Abstract
Reducing circulating LDL-cholesterol (LDL-c) reduces the risk of cardiovascular disease in people with hypercholesterolemia. Current approaches to reduce circulating LDL-c include statins, which inhibit cholesterol synthesis, and ezetimibe, which blocks cholesterol absorption. Both elevate serum PCSK9 protein levels in patients, which could attenuate their efficacy by reducing the amount of cholesterol cleared from circulation. To determine whether PCSK9 inhibition could enhance LDL-c lowering of both statins and ezetimibe, we utilized small interfering RNAs (siRNAs) to knock down Pcsk9, together with ezetimibe, rosuvastatin, and an ezetimibe/rosuvastatin combination in a mouse model with a human-like lipid profile. We found that ezetimibe, rosuvastatin, and ezetimibe/rosuvastatin combined lower serum cholesterol but induce the expression of Pcsk9 as well as the Srebp-2 hepatic cholesterol biosynthesis pathway. Pcsk9 knockdown in combination with either treatment led to greater reductions in serum non-HDL with a near-uniform reduction of all LDL-c subfractions. In addition to reducing serum cholesterol, the combined rosuvastatin/ezetimibe/Pcsk9 siRNA treatment exhibited a significant reduction in serum APOB protein and triglyceride levels. Taken together, these data provide evidence that PCSK9 inhibitors, in combination with current therapies, have the potential to achieve greater reductions in both serum cholesterol and triglycerides.
Keywords: cholesterol/metabolism, drug therapy, gene expression
Elevated circulating LDL cholesterol (LDL-c) is a risk factor for the development of atherosclerosis, and it is well documented that reducing the level of circulating LDL-c lowers the incidence of death from cardiovascular disease (as reviewed in Refs. 1–4). The level of circulating LDL-c is determined by the interplay between cholesterol synthesis, dietary cholesterol absorption, and cholesterol clearance (as reviewed in Refs. 5, 6). Current standard of care treatments to reduce serum LDL-c include statins, which inhibit cholesterol synthesis, and ezetimibe, which inhibits dietary and biliary cholesterol absorption (7–10). Both are effective means of reducing circulating LDL-c, although each inhibitor class induces a feedback mechanism leading to the induction of SREBP-2 (sterol-regulatory-element-binding protein-2). This leads to an increase in the expression of cholesterol biosynthesis genes, potentially blunting the efficacy of these treatments (11–17). In addition, induction of the SREBP-2 pathway potentially decreases LDL-c clearance through the increased expression of proprotein convertase subtilisin/kexin type 9 (Pcsk9) (18). PCSK9 increases circulating LDL-c by binding to and inducing the internalization and subsequent degradation of the LDL receptor (LDLr) (19–27). An increase in PCSK9 serum protein would thus reduce LDLr levels on the hepatic cell surface and consequently raise the amount of LDL-c in circulation (28–30). However, activation of the SREBP-2 pathway also leads to increased Ldlr expression. Therefore, the degree to which SREBP-2 pathway induction leads to decreased cholesterol clearance depends on the degree to which Pcsk9 expression is increased relative to Ldlr.
PCSK9 inhibitors are currently being explored as a means to lower circulating LDL-c (31–38), and it is well-documented that both statin- and ezetimibe-treated patients exhibit elevated serum PCSK9 protein levels (7, 11–14, 39–43). A greater reduction in LDL-c is observed for Pcsk9 knockout mice administered statins, providing evidence that PCSK9 inhibitors, in combination with inhibitors of liver cholesterol synthesis, should result in a greater reduction in circulating LDL-c (20). Here, we evaluated whether the combined inhibition of cholesterol absorption, cholesterol synthesis, and increased LDL-c clearance could lead to greater reductions in circulating LDL-c by small interfering RNA (siRNA)-mediated knockdown of Pcsk9 in combination with ezetimibe, rosuvastatin, or an ezetimibe/rosuvastatin treatment.
MATERIALS AND METHODS
siRNA design and synthesis
siRNAs were designed and synthesized as described previously (44, 45). The complementary strands were annealed. The duplex was ultrafiltered and lyophilized. Duplex purity was evaluated using LC/MS and tested for the presence of endotoxin by standard methods.
Preparation of siRNA-lipid nanoparticle complex
siRNA-lipid nanoparticles (LNPs) were made as described previously, except a 60:38:2 molar ratio of the cationic lipid CLinDMA (2-{4-[(3b)-cholest-5-en-3-yloxy]butoxy}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine; Merck and Co), cholesterol (Northern Lipids), and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol; NOF Corporation) was used (46).
In vivo
C57Bl/6 mice engineered to be hemizygous for Ldlr and the overexpression of hCetp driven by the endogenous Apoa1 promoter (B6-Ldlr<tm1>Tg (APOA1-CETP, Taconic) were used for these studies. Female mice ∼16–20 weeks of age were individually housed several days prior to the start of the study. At the start of the study, animals were switched to a low-fat Western diet (Lab Diets 5020 9F) containing 9% crude fat from breeder chow (Lab Diets 5001*) containing 4.5% crude fat. siRNAs were administered by intravenous injection. Both ezetimibe and rosuvastatin were formulated in 0.5% methyl cellulose and administered by oral gavage. Animals were euthanized by CO2 inhalation. Immediately after euthanasia, serum was collected using serum separator tubes and allowed to clot at room temperature for 30 min. Liver sections were excised, placed in either RNA Later (right medial lobe), 10% neutral buffered formalin (NBF, left medial lobe), or flash frozen (the remainder) and stored until further use.
RNA isolation, qRT-PCR, and ELISA
RNA isolation and quantitative real-time PCR were performed using Qiagen's RNeasy96 Universal Tissue Kit together with TaqMan Gene Expression reagents according to the supplied product protocol. An on-column DNase I treatment was performed, and samples were washed three times prior to elution in 100 μl RNase-free water. Reverse transcription was performed using the Cells to Ct kit (Ambion) in a 20 μl volume with 350 ng of RNA in 1× reverse transcriptase and buffer incubated at 37°C for 1 h. Changes in the expression of 361 genes (see supplementary Table I) involved in hepatic lipid metabolism were analyzed using a custom RT2 Profiler PCR Array (SA Biosciences) according to the supplied product protocol.
PCSK9 serum protein levels were measured by a dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) as described previously (47). Briefly, Immulon 4HBX clear 96-well polystyrene high binding plates (ThermoLabsystems) were incubated with a monoclonal antibody reactive to mouse PCSK9. The antibody solution was removed, and 200 μl of 1× blocking buffer [1% BSA (Sigma) in 1× TBST (Sigma)] was added. Serum samples (20 μl) were incubated at 37°C for 1 h, and wells were subsequently washed three times with 300 μl of TBST. A second monoclonal antibody reactive to mouse PCSK9 and biotinylated (biotinylated anti-mPCSK9 Fab B) was added, and samples were incubated for 1 h to detect PCSK9. Wells were subsequently washed three times with 300 μl TBST. Samples were next incubated with a 1:1000 dilution of streptavidin-europium solution (Perkin-Elmer) at room temperature for 20 min with gentle agitation. Wells were washed three times with 300 μl TBST, 100 μl/well of DELFIA Enhancer (Perkin Elmer) solution was added, and samples were incubated for 30 min at room temperature with gentle agitation prior to measurement with a europium plate reader.
Cholesterol and triglyceride analysis
Liver samples of approximately 250 mg were frozen and stored at −80°C until analysis. Individual samples were extracted according to the procedure of Folch, Lees, and Sloane-Stanley (48). Chromatography was performed as described by Burrier et al. (49) using an isocratic mobile phase containing 98.8% hexane and 1.2% isopropanol at a flow rate of 2 ml/min through a Zorbax Sil (4.6 × 25 cm) silica column (Agilent Technologies # 880952-701). Lipids in a 5 μl injection were detected by absorbance at 206 nm and quantitated by computer integration (System Gold, Beckman) of the area under the curve (AUC). Cholesterol, cholesteryl ester (CE), and triglyceride concentrations were determined by comparison to standard curves using Nonpolar Lipid Mix-B (Matreya, Inc.; Pleasant Gap, PA), C/N 1130.
For serum cholesterol analysis, lipase inhibitor (Sigma-Aldrich) was added at a 1:100 (v/v) ratio of inhibitor to serum, followed by mixing (600 rpm at 4°C for 1.5 min) and centrifugation (2,000 rpm at 4°C for 2 min). Serum total and HDL levels were measured using Wako's total and HDL kits according to the supplied product protocol. Non-HDL was calculated by subtracting HDL from total cholesterol measurements.
For LDL subfraction analysis, the lipoprotein profile of the mouse plasma was analyzed using gradient gel electrophoresis (Lipoprint LDL Subfraction System, Quantimetrix) according to the recommended protocol. The resolved lipoprotein bands were quantified based on lipid content using the software provided by the vendor.
Histology and hematology
Mouse liver samples (in 10% NBF) were processed to paraffin, sectioned, and stained with hematoxylin and eosin (H and E). The H and E-stained slides were reviewed by a board-certified veterinary pathologist for inflammation. Liver toxicity was measured by analyzing serum ALT (alanine aminotransferase), AST (aspartate aminotransferase), and LDH (lactate dehydrogenase) levels from 80 μl of serum using ACE Alera® Clinical Chemistry System (Alfa Wassermann, Inc.).
RESULTS
siRNA mediated knockdown of Pcsk9 reduced serum #8232non-HDL in combination with a maximum efficacious dose of ezetimibe
Wild-type mice typically have very low LDL-c levels, and a strain with elevated LDL-c levels was specifically chosen to provide a better window to assess reductions in LDL-c. This strain contains hemizygous mutations resulting in the partial knockdown of Ldlr (Ldlr+/−) and overexpression of the human CE transferase protein (Cetp) gene driven by the mouse Apoa1 promoter (Apoa1-hCetp+/−). These two mutations increased circulating LDL-c and led to a lipid profile that resembled the lipid profile observed in humans. (Tadin-Strapps et al., unpublished observations).
We first evaluated the efficacy of ezetimibe in this model by administering a maximum efficacious dose of ezetimibe (10 mg/kg/day), daily, for 7 days. Following 7 days of treatment, serum non-HDL, which serves as a close approximation of LDL-c in this model, decreased by 21% (P < 0.0001), whereas Pcsk9 liver mRNA and serum PCSK9 protein levels increased by 2- and 3-fold (P < 0.001 and P < 0.0001), respectively (see supplementary Figs. I, II; one-way ANOVA, Tukey posttest).
To investigate whether Pcsk9 knockdown could further reduce serum cholesterol when administered with ezetimibe, we utilized siRNAs designed against the mouse Pcsk9 mRNA transcript and formulated in a lipid nanoparticle (LNP) to achieve liver-targeted Pcsk9 knockdown. Mice were administered a maximum efficacious dose of ezetimibe (10 mg/kg/day) for 7 days, and on day 4, animals were administered a maximum tolerated dose (6 mg/kg) of a Pcsk9 siRNA [Pcsk9(1035)]. PCSK9 (liver mRNA and serum protein) levels were elevated 2-fold (P < 0.05 and P < 0.0001, respectively) following ezetimibe treatment. Both were reduced 4-fold (P < 0.0001) following the administration of the Pcsk9 siRNA (see supplementary Fig. IIA, B; one-way ANOVA, Tukey posttest). The combination of ezetimibe with Pcsk9(1035) led to a 50% (P < 0.0001) reduction in serum non-HDL relative to the vehicle/control siRNA treatment group and a 36% (P < 0.0001) and 33% (P < 0.0001) reduction relative to the ezetimibe and Pcsk9 siRNA individual treatments, respectively (see supplementary Fig. IIC; one-way ANOVA, Tukey posttest).
Both liver enzymes and pathological analysis of hematoxylin and eosin stained liver sections were evaluated to determine the impact that these treatments had on liver function. Liver enzyme levels (ALT, AST, and LDH) were within a range consistent with normal hepatic function, with no significant variation observed across groups (see supplementary Fig. IIIA–C). Additionally, hematoxlyin and eosin stained liver sections were reviewed by a board-certified pathologist and scored utilizing a subjective scoring system for the degree of inflammation. In all cases, inflammation was scored as minimal or mild and would not be expected to impede hepatic function (see supplementary Fig. IIID–E).
siRNA mediated knockdown of Pcsk9 led to greater reductions in both serum non-HDL and serum APOB protein levels in combination with ezetimibe, rosuvastatin, and ezetimibe/rosuvastatin treatments
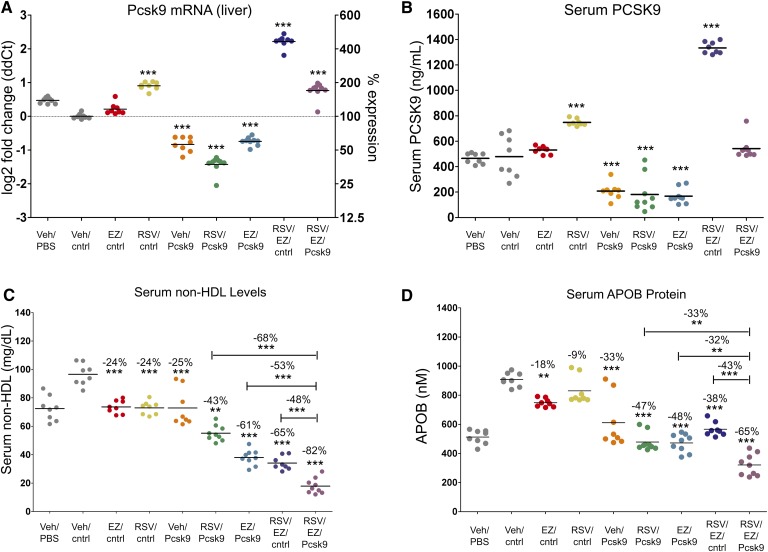
In addition to ezetimibe, statins have been reported to elevate serum PCSK9 protein levels, either alone or in combination with ezetimibe (39–41, 43). To determine whether Pcsk9 knockdown can further reduce serum cholesterol following ezetimibe, statin, or an ezetimibe/statin combination, we treated mice with ezetimibe (10 mg/kg/day), rosuvastatin (20 mg/kg/day), or the combination for 14 days, and on day 11, administered a 6 mg/kg dose of another Pcsk9 targeting siRNA [Pcsk9(1076)]. Both Pcsk9 liver mRNA and serum PCSK9 protein levels were significantly elevated, 2-fold (P < 0.0001) for the rosuvastatin and 4-fold (P < 0.0001) for the ezetimibe/rosuvastatin combination. In this experiment, only a slight but not significant elevation was observed for ezetimibe relative to the vehicle/control siRNA treatment (Fig. 1A, B; one-way ANOVA, Tukey posttest). Administration of the Pcsk9 siRNA reduced both PCSK9 liver mRNA and serum protein levels, and this was associated with an increase in hepatic LDLr (see supplementary Fig. IV). Administration of the Pcsk9 siRNA with either ezetimibe or rosuvastatin reduced PCSK9 below the levels observed for the vehicle/control siRNA treatment, whereas the ezetimibe/rosuvastatin combination induced PCSK9 to such an extent that coadministration of the Pcsk9 siRNA with the ezetimibe/rosuvastatin combination reduced PCSK9 back down to vehicle/control siRNA treatment levels (Fig. 1A, B; RSV/EZ/cntrl vs. RSV/EZ/Pcsk9).
Fig. 1.
Small interfering RNA (siRNA)-mediated knockdown of Pcsk9 led to greater reductions in both serum cholesterol and serum apolipoprotein B (APOB) protein levels in combination with ezetimibe, rosuvastatin, and ezetimibe/rosuvastatin treatments. (A) Pcsk9 mRNA expression and (B) serum PCSK9 protein levels following treatment. (C) siRNA-mediated knockdown of Pcsk9 resulted in a significant decrease in serum non-HDL and (D) serum APOB protein levels in combination with ezetimibe, rosuvastatin, and ezetimibe/rosuvastatin treatments. Vehicle/PBS and Vehicle/cntrl siRNA groups serve as negative controls. Individual animals (circles) and group means (bars) are shown. P values and the percent difference were calculated relative to the Vehicle/cntrl siRNA group unless otherwise indicated by a bar. * P < 0.05, ** P < 0.001, *** P < 0.0001.
Ezetimibe, rosuvastatin, and Pcsk9(1076) significantly reduced serum non-HDL levels by ∼25% (P < 0.0001) relative to the vehicle/control siRNA treatment (Fig. 1C; one-way ANOVA, Tukey posttest). For both ezetimibe and Pcsk9(1076), this was associated with a significant reduction in serum apolipoprotein B (APOB), 18% (P < 0.001) and 33% (P < 0.0001) for ezetimibe and Pcsk9(1076), respectively (Fig. 2D; one-way ANOVA, Tukey posttest). Pcsk9(1076), in combination with either ezetimibe or rosuvastatin, led to even greater reductions in both serum non-HDL (43–65%,) and APOB (38–48%) relative to the individual treatments (Fig. 1C, D). The combination of Pcsk9(1076) with the ezetimibe/rosuvastatin treatment led to significantly lower serum non-HDL cholesterol levels relative to the other combination groups (Fig. 1C; one-way ANOVA, Tukey post test) [compare RSV/EZ/Pcsk9(1076) to EZ/Pcsk9(1076) where differences of 53% (P < 0.0001) were observed, RSV/EZ/Pcsk9(1076) to RSV/Pcsk9(1076) where differences of 68% (P < 0.0001) were observed, and RSV/EZ/Pcsk9(1076) to EZ/RSV where differences of 48% (P < 0.0001) were observed]. In addition, significant reductions in serum APOB were observed for the RSV/EZ/Pcsk9(1076) group relative to EZ/Pcsk9(1076) (32%, P < 0.05), RSV/Pcsk9(1076) (33%, P < 0.0001), and EZ/RSV (43%, P < 0.0001) groups (Fig. 1D; one-way ANOVA, Tukey posttest).
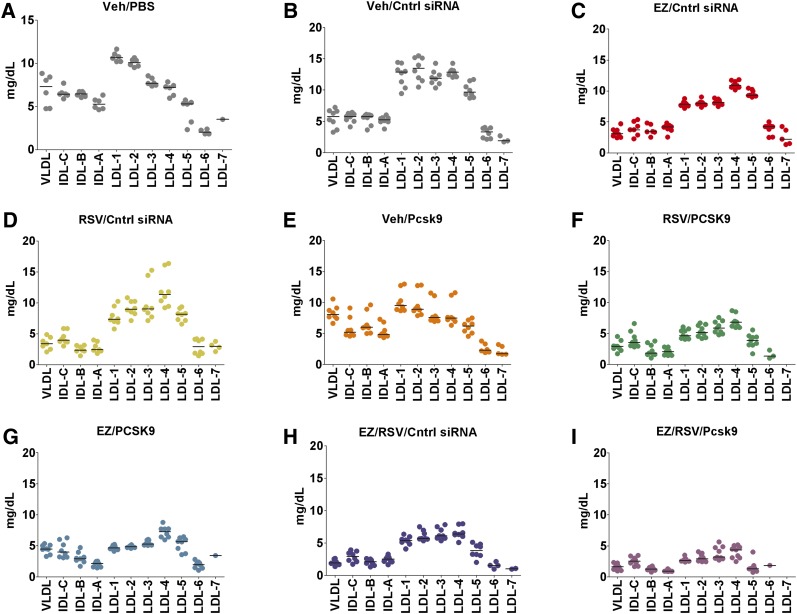
Fig. 2.
All LDL-cholesterol (LDL-c) subfractions are reduced following combination treatment. A–I: Serum lipoproteins are resolved to discrete bands consisting of VLDL, IDL bands C, B, A, LDL subfractions 1 to 7, and an HDL band using a lipoprint system. Each subfraction represents a percentage based on cholesterol dye staining, which is converted to mg/dl based on the known amount of total cholesterol previously measured for each sample. Individual animals (circles) and group means (bars) are shown.
The observed changes to PCSK9, LDLr, non-HDL, and APOB were confirmed with the Pcsk9(1035) sequence for the triple combination relative to EZ/Pcsk9(1035), RSV/Pcsk9(1035), and EZ/RSV treatments (see supplementary Figs. V–VII; compare RSV/EZ/Pcsk9(1035) to EZ/Pcsk9(1035) where differences of 65% for non-HDL (P < 0.0001) and 27% for APOB (P < 0.05) were observed, RSV/EZ/Pcsk9(1035) to RSV/Pcsk9(1035) where differences of 74% for non-HDL (P < 0.0001) and 45% for APOB (P < 0.0001) were observed, RSV/EZ/Pcsk9(1035) to RSV/EZ where differences of 55% for non-HDL (P < 0.0001) and 47% for APOB (P < 0.0001) were observed). Taken together, these data demonstrate that siRNA-mediated knockdown of Pcsk9 negates the ezetimibe/rosuvastatin-induced increase in PCSK9 (liver mRNA and serum protein) leading to greater cholesterol lowering.
A near-uniform reduction in all LDL-c subfractions was observed following combination treatments
Having a disproportionate number of small LDL particles has been demonstrated to be an independent risk factor for coronary heart disease, and it has been proposed that small LDL particles may reside longer within circulation and may be more prone to uptake by macrophages in the atheroma (50). Here, we analyzed the size distribution of LDL particles using a gradient gel electrophoresis system (LDL Lipoprint) along with a human serum control (Liposure). The gel system can resolve serum lipoproteins to discrete bands consisting of VLDL, IDL bands C, B, A, LDL subfractions 1 to 7, and an HDL band. For normal human serum, LDL is distributed among subfractions 1 to 4, peaking at subfraction 1 (51). In our mouse model with a human-like serum lipid profile, LDL has a broader distribution centered at subfraction 2 (Fig. 2A, B). All combinations of rosuvastatin, ezetimibe, and the Pcsk9 siRNA that were evaluated resulted in a near-uniform reduction in LDL subfractions, suggesting that the combination treatments do not disproportionately lower a particular LDL subfraction (Fig. 2).
The rosuvastatin/ezetimibe/Pcsk9 siRNA treatment reduced serum triglycerides
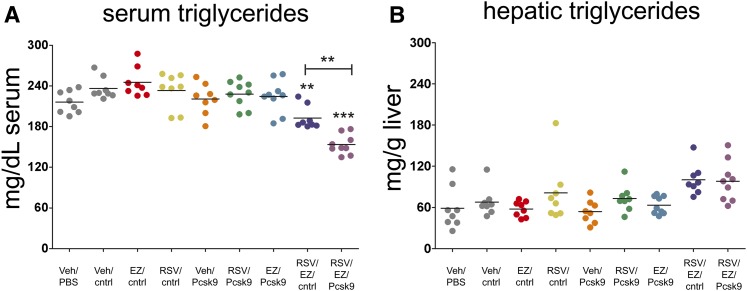
In addition to cholesterol, elevated serum triglyceride levels have been associated with atherosclerosis (52, 53). We next measured serum triglyceride levels and observed a significant reduction in serum triglycerides for the ezetimibe/rosuvastatin/Pcsk9(1076) siRNA combination relative to the other groups (Fig. 3A; P < 0.001; one-way ANOVA, Tukey posttest). Hepatic triglyceride levels were also measured to determine whether the observed reduction in serum triglycerides for the rosuvastatin/ezetimibe/Pcsk9 siRNA combination treatment led to their accumulation within the liver. Hepatic triglyceride levels trended upward but did not reach significance (one-way ANOVA, Tukey posttest) for the rosuvastatin/ezetimibe/Pcsk9 siRNA treatment relative to controls (Fig. 3B; compare RSV/EZ/Pcsk9 to Veh/cntrl).
Fig. 3.
Serum triglycerides are reduced following ezetimibe/rosuvastatin/Pcsk9 siRNA treatment. (A) Significant reductions in serum triglycerides are observed for the ezetimibe/rosuvastatin/Pcsk9(1076) siRNA treatment relative to the negative control group (RSV/EZ/Pcsk9 vs. Veh/cntrl). (B) A trend toward increased hepatic triglycerides observed for the ezetimibe/rosuvastatin/Pcsk9(1076) siRNA treatment, which failed to reach significance. Individual animals (circles) and group means (bars) are shown. The P values between each group relative to Veh/cntrl are indicated. * P < 0.05, ** P < 0.001, *** P < 0.0001.
Ezetimibe and the ezetimibe/rosuvastatin combination activated hepatic Srebp-2, which increased the expression of the cholesterol biosynthesis pathway
Both ezetimibe and statins have been reported to influence the expression of numerous genes involved in hepatic lipid metabolism (12, 16, 17). We analyzed the expression of 361 genes involved in hepatic lipid metabolism using qRT-PCR and found that many genes within the Srebp-2 pathway were induced following ezetimibe treatment (2.5-fold average induction relative to control); these were induced further (11-fold average induction relative to control) following the ezetimibe/rosuvastatin combination treatment (Table 1, and see supplementary Table I). In contrast to the ezetimibe and ezetimibe/rosuvastatin treatments, Pcsk9 knockdown did not result in a significant increase in the expression of Srebp-2 pathway genes (see supplementary Table I), even though serum PCSK9 protein levels were significantly (P < 0.0001) reduced (Fig. 1B, see supplementary Fig. IIB; Vehicle/Pcsk9 siRNA vs. Vehicle/cntrl siRNA), and there was a significant decrease (21–25%, P < 0.0001) in serum non-HDL (Fig. 1C, and see supplementary Fig. IIC; Veh/Pcsk9 siRNA vs. Veh/Cntrl). Consistent with Pcsk9 knockdown resulting in only minor changes to the hepatic gene signature, the level of Srebp-2 pathway induction was similar for the ezetimibe and the ezetimibe/Pcsk9 siRNA combination treatments. This was also found for the ezetimibe/rosuvastatin treatment relative to ezetimibe/rosuvastatin/Pcsk9 siRNA treatment (Table 1).
TABLE 1.
The Srebp-2 pathway is induced following ezetimibe and ezetimibe/rosuvastatin combination treatments
| Pcsk9 |
EZ |
EZ/Pcsk9 |
RSV/EZ |
RSV/EZ/Pcsk9 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes involved in the Srebp2 pathway | Gene | Fold reg | P | Fold reg | P | Fold reg | P | Fold reg | P | Fold reg | P |
| Hmgcr | 1.5 | 0.02 | 3.2 | 0.000078 | 2.4 | 0.000091 | 18.7 | 0.00008 | 13.0 | 0.000055 | |
| Hmgcs1 | −1.0 | 0.8 | 2.8 | 0.000006 | 2.4 | 0.000001 | 9.9 | 0 | 8.7 | 0 | |
| Mvk | −1.3 | 0.02 | 1.8 | 0.000995 | 2.2 | 0.000004 | 9.5 | 0 | 8.9 | 0.000054 | |
| Pmvk | −1.1 | 0.06 | 1.9 | 0 | 1.4 | 0.000023 | 6.0 | 0 | 4.6 | 0 | |
| Mvd | −1.0 | 0.9 | 2.5 | 0.000013 | 2.3 | 0.000006 | 10.4 | 0 | 10.8 | 0.000004 | |
| Idi1 | −1.2 | 0.05 | 3.3 | 0 | 2.7 | 0.000001 | 20.8 | 0 | 20.3 | 0.000001 | |
| Fdps | −1.2 | 0.2 | 2.2 | 0 | 1.7 | 0.000003 | 3.3 | 0 | 5.4 | 0 | |
| Fdft1 | 1.1 | 0.28 | 2.1 | 0 | 1.9 | 0 | 8.4 | 0 | 6.8 | 0 | |
| Cyp51a1 | −1.2 | 0.02 | 2.9 | 0 | 2.3 | 0 | 14.2 | 0 | 17.2 | 0 | |
| Dhcr7 | 1.2 | 0.01 | 2.0 | 0.000001 | 2.2 | 0 | 5.8 | 0 | 5.7 | 0 | |
The expression of selected Srebp-2 pathway genes were analyzed using qRT-PCR. The fold regulation was calculated for the treatment group relative to the Veh/cntrl siRNA group. P values were calculated using a two-tailed t-test between control and treatment groups. The analysis software contains a 6-digit cut-off. Therefore, p-values below 6-digits are represented with a 0.
Both CEs and free cholesterol (FC) were measured by HPLC to determine whether the observed induction of the Srebp-2 pathway led to the accumulation of cholesterol within the liver. The hepatic levels of CE and FC were not elevated, despite increased expression of Srebp-2 pathway genes following ezetimibe and ezetimibe/rosuvastatin treatment, and in fact, a modest decrease in CE was observed for most groups relative to controls (see supplementary Fig. VIII).
DISCUSSION
Here, we report that treatment with two distinct Pcsk9-targeting siRNAs results in a modest decrease in serum non-HDL and APOB protein levels in a mouse model with a human-like serum lipid profile (Apoa1 promoter-driven hCetp+/− and Ldlr+/− mice). The results reported here are consistent with the reported decrease in serum cholesterol observed following monoclonal antibody-, siRNA-, and antisense-mediated inhibition of PCSK9 as well as for Pcsk9−/− mice (20, 34–38, 47). Collectively, these data demonstrate the effectiveness of inhibiting PCSK9 in reducing serum cholesterol using multiple methods in several models, including wild-type mice, diet-induced hyperlipidemic mice, and mice engineered to exhibit a healthy human lipid profile. It has been proposed that serum cholesterol levels in hCetp+/− Ldlr+/− mice may be more sensitive to LDL-c-lowering treatments relative to wild-type mice, since the lipid profile is no longer dominated by HDL. hCetp+/− Ldlr+/− mice, therefore, represent an attractive alternative to mouse models of hyperlipidemia, and the fact that they are heterozygous for Ldlr expression may make this model more representative of the response expected from familial hypercholesterolemia heterozygotes.
It is worth noting that HDL was reduced in some experiments following Pcsk9 siRNA, ezetimibe, rosuvastatin, or combination treatment. Reductions in HDL have been reported for siRNA-mediated knockdown (KD) of ApoB, and it remains possible that the observed reductions in HDL are due to a reduction in serum APOB following Pcsk9 siRNA, ezetimibe, rosuvastatin, or combination treatment. The mechanism behind APOB-mediated changes in HDL is unknown, although it has been proposed that APOB could influence mature HDL particles indirectly through the reduction of HDL components supplied by APOB containing particles (54).
Both rosuvastatin and ezetimibe led to a decrease in serum cholesterol but induced the expression of the Srebp-2 pathway in our model, which is also consistent with several previous reports (12, 16, 17, 55–57). Induction of the Srebp-2 pathway most likely serves as a homeostatic response, probably blunting the efficacy of these treatments. Interestingly, siRNA-mediated knockdown of Pcsk9 did not induce the Srebp-2 pathway, although we cannot rule out the possibility that prolonged knockdown of Pcsk9 (>3 days) would not have this effect.
In addition, ezetimibe and rosuvastatin induced PCSK9 (liver mRNA and serum protein), consistent with both preclinical and clinical data for both statins and ezetimibe (7, 11–14, 39–43). Our findings demonstrate that knocking-down Pcsk9 can counter this response leading to greater reductions in serum non-HDL and APOB protein levels. These data provide evidence that PCSK9 inhibitors could serve as a follow-on therapy to counter the feedback mechanism leading to the induction of Pcsk9, which could be of value to patients who are unable to reduce their LDL-c to their target goal using current treatment strategies. The combined ezetimibe/rosuvastatin/Pcsk9 siRNA treatment additionally led to a reduction in serum triglycerides. Taken together, these data suggest that inhibiting PCSK9, in combination with these two cholesterol-lowering therapies, can improve the lipid profile for dislipidemic patients.
It is also worth noting that although the HDL-to-LDL ratio in our model is comparable to the ratio observed for a healthy human, the distribution of the LDL subfraction is broader, with a greater percentage of LDL consisting of the smaller, pro-atherogenic, LDL subparticles. Following ezetimibe, rosuvastatin, Pcsk9 siRNA, or combination treatments, we observed a near-uniform reduction in both large and small LDL subfractions. In the clinic, pravastatin and simvastatin have generally been reported to only moderately impact the distribution of the LDL subfraction, whereas fluvastatin, atorvastatin, and rosuvastatin have all been shown to shift the distribution in some instances (58–61). Here, we observe no appreciable shift following rosuvastatin treatment, which could be due either to differences between our model and humans or differences in the duration of treatment (14 days in our study compared with 8 weeks in the clinic). The impact of ezetimibe on LDL subparticle distribution varies depending on the clinical study (59, 60, 62). Such variability between studies makes meaningful comparisons to our data set in mice difficult.
Finally, it is important to note that these data demonstrate the utility of siRNAs as tools for target validation. Given the speed with which siRNAs can be designed, synthesized, and validated against a target of interest, we believe that siRNAs will become a frequently used tool to help identify potential product combinations at an earlier phase of drug development.
Supplementary Material
Acknowledgments
The authors would like to thank Duncan Brown for siRNA design, Dipali Ruhela and her group for providing the LNP formulations, Michael Krimm for liver enzyme analysis, and Premier Laboratory for histological services and support with pathology review.
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- APOB
- apolipoprotein B
- AST
- aspartate aminotransferase
- CE
- cholesteryl ester
- DELFIA
- dissociation-enhanced lanthanide fluorescence immunoassay
- FC
- free cholesterol
- H and E
- hematoxylin and eosin
- LDH
- lactate dehydrogenase
- LDL-c
- LDL cholesterol
- LDLr
- LDL receptor
- LNP
- lipid nanoparticle
- NBF
- neutral buffered formalin
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- siRNA
- small interfering RNA
- SREBP-2
- sterol-regulatory-element-binding protein-2
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight figures and two tables.
REFERENCES
- 1.Steinberg D. 2006. Thematic review series: the Pathogenesis of Atherosclerosis. An interpretive history of the cholesterol controversy, part V: The discovery of the statins and the end of the controversy. J. Lipid Res. 47: 1339–1351. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg D. 2005. Thematic review series: the pathogenesis of atherosclerosis. An interpretive history of the cholesterol controversy, part II: the early evidence linking hypercholesterolemia to coronary disease in humans. J. Lipid Res. 46: 179–190. [DOI] [PubMed] [Google Scholar]
- 3.Steinberg D. 2005. Thematic review series: the pathogenesis of atherosclerosis: an interpretive history of the cholesterol controversy, part III: mechanistically defining the role of hyperlipidemia. J. Lipid Res. 46: 2037–2051. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg D. 2004. Thematic review series: the pathogenesis of atherosclerosis. An interpretive history of the cholesterol controversy: part I. J. Lipid Res. 45: 1583–1593. [DOI] [PubMed] [Google Scholar]
- 5.Maxfield F. R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438: 612–621. [DOI] [PubMed] [Google Scholar]
- 6.Gylling H. 2004. Cholesterol metabolism and its implications for therapeutic interventions in patients with hypercholesterolaemia. Int. J. Clin. Pract. 58: 859–866. [DOI] [PubMed] [Google Scholar]
- 7.Altmann S. W., Davis H. R., Jr, Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204. [DOI] [PubMed] [Google Scholar]
- 8.Endo A., Kuroda M., Tsujita Y. 1976. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogenesis produced by Penicillium citrinium. J. Antibiot. (Tokyo). 29: 1346–1348. [DOI] [PubMed] [Google Scholar]
- 9.Endo A., Kuroda M., Tanzawa K. 1976. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 72: 323–326. [DOI] [PubMed] [Google Scholar]
- 10.Tsujita Y., Kuroda M., Tanzawa K., Kitano N., Endo A. 1979. Hypolipidemic effects in dogs of ML-236B, a competitive inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Atherosclerosis. 32: 307–313. [DOI] [PubMed] [Google Scholar]
- 11.Davis H. R., Jr, Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., et al. 2004. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J. Biol. Chem. 279: 33586–33592. [DOI] [PubMed] [Google Scholar]
- 12.Halleck M., Davis H. R., Kirschmeier P., Levitan D., Snyder R. D., Treinen K., Macdonald J. S. 2009. An assessment of the carcinogenic potential of ezetimibe using nonclinical data in a weight-of-evidence approach. Toxicology. 258: 116–130. [DOI] [PubMed] [Google Scholar]
- 13.Sudhop T., Reber M., Tribble D., Sapre A., Taggart W., Gibbons P., Musliner T., von Bergmann K., Lutjohann D. 2009. Changes in cholesterol absorption and cholesterol synthesis caused by ezetimibe and/or simvastatin in men. J. Lipid Res. 50: 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudhop T., Lutjohann D., Kodal A., Igel M., Tribble D. L., Shah S., Perevozskaya I., von Bergmann K. 2002. Inhibition of intestinal cholesterol absorption by ezetimibe in humans. Circulation. 106: 1943–1948. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 16.Roglans N., Verd J. C., Peris C., Alegret M., Vázquez M., Adzet T., Diaz C., Hernández G., Laguna J. C., Sánchez R. M. 2002. High doses of atorvastatin and simvastatin induce key enzymes involved in VLDL production. Lipids. 37: 445–454. [DOI] [PubMed] [Google Scholar]
- 17.Scharnagl H., Schinker R., Gierens H., Nauck M., Wieland H., Marz W. 2001. Effect of atorvastatin, simvastatin, and lovastatin on the metabolism of cholesterol and triacylglycerides in HepG2 cells. Biochem. Pharmacol. 62: 1545–1555. [DOI] [PubMed] [Google Scholar]
- 18.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxwell K. N., Breslow J. L. 2004. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA. 101: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 102: 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. 2006. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 116: 2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J. C., Boerwinkle E., Mosley T. H., Jr, Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 23.Hallman D. M., Srinivasan S. R., Chen W., Boerwinkle E., Berenson G. S. 2007. Relation of PCSK9 mutations to serum low-density lipoprotein cholesterol in childhood and adulthood (from The Bogalusa Heart Study). Am. J. Cardiol. 100: 69–72. [DOI] [PubMed] [Google Scholar]
- 24.Hooper A. J., Marais A. D., Tanyanyiwa D. M., Burnett J. R. 2007. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 193: 445–448. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z., Tuakli-Wosornu Y., Lagace T. A., Kinch L., Grishin N. V., Horton J. D., Cohen J. C., Hobbs H. H. 2006. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. 2008. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA. 105: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. 2007. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 282: 18602–18612. [DOI] [PubMed] [Google Scholar]
- 28.Davis H. R., Jr, Pula K. K., Alton K. B., Burrier R. E., Watkins R. W. 2001. The synergistic hypocholesterolemic activity of the potent cholesterol absorption inhibitor, ezetimibe, in combination with 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors in dogs. Metabolism. 50: 1234–1241. [DOI] [PubMed] [Google Scholar]
- 29.Kastelein J. J., Akdim F., Stroes E. S., Zwinderman A. H., Bots M. L., Stalenhoef A. F., Visseren F. L., Sijbrands E. J., Trip M. D., Stein E. A., et al. 2008. Simvastatin with or without ezetimibe in familial hypercholesterolemia. N. Engl. J. Med. 358: 1431–1443. [DOI] [PubMed] [Google Scholar]
- 30.Grefhorst A., McNutt M. C., Lagace T. A., Horton J. D. 2008. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J. Lipid Res. 49: 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan J. C. Y., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., et al. 2009. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc. Natl. Acad. Sci. USA. 106: 9820–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan L., Pang L., Zhang R., Murgolo N. J., Lan H., Hedrick J. A. 2008. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 375: 69–73. [DOI] [PubMed] [Google Scholar]
- 33.Ni Y. G., Condra J. H., Orsatti L., Shen X., Di Marco S., Pandit S., Bottomley M. J., Ruggeri L., Cummings R. T., Cubbon R. M., et al. 2010. A proprotein convertase subtilisin-like/kexin type 9 (PCSK9) C-terminal domain antibody antigen-binding fragment inhibits PCSK9 internalization and restores low density lipoprotein uptake. J. Biol. Chem. 285: 12882–12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta N., Fisker N., Asselin M. C., Lindholm M., Rosenbohm C., Orum H., Elmen J., Seidah N. G., Straarup E. M. 2010. A locked nucleic acid antisense oligonucleotide (LNA) silences PCSK9 and enhances LDLR expression in vitro and in vivo. PLoS ONE. 5: e10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., et al. 2009. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc. Natl. Acad. Sci. USA. 106: 9820–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duff C. J., Scott M. J., Kirby I. T., Hutchinson S. E., Martin S. L., Hooper N. M. 2009. Antibody-mediated disruption of the interaction between PCSK9 and the low-density lipoprotein receptor. Biochem. J. 419: 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank-Kamenetsky M., Grefhorst A., Anderson N. N., Racie T. S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., et al. 2008. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl. Acad. Sci. USA. 105: 11915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham M. J., Lemonidis K. M., Whipple C. P., Subramaniam A., Monia B. P., Crooke S. T., Crooke R. M. 2007. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J. Lipid Res. 48: 763–767. [DOI] [PubMed] [Google Scholar]
- 39.Welder G., Zineh I., Pacanowski M. A., Troutt J. S., Cao G., Konard R. J. 2010. High-dose atorvastatin causes a rapid sustained increase in human serum PCSK9 and disrupts its correlation with LDL cholesterol. J. Lipid Res. 51: 2714–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubuc G., Tremblay M., Pare G., Jacques H., Hamelin J., Benjannet S., Boulet L., Genest J., Bernier L., Seidah N. G., et al. 2010. A new method for measurement of total plasma PCSK9: clinical applications. J. Lipid Res. 51: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong B., Wu M., Li H., Kraemer F. B., Adeli K., Seidah N. G., Park S. W., Liu J. 2010. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 51: 1486–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamon-Fava S., Diffenderfer M. R., Barrett P. H., Buchsbaum A., Matthan N. R., Lichtenstein A. H., Dolnikowski G. G., Horvath K., Asztalos B. F., Zago V., et al. 2007. Effects of different doses of atorvastatin on human apolipoprotein B-100, B-48, and A-I metabolism. J. Lipid Res. 48: 1746–1753. [DOI] [PubMed] [Google Scholar]
- 43.Careskey H. E., Davis R. A., Alborn W. E., Troutt J. S., Cao G., Konrad R. J. 2008. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 49: 394–398. [DOI] [PubMed] [Google Scholar]
- 44.Majercak J., Ray W. J., Espeseth A., Simon A., Shi X. P., Wolffe C., Getty K., Marine S., Stec E., Ferrer M., et al. 2006. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 103: 17967–17972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wincott F., DiRenzo A., Shaffer C., Grimm S., Tracz D., Workman C., Sweedler D., Gonzalez C., Scaringe S., Usman N. 1995. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 23: 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strapps W. R., Pickering V., Muiru G. T., Rice J., Orsborn S., Polisky B. A., Sachs A., Bartz S. R. 2010. The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res. 38: 4788–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni Y. G., Di Marco S., Condra J. H., Peterson L. B., Wang W., Wang F., Pandit S., Hammond H. A., Rosa R., Cummings R. T., et al. 2011. A PCSK9-binding antibody that structurally mimics the EGF(A) domain of LDL-receptor reduces LDL cholesterol in vivo. J. Lipid Res. 52: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 49.Burrier R. E., Smith A. A., McGregor D. G., Hoos L. M., Zilli D. L., Davis H. R., Jr 1995. The effect of acyl CoA:cholesterol acyltransferase inhibition on the uptake, esterification and secretion of cholesterol by the hamster small intestine. J. Pharmacol. Exp. Ther. 272: 156–163. [PubMed] [Google Scholar]
- 50.Coresh J., Kwiterovich P. O., Jr, Smith H. H., Bachorik P. S. 1993. Association of plasma triglyceride concentration and LDL particle diameter, density, and chemical composition with premature coronary artery disease in men and women. J. Lipid Res. 34: 1687–1697. [PubMed] [Google Scholar]
- 51.Hoefner D. M., Hodel S. D., O'Brien J. F., Branum E. L., Sun D., Meissner I., McConnell J. P. 2001. Development of a rapid, quantitative method for LDL subfractionation with use of the Quantimetrix Lipoprint LDL System. Clin. Chem. 47: 266–274. [PubMed] [Google Scholar]
- 52.Austin M. A., Hokanson J. E., Edwards K. L. 1998. Hypertriglyceridemia as a cardiovascular risk factor. Am. J. Cardiol. 81: 7B–12B. [DOI] [PubMed] [Google Scholar]
- 53.Hokanson J. E., Austin M. A. 1996. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J. Cardiovasc. Risk. 3: 213–219. [PubMed] [Google Scholar]
- 54.Lieu H. D., Withycombe S. K., Walker Q., Rong J. X., Walzem R. L., Wong J. S., Hamilton R. L., Fisher E. A., Young S. G. 2003. Eliminating atherogenesis in mice by switching off hepatic lipoprotein secretion. Circulation. 107: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 55.Harada L. M., Carrilho A. J., Oliveira H. C., Nakandakare E. R., Quintao E. C. 2006. Regulation of hepatic cholesterol metabolism in CETP/LDLr mice by cholesterol feeding and by drugs (cholestyramine and lovastatin) that lower plasma cholesterol. Clin. Exp. Pharmacol. Physiol. 33: 1209–1215. [DOI] [PubMed] [Google Scholar]
- 56.Davis H. R., Jr, Hoos L. M., Tetzloff G., Maguire M., Zhu L. J., Graziano M. P., Altmann S. W. 2007. Deficiency of Niemann-Pick C1 Like 1 prevents atherosclerosis in ApoE−/− mice. Arterioscler. Thromb. Vasc. Biol. 27: 841–849. [DOI] [PubMed] [Google Scholar]
- 57.Davis H. R., Jr, Compton D. S., Hoos L., Tetzloff G. 2001. Ezetimibe, a potent cholesterol absorption inhibitor, inhibits the development of atherosclerosis in ApoE knockout mice. Arterioscler. Thromb. Vasc. Biol. 21: 2032–2038. [DOI] [PubMed] [Google Scholar]
- 58.Rizzo M., Berneis K. 2006. The clinical relevance of low-density-lipoproteins size modulation by statins. Cardiovasc. Drugs Ther. 20: 205–217. [DOI] [PubMed] [Google Scholar]
- 59.Berneis K., Rizzo M., Berthold H. K., Spinas G. A., Krone W., Gouni-Berthold I. 2010. Ezetimibe alone or in combination with simvastatin increases small dense low-density lipoproteins in healthy men: a randomized trial. Eur. Heart J. 31: 1633–1639. [DOI] [PubMed] [Google Scholar]
- 60.Mihalis Kalogirou V. T., Saougos V., Lagos K., Tselepis A. D., Elisaf M. 2007. Clinical research effect of ezetimibe on lipoprotein subfraction concentrations: the role of atorvastatin pretreatment. Arch. Med. Sci. 3: 344–350. [Google Scholar]
- 61.Caslake M. J., Stewart G., Day S. P., Daly E., McTaggart F., Chapman M. J., Durrington P., Laggner P., Mackness M., Pears J., et al. 2003. Phenotype-dependent and -independent actions of rosuvastatin on atherogenic lipoprotein subfractions in hyperlipidaemia. Atherosclerosis. 171: 245–253. [DOI] [PubMed] [Google Scholar]
- 62.Farnier M., Freeman M. W., Macdonell G., Perevozskaya I., Davies M. J., Mitchel Y. B., Gumbiner B. 2005. Efficacy and safety of the coadministration of ezetimibe with fenofibrate in patients with mixed hyperlipidaemia. Eur. Heart J. 26: 897–905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.