions of DGAT1 and DGAT2. Adipocytes with a single deletion of either enzyme were capable of TG synthesis and LD formation. In contrast, adipocytes with deletions of both DGATs were severely lacking in TG and did not have LDs, indicating that DGAT1 and DGAT2 account for nearly all TG synthesis in adipocytes and appear to be required for LD formation during adipogenesis. DGAT enzymes were not absolutely required for LD formation in mammalian cells, however; macrophages deficient in both DGAT enzymes were able to form LDs when incubated with cholesterol-rich lipoproteins. Although adipocytes lacking both DGATs had no TG or LDs, they were fully differentiated by multiple criteria. Our findings show that DGAT1 and DGAT2 account for the vast majority of TG synthesis in mice, and DGAT function is required for LDs in adipocytes, but not in all cell types.
Keywords: neutral lipids, diacylglycerol, sterol esters, lipases, phospholipids, adipogenesis, macrophages, perilipin
Lipid droplets (LDs) are found in nearly all eukaryotic cells, and each consists of a neutral lipid core enveloped by a phospholipid monolayer and surface proteins (1). The main lipids found in the cores of LDs are triacylglycerols (TGs) and sterol esters (SEs). Whether TGs are required for the formation of LDs is unknown. The major known enzymes that catalyze TG synthesis in mammals are the acyl CoA:diacylglycerol acyltransferases (DGAT; Fig. 1A) (2), which catalyze the covalent addition of a fatty acyl chain to diacylglycerol. Genetic deletion of DGAT1 in mice revealed that this enzyme is not essential and that DGAT1 knockout (DGAT1 KO) mice have reductions in TG levels in many tissues, including adipose tissue, when fed a high-fat diet (3). Deletion of DGAT2 revealed that this enzyme is essential: mice lacking DGAT2 have severe reductions in TG levels and die shortly after birth (4). Nevertheless, newborn DGAT2 KO mice do have some TG, which may be due to DGAT1 activity. Given that enzymes in both the DGAT1 (MBOAT, 16 family members) and DGAT2 (7 members) families possess many different lipid acyltransferase activities (5, 6), and that several of these enzymes have unknown functional activities, it is unclear whether TG is normally made by enzymes other than DGAT1 and DGAT2. Furthermore, it is unclear whether cells lacking DGAT enzymes are competent to form LDs.
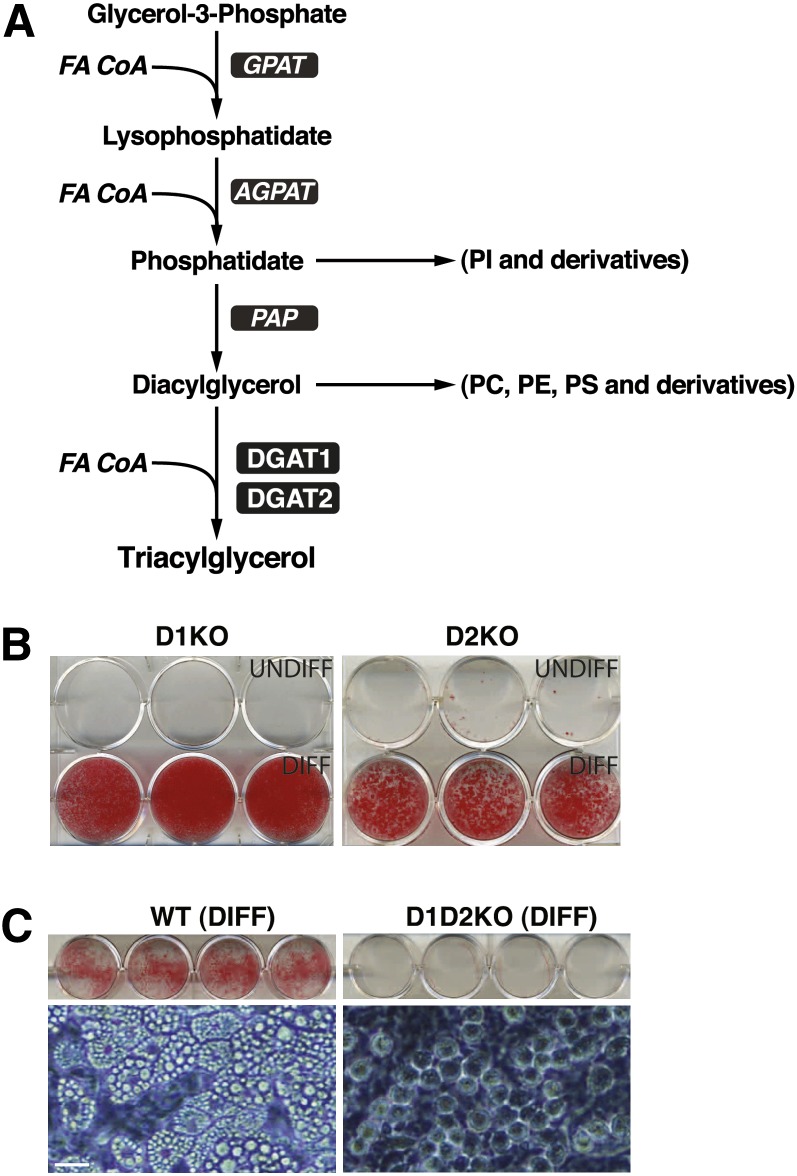
Fig. 1.
Lack of lipid accumulation in MEFs deficient in both DGAT1 and DGAT2. A: The Kennedy pathway of TG synthesis. FA CoA, fatty acyl CoA; GPAT, glycerol-phosphate acyltransferase; AGPAT, acylglycerolphosphate acyltransferase; PAP, phosphatidic acid phosphatase. B: Single deletions of DGAT1 (D1KO) or DGAT2 (D2KO) do not affect adipogenesis as assessed by lipid accumulation (Oil-Red-O staining). UNDIFF, undifferentiated; DIFF, differentiated. C: Differentiation of MEFs deficient in both DGATs (D1D2KO) results in cells that lack lipid staining (top) and appear to lack LDs (bottom, phase-contrast microscopy image). Scale bar, 20 μm. Experiment was performed six times, with representative results shown. Exogenous FAs were not added during differentiation.
In this study, we sought to determine the requirements for DGAT enzymes for TG synthesis and LD formation in mammals, focusing on adipocytes, the major cell type for TG storage. To do so, we utilized a murine embryonic fibroblast (MEF) differentiation paradigm combined with genetic deletions of DGAT genes. The MEF differentiation system has been utilized extensively to study the function of genes in adipocyte formation (7–11) and is a well-accepted in vitro model. In particular, this model is useful for examining the role of essential genes, such as DGAT2 (4), in which early postnatal lethality precludes studies of adipogenesis in vivo. The adipocyte paradigm is also very useful for studying the enzymes of TG synthesis because they are present at high levels in adipocytes. Here we focused on the ability of the differentiated adipocytes with DGAT deletions to synthesize TG and form LDs. We also examined these processes in macrophages lacking both DGAT1 and DGAT2 derived from fetal tissues.
MATERIALS AND METHODS
All materials were from Sigma unless otherwise indicated.
Mice
DGAT1 and DGAT2 KO mice were generated as described (3, 4). To generate mice deficient in both DGAT1 and DGAT2 (D1D2KO), Dgat1Dgat2+/− mice were interbred. Wild-type (WT) controls were from a WT litter with identical plug date. Adiponectin-null mice were as described (12). All mice were C57BL/6J background. All experiments were in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and were approved by the University of California San Francisco Institutional Animal Care and Use Committee.
Cells
MEFs.
Timed pregnant mice were euthanized at E14, and MEFs were prepared and differentiated according to the standard protocol (13). Briefly, embryos were dissected away from the uterus and embryonic membranes. The viscera and head were removed, and the head was used for genotyping. The carcass was minced and incubated at 37°C for 45 min in 0.05% trypsin. The cells were triturated and then plated in high-glucose DMEM (Invitrogen) and 10% FBS (Hyclone) supplemented with antibiotics. Two days after confluence, cells were treated with adipogenic cocktail (5 μg/ml insulin, 500 μM 3-Isobutyl-1-methylxanthine, 1 μM dexamethasone, 10 μM troglitazone). On day 4, cells were treated with insulin and troglitazone. On day 6, cells were switched back to media with no supplements. Experiments were performed on day 8 or day 9 unless otherwise indicated. Highest-yield differentiation of MEFs occurred with minimal passaging of cells.
Macrophages.
E14 livers were incubated at 37°C in 0.05% trypsin for 15 min, then triturated, and plated on noncoated plasticware in RPMI+10% FBS with 10 ng/ml recombinant macrophage colony-stimulating factor (M-CSF; R and D Systems). Cells were used 6 days after plating. Acetylated LDL (Kalen Biomedical) was used at 25 μg/ml to induce SE droplets.
3T3-L1 cells were purchased from American Type Culture Collection and were differentiated as for MEFs, but without troglitazone.
Statistical analyses
Unless otherwise indicated, P values were determined by Student's t-test.
Assays
ORO staining.
Cells were washed with PBS, fixed with 4% paraformaldehyde, washed with water, dried, and stained with 0.3% Oil Red O (ORO) in 60% isopropanol for 15 min. Stained cells were washed with 70% ethanol and water and dried, and the tissue culture plates were scanned on a flatbed scanner.
Cellular lipid analysis.
TLC plates were dipped in 10% cupric sulfate in 8.4% phosphoric acid and charred at 140°C for 10 min.
Tissue TG was quantified by densitometry of the charred plate with ImageJ.
Cellular lipid synthesis.
Adipocytes were treated with 0.125 μCi/ml of [14C]oleic acid and 250 μM cold oleic acid for 2 h. Cells were washed twice with PBS, and lipids were extracted with hexane-isopropanol (3:2), dried down, run on TLC, and quantified using a Bioscan AR-2000 instrument. Macrophages were pretreated with acetylated LDL for 24 h and labeled with 0.125 μCi/ml of [14C]oleic acid tracer during the last 2 h. Neutral lipids were run on hexane-ethyl ether-glacial acetic acid (80:20:1) and polar lipids on methyl acetate-2-propanol-chloroform-methanol-0.25% KCl (25:25:25:10:9) solvent system.
DGAT activity assays.
DGAT assays were performed as described (5). Briefly, 50 μg of protein lysate (in 250 mM sucrose, 50 mM Tris-HCl, pH 7.4, supplemented with protease inhibitors; Complete Roche) were incubated for 5 min at 37°C with 200 μM diacylglycerol (dissolved in acetone) and 25 μM [14C]oleoyl-CoA. The reaction was stopped with 4 ml of 3:1 chloroform-methanol, 750 μl of water was added, and the organic phase was extracted, dried, and subjected to TLC (hexane-diethylether-acetic acid, 80:20:1 as solvent). TG product was quantified as for TG synthesis.
TG hydrolase activity assays.
Cells were washed two times with PBS and collected by scraping. Cells were disrupted by sonication (Virsonic 425,Virtis) at 4°C in buffer A (50 mM Tris-HCl, pH 7.4, 250 mM sucrose, 1 mM EDTA, 1 mM dithiothreitol, 20 µg/ml leupeptin, 1 µg/ml pepstatin, 2 µg/ml antipain). The cell lysate was centrifuged at 15,000 g for 15 min at 4°C, and the infranant was used for TG hydrolase activity. Protein content was determined using the Bradford method (Biorad Laboratories)
The TG hydrolase assay was performed as described (14). The reaction contained 40 µg protein and was incubated with 100 µl substrate in a total volume of 200 µl at 37°C for 60 min. In some reactions, the hormone-sensitive lipase (HSL) inhibitor (NNC 0076-0000-0079; kind gift of Novo-Nordisk) was used at 25 μM. The reaction was terminated by addition of 3.25 ml methanol-chloroform-heptane (10:9:7 v/v/v) and 1 ml of potassium carbonate, 0.1 M boric acid, pH 10.5. After centrifugation at 800 g for 15 min, the radioactivity in 1 ml of the upper aqueous phase was determined by liquid scintillation counting. The substrate for TG hydrolase was prepared by emulsifying 33 nM triolein/assay (glyceroltri[9,10(n)-3H] oleate 40,000 cpm/nmol; GE Healthcare) and 45 μM phosphatidylcholine (PC)-phosphatidylinositol (3:1) by sonication in 100 mM potassium phosphate buffer, pH 7.4 and 2% FA-free BSA. Glycerol release was determined using Free Glycerol Reagent (Sigma).
Glucose transport assays were performed as described (15), with modifications. On day 7, the culture medium for adipocytes was changed to low glucose (5 mM). On day 10, cells were cultured in serum-free Krebs-Ringer phosphate buffer (128 mM NaCl, 12.5 mM NaH2PO4/Na2HPO4, pH 7.4, 5 mM KCl, 1.3 mM MgSO4, 1.37 mM CaCl2) containing 0.5% BSA for 4 h. The insulin-treated group was treated with 100 nM insulin for 15 min. 3H-2-deoxylgucose (1 μCi/ml, final total 2-deoxylgucose concentration 100 μM) was then added for 5 min, cells were washed four times with ice cold PBS and lysed, and the lysate was subjected to scintillation counting. Separate groups of cells for each genotype were pretreated with 30 μM cytochalasin B, and the cytochalasin-independent counts were subtracted from totals to account for nonspecific transport.
Western blotting
Western blots were incubated with anti- adipose triglyceride lipase (ATGL) polyclonal antibody (1:1000; Cell Signaling), anti-total-HSL polyclonal antiserum (1:2000; Cell Signaling), anti-phospho-serine563-HSL (1:1000; Cell Signaling), anti-comparative gene identification 58 (CGI-58) polyclonal antiserum (1:10000), β-actin polyclonal antibody (1:2000), or anti-perilipin antibody (1:1000; Affinity Bioreagents). For all immunoblots, the antibody binding was subjected to appropriate HRP-conjugated secondary antibodies, detected with ECL plus (GE Healthcare), and exposed to X-ray film (Hyperfilm ECL; GE Healthcare).
Immunofluorescence
Cells were washed with PBS, fixed in 4% paraformaldehyde for 10 min, washed with PBS, and blocked in 1% goat serum. Cells were incubated in primary antibody for 1 h at room temperature and then with appropriate secondary antibody for 1 h at room temperature. Cells were stained for LDs with BODIPY493/503 (1 μg/ml; Molecular Probes) and for nuclei with Hoechst 33342 (Molecular Probes). Images were acquired with MicroManager on a Nikon Eclipse Ti 2000 confocal microscope. Primary perilipin antibody (kind gift of Dawn Brasaemle) was used at 1:5000. Alexa 564-conjugated secondary antibody (Invitrogen) was used at 1:500.
Electron microscopy
Cells were fixed in 1.5% glutaraldehyde, 4% polyvinylpyrrolidone, and 0.5% calcium chloride in 0.1 M sodium cacodylate buffer, pH 7.4. The imidazole-buffered osmium tetroxide procedure (16) was used to stain for lipids. Tissue was en block-stained in aqueous uranyl acetate, dehydrated, infiltrated, and embedded in LX-112 resin (Ladd Research Industries). Samples were ultrathin sectioned on the Reichert Ultracut S ultramicrotome and stained with 0.8% lead citrate. Grids were examined on a JEOL JEM-1230 electron microscope (JEOL USA) and photographed using the Gatan Ultrascan 1000 digital camera (Gatan). Mitochondrial area determinations were performed as described (16). Caveolae density was determined by counting the number of caveolae per linear micron in a ribbon whose width was three times the caveolae diameter and parallel to the cell surface.
Mass spectrometric analysis of TG molecular species
TG analyses were as described (17, 18). Briefly, TGs were extracted into chloroform by the Bligh and Dyer method, the chloroform layer was evaporated to dryness under nitrogen, and the lipid extract was reconstituted with 1:1 CHCl3/MeOH. Lipid extracts were flushed with nitrogen, capped, and stored at −20°C before ESI/MS analysis. A TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with an automated nanospray apparatus (Nanomate HD; Advion Bioscience) and Xcalibur system software were utilized (19). Each lipid extract was diluted to <50 pmol of total lipids/μl with CHCl3-MeOH-isopropanol (1:2:4 v/v/v) before infusion into the mass spectrometer by the Nanomate apparatus. For all analyses, the first and third quadrupoles were used as independent mass analyzers with mass resolution settings of 0.7 Th, and the second quadrupole served as a collision cell for MS/MS. Typically, spectra were acquired using a 2 min period of signal averaging in the profile mode for each full mass spectra scan. For MS/MS, the collision gas pressure was set at 1.0 mTorr, with the collision energy set at 35 eV. For each tandem mass spectrum, a 2−5 min period of signal averaging in the profile mode was employed. All full MS scans and MS/MS scans were automatically acquired using a customized sequence subroutine operated under Xcalibur software. Data from biological samples were normalized to the protein content, and all data are presented as the mean ± standard error.
Transcriptional analysis
Total RNA was purified from cultured cells with Trizol (Invitrogen). Contaminating genomic DNA was removed with Turbo DNA-free DNase (Ambion). Sample amplification and labeling was performed according to the manufacturer's specifications with Affymetrix WT cDNA Synthesis and Amplification Kits and WT Terminal Labeling Kits. Biotinylated single stranded DNAs were hybridized to Affymetrix Mouse Gene 1.0 sense target arrays, stained, and scanned according to the manufacturer's instructions. Data analysis was performed in R with Bioconductor software packages (20). Data were read and preprocessed using the affy package implementation of robust multichip analysis. These data were subsequently analyzed and filtered using the limma package (21). Filtering cutoffs were log2-fold change >1 and nominal P value <0.001. Heatmaps were generated using the gplots package, and clusters were calculated using the HOPACH package (22). For quantitative PCR analysis, mRNA was reverse-transcribed with iScript (Biorad), and template was amplified using SYBRGreen master mix (Applied Biosystems) on an Applied Biosystems 7900 thermal cycler. The ΔΔCt method was used with normalization to cyclophilin. Primer sequences for all genes were from http://mouseprimerdepot.nci.nih.gov/.
ELISA
Adiponectin and high-molecular weight adiponectin were measured using an ELISA kit from ALPCO. The protocol was modified to adapt the serum-based ELISA to the lower levels of adiponectin found in conditioned media. Specifically, 50 μl conditioned medium was incubated with 100 μl protease solution, which was then neutralized by 100 μl sample pretreatment buffer. The sample was then diluted 25 times in dilution buffer, and the rest of the assay was performed according to the manufacturer's instructions. The modification was validated by comparing results obtained with the two different dilutions on concentrated samples. The dilution paradigm was also validated using serum from adiponectin-null mice.
RESULTS
Single deletion of either DGAT does not affect ability of adipocytes to make LDs or TGs
We first determined whether deletion of individual DGAT enzymes affected TG synthesis and LD formation in adipocytes differentiated from MEFs. As expected, given that DGAT1 KO (D1KO) mice have white adipose tissue (WAT), the deletion of DGAT1 did not impair TG synthesis or LD formation (Fig. 1B). Similarly, adipocytes with a single deletion of DGAT2 (D2KO) had as much or more TG than WT adipocytes (Fig. 1B and data not shown). For either single knockout, a high proportion of cells differentiated, and differentiation resulted in cells with readily apparent LDs (by light microscopy) that stained with Oil-Red O (ORO; Fig. 1B). TG content and TG synthetic rates in intact cells in D1KO and D2KO adipocytes were not impaired compared with WT adipocytes (not shown).
We next sought to analyze cells that lacked both DGAT enzymes. We isolated primary MEFs derived from D1D2KO E14 embryos and treated them with adipogenic factors. A high proportion of cells changed morphology, consistent with differentiation. In contrast to single DGAT deletions, however, D1D2KO differentiated cells lacked ORO staining and appeared to lack LDs by light microscopy (Fig. 1C).
Adipocytes lacking DGAT1 and DGAT2 are deficient in DGAT activity and TG synthesis
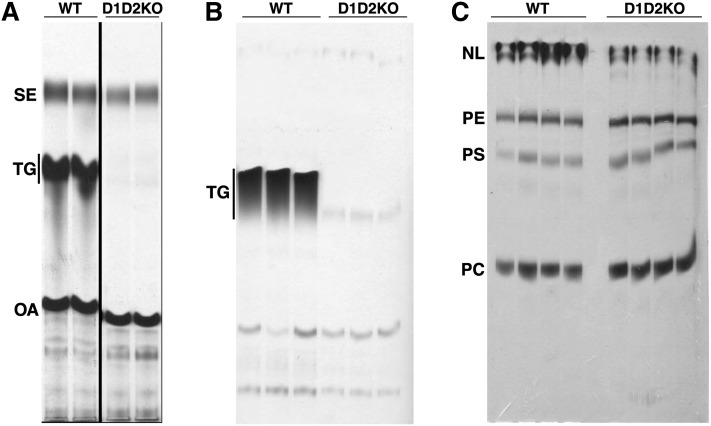
We next assessed TG synthesis in the D1D2KO adipocytes. DGAT activity in whole-cell lysates was essentially absent (Fig. 2A). Corroborating this result, TG synthesis in intact cells incubated with [14C]oleic acid showed that radiolabel incorporation into TG in D1D2KO adipocytes was ∼5% that of WT cells (Fig. 2B). This was not due to a defect in FA uptake, inasmuch as the cellular uptake of [14C]oleic acid (after 5 min) was only modestly decreased (∼30% reduction) in D1D2KO cells (data not shown). In contrast to the diminished TG incorporation, there was increased accumulation of [14C]oleic acid into PC in D1D2KO adipocytes (Fig. 2C; 173 ± 15% of WT levels, P = 0.005). The incorporation into phosphatidylserine was also increased (138 ± 14% of WT levels, P = 0.01) in D1D2KO adipocytes. There was a trend for increased phosphatidylethanolamine (PE) synthesis (112 ± 6% of WT levels, P = 0.27). Thus, in the absence of DGAT enzymes, TG synthesis was nearly absent, and there was some increased flux of exogenous FAs into phospholipids.
Fig. 2.
MEFs deficient in both DGATs (D1D2KO) have greatly reduced TG synthesis. A: DGAT activity is nearly absent in D1D2KO differentiated cells. Shown is an autoradiograph of radiolabeled lipids, separated by TLC, from in vitro DGAT assays. SE, sterol esters; TG, triacylglycerol; OA, oleic acid. B: Lack of TG synthesis in intact D1D2KO differentiated cells. Cells were incubated with [14C]oleic acid, and lipids were analyzed by TLC and autoradiography. C: Increased incorporation of labeled oleic acid into PC in intact D1D2KO differentiated cells. NL, neutral lipids; PE, phosphatidylethanolamine; PS, phosphatidylserine; PC, phosphatidylcholine; WT, wild-type. For B and C, cells were differentiated without exogenous FA and then loaded with 250 μM exogenous oleic acid during labeling. Experiments were performed three times, with representative results shown.
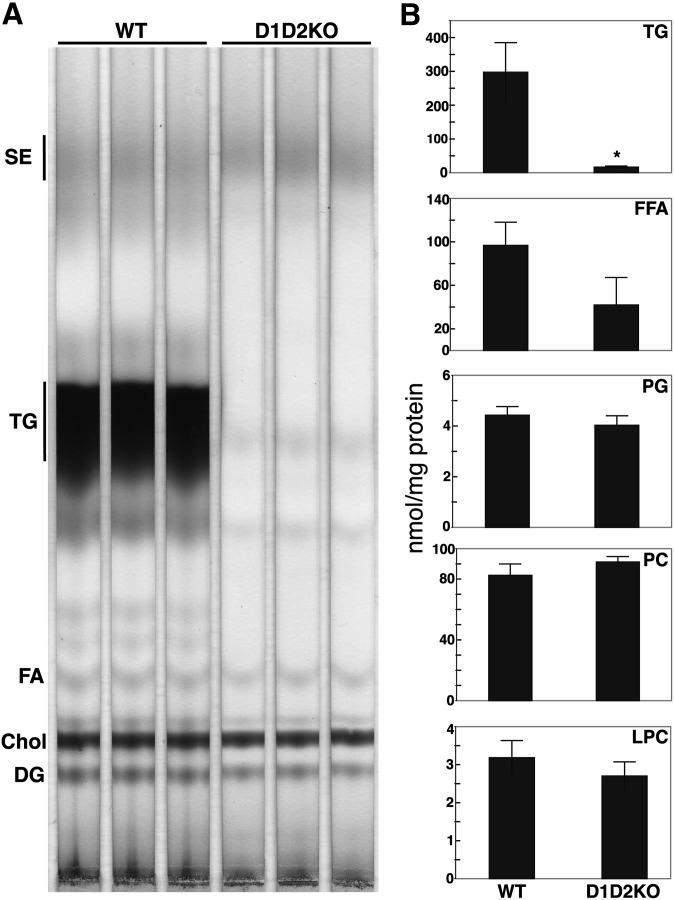
We also measured levels of TG mass in the D1D2KO adipocytes. TLC analysis of cellular lipids revealed that WT cells had large amounts of TG, whereas TG was nearly absent in D1D2KO adipocytes (Fig. 3A). As measured by MS, the total TG species were reduced ∼95% in D1D2KO cells (Fig. 3B; 16 ± 2 vs. 298 ± 88 nmol/mg protein for WT). The levels of the most common, longer chain TG species were reduced to a greater extent (e.g., to 1.4% of WT for C48:2 species, 1.7% for C48:1, and 2.1% for C50:2) than other species in D1D2KO adipocytes. Intracellular free FA levels were lower in D1D2KO cells (62% reduction; P < 0.05 for the most abundant species, 16:1, and a trend for decrease in overall intracellular FA levels, P = 0.06), whereas levels of PC, lysophosphatidylcholine, and phosphatidylglycerol were similar in D1D2KO and WT cells (Fig. 3B). Thus, by measurements of both DGAT activity and mass, TG synthesis is profoundly reduced in the absence of DGAT enzymes, indicating that the two DGAT enzymes account for nearly all of the TG synthesis in murine adipocytes.
Fig. 3.
D1D2KO adipocytes have reduced TG levels. A: Charred TLC plate of lipid extracts from WT and D1D2KO differentiated cells. B: Quantification of mass spectrometry analysis of lipids from WT and D1D2KO adipocytes showing content of TG, free fatty acid (FFA), phosphatidylglycerol (PG), phosphatidylcholine, (PC), lysophosphatidylcholine (LPC). * P < 0.05, n = 3. Exogenous FAs were not added during differentiation. The experiment was performed three times, with mass spectrometry performed on a single sample set. Error bars represent SD.
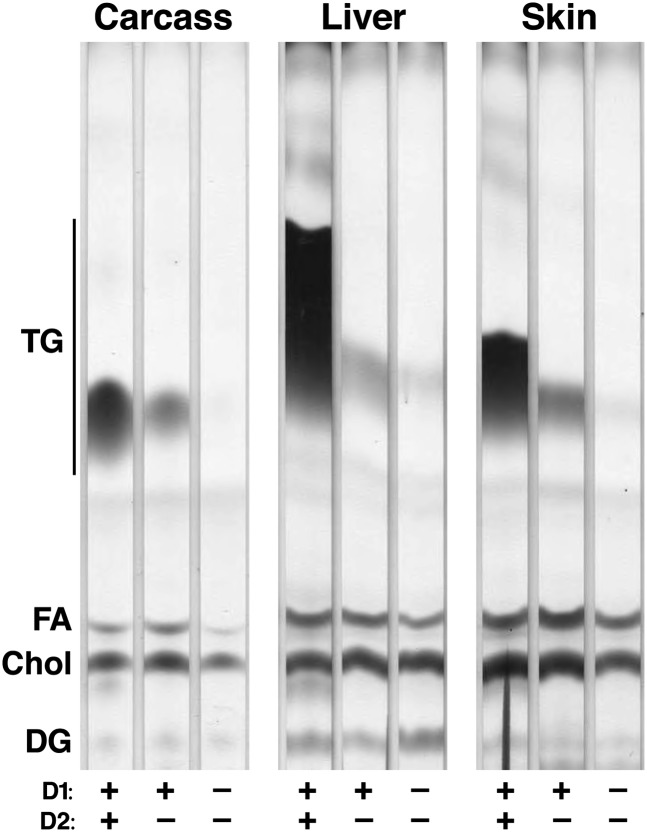
We also assessed TG content in tissues of newborn D1D2KO mice. These mice were born at expected Mendelian ratios, indicating no requirements for the DGAT enzymes for embryonic viability. Like DGAT2-null mice (4), D1D2KO mice died within hours after birth. As described previously, TG levels were lower in tissues of D2KO mice (10–30% of WT in D2KO liver, skin, and carcass). Levels were even lower in the D1D2KO mice (1–3% of WT; Fig. 4). Thus, DGAT1 and DGAT2 appear to together account for the vast majority of TG synthesis in most murine tissues.
Fig. 4.
D1D2KO newborn mice are severely depleted in TG. Lipid extracts from carcass, liver, and skin of newborn WT, D2KO, and D1D2KO mice were analyzed by TLC. The experiment was performed three times, with representative results shown.
Adipocytes lacking both DGAT enzymes lack LDs
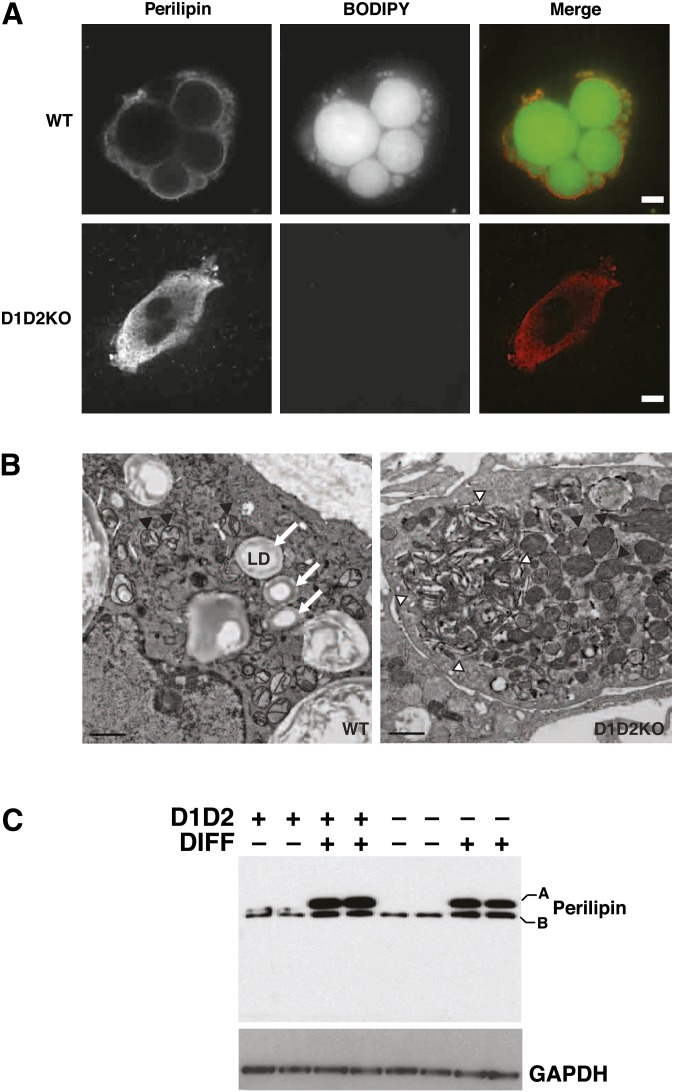
We next asked whether D1D2KO adipocytes, lacking TGs, had LDs. Whereas WT adipocytes had large LDs that were detected by staining with BODIPY 493/503, D1D2KO adipocytes completely lacked LDs (Fig. 5A). Both WT and D1D2KO adipocytes expressed the LD marker protein perilipin (Fig. 5A, C). In WT cells, perilipin mostly surrounded LDs, whereas in D1D2KO cells, perilipin staining appeared reticular, consistent with an endoplasmic reticulum localization.
Fig. 5.
D1D2KO adipocytes express perilipin but lack LDs and accumulate membranous aggregates. A: BODIPY493/503 (BODIPY, green) and perilipin (red) staining of WT and D1D2KO differentiated cells. Scale bar, 5 μm. B: WT and D1D2KO adipocytes visualized by electron microscopy. Lipid droplets are indicated by arrows. Mitochondria are indicated by black arrowheads. A large membranous aggregate is visible (outlined by white arrowheads) in the D1D2KO adipocyte. Aggregates were visible in all D1D2KO adipocytes. Scale bar, 1 μm. C: Western blot of perilipin in undifferentiated (DIFF–) and differentiated (DIFF+) WT (D1D2+) and D1D2KO (D1D2–) cells. A and B indicate the different isoforms of perilipin.
Electron microscopic examination of D1D2KO adipocytes confirmed the total absence of LDs (Fig. 5B). D1D2KO adipocytes exhibited some unique features, including abnormal whorls of electron-dense material that appeared to be collections of membranes (Fig. 5B, right panel arrowheads) and more-abundant mitochondria (2.5-fold increase vs. WT, P < 10−4). By comparison, the numbers of plasma membrane caveolae, a feature of differentiated adipocytes, were similar in WT and D1D2KO cells (data not shown). Thus, D1D2KO adipocytes showed no evidence of LDs by either light or electron microscopy.
DGAT enzymes are not required for forming SE-containing LDs in macrophages
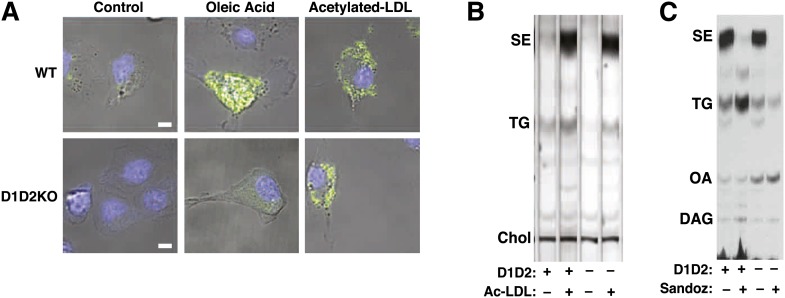
Because DGAT enzymes were required for LDs in differentiated adipocytes, we asked whether DGAT enzymes were required more generally for LD formation in mammalian cells. To address this question, we turned to cultured macrophages, in which LDs often contain large amounts of cholesteryl esters, synthesized by ACAT1 (23). We examined LD formation in D1D2KO murine macrophages that were cultured in the presence of cholesterol-rich acetylated LDL. Because D1D2KO mice die just after birth, we could not obtain macrophages from bone marrow or the peritoneum. We therefore prepared macrophages from fetal livers by culturing cells in the presence of M-CSF (24). Macrophages of WT and D1D2KO mice were treated with vehicle, oleic acid (to induce TG-containing droplets), or acetylated LDL (to induce cholesteryl ester-containing droplets). In WT macrophages, a small number of LDs were found in vehicle-treated cells, and both the size and number of droplets increased dramatically with oleic acid or acetylated-LDL treatments (Fig. 6A, top panels). In contrast, D1D2KO macrophages had no visible droplets in vehicle-treated cells and did not form droplets with oleic acid treatment (Fig. 6A, bottom panels). However, D1D2KO macrophages did form LDs when they were incubated with acetylated LDL. Analysis of lipid extracts from these cells revealed that both WT and D1D2KO macrophages treated with acetylated LDL had much greater amounts of SEs than control-treated cells (Fig. 6B). Interestingly, D1D2KO macrophages also contained significant amounts of TG (∼60% of WT) when incubated with acetylated LDL (Fig. 6B).
Fig. 6.
D1D2KO macrophages can make SE-containing LDs and can synthesize TG. A: WT and D1D2KO macrophages untreated (Control) or treated with oleic acid (500 μM) or acetylated LDL (25 μg/ml). Scale bar, 5 μm. Blue, Hoechst 33342; green, BODIPY 493/503. B: Analysis of lipids in conditions for A by TLC. C: TLC of WT and D1D2KO acetylated LDL-treated macrophages in the presence or absence of Sandoz 58-035 that have been incubated with [14C]oleic acid tracer without other exogenous oleic acid. The experiment was performed three times, with representative results shown. DAG, diacylglycerol.
To ensure that the intracellular cholesteryl esters and TG were synthesized by the macrophages (and not merely internalized), we performed similar acetylated-LDL loading experiments in the presence of [14C]oleic acid. We found that this FA tracer was incorporated into both cholesteryl esters and TG of WT and D1D2KO macrophages (Fig. 6C). The accumulation of SEs was mediated by ACAT, as it was inhibited in both WT and D1D2KO macrophages by the ACAT inhibitor Sandoz 58-035. In the presence of the ACAT inhibitor, WT macrophages shunted the [14C]oleic acid to TG synthesis; this was not found in the absence of DGAT enzymes.
Adipogenesis is intact in differentiated fibroblasts lacking both DGAT1 and DGAT2
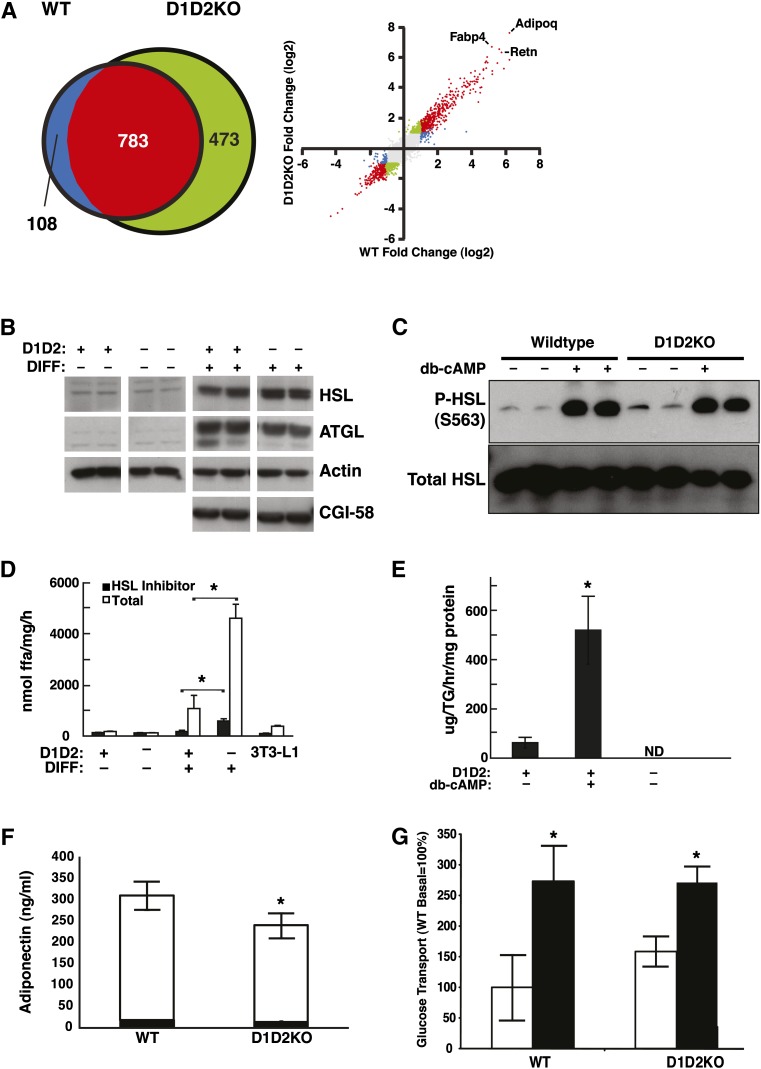
To verify that adipogenesis was intact in D1D2KO cells, we analyzed these cells for transcriptional changes, protein expression, and other functional activities that are characteristic of adipocytes. Microarray analyses revealed that mRNAs whose levels were altered as a result of adipogenic differentiation were highly similar in WT and D1D2KO cells (n = 891 genes for WT and 1,256 genes for D1D2KO; Fig. 7A). Nearly 90% of the genes changed in the WT differentiation program were also changed in D1D2KO cells (Fig. 7A). Overall, the changes in gene expression for WT and D1D2KO differentiation were highly correlated (Fig. 7A; R2 = 0.87). In both genotypes, GO Term analysis and gene set enrichment analysis with the Kyoto Encyclopedia of Genes and Genomes pathway annotation identified many adipocyte-related gene sets, such as FA metabolism and peroxisome proliferator-activated receptor γ (PPAR-γ) signaling, enriched in differentiated compared with undifferentiated cells (see supplementary Table I and supplementary Fig. I).
Fig. 7.
D1D2KO differentiated cells have the cellular physiology of adipocytes. A: Results of microarray analyses of mRNA from WT and D1D2KO cells. Left: Venn diagram of genes regulated differently during differentiation in WT and D1D2KO adipocytes. Right: Scatterplot of induction values (differentiated/undifferentiated) for individual genes in the microarray. Points are color-coded based on meeting criteria for induction with differentiation (2-fold change and P < 0.001): red, meets criteria for both WT and D1D2KO; green, meets criteria for D1D2KO only; blue, meets criteria for WT only; gray, meets criteria for neither. Individual positions of three highly induced adipocyte markers (Adipoq, Fabp4, and Retn) are indicated. B: Expression of HSL, ATGL, and CGI-58 in WT (D1D2+) and D1D2KO (D1D2–) undifferentiated (DIFF–) and differentiated (DIFF+) MEFs. C: Intact phosphorylation of HSL at serine 563 (S563) in D1D2KO adipocytes induced by dibutyryl cAMP (db-cAMP). D: TG hydrolase activity in lysates of D1D2KO undifferentiated and differentiated MEFs and 3T3-L1 adipocytes. E: Glycerol release from WT and D1D2KO adipocytes untreated or treated with db-cAMP. F: Intact total (white bar) and HMW adiponectin (black bar) secretion in WT and D1D2KO differentiated MEFs. (G) Basal and insulin-stimulated glucose transport in WT and D1D2KO adipocytes. White bars, basal; black bars, insulin-stimulated. ND, none detected. * P < 0.05, n = 4. B–G, experiments were performed three times with representative results shown. Error bars represent SD.
To validate the microarray findings, we examined ∼20 adipocyte differentiation markers by quantitative PCR and found that they were induced in both WT and D1D2KO cells with differentiation (see supplementary Table II). Indeed, the levels of induction of many adipocyte markers were higher in D1D2KO cells than in WT cells. Numerous genes of brown adipose tissue were significantly increased with adipogenesis for both genotypes, probably reflecting the use of a PPAR-γ agonist in the differentiation cocktail (25).
D1D2KO differentiated cells expressed many proteins that are hallmarks of adipocytes. Immunoblotting revealed that the major adipocyte lipases, HSL and ATGL, were expressed in both WT and D1D2KO differentiated cells, as was the ATGL cofactor CGI-58 (Fig. 7B). Notably, the expression of HSL was 90% higher in D1D2KO differentiated cells than in WT cells (Fig. 7B).
Hormone-mediated fat breakdown, or lipolysis, is a robust process in adipocytes. The signal transduction pathway for lipolysis consists of adrenergic receptor-mediated increases in cAMP, activation of protein kinase A (PKA), and phosphorylation of HSL. Both WT and D1D2KO differentiated cells showed phosphorylation of HSL at S563, a well-characterized PKA site that results in activation of HSL activity (26), in response to dibutyryl cAMP, a PKA activator (Fig. 7C). In addition, lysates of D1D2KO differentiated cells possessed lipase activities that were higher than with WT cells (P < 0.05; Fig. 7D). This was the case for both total lipase activity, which was increased 4–5-fold, and for non-HSL activity, as assessed by the use of the HSL inhibitor NNC 0076-0000-0079. As expected, because D1D2KO adipocytes lack stores of TG, they did not release glycerol, a terminal product of lipolysis, during basal conditions or in response to dibutyryl cAMP (Fig. 7E).
D1D2KO differentiated cells also secreted the adipocyte-specific hormone adiponectin, including the high-molecular-weight isoform (Fig. 7F). The amount of total adiponectin secreted by D1D2KO differentiated cells was modestly decreased (Fig. 7F; ∼23% reduction, white bars, P < 0.05). However, the amounts of high-molecular-weight adiponectin, which is most correlated with the beneficial metabolic effects (27), were similar in media from WT and D1D2KO differentiated cells (Fig. 7F, dark bars).
Another hallmark of adipocytes is the ability to take up glucose in the presence of insulin. The levels of basal and insulin-stimulated glucose uptake (assayed by using 3H-2-deoxyglucose) were similar in WT and D1D2KO adipocytes (Fig. 7G), and there was a trend for increased basal glucose uptake in D1D2KO adipocytes. Glucose uptake increased from basal levels in response to insulin in adipocytes of both genotypes. As a whole, these results clearly showed that adipogenesis was intact in differentiated D1D2KO MEFs and that these cells had all the hallmarks of adipocytes except TG accumulation.
DISCUSSION
In this study, we show that DGAT1 and DGAT2 account for nearly all TG synthesis in adipocytes and other murine tissues and that in the absence of DGAT enzymes, adipocytes lack TG and LDs. We also found that DGAT enzymes and DGAT-mediated TG synthesis and accumulation are not required for adipocyte differentiation in vitro. Although DGAT enzymes appear to be essential for LDs in adipocytes, they are not absolutely required for LD formation, inasmuch as macrophages lacking both DGATs were capable of forming LDs upon cholesterol loading with acetylated LDL.
Our results underscore the dominant roles that DGAT1 and DGAT2 enzymes play in murine and probably mammalian TG synthesis. We found that DGAT1 and DGAT2 account for the vast majority of TG normally found in murine adipocytes derived from MEFs. The inability of D1D2KO adipocytes to synthesize TG was probably not due to decreased availability of intracellular substrates, inasmuch as uptake of glucose used for the production of FAs was normal and the D1D2KO MEFs had detectable intracellular free FAs. DGAT1 and DGAT2 also account for nearly all TG in tissues of newborn pups, which lack WAT. Previous single deletions of DGATs in mice indicated that DGAT1 contributed modestly to total TG synthesis (3) and that DGAT2 played a major role, inasmuch as TG content was greatly reduced in newborn DGAT2 KO mice (4). In the current study, the double deletion of DGAT1 and DGAT2 resulted in newborn pups with even more severely reduced TG content (∼1–3% of WT; Fig. 4). Like D2KO mice, D1D2KO mice died within hours after birth. Thus, mice lacking both DGAT enzymes have apparently normal embryonic development. Of note, the single deletion of DGAT2 did not impair the ability of MEF-derived adipocytes to synthesize TG. We suspect that DGAT1 is able to compensate for DGAT2 deficiency in adipocytes, because we previously showed in D2KO hepatocytes that DGAT1 can compensate, provided that substrates are abundant (4), and this is probably the case during adipogenesis.
Our study also reveals that some TG can be synthesized in the absence of DGAT1 and DGAT2. Small amounts of TG were present in D1D2KO adipocytes and, to a greater degree, in cholesterol-loaded D1D2KO macrophages. This TG may be from either an alternative DGAT activity or a non-DGAT enzymatic activity. DGAT2 is not the only DGAT2 family member to possess DGAT activity in vitro (6, 28). However, there was no detectable DGAT activity in lysates from D1D2KO macrophages (data not shown). Potential non-DGAT activities include diacylglycerol transacylase, which converts two molecules of diacylglycerol (DG) to the products TG and monoacylglycerol (29), and phospholipid diacylglycerol acyltransferase (PDAT), which catalyzes the transfer of a fatty acyl moiety from a phospholipid to DG to form TG and lysophospholipid products. DG transacylase activity has been reported in rodent intestine (29, 30). To date, PDAT activity has been reported only in yeast and plants (31). Whatever the source of this TG synthesis, the near absence of TG in D1D2KO newborn mice suggests that its contributions are normally relatively small.
Interestingly, we did not see accumulation of DG in D1D2KO adipocytes. This is probably because DG can flux into a variety of other lipid pathways. For example, DG serves as a substrate for de novo synthesis of phospholipids such as PC and PE (Fig. 1). Indeed, we observed a 73% increase in PC synthesis in D1D2KO adipocytes, supporting this hypothesis. Despite the increase in PC synthesis with oleate loading, we did not find an increase in steady-state PC levels, suggesting that PC degradation is also increased to maintain homeostasis, as has been described previously (32). The lack of DG accumulation in D1D2KO adipocytes may also be due to lipases. In particular, HSL functions predominantly as a DG lipase (33), and HSL activity was prominent in the D1D2KO adipocytes.
In the absence of DGAT enzymes, adipocytes did not have LDs, suggesting that TGs are the major neutral lipid required for LD formation in this cell type. On the other hand, D1D2KO macrophages formed SE-containing LDs when they were loaded with cholesterol. Thus, DGAT1 and DGAT2 are not specifically required for LD formation in all cell types, and other enzymatic activities can contribute to LD formation. In agreement with this result, the yeast Saccharomyces cerevesiae has four major enzymes of neutral lipid synthesis (Lro1p and Dga1p for TG, and Are1p and Are2p for SE), and deletion of all four genes is required to eliminate LD formation (34, 35). Whether TGs are absolutely required for LD formation remains an open question. Add-back studies in yeast, in which individual enzymes were expressed in the quadruple mutant, revealed a strong dependency of LD formation on TG (versus SE) synthesis (35). Our data are consistent with this result; we always found some TG when LDs were present in either adipocytes or macrophages. However, our results do not exclude the possibility that cholesteryl esters alone may be sufficient to drive LD formation. This cannot be tested in mammalian cells, however, until the all sources of TG synthesis activity are identified and eliminated. It is also possible that a contributing factor to the lack of LDs in D1D2KO adipocytes is an inability to accumulate TG due to increased lipase activity in addition to very low TG synthesis rates.
Our results demonstrating intact adipogenesis in D1D2KO cells, which lack the enzymes catalyzing the final step in TG synthesis, contrast with specific proximal defects in the de novo (Kennedy) pathway of TG synthesis. More-proximal blocks at the level of 1-acyl-sn-glycerol-3-phosphate acyltransferase 2 (AGPAT2) in humans and mice (36, 37) or lipin 1 in mice (38) lead to lipoatrophy and associated loss of adipokines, such as leptin and adiponectin, and insulin resistance. In addition, lipin 1-deficient (fld) MEFs fail to differentiate in vitro, as evidenced by lack of induction of adipogenic markers (39). Knockdown of AGPAT2 via small interfering RNA in OP9 cells (40) or small hairpin RNA-mediated knockdown of glycerol-3-phosphate acyltransferase in 3T3-L1 cells (41) also inhibits the adipogenic program. What could account for the differences in adipocyte differentiation in proximal and distal steps in the pathway? One possible explanation is that in addition to generating lipid species used for storage, Kennedy pathway intermediates also act as lipid signaling molecules to stimulate adipocyte differentiation through a “feed-forward” mechanism. In this way, increased substrate flux through the pathway might promote both the storage of TG and the differentiation of cells into adipocytes to store these substrates. Such intermediary substrates might act as ligands for PPAR-γ. The existence of endogenous PPAR-γ ligands is controversial (10, 42, 43), but collectively, these studies support the existence of such lipid signaling molecules.
A limitation of our study is that we could not examine TG and LDs in adipocytes in vivo in adult mice lacking both DGAT1 and DGAT2. An in vivo assessment would require crossing DGAT1 KO mice with a conditional knockout of DGAT2 in the WAT, and the latter mice are not currently available. Nevertheless, the MEF model that we employed has been extensively utilized to examine the functional roles of other genes in adipogenesis (8–11, 44), and it is a well-accepted means to examine adipogenesis and adipocyte-related processes. When DGAT2 tissue-specific knockout mice become available, our findings concerning adipocytes can then be tested in WAT.
Our study agrees with a previous study that tested the relationship of TG synthesis and adipocyte differentiation through biochemical depletion of FA synthesis. Culturing of preadipocytes in biotin-deficient medium diminished FA synthesis and TG formation (45), inasmuch as biotin is a cofactor for acetyl CoA carboxylase, a key enzyme in FA synthesis. Under these conditions of TG depletion, preadipocytes displayed some signs of adipocyte differentiation, including morphological changes as assessed by light microscopy and biochemically as assessed by lipoprotein lipase enzyme activity (45). It is difficult to directly compare this system with the genetic deletions we employed, however, because rendering media biotin-deficient requires extensive dialysis of serum, which depletes the medium of many low-molecular-weight molecules, including FAs, and therefore probably has more-profound effects on cell metabolism.
In summary, our results show that the two DGAT enzymes account for nearly all TG synthesis in murine adipocytes. In the absence of TG, D1D2KO adipocytes do not make LDs, but rather form membrane aggregrates, visible by electron microscopy. Despite the absence of TG and LDs, D1D2KO adipocytes are otherwise similar to WT adipocytes with respect to adipocyte differentiation, with similar mRNA expression, expression of several adipocyte-specific proteins, TG lipase activity, and secretion of the adipokine adiponectin. Our finding that DGAT-deficient MEFs are competent to undergo adipogenesis, in contrast to specific defects in the proximal steps in TG synthesis, implies that DG or a DG-derived lipid might be an adipogenic signal. Finally, unlike adipocytes, macrophages lacking DGAT enzymes can form TG and LD, implying that non-DGAT enzymes contribute to TG synthesis under certain conditions.
Supplementary Material
Acknowledgments
The authors thank members of the Farese laboratory for discussions, Tobias Walther and Dawn Brasaemle for comments on the manuscript, Caroline Mrejen for helpful discussions on microarray and electron microscopy experiments, Collin Shaw and Craig LaMarca for helpful discussions on HMW adiponectin measurements, Jennifer Gregg and the Gladstone Genomics Core for microarray hybridization, Jinny Wong of the Gladstone Electron Microscopy core for electron microscopy, Dawn Brasaemle for anti-perilipin antibody, Daryl Jones for assistance in manuscript preparation, and Gary Howard for editorial assistance.
Footnotes
Abbreviations:
- AGPAT
- 1-acyl-sn-glycerol-3-phosphate acyltransferase
- ATGL
- adipose triglyceride lipase
- CGI-58
- comparative gene identification 58
- DG
- diacylglycerol
- DGAT
- diacylglycerol acyltransferase
- HSL
- hormone-sensitive lipase
- KO
- knock-out
- LD
- lipid droplet
- M-CSF
- macrophage colony-stimulating factor
- MEF
- murine embryonic fibroblast
- ORO
- Oil Red O
- PC
- phosphatidylcholine
- PDAT
- phospholipid diacylglycerol acyltransferase
- PE
- phosphatidylethanolamine
- PKA
- protein kinase A
- PPAR
- peroxisome proliferator-activated receptor
- SE
- sterol ester
- TG
- triacylglycerol
- WAT
- white adipose tissue
- WT
- wild-type
This work was supported by University of California San Francisco Diabetes, Endocrinology and Metabolism Training Grant NIDDK DK07418-27, National Institutes of Health career physician-scientist award (KO8-DK-081680 to C.H.), National Institutes of Health Grants RO1-DK-056084 (R.F.) and P01-HL-057278 (R.G.), animal facilities National Institutes of Health/National Center for Research Resources Grant CO6 RR-018928, and the J. David Gladstone Institutes. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and one figure.
REFERENCES
- 1.Farese R. V., Jr., Walther T. C. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr 2008. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49: 2283–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr 2000. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25: 87–90. [DOI] [PubMed] [Google Scholar]
- 4.Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V., Jr 2004. Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J. Biol. Chem. 279: 11767–11776. [DOI] [PubMed] [Google Scholar]
- 5.Yen C. L., Monetti M., Burri B. J., Farese R. V., Jr 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 1502–1511. [DOI] [PubMed] [Google Scholar]
- 6.Yen C. L., Brown C. H. t., Monetti M., Farese R. V., Jr 2005. A human skin multifunctional O-acyltransferase that catalyzes the synthesis of acylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen E. D., MacDougald O. A. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7: 885–896. [DOI] [PubMed] [Google Scholar]
- 8.Nofsinger R. R., Li P., Hong S. H., Jonker J. W., Barish G. D., Ying H., Cheng S. Y., Leblanc M., Xu W., Pei L., et al. 2008. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc. Natl. Acad. Sci. USA. 105: 20021–20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park K. W., Waki H., Villanueva C. J., Monticelli L. A., Hong C., Kang S., MacDougald O. A., Goldrath A. W., Tontonoz P. 2008. Inhibitor of DNA binding 2 is a small molecule-inducible modulator of peroxisome proliferator-activated receptor-gamma expression and adipocyte differentiation. Mol. Endocrinol. 22: 2038–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walkey C. J., Spiegelman B. M. 2008. A functional peroxisome proliferator-activated receptor-gamma ligand-binding domain is not required for adipogenesis. J. Biol. Chem. 283: 24290–24294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z., Yu S., Hsu C. H., Eguchi J., Rosen E. D. 2008. The orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II is a critical regulator of adipogenesis. Proc. Natl. Acad. Sci. USA. 105: 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streeper R. S., Koliwad S. K., Villanueva C. J., Farese R. V., Jr 2006. Effects of DGAT1 deficiency on energy and glucose metabolism are independent of adiponectin. Am. J. Physiol. Endocrinol. Metab. 291: E388–E394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Fu M., Cui T., Xiong C., Xu K., Zhong W., Xiao Y., Floyd D., Liang J., Li E., et al. 2004. Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proc. Natl. Acad. Sci. USA. 101: 10703–10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 15.Houstis N., Rosen E. D., Lander E. S. 2006. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 440: 944–948. [DOI] [PubMed] [Google Scholar]
- 16.Angermuller S., Fahimi H. D. 1982. Imidazole-buffered osmium tetroxide: an excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 14: 823–835. [DOI] [PubMed] [Google Scholar]
- 17.Han X., Gross R. W. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100. [DOI] [PubMed] [Google Scholar]
- 18.Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 19.Han X., Yang K., Gross R. W. 2008. Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 22: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wettenhall J. M., Smyth G. K. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 20: 3705–3706. [DOI] [PubMed] [Google Scholar]
- 22.Pollard K. S., van der Laan M. J. 2002. A method to identify significant clusters in gene expression data. 6th World Multiconf Systemics, Cybernetics Informatics (SCI2002) II: 318–325. [Google Scholar]
- 23.Meiner V. L., Cases S., Myers H. M., Sande E. R., Bellosta S., Schambelan M., Pitas R. E., McGuire J., Herz J., Farese R. V., Jr 1996. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc. Natl. Acad. Sci. USA. 93: 14041–14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz A., Vanhille L., Mohideen P., Kelly L. M., Otto C., Bakri Y., Mossadegh N., Sarrazin S., Sieweke M. H. 2006. Development of macrophages with altered actin organization in the absence of MafB. Mol. Cell. Biol. 26: 6808–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovic N., Walden T. B., Shabalina I. G., Timmons J. A., Cannon B., Nedergaard J. 2010. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285: 7153–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthonsen M. W., Ronnstrand L., Wernstedt C., Degerman E., Holm C. 1998. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J. Biol. Chem. 273: 215–221. [DOI] [PubMed] [Google Scholar]
- 27.Pajvani U. B., Hawkins M., Combs T. P., Rajala M. W., Doebber T., Berger J. P., Wagner J. A., Wu M., Knopps A., Xiang A. H., et al. 2004. Complex distribution, not absolute amount of adiponectin, correlates with thiazolidinedione-mediated improvement in insulin sensitivity. J. Biol. Chem. 279: 12152–12162. [DOI] [PubMed] [Google Scholar]
- 28.Turkish A. R., Henneberry A. L., Cromley D., Padamsee M., Oelkers P., Bazzi H., Christiano A. M., Billheimer J. T., Sturley S. L. 2005. Identification of two novel human acyl-CoA wax alcohol acyltransferases: members of the diacylglycerol acyltransferase 2 (DGAT2) gene superfamily. J. Biol. Chem. 280: 14755–14764. [DOI] [PubMed] [Google Scholar]
- 29.Lehner R., Kuksis A. 1993. Triacylglycerol synthesis by an sn-1,2(2,3)-diacylglycerol transacylase from rat intestinal microsomes. J. Biol. Chem. 268: 8781–8786. [PubMed] [Google Scholar]
- 30.Buhman K. K., Smith S. J., Stone S. J., Repa J. J., Wong J. S., Knapp F. F., Jr., Burri B. J., Hamilton R. L., Abumrad N. A., Farese R. V., Jr 2002. DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. J. Biol. Chem. 277: 25474–25479. [DOI] [PubMed] [Google Scholar]
- 31.Banas A., Dahlqvist A., Stahl U., Lenman M., Stymne S. 2000. The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem. Soc. Trans. 28: 703–705. [PubMed] [Google Scholar]
- 32.Walkey C. J., Kalmar G. B., Cornell R. B. 1994. Overexpression of rat liver CTP:phosphocholine cytidylyltransferase accelerates phosphatidylcholine synthesis and degradation. J. Biol. Chem. 269: 5742–5749. [PubMed] [Google Scholar]
- 33.Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. 2002. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 277: 4806–4815. [DOI] [PubMed] [Google Scholar]
- 34.Sandager L., Gustavsson M. H., Stahl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277: 6478–6482. [DOI] [PubMed] [Google Scholar]
- 35.Sorger D., Athenstaedt K., Hrastnik C., Daum G. 2004. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 279: 31190–31196. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal A. K., Arioglu E., De Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., Garg A. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31: 21–23. [DOI] [PubMed] [Google Scholar]
- 37.Cortes V. A., Curtis D. E., Sukumaran S., Shao X., Parameswara V., Rashid S., Smith A. R., Ren J., Esser V., Hammer R. E., et al. 2009. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metab. 9: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterfy M., Phan J., Xu P., Reue K. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- 39.Phan J., Peterfy M., Reue K. 2004. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 279: 29558–29564. [DOI] [PubMed] [Google Scholar]
- 40.Gale S. E., Frolov A., Han X., Bickel P. E., Cao L., Bowcock A., Schaffer J. E., Ory D. S. 2006. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 281: 11082–11089. [DOI] [PubMed] [Google Scholar]
- 41.Shan D., Li J. L., Wu L., Li D., Hurov J., Tobin J. F., Gimeno R. E., Cao J. 2010. GPAT3 and GPAT4 are regulated by insulin-stimulated phosphorylation and play distinct roles in adipogenesis. J. Lipid Res. 51: 1971–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J. B., Wright H. M., Wright M., Spiegelman B. M. 1998. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA. 95: 4333–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzameli I., Fang H., Ollero M., Shi H., Hamm J. K., Kievit P., Hollenberg A. N., Flier J. S. 2004. Regulated production of a peroxisome proliferator-activated receptor-gamma ligand during an early phase of adipocyte differentiation in 3T3–L1 adipocytes. J. Biol. Chem. 279: 36093–36102. [DOI] [PubMed] [Google Scholar]
- 44.Rosen E. D., Walkey C. J., Puigserver P., Spiegelman B. M. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14: 1293–1307. [PubMed] [Google Scholar]
- 45.Kuri-Harcuch W., Wise L. S., Green H. 1978. Interruption of the adipose conversion of 3T3 cells by biotin deficiency: differentiation without triglyceride accumulation. Cell. 14: 53–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.