Abstract
Traits related to fatness, important as economic factors in pork production, are associated with serious diseases in humans. Genetical genomics is a useful approach for studying the effects of genetic variation at the molecular level in biological systems. Here we applied a whole-genome association analysis to hepatic gene expression traits, focusing on transcripts with expression levels that correlated with fatness traits in a porcine model. A total of 150 crossbred pigs [Pietrain×(German Large White × German Landrace)] were studied for transcript levels in the liver. The 24K Affymetrix expression microarrays and 60K Illumina single nucleotide polymorphism (SNP) chips were used for genotyping. A total of 663 genes, whose expression significantly correlated with the trait “fat area,” were analyzed for enrichment of functional annotation groups as defined in the Ingenuity Pathways Knowledge Base (IPKB). Genes involved in metabolism of various macromolecules and nutrients as well as functions related to dynamic cellular processes correlated with fatness traits. Regions affecting the transcription levels of these genes were mapped and revealed 4,727 expression quantitative trait loci (eQTL) at P < 10−5, including 448 cis-eQTL. In this study, genome-wide association analysis of trait-correlated expression was successfully used in a porcine model to display molecular networks and list genes relevant to fatness traits.
Keywords: expression quantitative trait loci, obesity, fat area, trait-associated expression, functional genomics, genome-wide association, microarrays, single nucleotide polymorphism chip
The metabolic activities of the liver are essential for providing fuel to the peripheral organs. Most components absorbed by the intestine pass through the liver, which enables it to regulate the levels of many metabolites in the blood. Storage, utilization, and partitioning of nutrients - including lipids - largely depend on the liver. Thus, genes active in the liver could potentially affect many traits related to body composition, leanness, and obesity. Traits related to fatness are important not only as economic factors in pork production but also because of their association with serious diseases in humans (1, 2). Pigs share many similarities with humans in physiology and genomes and, therefore, provide a good model to study the genetic determination of complex traits (3). The study of a porcine model is beneficial because a number of fatness traits (e.g., “fat area”) can be determined quantitatively with great accuracy and reproducibility post mortem, and RNA samples are available for transcriptomic approaches. In humans, indices for obesity are obtained intra vitam (i.e., body mass index or body fat percentage are estimated based on statistical models of measures of near-infrared interactance, body average density measurement, bioelectrical impedance analysis, or anthropometric methods) (4).
Functional genomics provides an insight into the molecular processes underlying phenotypic differences. The analysis of trait-correlated expression levels reveals genes belonging to pathways or networks relevant for the control of quantitative traits. However, holistic expression profiling does not often discriminate differential expression of the genes as either an effect or a cause of variation.
Genome-wide association studies (GWAS) in large natural populations of unrelated individuals have resulted in the association of many genes with complex traits (http://www.genome.gov/gwastudies). Unfortunately, for a considerable proportion of these genes and their proteins, it is not yet clear what their downstream effects and functions are.
Genetical genomics is a useful approach for studying the effect of genetic variation at the molecular level in biological systems. Genome-wide studies of gene expression are starting to provide information on genetic variation that affects gene expression levels (5). Recent studies in a variety of organisms have shown that levels of gene expression are often highly heritable (6–9) and that for many genes, it is possible to map cis- and trans-acting factors using linkage (8–12) or association mapping (7, 13–16).
We aimed to identify candidate genes for the fatness trait “fat area” by combining (i) genome-wide expression profiling and genotyping, (ii) analyzing correlation of transcript abundance with organismal phenotypic characteristics (fatness/obesity), and (iii) studying subsequent association of genomic variation with variation of expression of transcripts showing trait-correlated expression to determine expression quantitative trait loci (eQTL). Performance-tested animals of commercial crossbred herds were analyzed using 24K expression and 60K genotyping microarrays.
MATERIAL AND METHODS
Animals and tissue collection
Animal care and tissue collection processes followed the guidelines of the German Law of Animal Protection, and the experimental protocol was approved by the Institutional Animal Care Committee. This study was based on trait measurements, genotyping records, expression profiles, and genome-wide association analyses done with 150 performance-tested pigs from commercial herds of the crossbreed Pietrain×(German Large White × German Landrace) [Pi×(DE×DL)]. These animals represented 70 full-sibling families derived from 39 sires and 67 dams. The carcass and fat traits data were collected according to the guidelines of the Zentral Verband der Deutschen Schweineproduktion e.V. (ZDS) (17). Fat area was measured between the 13th and 14th rib on the M. longissimus dorsi (mean and standard error, 15.2 ± 3.2 cm2).
Whole-genome scan and quality control
Illumina bead array technology was used to carry out all genotyping reactions in accordance with the manufacturer's protocol for the single nucleotide polymorphism (SNP) Infinium HD assay (http://www.illumina.com). Genotyping was performed using the PorcineSNP60 BeadChip of Illumina (Illumina, Inc., San Diego, CA). In brief, 200 ng of DNA were used for genome-wide amplification and subsequent fragmentation. DNA was hybridized to the 62,163 locus-specific 50mers covalently linked to the beads distributed on the surface of the microarray. Single-base extension of the oligos on the BeadChip was then performed using the captured DNA as a template, incorporating detectable labels on the BeadChip. The signals of each wavelength were determined using an Illumina iScan that converted the images to intensity data. The intensity data for each SNP were normalized and assigned a cluster position and genotype with the GenomeStudio software (Illumina, Inc.), and a quality score for each genotype was generated. A total of 150 samples were genotyped for 62,163 SNPs. Quality of the data was evaluated, and samples with call rates <95% were removed. Markers were excluded if they had low minor allele frequency (MAF) <5%. The deviation from Hardy-Weinberg equilibrium was not considered because a three-way crossbreed pig population was used, where deviation from Hardy Weinberg equilibrium can be expected due to discordant allele frequencies in the parental breeds. The average call rate for all samples was 99.8% ± 0.2. In total, 11,505 SNPs were removed because of low call rates (GenCall score < 0.4) or MAF.
Whole-genome expression profiling
Gene expression profiling of the liver was conducted with the same 150 animals. In brief, total RNA of the liver was isolated using TRI Reagent (Sigma, Taufkirchen, Germany) and used for target preparation for microarray hybridization. According to Affymetrix protocols, 500 ng of total RNA were reversely transcribed into cDNA, transcribed into cRNA, and labeled using the Affymetrix One cycle synthesis and labeling kit (Affymetrix, UK) to prepare antisense biotinylated RNA targets. Liver expression patterns were produced using 150 GeneChip Porcine Genome Arrays (Affymetrix). The quality of hybridization was assessed in all samples following the manufacturer's recommendations. Data were analyzed with the Affymetrix GCOS 1.1.1 software, using global scaling to a target signal of 500 for background correction and probe summarization. Data were then imported into the Expression Console software (Affymetrix) for subsequent analysis. First the data were processed with the MAS5.0 algorithm to generate probe cell intensity values and to evaluate presence and absence of transcripts. Using default settings with detection of P < 0.04 for “present”’, P ≥ 0.04 and P ≤ 0.06 for “marginal”, and P < 0.06 for “absent,” only “present” calls were used. Then quantitative expression levels of the “present” transcripts were estimated, and normalization was performed using probe logarithmic intensity error (PLIER). PLIER data were used for further statistical analysis of expression levels. The microarray data related to all samples were deposited in the Gene Expression Omnibus public repository (GEO accession number GSE25445).
Correlation between traits and expression levels
Organismal phenotypes and expression levels were adjusted for systematic effects by ANOVA performed with the “Mixed” procedure of SAS software (SAS version 9.1, SAS Institute, Cary, NC) before analyzing their correlation. “Gender” was used as a fixed effect, “sire” and “slaughter day” as random effects, and “carcass weight” as a covariate. Subsequently, Pearson correlation coefficients were calculated between the residuals of log2-transformed expression intensities and the trait “fat area” (FA) ( i.e., the area of the subcutaneous back fat on M. longissimus dorsi between the 13th and 14th rib. Genes that showed correlation at P ≤ 0.05 were analyzed further.
Pathway analysis and network generation
On the basis of BLAST comparison of the Affymetrix porcine target sequences with the porcine genome sequence (Ensembl_Sscrofa_9, released April 2009), a total of 20,689 of the 24,123 probe sets on the Affymetrix Porcine GeneChip were annotated (18) for use in this study. The list of significant trait-correlated transcripts was analyzed, referring to predefined pathways and functional categories of the Ingenuity Pathways Knowledge Base (IPKB) using Ingenuity Pathways Analysis (IPA). Canonical pathways were also identified from the IPA library, which were most significant to the input data. The significance of the association between the dataset and the predefined pathways and functional categories was measured by Fischer's exact test, providing a P value to determine the probability that the association between the genes in the dataset and the pathway was explained by chance alone.
Furthermore, identifiers of genes with correlated expression were assigned to the corresponding gene objects of the IPKB. These genes, called “focus genes,” were overlaid onto a global molecular network developed from information contained in the IPKB. Networks of these focus genes were then algorithmically generated based on their connectivity. The functional analysis of members of a network identified the biological functions (and/or diseases) that were most significant to the genes in the network. Fischer's exact test was used to calculate a P value for the networks. A score was determined according to the IPA 7 Feature Manual that reflected the relevance of the network based on the number of regulated genes within a network and the size of the network.
Genome-wide association analysis
For the association analysis, log2-transformed expression levels of 150 individuals were used as traits for whole-genome association analysis with SNPs genotyped by a mixed model ANOVA using JMP Genomics (SAS Institute). “Genotype” and “gender” were used as fixed effects, “sire” and “slaughter day” as random effects, and “carcass weight” as a covariate. The sequences flanking the SNPs represented on the Illumina SNP chip, which were significantly associated with the level of transcription of any probe set of the Affymetrix expression microarrays, were assigned to the porcine genome sequences (Ensembl_Sscrofa_9, released April 2009). Annotation and localization of SNP sites and probe sets allowed discriminating cis- and trans-regulation. We defined an eQTL as “cis” if an associated SNP was located within an area less than 10 Mb from the probe set/gene. All other eQTL were considered as “trans.” Results are reported at thresholds of P < 1.4 × 10−9 (corresponding to a Bonferroni-corrected level of significances of P ≤ 0.05) and P < 10−5. A significance threshold of P < 10−5 was used, representing a tradeoff between avoidance of false-positive associations while taking into account the likely higher dependence among the expression traits (correlated expression) and among the SNP genotypes (linkage disequilibrium).
RESULTS
Expression profiling and genotyping
To characterize the genetic architecture of hepatic gene expression relevant to fatness traits, 150 performance-tested pigs were genotyped using the PorcineSNP60K BeadChip (Illumina) and expression-profiled using the 24K expression microarray. Out of 62,163 SNPs of the 60K SNP-Chip, a total of 52,727 SNPs with genotyping call rates greater than 95% and MAF greater than 5% were selected for genome-wide association analysis. The GeneChip Porcine Genome Arrays (Affymetrix) contained 24,123 probe sets representing 20,689 known genes (18), of which 10,945 probe sets were used for further analysis that showed consistent expression according to MAS5 analysis. Analysis of correlation between residuals derived from the mixed model analysis of the trait FA and the expression levels of 10,945 probe sets, respectively, revealed 663 probe sets that showed significant correlation with FA (supplementary Table I). The correlation coefficients at P ≤ 0.05 ranged |0.16-0.33|. FA was positively correlated with 213 genes and negatively correlated with 450 genes. These genes were functional candidates for traits related to fatness.
Detection of transcripts with fat area-correlated expression
We tested the list of significant positively and negatively correlated functional candidate genes for the enrichment in functional annotation groups as defined in the IPKB. Genes with expression levels that correlated positively with FA mainly belonged to categories related to metabolism of various macromolecules and nutrients, whereas genes with negative correlations belonged to functions related to dynamic cellular processes (Table 1).
TABLE 1.
Molecular and cellular functions of genes showing expression levels correlated positively or negatively with fat area
| Molecular and Cellular Functions | Number of Genes | P |
|---|---|---|
| Positive correlation | ||
| Lipid metabolism | 22 | 9.10E-03 |
| Small molecule biochemistry | 39 | 9.10E-03 |
| Drug metabolism | 12 | 9.10E-03 |
| Vitamin and mineral metabolism | 9 | 9.10E-03 |
| Cell death | 20 | 7.46E-03 |
| Negative correlation | ||
| Cell death | 103 | 4.69E-04 |
| Cellular development | 91 | 7.64E-04 |
| Cellular growth and proliferation | 117 | 9.07E-04 |
| Cell morphology | 48 | 1.84E-03 |
| Cell cycle | 34 | 2.48E-03 |
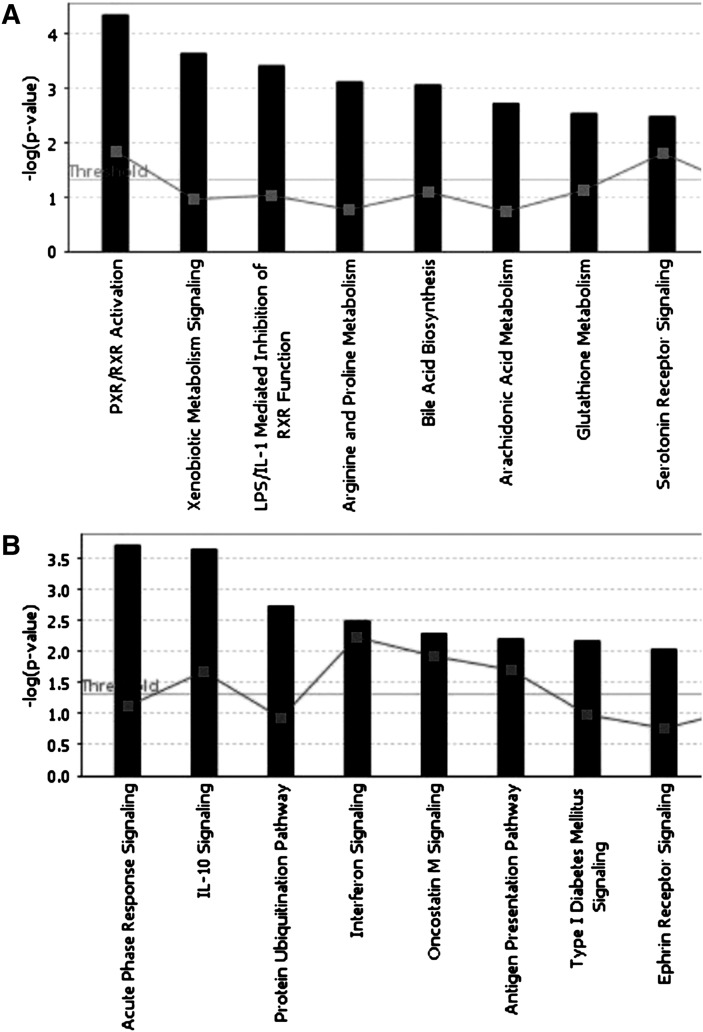
To further refine the functional annotation of these gene sets with FA-correlated expression, their assignment to canonical pathways was explored using the IPKB. The positively and negatively correlated expression levels of functional candidate genes were uploaded as gene datasets for analysis. Significantly enriched canonical pathways of the transcription profiles with positive correlation to FA belonged to pregnane X receptor (PXR) / retinoid X receptor (RXR) activation (SCD, GSTM2, CYP1A2, FOXO1, CYP3A4, and CYP7A1); xenobiotic metabolism signaling (PNPLA7, ALDH4A1, GSTT1, MAOB, GSTM2, CYP1A2, CYP3A4, FMO1, CES1, including EG:1066, and CITED2); lipopolysaccharide (LPS) / interleukin (IL)-1-mediated inhibition of RXR function (ALDH4A1, GSTT1, MAOB, GSTM2, SLC10A1, CYP3A4, CYP7A1, and FMO1); arginine and proline metabolism (CKB, ALDH4A1, MAOB, ASS1, and SAT2); bile acid biosynthesis (ALDH4A1, CYP3A4, CYP7A1, and ADHFE1); arachidonic acid metabolism (GSTT1, GPX3, CYP4F2, CYP1A2, CYP3A4, and PRDX6); glutathione metabolism (GSTT1, GPX3, GSTM2, and PRDX6); and serotonin receptor signaling (MAOB, PCBD1, and QDPR) (Fig. 1A). For the negatively correlated genes, the list indicated enrichment of genes belonging to the canonical pathways of acute phase response signaling (MTOR, SOD2, RIPK1, NFKBIA, MYD88, IL1RN, CRP, STAT3, LBP, SERPINE1, MAP2K1, and FGG); IL-10 signaling (CCR1, NFKBIA, SP1, IL1RN, IL10RB, STAT3, and LBP); protein ubiquitination (UBD, PSMB9, UCHL3, PSMA6, USP15, HLA-A, UBE2A, PSMD10, PSMA5, PSMB8, and HLA-C); interferon signaling (PTPN2, PSMB8, IFNAR1, and IRF1); oncostatin M signaling (EPAS1, OSMR, STAT3, and MAP2K1); antigen presentation pathway (PSMB9, HLA-A, PSMB8, and HLA-C); type I diabetes mellitus signaling (CD247, RIPK1, NFKBIA, HLA-A, MYD88, IRF1, and HLA-C); and ephrin receptor signaling (GNB1, CDC42, CXCR4, CXCL12, ARPC5, GNA11, ATF4, STAT3, and MAP2K1) (Fig. 1B).
Fig. 1.
Global canonical pathway analysis of gene expression patterns (A) positively correlated with fat area and (B) negatively correlated with fat area. The datasets were analyzed using the IPA software. Significance is expressed as a P value calculated using Fisher's exact test.
Whole-genome association analyses for abundance of transcripts with FA-correlated expression (eQTL)
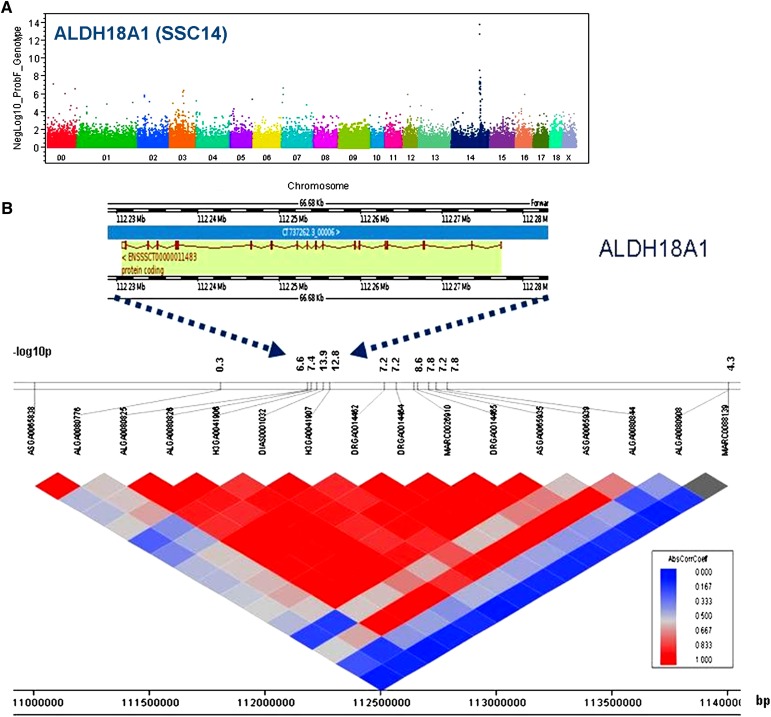
Association mapping of gene expression traits allowed us to identify eQTL (i.e., genomic regions that have a regulatory effect on the expression traits). Two types of eQTL could be distinguished: those that mapped near (less than 10 Mb from) the gene that encodes the transcript (“cis”) and those that mapped elsewhere in the genome (“trans”). To link eQTL to an organismal phenotype, we focused on the transcripts that showed expression levels correlated with the classical trait FA, and only these were subjected to association analysis using 52,727 SNPs. The association P values were adjusted to control for multiple testing using the stringent, conservative Bonferroni correction method to constrain the study-wise significance level. In total, 52,727 SNPs were tested for association to each of the 663 expression traits. Therefore, the Bonferroni-adjusted P threshold was computed as 0.05/(52,727 × 663) = 1.43 × 10−9. At this threshold, 555 eQTL were identified, which were composed of 96 cis- and 364 trans-eQTL. However, the Bonferroni adjustment method is likely too stringent, given the dependence among expression traits and SNP genotypes. For example, the expression level of ALDH18A1 is associated with a number of SNPs in the surrounding sequences of this gene where strong linkage disequilibrium (LD) exists (Fig. 2). Because the Bonferroni adjustment might be overly conservative, we set a more relaxed threshold at nominal values of P < 1.0 × 10−5, which is used in many studies (http://www.genome.gov/gwastudies) (19, 20). At this threshold, 663 probe sets revealed 4,727 eQTL with a mean negative log of the P value of 6.98 (supplementary Table II). For 3,745 of these, there was information on the genomic localization of both the SNP marker and the transcript represented by the probe set. This allowed assignment of either cis or trans regulation. In fact, 448 cis-eQTL and 3,297 trans-eQTL were detected with mean negative log of the P values of 7.81 and 6.90, respectively. For the rest of the eQTL (982), either the SNP marker or the corresponding probe set could not be localized. The 448 cis-eQTL corresponded to 71 genes, and the 3,297 trans-eQTL were related to 408 genes (Table 2). Functional candidate genes with either a positive or negative correlation with FA and with either a cis- or trans-eQTL are shown in Tables 3–6.
Fig. 2.
Significant association of genetic variation with expression of ALDH18A1. A: The expression level of ALDH18A1 is significantly associated with many SNPs in the same region. The P values (−log10 transformed) on the vertical axis are plotted against the genomic position of SNPs on the horizontal axis. B: ALDH18A1 located at SSC14 position 112 Mb. The P values (−log10 transformed) of SNPs in the same region are listed (indicated by blue arrows). The linkage disequilibrium pattern around the gene is shown. The scale of correlation coefficient varies from red to blue (high to low). SNP positions are located under the linkage disequilibrium pattern.
TABLE 2.
Numerical summary of the whole-genome association study for eQTL of genes showing expression levels correlated with fat area
| Number of eQTL |
Number of cis-eQTL |
Number of trans-eQTL |
|||||
|---|---|---|---|---|---|---|---|
| Total Probe Set | Number of SNP | P < 10 | P < 10 | P < 10 | P < 10 | P < 10 | P < 10 |
| 663 | 52,727 | 4,727 | 555 | 448 | 76 | 3,297 | 364 |
TABLE 3.
SNP markers showing association with the expression levels of genes positively correlated with fat area and their positions indicating cis-regulation
| Gene Symbol | Gene Location(Mb, SSC) | SNP | SNP Location(Mb, SSC) | Association−log10 (P) | R2a | Pb |
|---|---|---|---|---|---|---|
| CA7 | 103.0 (4) | ALGA0111689 | 108.0 (4) | 5.1 | 0.24 | 3.22E-03 |
| CYP1A2 | 64.3 (7) | MARC0013048 | 62.6 (7) | 5.9 | 0.19 | 1.75E-02 |
| CYP4F2 | 42.8 (2) | ALGA0013037 | 39.3 (2) | 5.5 | 0.20 | 1.56E-02 |
| FAM134B | 5.15 (16) | H3GA0045908 | 5.3 (16) | 12.5 | 0.29 | 3.68E-04 |
| FOXO1 | 15.1 (11) | ALGA0119631 | 22.7 (11) | 5.2 | 0.23 | 4.55E-03 |
| FOXRED1 | 51.7 (9) | ASGA0043161 | 51.8 (9) | 5.9 | 0.19 | 2.20E-02 |
| GSTT1 | 50.8 (14) | H3GA0040142 | 49.2 (14) | 6.2 | 0.18 | 2.39E-02 |
| HDHD2 | 291.0 (1) | MARC0011129 | 290.0 (1) | 9.7 | 0.20 | 1.43E-02 |
| PLIN4 | 48.7 (2) | ASGA0090304 | 48.6 (2) | 7.5 | 0.22 | 7.95E-03 |
| XYLB | 19.0 (13) | ALGA0068858 | 19.1 (13) | 17.5 | 0.18 | 2.49E-02 |
Correlation between expression and fat area.
P of correlation (R2).
TABLE 4.
SNP markers showing association with the expression levels of genes negatively correlated with fat area and their positions indicating cis-regulation
| Gene Symbol | Gene Location(Mb, SSC) | SNP | SNP Location(Mb, SSC) | Association−log10 (P) | R2a | Pb |
|---|---|---|---|---|---|---|
| ALDH18A1 | 112.0 (14) | DIAS0001032 | 112.0 (14) | 13.8 | −0.29 | 2.80E-04 |
| ANO10 | 22.6 (13) | M1GA0017465 | 22.9 (13) | 6.2 | −0.20 | 1.26E-02 |
| ANXA5 | 87.9 (8) | ALGA0048900 | 87.9 (8) | 18.8 | −0.33 | 4.00E-05 |
| B4GALT5 | 53.5 (17) | M1GA0022228 | 53.5 (17) | 8.5 | −0.29 | 3.72E-04 |
| BRD2 | 29.5 (7) | M1GA0009853 | 32.8 (7) | 6.4 | −0.22 | 7.55E-03 |
| C1orf57 | 59.3 (14) | H3GA0040407 | 59.2 (14) | 18.7 | −0.21 | 9.43E-03 |
| ENSA | 103.0 (4) | H3GA0013772 | 104.0 (4) | 6.6 | −0.21 | 1.20E-02 |
| H2AFY | 125.0 (2) | CASI0010120 | 125.0 (2) | 6.0 | −0.20 | 1.35E-02 |
| HS2ST1 | 34.9 (9) | ASGA0042539 | 35.2 (9) | 9.5 | −0.21 | 9.70E-03 |
| MRPL44 | 118.0 (15) | ASGA0070945 | 118.0 (15) | 7.9 | −0.21 | 1.09E-02 |
Correlation between expression and fat area.
P of correlation (R2).
TABLE 5.
SNP markers showing association with the expression levels of genes positively correlated with fat area and their positions indicating trans-regulation
| Gene Symbol | Gene Location(Mb, SSC) | SNP | SNP Location(Mb, SSC) | Association−log10 (P) | R2a | Pb |
|---|---|---|---|---|---|---|
| APCDD1 | (6) | H3GA0010071 | 87.1 (3) | 6.3 | 0.31 | 1.10E-04 |
| CA7 | 103.0 (4) | ALGA0016578 | 128.0 (2) | 5.0 | 0.24 | 3.22E-03 |
| CYP7A1 | 77.2 (4) | H3GA0001640 | 51.5 (1) | 5.4 | 0.25 | 2.25E-03 |
| FAM134B | 5.2 (16) | H3GA0051448 | 6.1 (X) | 5.1 | 0.29 | 3.68E-04 |
| FOXO1 | 15.1 (11) | ASGA0064852 | 87.2 (14) | 6.0 | 0.23 | 4.55E-03 |
| GPX3 | 69.4 (16) | ALGA0019285 | 53.5 (3) | 5.0 | 0.24 | 2.75E-03 |
| HYI | 120.0 (6) | H3GA0005137 | 284.0 (1) | 5.7 | 0.23 | 3.85E-03 |
| LOC100156168 | 86.8 (1) | ALGA0008875 | 255.0 (1) | 6.1 | 0.28 | 5.93E-04 |
| XPO5 | 0.01 (7) | DRGA0005753 | 40.7 (5) | 5.1 | 0.25 | 2.50E-03 |
| ZADH2 | 156.0 (1) | SIRI0000493 | 136.0 (4) | 5.2 | 0.26 | 1.34E-03 |
Correlation between expression and fat area.
P of correlation (R2).
TABLE 6.
SNP markers showing association with the expression levels of genes negatively correlated with fat area and their positions indicating trans-regulation
| Gene Symbol | Gene Location(Mb, SSC) | SNP | SNP Location(Mb, SSC) | Association−log10 (P) | R2a | Pb |
|---|---|---|---|---|---|---|
| AGPAT9 | 116.0 (8) | ASGA0058243 | 63.5 (13) | 6.9 | −0.28 | 4.54E-04 |
| ALDH18A1 | 112.0 (14) | ALGA0038327 | 7.3 (7) | 6.7 | −0.29 | 2.80E-04 |
| ANXA5 | 87.9 (8) | ALGA0068733 | 17.1 (13) | 6.2 | −0.33 | 4.00E-05 |
| CCL2 | 38.4 (12) | MARC0091801 | 17.6 (9) | 8.9 | −0.27 | 7.06E-04 |
| CD53 | 114.0 (4) | H3GA0055035 | 31.8 (3) | 7.2 | −0.27 | 1.00E-03 |
| IER5 | 115.0 (9) | ALGA0015640 | 112.0 (2) | 9.2 | −0.30 | 2.44E-04 |
| LOC728835 | 37.2 (12) | ALGA0003946 | 67.7 (1) | 5.4 | −0.27 | 8.65E-04 |
| PLIN2 | 213.0 (1) | DRGA0009341 | 37.6 (9) | 5.8 | −0.30 | 1.72E-04 |
| RIPK1 | 2.9 (7) | ALGA0015662 | 112.0 (2) | 5.4 | −0.32 | 7.67E-05 |
| TRAFD1 | 39.5 (14) | H3GA0001667 | 53.5 (1) | 5.4 | −0.28 | 6.26E-04 |
Correlation between expression and fat area.
P of correlation (R2).
Biologically relevant pathways and network generation
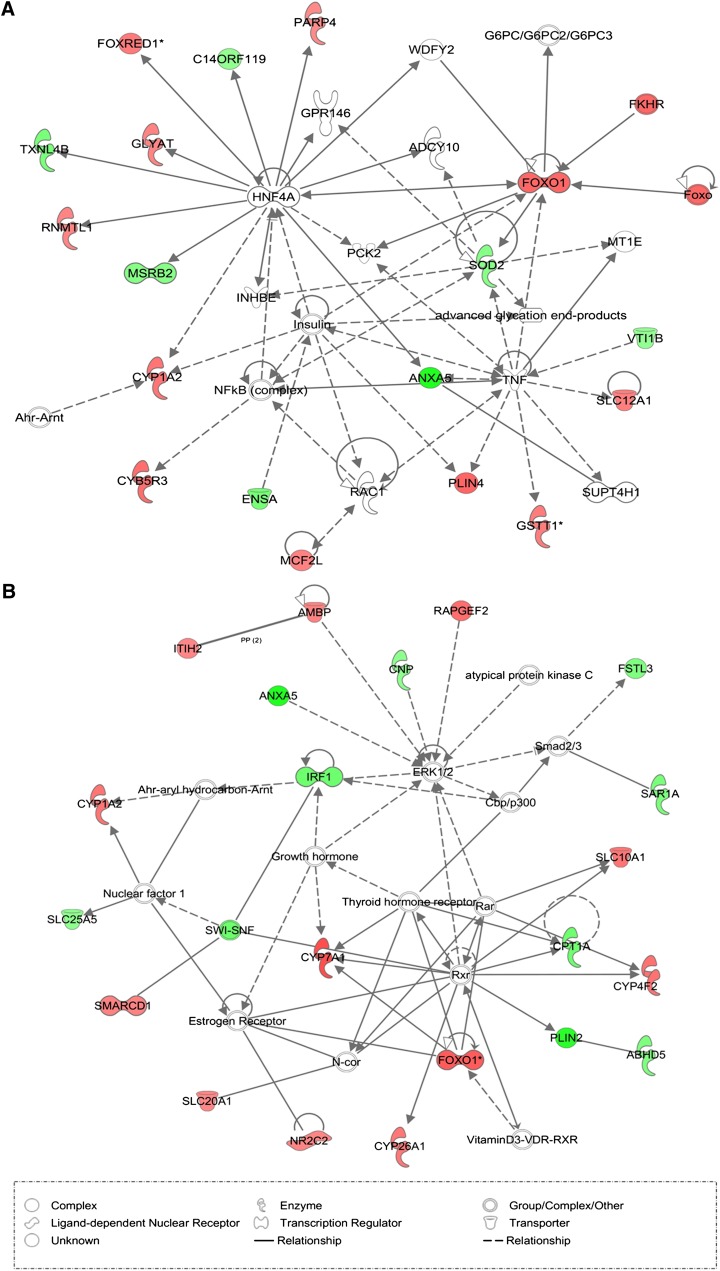
To determine the biologically relevant networks beyond canonical pathways, pathway analysis was performed for gene sets with cis- (71 genes) and trans- (408 genes) eQTL using the IPKB. The networks describe functional relationships between gene products based on known interactions reported in the literature. Fig. 3 exemplarily shows the highest ranking networks deduced from the list of functional candidate genes having either cis-eQTL or trans-eQTL. The network of cis-regulated genes comprised 19 focus genes belonging to lipid metabolism, small molecule biochemistry, and cell death (Fig. 3A). The second network of genes with trans-regulation contained 21 focus genes associated with lipid metabolism, small molecule biochemistry, and genetic disorder (Fig. 3B).
Fig. 3.
Pathway analysis based on the IPKB. The high-scoring gene networks significantly correlated with fat area and (A) cis-regulation or (B) trans-acting eQTL. Both networks belong to lipid metabolism, small molecule biochemistry, and cell death or genetic disorder. Color shading corresponds to the type of correlation, with red for positive correlation and green for negative correlation. The nodes without color are not from the list of correlated genes. The shape of the node indicates the major function of the protein. A line denotes binding of the products of two genes while “acts on” is denoted by a line with an arrow and “direct interaction” by a line without an arrow. A dotted line denotes an indirect interaction.
DISCUSSION
To get more insight into the genetic control of traits related to obesity and to address candidate genes for these organismal phenotypes, the genetics of expression was studied in porcine liver, a metabolically active tissue that is critical to a number of core biological processes and that plays a role not only in carcass traits in pigs but also in body composition, obesity and a number of common metabolic diseases in humans. We achieved detecting functional candidate genes for these traits by exploiting trait-correlated expression and assigning eQTL to these genes through genome-wide SNP-array based association analysis. Ghazalpour et al. (21) found that the mapping resolution for eQTL was significantly greater in the outbred population than in F2 intercrosses and showed how this precise mapping could resolve previously identified loci (in F2 intercross studies), which affect many different transcript levels (known as eQTL ‘hotspots), into distinct regions (21). The success of these studies prompted us to investigate the potential use of a commercial pig herd (outbred) for eQTL studies compared with the F2 population from our previous studies (22–24). Here, a whole-genome association study for hepatic gene expression profiles was carried out in a commercial herd of Pi×(DE×DL).
The application of global gene expression analysis to studies of cell and tissues has provided a wealth of data relevant to complex traits. Accounting for 10,945 probe sets representing genes found expressed in liver, more than 70,000 eQTL can be expected at P < 10−5. However, only 6% of liver transcripts out of the 10,945 probe sets were found to correlate with the trait “fat area.” Among the carcass traits related to fatness that are measured during performance testing in commercial breeding schemes, “fat area” is highly heritable and can be considered an indicator of obesity and body composition (25). FA shows high genetic correlation with plasma leptin levels, which in turn are correlated with adipositas and habitus (25–27). Finding statistically significant correlations between a trait and particular genes suggest a biologic relationship between them (22, 28, 29). IPA essentially evaluates the enrichment of particular biological processes and molecular functions of gene sets by examining information collected by databases such as Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and IPKB. In this study, many genes involved in lipid metabolism were positively correlated with FA, whereas genes with negative correlation were associated with cellular development, cellular growth, and proliferation. These findings were in agreement with our previous conclusion that the genetic differences between obese and lean pig breeds were associated with upregulation of lipid metabolic pathways in obese pigs and upregulation of cell growth and/or maintenance and cell proliferation pathways in lean pigs (30).
Canonical pathway analysis
Canonical pathway analysis highlighted pathways involving signaling via PXR and RXR. These pathways play an important regulatory role in metabolic pathways (glucose, fatty acid, and cholesterol metabolism) (31). Individual genes from the lists that were detected here and assigned to the pathways of “PXR/RXR activation,” “xenobiotic metabolism signaling,” and “LPS/IL-1-mediated inhibition of RXR function” point to lipid metabolism instead of any function related to detoxification. Several of the genes in these pathways have been shown to influence adipose tissue mass in other studies. For example, forkhead box protein O1 (FOXO1) plays an important role in modulating metabolic functions. It induces the expression of gluconeogenic genes and contributes to hyperglycemia during hepatic insulin resistance (32, 33). The other two pathways (bile acid biosynthesis and arachidonic acid metabolism) are involved in cholesterol metabolism. Similar pathways were also found positively correlated with fat mass in mice (34, 35). The conversion of cholesterol to bile acid is the major pathway for cholesterol catabolism. In bile acid pathways, cholesterol serves as a substrate that is converted into bile acid through the activation of cholesterol 7 α hydroxylase (CYP7A1), the transcript for which is significantly increased in pigs with a high fat area. This protein is the first rate-limiting enzyme in bile acid synthesis in the liver. FoxO1 has been shown to stimulate rat Cyp7a1 gene transcription, and mutations in the FoxO1 binding site attenuated rat Cyp7a1 promoter activity (36). In our study, both FOXO1 and CYP7A1 transcripts were significantly increased in pigs with a high fat area. The aldehyde dehydrogenase (ALDH)4A1 gene family, which is involved in arginine and proline metabolism, was also found to be significantly correlated with FA. The increased expression of aldehyde dehydrogenase in mice with higher subcutaneous fat-pad mass has previously been reported (37). Another pathway with a significant correlation with high FA was glutathione metabolism. Glutathione is a major endogenous cellular anti-oxidant. We found that four of the genes in this pathway were significantly overexpressed in pigs with high FA. Glutathione peroxidase (GPX)3 is involved in cellular protection against oxidative damage through the reduction of peroxides (38). The cytosilic isoform of GPX3 has been associated with obesity (39), and there is evidence for causal relationship of GPX3 to obesity and for the idea that GPX3 overexpression modifies insulin resistance (40). Recently, transgenic and knockout mouse models of GPX3 resulted in significant changes in obesity-related traits (41). PRDX6 is an antioxidant enzyme with calcium-independent phospholipase A2 activity (42). PRDX6 was identified in a recent study as a possible candidate gene underlying a novel obesity locus on chromosome 1q24 in an isolated population of Cilento, Italy (43). Recently, a study of 21,000 individuals identified PRDX6 associated with body mass index (BMI) as an indicator of obesity (44). Using the porcine model providing reliable phenotypes and samples, we were able to associate canonical pathways rather than only single genes with the organismal phenotype related to obesity. For some elements of these canonical pathways there are supporting data from functional studies in rodent models and epidemiological studies in human.
In this study, genes whose expression was found to be negatively correlated with fatness were involved in acute phase response signaling, protein ubiquitination, IL-10 signaling, and the antigen presentation pathway. These pathways, indicating a higher cellular turnover, activity, and reactivity, were also found to correlate with leptin-mediated weight loss in ob/ob mice after leptin administration (45).
Whole-genome association analyses for abundance of transcripts with ldquofat areardquo-correlated expression (eQTL)
We present here the first eQTL study based on whole-genome association analysis in a commercial crossbred population with focus on the FA trait. In the first step, we analyzed the correlation between gene expression and the phenotype related to fatness, which revealed biologically meaningful relationships. In the second step, eQTL were identified for transcripts that showed trait-correlated expression, which informed us about the genomic location of putative regulatory loci. This strategy reduced the number of several thousand eQTL that were not associated with FA. A genome-wide association screen for expression SNPs (eSNP) was performed on 150 pigs from commercial herds. The eSNPs were assessed using a mixed model analysis with “genotype” and “sex” as fixed effects, “sire” and “slaughter days” as random effects, and “carcass weight” as a covariate. For the identification of sequence variants having cis- and trans-regulatory effects on expression traits, SNPs located within a 10 Mb window at the location of the probe set were defined as cis-eQTL. The trans-acting eQTL represent transcripts whose abundance is regulated by loci remote from the genomic locus of each of these genes. In our study, the proportion of trans-eQTL was higher (70%) compared with cis-eQTL (10%) at P < 1.0 × 10−5; the remaining eQTL could not be assigned cis or trans. Most studies in model organisms have shown that the number of traits with significant trans-eQTL is fewer than the number of expression traits with significant cis-eQTL (9–11, 46). Here, eQTL analysis was focused only on biologically meaningful relationship with the traits.
Biologically relevant pathways and network generation of cis- and trans-regulated genes
A network of cis-regulated genes comprised 19 focus genes with a score of 40 belonging to lipid metabolism, small molecule biochemistry, and cell death (Fig. 2A). In this network, 10 probe sets representing cis-regulated genes had hepatocyte nuclear factor 4 alpha (HNF4A), a common transcription factor. HNF4A, the protein encoded by this gene, is a nuclear transcription factor that binds DNA as a homodimer to the promoters of several genes involved in hepatic pathways. The 10 commonly cis-regulated target genes of HNF4A are likely to exhibit polymorphisms affecting their own expression. Moreover, these polymorphic sites may be located in the target region of the common transcription factor HNF4A. Thus the coregulation of the 10 probe sets representing cis-regulated genes is likely due to differences in the affinity of allelic variants of the HNF4A binding sites in their promoters. Another central component of the network was FOXO1. The transcript of this molecule not only significantly correlated with FA, as described above, but was also cis- and trans-regulated. Another node within the network is the tumor necrosis factor (TNF) interacting with GSTT1, PLIN4, ANXA5, SOD2, VTI1B, and SLC12A1, some of which were previously associated with fat traits in human (47, 48). Perilipins (PLIN) are proteins localized at the surface of the lipid droplets in adipocytes, steroid-producing cells, and play a key role in the cellular regulation of triglyceride deposition and mobilization (49). PLIN polymorphisms have been associated with obesity-related phenotypes (47, 50). Mitochondrial superoxide dismutase-2 (SOD-2) is an endogenous anti-oxidant enzyme. Gene variants of SOD-2 have been shown to be associated with coronary heart disease in diabetes mellitus (48). For many genes mentioned here, functional or epidemiological association to fatness traits has been shown in rodents and humans. Using the porcine model, we were able to provide functional and genetic links between these genes and to derive networks.
For the network of genes with trans-eQTL, central but not differentially regulated molecules were the retinoid receptor, estrogen receptor, thyroid hormone receptor, and growth hormone. This finding is consistent with published eQTL studies, which have indicated that trans-eQTL intervals encode many different gene classes, including receptors and their ligands, which can affect their target transcript via a context-dependent mechanism (51–53). Together, the central components of the trans-networks typically represented hierarchically high positions in signaling cascades, whereas downstream components of signaling pathways were represented in the cis-networks.
The correlation between expression levels and the organismal phenotype tended to be higher for trans-regulated genes. This correlation comprised genetic causal effects that the transcript may have on the phenotype as well as common dependence on environmental factors. Marker association with expression levels tended to be more significant for cases with cis-regulation, where genetic effects were expected to be prevailing. As we noticed in our previous study, eQTL mapping with its potential to categorize cis- and trans-effects provided a way to discriminate between effect and cause of trait-associated differential expression (22–24, 54, 55). This finding was also supported by other studies in model organisms; environmentally induced changes in gene expression seem to act through prominent trans-effects (56, 57).
The number of GWAS in humans and mice continues to rise rapidly. In the GWA studies to date, 10-15% of the top hits have affected a known eQTL in a public data set (58). Recently, a study has shown that trait-associated SNPs are more likely to be eQTLs (20). These findings are robust across a range of thresholds for establishing eQTL (P range from 10−4 to 10−8).
To our knowledge, this is the first report of a comprehensive scan for eQTL associated with obesity in pigs. Many of the candidate genes previously identified in humans and mice have been confirmed here in the porcine model. Moreover, lists of novel candidate genes for fat traits were identified for the first time here in the pig model. These candidate genes for fat traits are valuable for human research due to the many similarities in physiology and genome between pigs and humans. The application of this information can facilitate the detection of trait-associated SNPs and a better understanding of the biology of complex traits.
Supplementary Material
Acknowledgments
The authors thank Annette Jugert and Joana Bittner for their excellent technical help.
Footnotes
Abbreviations:
- ALDH
- aldehyde dehydrogenase
- BMI
- body mass index
- CYP7A1
- cholesterol 7 α hydroxylase
- eQTL
- expression quantitative trait loci
- eSNP
- expression SNP
- FA
- fat area
- FOXO1
- forkhead box protein O1
- GPX
- glutathione peroxidase
- GWAS
- genome-wide association studies
- HNF4A
- hepatocyte nuclear factor 4 alpha
- IL
- interleukin
- IPA
- Ingenuity Pathways Analysis
- IPKB
- Ingenuity Pathways Knowledge Base
- LD
- linkage disequilibrium
- LPS
- lipopolysaccharide
- MAF
- minor allele frequency
- Pi×(DE×DL)
- Pietrain×(German Large White × German Landrace) pig
- PLIN
- perilipin
- PXR
- pregnane X receptor
- RXR
- retinoid X receptor
- SNP
- single nucleotide polymorphism
- SOD-2
- superoxide dismutase-2
- TNF
- tumor necrosis factor
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables.
REFERENCES
- 1.Reilly J. J., Kelly J. Long-term impact of overweight and obesity in childhood and adolescence on morbidity and premature mortality in adulthood: systematic review. Int. J. Obes. Epub ahead of print. October 26, 2010; doi:10.1038/ijo.2010.222. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty S., Zawieja S., Wang W., Zawieja D. C., Muthuchamy M. 2010. Lymphatic system: a vital link between metabolic syndrome and inflammation. Ann. N. Y. Acad. Sci. 1207 (Suppl. 1): E94–E102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bode G., Clausing P., Gervais F., Loegsted J., Luft J., Nogues V., Sims J. 2010. The utility of the minipig as an animal model in regulatory toxicology. J. Pharmacol. Toxicol. Methods. 62: 196–220. [DOI] [PubMed] [Google Scholar]
- 4.Goh V. H., Tain C. F., Tong T. Y., Mok H. P., Wong M. T. 2004. Are BMI and other anthropometric measures appropriate as indices for obesity? A study in an Asian population. J. Lipid Res. 45: 1892–1898. [DOI] [PubMed] [Google Scholar]
- 5.Gilad Y., Rifkin S. A., Pritchard J. K. 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung V., Conlin L., Weber T., Arcaro M., Jen K., Morley M., Spielman R. 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 33: 422–425. [DOI] [PubMed] [Google Scholar]
- 7.Dixon A. L., Liang L., Moffatt M. F., Chen W., Heath S., Wong K. C., Taylor J., Burnett E., Gut I., Farrall M., et al. 2007. A genome-wide association study of global gene expression. Nat. Genet. 39: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 8.Göring H. H., Curran J. E., Johnson M., Dyer T. D., Charlesworth J., Cole S. A., Jowett J. B., Abraham L. J., Rainwater D. L., Comuzzie A. G., et al. 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 39: 1208–1216. [DOI] [PubMed] [Google Scholar]
- 9.Emilsson V., Thorleifsson G., Zhang B., Leonardson A., Zink F., Zhu J., Carlson S., Helgason A., Walters G. B., Gunnarsdottir S., et al. 2008. Genetics of gene expression and its effect on disease. Nature. 452: 423–428. [DOI] [PubMed] [Google Scholar]
- 10.Brem R. B., Yvert G., Clinton R., Kruglyak L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science. 296: 752–755. [DOI] [PubMed] [Google Scholar]
- 11.Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., Colinayo V., Ruff T. G., Milligan S. B., Lamb J. R., Cavet G., et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature. 422: 297–301. [DOI] [PubMed] [Google Scholar]
- 12.Morley M., Molony C., Weber T., Devlin J., Ewens K., Spielman R., Cheung V. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature. 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung V., Spielman R., Ewens K., Weber T., Morley M., Burdick J. 2005. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 437: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stranger B., Nica A., Forrest M., Dimas A., Bird C., Beazley C., Ingle C., Dunning M., Flicek P., Koller D., et al. 2007. Population genomics of human gene expression. Nat. Genet. 39: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stranger B., Forrest M., Dunning M., Ingle C., Beazley C., Redon R., Bird C., de Grassi A., Lee C., Tyler-Smith C., et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T. J., Sladek R., Majewski J. 2008. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 40: 225–231. [DOI] [PubMed] [Google Scholar]
- 17.Zentral Verband der Deutschen Schweinproduktion (ZDS). 2004. Richtlinien für die Stationsprüfung auf Mastleistung, Schlachtkörperwert und Fleischbeschaffenheit beim Schwein. Bonn, Germany. [Google Scholar]
- 18.Naraballobh W., Chomdej S., Murani E., Wimmers K., Ponsuksili S. 2010. Annotation and in silico localization of the Affymetrix GeneChip Porcine Genome Array. Arch. Tierz. 53: 230–238. [Google Scholar]
- 19.Schadt E. E., Molony C., Chudin E., Hao K., Yang X., Lum P. Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolae D. L., Gamazon E., Zhang W., Duan S., Dolan M. E., Cox N. J. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6: e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghazalpour A., Doss S., Kang H., Farber C., Wen P. Z., Brozell A., Castellanos R., Eskin E., Smith D. J., Drake T. A., et al. 2008. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet. 4: e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponsuksili S., Jonas E., Murani E., Phatsara C., Srikanchai T., Walz C., Schwerin M., Schellander K., Wimmers K. 2008. Trait-correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 9: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponsuksili S., Murani E., Schwerin M., Schellander K., Wimmers K. 2010a. Identification of expression QTL (eQTL) of genes expressed in porcine M. longissimus dorsi and associated with meat quality traits. BMC Genomics. 11: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponsuksili S., Murani E., Phatsara C., Schwerin M., Schellander K., Wimmers K. 2010b. Expression quantitative trait loci analysis of genes in porcine muscle by quantitative real-time RT-PCR compared to microarray data. Heredity. 105: 309–317. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki K., Inomata K., Katoh K., Kadowaki H., Shibata T. 2009. Genetic correlations among carcass cross-sectional fat area ratios, production traits, intramuscular fat, and serum leptin concentration in Duroc pigs. J. Anim. Sci. 87: 2209–2215. [DOI] [PubMed] [Google Scholar]
- 26.Hoque M. A., Katoh K., Suzuki K. 2009. Genetic associations of residual feed intake with serum insulin-like growth factor-I and leptin concentrations, meat quality, and carcass cross sectional fat area ratios in Duroc pigs. J. Anim. Sci. 87: 3069–3075. [DOI] [PubMed] [Google Scholar]
- 27.Ranganathan S., Maffei M., Kern P. A. 1998. Adipose tissue ob mRNA expression in humans: discordance with plasma leptin and relationship with adipose TNFalpha expression. J. Lipid Res. 39: 724–730. [PubMed] [Google Scholar]
- 28.Segal E., Friedman N., Kaminski N., Regev A., Koller D. 2005. From signatures to models: understanding cancer using microarrays. Nat. Genet. 37 (Suppl.): S38–S45. [DOI] [PubMed] [Google Scholar]
- 29.Ponsuksili S., Murani E., Phatsara C., Schwerin M., Schellander K., Wimmers K. 2009. Porcine muscle sensory attributes associate with major changes in gene networks involving CAPZB, ANKRD1, and CTBP2. Funct. Integr. Genomics. 9: 455–471. [DOI] [PubMed] [Google Scholar]
- 30.Ponsuksili S., Murani E., Walz C., Schwerin M., Wimmers K. 2007. Pre- and postnatal hepatic gene expression profiles of two pig breeds differing in body composition: insight into pathways of metabolic regulation. Physiol. Genomics. 29: 267–279. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja H. S., Szanto A., Nagy L., Davies P. J. 2003. The retinoid X receptor and its ligands: versatile regulators of metabolic function, cell differentiation and cell death. J. Biol. Regul. Homeost. Agents. 17: 29–45. [PubMed] [Google Scholar]
- 32.Schmoll D., Walker K. S., Alessi D. R., Grempler R., Burchell A., Guo S., Walther R., Unterman T. G. 2000. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 275: 36324–36333. [DOI] [PubMed] [Google Scholar]
- 33.Hall R. K., Yamasaki T., Kucera T., Waltner-Law M., O'Brien R., Granner D. K. 2000. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 275: 30169–30175. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons G. F. 2003. Regulation of fatty acid and cholesterol synthesis: co-operation or competition? Prog. Lipid Res. 42: 479–497. [DOI] [PubMed] [Google Scholar]
- 35.Ghazalpour A., Doss S., Sheth S. S., Ingram-Drake L. A., Schadt E. E., Lusis A. J., Drake T. A. 2005. Genomic analysis of metabolic pathway gene expression in mice. Genome Biol. 6: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T., Ma H., Chiang J. Y. 2008. TGFbeta1, TNFalpha, and insulin signaling crosstalk in regulation of the rat cholesterol 7 alpha-hydroxylase gene expression. J. Lipid Res. 49: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demozay D., Rocchi S., Mas J. C., Grillo S., Pirola L., Chavey C., Van Obberghen E. 2004. Fatty aldehyde dehydrogenase: potential role in oxidative stress protection and regulation of its gene expression by insulin. J. Biol. Chem. 279: 6261–6270. [DOI] [PubMed] [Google Scholar]
- 38.Ishibashi N., Mirochnitchenko O. 2002. Chemokine expression in transgenic mice overproducing human glutathione peroxidases. Methods Enzymol. 353: 460–476. [DOI] [PubMed] [Google Scholar]
- 39.McClung J. P., Roneker C. A., Mu W., Lisk D. J., Langlais P., Liu F., Lei X. G. 2004. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc. Natl. Acad. Sci. USA. 101: 8852–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y. S., Kim A. Y., Choi J. W., Kim M., Yasue S., Son H. J., Masuzaki H., Park K. S., Kim J. B. 2008. Dysregulation of adipose glutathione peroxidase 3 in obesity contributes to local and systemic oxidative stress. Mol. Endocrinol. 22: 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X., Deignan J. L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R. R., et al. 2009. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 41: 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manevich Y., Reddy K. S., Shuvaeva T., Feinstein S. I., Fisher A. B. 2007. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J. Lipid Res. 48: 2306–2318. [DOI] [PubMed] [Google Scholar]
- 43.Ciullo M., Nutile T., Dalmasso C., Sorice R., Bellenguez C., Colonna V., Persico M. G., Bourgain C. 2008. Identification and replication of a novel obesity locus on chromosome 1q24 in isolated populations of Cilento. Diabetes. 57: 783–790. [DOI] [PubMed] [Google Scholar]
- 44.Naukkarinen J., Surakka I., Pietiläinen K. H., Rissanen A., Salomaa V., Ripatti S., Yki-Järvinen H., van Duijn C. M., Wichmann H. E., Kaprio J., et al. 2010. Use of genome-wide expression data to mine the “Gray Zone” of GWA studies leads to novel candidate obesity genes. PLoS Genet. 6: e1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma A., Bartell S. M., Baile C. A., Chen B., Podolsky R. H., McIndoe R. A., She J. X. 2010. Hepatic gene expression profiling reveals key pathways involved in leptin-mediated weight loss in ob/ob mice. PLoS ONE. 5: e12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehrabian M., Allayee H., Stockton J., Lum P. Y., Drake T. A., Castellani L. W., Suh M., Armour C., Edwards S., Lamb J., et al. 2005. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat. Genet. 37: 1224–1233. [DOI] [PubMed] [Google Scholar]
- 47.Deram S., Nicolau C. Y., Perez-Martinez P., Guazzelli I., Halpern A., Wajchenberg B. L., Ordovas J. M., Villares S. M. 2008. Effects of perilipin (PLIN) gene variation on metabolic syndrome risk and weight loss in obese children and adolescents. J. Clin. Endocrinol. Metab. 93: 4933–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones D. A., Prior S. L., Tang T. S., Bain S. C., Hurel S. J., Humphries S. E., Stephens J. W. 2010. Association between the rs4880 superoxide dismutase 2 (C>T) gene variant and coronary heart disease in diabetes mellitus. Diabetes Res. Clin. Pract. 90: 196–201. [DOI] [PubMed] [Google Scholar]
- 49.Tai E. S., Ordovas J. M. 2007. The role of perilipin in human obesity and insulin resistance. Curr. Opin. Lipidol. 18: 152–156. [DOI] [PubMed] [Google Scholar]
- 50.Sone Y., Yamaguchi K., Fujiwara A., Kido T., Kawahara K., Ishiwaki A., Kondo K., Morita Y., Tominaga N., Otsuka Y. 2010. Association of lifestyle factors, polymorphisms in adiponectin, perilipin and hormone sensitive lipase, and clinical markers in japanese males. J. Nutr. Sci. Vitaminol. (Tokyo). 56: 123–131. [DOI] [PubMed] [Google Scholar]
- 51.Yvert G., Brem R. B., Whittle J., Akey J. M., Foss E., Smith E. N., Mackelprang R., Kruglyak L. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 35: 57–64. [DOI] [PubMed] [Google Scholar]
- 52.Park C. C., Ahn S., Bloom J., Lin A., Wang R., Wu T., Sekar A., Khan A., Farr C., Lusis A. J., et al. 2008. Fine mapping of regulatory loci for mammalian gene expression using radiation hybrids. Nat. Genet. 40: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papeta N., Chan K. T., Prakash S., Martino J., Kiryluk K., Ballard D., Bruggeman L. A., Frankel R., Zheng Z., Klotman P. E., et al. 2009. Susceptibility loci for murine HIV-associated nephropathy encode trans-regulators of podocyte gene expression. J. Clin. Invest. 119: 1178–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wimmers K., Murani E., Ponsuksili S. 2010. Expression QTL and their applications in genetic improvement in farm animals. CAB Reviews. 5: 1–8. [Google Scholar]
- 55.Wimmers K., Murani E., Ponsuksili S. 2010. Functional genomics and genetical genomics approaches towards elucidating networks of genes affecting meat performance in pigs. Brief. Funct. Genomics. 9: 251–258. [DOI] [PubMed] [Google Scholar]
- 56.Li Y., Álvarez O. A., Gutteling E. W., Tijsterman M., Fu J., Riksen J., Hazendonk E., Prins P., Plasterk R. H., Jansen R. C., et al. 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith E. N., Kruglyak L. 2008. Gene-environment interaction in yeast gene expression. PLoS Biol. 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10: 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.