Abstract
Diacylglycerol (DAG) acyl transferase 1 (Dgat1) knockout (−/−) mice are resistant to high-fat-induced obesity and insulin resistance, but the reasons are unclear. Dgat1−/− mice had reduced mRNA levels of all three Ppar genes and genes involved in fatty acid oxidation in the myocardium of Dgat1−/− mice. Although DGAT1 converts DAG to triglyceride (TG), tissue levels of DAG were not increased in Dgat1−/− mice. Hearts of chow-diet Dgat1−/− mice were larger than those of wild-type (WT) mice, but cardiac function was normal. Skeletal muscles from Dgat1−/− mice were also larger. Muscle hypertrophy factors phospho-AKT and phospho-mTOR were increased in Dgat1−/− cardiac and skeletal muscle. In contrast to muscle, liver from Dgat1−/− mice had no reduction in mRNA levels of genes mediating fatty acid oxidation. Glucose uptake was increased in cardiac and skeletal muscle in Dgat1−/− mice. Treatment with an inhibitor specific for DGAT1 led to similarly striking reductions in mRNA levels of genes mediating fatty acid oxidation in cardiac and skeletal muscle. These changes were reproduced in cultured myocytes with the DGAT1 inhibitor, which also blocked the increase in mRNA levels of Ppar genes and their targets induced by palmitic acid. Thus, loss of DGAT1 activity in muscles decreases mRNA levels of genes involved in lipid uptake and oxidation.
Keywords: heart, triglyceride, diacylglycerol acyl transferase, ceramide, exercise, muscle hypertrophy, peroxisome proliferator-activated receptor
Triglyceride (TG) is the major energy storage form in tissues. The final step of TG synthesis, the conversion of diacylglycerol (DAG) to TG, is catalyzed by DAG acyltransferases (DGAT). Dgat1 belongs to a family of membrane-bound O-acyltranferase (MBOAT), which includes acyl CoA:cholesterol acyltransferase (Acat) 1 and Acat2 (1–3). Dgat1 is widely expressed in all tissues, with high expression in white adipose tissue, skeletal muscle, heart, and intestine (1, 2). Dgat2 is a member of the monoacylglycerol acyltransferase family and is primarily expressed in the liver and adipose tissue (2); DGAT2 is thought to be the primary source of DGAT activity in the liver (4).
The effects of Dgat1 overexpression in tissues have suggested that conversion of DAG to TG can be a detoxifying process (5, 6). Overexpression of Dgat1 in skeletal muscle increased tissue content of TG but improved insulin sensitivity (5). These data support the hypothesis that conversion of intermediary lipids to TG via DGAT1 is beneficial. In fact, this alteration in lipid metabolism appears to mimic that found with athletic training, in which TG stores increase, fatty acid oxidation increases, and insulin sensitivity improves (7, 8). Overexpression of DGAT1 in cardiomyocytes increased TG and decreased levels of toxic lipids, such as DAG, ceramide, and free fatty acids (6). When a cardiomyocyte-expressing human DGAT1 transgene was crossed onto a heart failure model overexpressing acyl CoA synthetase 1, heart function and survival rate were significantly improved (6). Similarly, overexpression of DGAT1 in macrophages alleviated fatty acid-induced inflammation and high-fat feeding-induced insulin resistance (9).
Despite these seemingly beneficial actions of DGAT1 overexpression, its deletion does not lead to an opposite phenotype. Dgat1 knockout (Dgat1−/−) mice have reduced diet-induced obesity and less insulin resistance (10). Greater insulin sensitivity was also found in skeletal muscle in these mice (22); however, the reasons remain unclear. Other experiments showed that the transplantation of Dgat1−/− white adipose tissue decreases adiposity and enhances glucose disposal in wild-type (WT) mice; adiponectin secreted by adipose tissue was postulated to contribute to the increased energy expenditure and insulin sensitivity in Dgat1−/− mice (11, 12). This observation has spurred pharmaceutical development of DGAT1 inhibitors. However, the effect of loss of DGAT1 activity in other important organs, such as the heart, has not been reported. We hypothesized that Dgat1−/− mice would have reduced conversion of DAG to triglyceride. This might lead to accumulation of DAG, ceramide, and fatty acids in cardiac muscle, which could lead to cardiomyopathy, especially in the setting of high-fat diets.
In this study, we describe the effects of acute and chronic loss of DGAT1 activity in muscles. We show that loss of this enzyme modulates counter-regulatory pathways that reduce muscle lipid and increase glucose uptake. This finding suggests that DGAT inhibitors may be of benefit in the treatment of reperfusion injury.
MATERIALS AND METHODS
Mice and diet
Animal protocols were in compliance with accepted standards of animal care and were approved by the Columbia University Institutional Animal Care and Use Committee. Male mice were used in experiments unless otherwise indicated. WT, C57BL/6J, and Dgat1−/− C57BL/6J mice (10) were purchased from the Jackson Laboratory. Mice were housed in a barrier facility with 12-h light/12-h dark cycles and had ad libitum access to either chow diet (5053 PicoLab Rodent Diet 20; Purina Mills) or high-fat diet (45% fat primarily from lard, 20% protein, 35% carbohydrates, catalog no. D12451: Research Diets, Inc.).
Lipid measurements
To measure TG, DAG, and ceramide, lipids were first extracted from muscles using chloroform/methanol/HCl (v/v/v, 2:1:0.01) (5). Butylated hydroxytoluene (0.01%) was included in the extraction solution as an antioxidant, and [3H]triolein (0.25 μCi) was used as an internal control for TG recovery. TG and fatty acid concentrations in lipid extracts were determined enzymatically with colorimetric kits (Sigma-Aldrich). DAG and ceramide levels were measured using a DAG kinase-based method. Lipids extracted from muscle specimens were dried under a stream of N2, redissolved in 7.5% octyl-β-D-glucoside containing 5 mM cardiolipin and 1 mM diethylenetriamine pentaacetate, in which DAG and ceramide are quantitatively phosphorylated to form 32P-labeled phosphatidic acid and ceramide-1-phosphate, respectively, which were then quantified. The reaction was carried out at room temperature for 30 min in 100 mM imidazole HCl, 100 mM NaCl, 25 mM MgCl2, 2 mM EGTA (pH 6.6), 2 mM DTT, 10 μg/100 μl DAG kinase (Sigma-Aldrich), 1 mM ATP, and 1 μCi/100 μl [γ-32P]ATP. The reaction was stopped by addition of chloroform/methanol (v/v, 2:1) and 1% HClO4, and lipids were extracted and washed twice with 1% HClO4. Lipids were resolved by TLC (Partisil K6 adsorption TLC plates, Whatman catalog no. LK6D); mobile phase contained chloroform/methanol/acetic acid (v/v/v, 65:15:5). The bands corresponding to phosphatidic acid and ceramide-1-phosphate were identified with known standards, and silicon was scraped into a scintillation vial for radioactivity measurement. [3H]triolein bands from the same TLC plates were identified and quantified in the same way and were used as controls for lipid recovery.
DGAT activity assay
DGAT activity was measured in membrane fractions isolated from muscle specimens using [14C]labeled palmitoyl-CoA as previously described (5). The amounts of both DAG and fatty acyl-CoA in the reaction mixture were in excess, and the reaction rate was of the first order with respect to the amount of input DGAT activity to be assayed.
Real-time PCR
Total RNA was extracted using a Trizol kit from Invitrogen. An amount of 1 μg of RNA was initially treated with DNase I (Invitrogen). The RNA samples were then reverse-transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time amplification was performed using iQ SYBR Green Supermix (Bio-Rad). Primers used for PCR amplification are listed in supplementary Table I. Analysis was performed using iCycler iQ Real-Time Detection System software (Bio-Rad).
Dual energy X-ray absorptiometry
The dual energy X-ray absorptiometry (DEXA) machine was calibrated before use. WT, Dgat1+/−, and Dgat1−/− mice were anesthetized with intraperitoneal injection of ketamine/xylazine (86.98 mg/kg and 13.4 mg/kg, respectively), and whole body measurements, excluding the head, were made (DEXA, PIXImus Lunar-GE). Measurements included body weight, lean mass, fat mass, and percentage of fat/body weight. One person performed all scans, and mice were always in the prostrate position on the imaging-positioning tray.
Exercise protocol
Mice were forced to swim in tanks. The swimming protocol was a modification of the procedure used by Ryder et al. (13). Water temperature was maintained at 34-35°C. Mice swam for 10 min sessions twice a day separated by a 10 min break. Sessions were increased by 10 min each day until the fourth day; mice were then allowed to swim for six 30 min intervals separated by 10-15 min rest periods. After the last swim interval, mice were dried and put back in their cages for 18-20 h.
Western blotting
An amount of 10-30 mg of tissue was homogenized in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, and 1 protease inhibitor cocktail tablet (Roche)/25-50 ml. Cell lysates (50 μg per sample) obtained after centrifugation at 15,000 g for 10 min at 4°C were resolved by SDS-PAGE and then transferred onto nitrocellulose membranes. Immunoblotting was carried out using the following primary antibodies: phospho-AKT (p-AKT), phospho-mTOR (p-mTOR), total AKT, total mTOR, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling), and PPARα, β, and γ (Santa Cruz).
Echocardiography
Two-dimensional echocardiography was performed using a high-resolution imaging system with a 30-MHz imaging transducer (Vevo 770; VisualSonics) in unconscious 3-4-month-old mice. The mice were anesthetized with 1.5-2% isoflurane and thereafter maintained on 1-1.5% isoflurane throughout the procedure. Two-dimensional echocardiographic images were obtained and recorded in a digital format. Images were analyzed offline by a researcher blinded to the murine genotype. Left ventricular end-diastolic dimension (LVDd) and left ventricular end-systolic dimension (LVDs) were measured. Percentage fractional shortening, which quantifies contraction of the ventricular wall and is an indication of muscle function, was calculated as % FS = ([LVDd − LVDs] / LVDd) × 100.
In vitro fatty acid oxidation and in vivo glucose and [14C]triolein-VLDL uptake
Cardiac muscle fatty acid oxidation was measured with heart slices as described previously (6). Hearts were removed and immediately sliced across the ventricles forming rings ∼0.1-0.2 mm thick and usually 20-30 mg wet weight. The cardiac muscles were first washed for 1 h at 37°C in Krebs-Henseleit bicarbonate buffer [10 mM HEPES (pH 7.35), 24 mM NaHCO3, 114 mM NaCl, 5 mM KCl, 1 mM MgCl2, and 2.2 mM CaCl2] containing 8 mM glucose, 32 mM mannitol, and 0.1% BSA. The reaction was initiated by adding to the mix preconjugated BSA/fatty acid solution (final concentrations: 0.2 mM palmitate with 1 μCi/ml of 9,10-[3H]palmitate and 1.25% BSA). The fatty acid oxidation product 3H2O was determined in muscles incubated for 2 h at 37°C. An aliquot of the completed reaction mixture was transferred into an open microtube, which was placed inside a scintillation vial containing 0.5 ml of distilled water. The scintillation vial was then capped and incubated at 50°C for 18-24 h to reach vapor-phase equilibrium. After the transfer of 3H2O from the reaction mixture to the scintillation vial (by water equilibration), the 3H2O in the scintillation vial was scintillation-counted. The transfer and counting efficiency was calibrated using a standard solution containing 0.1 μCi of 3H2O for re-equilibration.
Glucose uptake was measured as described (12). Basal glucose uptake was measured following an intravenous administration of 3 μCi of 2-deoxy-D-[1-3H]glucose (PerkinElmer Life Sciences) in 4 h fasted mice. Blood was collected 0.5, 15, 30, and 60 min following injection. At 60 min, tissues were excised after the body cavity was perfused with 10 ml of PBS by cardiac puncture.
VLDL was labeled using cholesteryl ester transfer protein to incorporate [14C]triolein into VLDL as described previously (14, 15). After intravenous injection, plasma counts at 0.5, 5, 10, and 15 min were measured, and the mice were perfused with PBS as above. Radioactivity in the tissues was measured in a scintillation counter, and tissue uptake was normalized to recovered counts in the liver.
Acute treatment with DGAT1 inhibitor (DGAT1i)
Male 8-week-old mice fed a chow diet or male 14-week-old mice after 6 weeks of high-fat (Harlan TD 88137) feeding were gavaged with 200 μl of vehicle (0.5% hydroxypropylmethyl cellulose/0.1% polysorbate 80) or DGAT1i (5 mg/kg, Compound 2 (10, 16, 17)) upon food removal at 8 AM. This DGAT1i is a potent inhibitor of human DGAT1 and is selective over DGAT2 and ACAT1 (16, 17). Importantly, this DGAT1i potently inhibits mouse microsomal DGAT1 activity (16). At the doses utilized in this study, this DGAT1i produces good blood levels in C57BL/6 mice. At 8 h postgavage, mice were sacrificed, and tissues collected for lipid measurement and RNA isolation.
Palmitic acid and DGAT1i treatment of myocytes
AC16 human cardiomyocytes and C2C12 myoblasts were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C. C2C12 myoblasts were maintained below 60% confluence to prevent confluence-induced differentiation. For experiments, cells were allowed to become confluent, and the media were supplemented with 2% horse serum instead of FBS for myotube formation. After 4-5 days of differentiation, myotubes were formed and used for experiments. AC16 human cardiomyocytes were changed to 2% FBS 24 h before experiments. Each experiment was performed on three separate occasions. Palmitic acid was administered to cells as a conjugate with 1.25% (w/v) fatty acid-free BSA at the final concentration of 0.5 mM for 12 h followed by 4 h of incubation in the presence or absence of DGAT1i (5 mM). Both cells were washed two times with ice-cold PBS prior to RNA isolation and quantitative real-time PCR.
To determine palmitic acid uptake, AC16 cardiomyocytes were pretreated with 5 mM DGAT1i for 4 h and then incubated for 15 min with 0.5 mM palmitic acid with 1 μCi/ml of 9,10-[3H]palmitic acid and 1.25% BSA. Cells were washed three times with ice-cold PBS prior to 0.1N Na2OH addition. An aliquot of homogenate was taken for radioactivity measurement.
Statistics
All data are presented as mean ± SD, unless indicated otherwise. Comparisons between two groups were performed using unpaired two-tailed Student's t-test with Statistica version 6.0 (StatSoft). A P value less than 0.05 was used to determine statistical significance.
RESULTS
Dgat1−/− mice have decreased heart and skeletal muscle DGAT activity
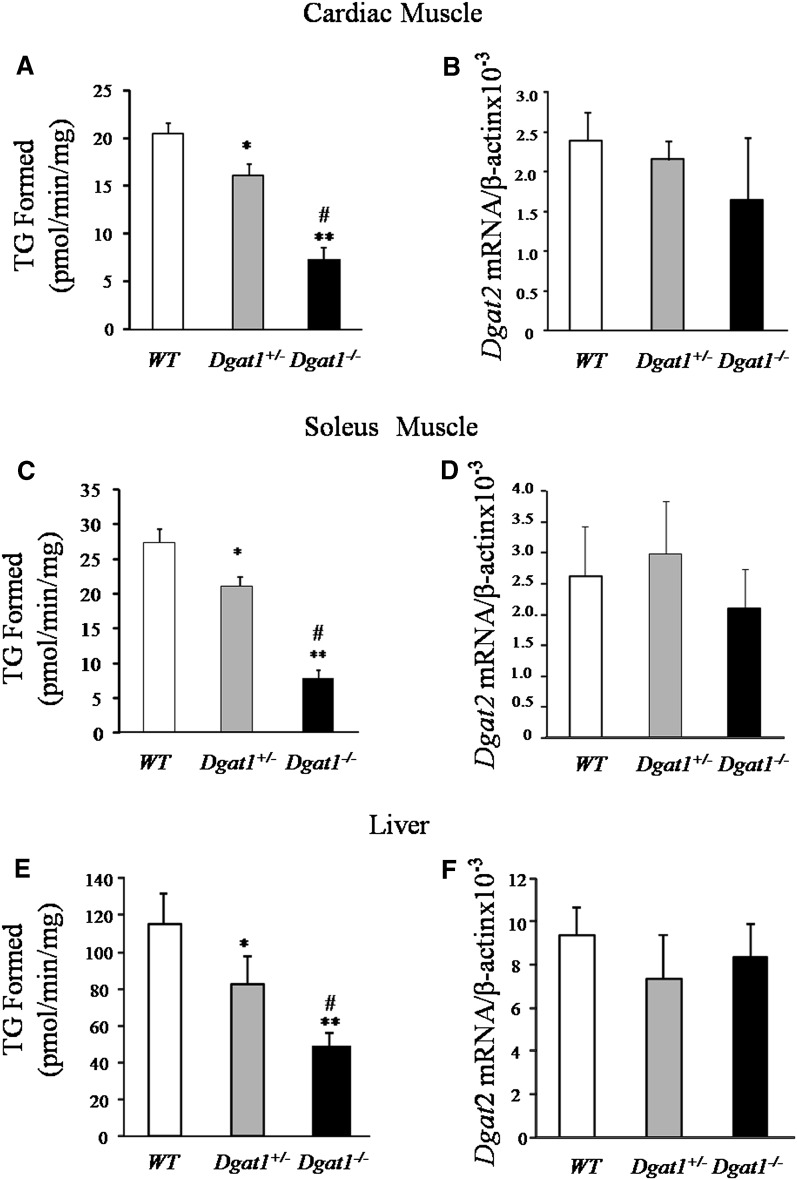
To determine if loss of Dgat1 in heart and skeletal muscle reduces or eliminates DGAT function, we measured DGAT activity in the heart and skeletal muscle tissue of Dgat1+/− and Dgat1 −/−mice. DGAT activity was decreased ∼20% and ∼65% in hearts of Dgat1+/− and Dgat1−/− mice, respectively (Fig. 1A). Dgat2 mRNA did not change in these tissues (Fig. 1B). In skeletal muscle, DGAT activity was also reduced (Fig. 1C), but Dgat2 mRNA did not change significantly (Fig. 1D). Dgat1−/− mouse liver had a ∼57% decrease in total DGAT activity, but DGAT activity was 4-fold greater than skeletal muscle both under WT and Dgat1−/− conditions. Therefore, Dgat1−/− liver had an activity level that was greater than that found in WT soleus muscle (Fig. 1E). These animals also had no compensatory increase in Dgat2 mRNA (Fig. 1F). Plasma levels of lipids and glucose in these mice are shown in supplementary Table II.
Fig. 1.
Total DGAT activity and Dgat2 mRNA levels in muscle and liver from Dgat1−/− mice. A, C, and E: Total DGAT activity levels in membrane fractions of heart, soleus muscle, and liver from WT, Dgat1+/−, and Dgat1−/− mice (n = 5, *P < 0.05 vs. WT, **P < 0.01 vs. WT, #P < 0.05 vs. Dgat1+/−). B, D, and E: Realtime-PCR quantification of Dgat2 mRNA levels in heart, soleus muscle, and liver of WT, Dgat1+/−, and Dgat1−/− mice (n = 5). DGAT, diacylglycerol acyl transferase; TG, triglyceride; WT, wild type.
DAG and ceramide levels were decreased in heart from Dgat1−/− mice
Surprisingly the content of TG was not significantly decreased in cardiac muscle in Dgat1−/− mice (Table 1). Also unlike what we expected, the contents of TG precursors DAG and ceramide were not increased but, in fact, were decreased ∼20% in Dgat1−/− hearts (Table 1). We then fed the mice a high-fat diet. High-fat-diet control mice had an increase in TG levels (∼15%), DAG levels (26%), and FFA levels (37%) in the heart (Table 1). As with chow, the contents of DAG and ceramide were reduced ∼16% and ∼22%, respectively, in hearts of high-fat-diet Dgat1−/− mice.
TABLE 1.
Lipid content in chow diet and high-fat diet WT and Dgat1 hearts (n=5–7)
| Chow Diet |
High-Fat Diet |
|||||
|---|---|---|---|---|---|---|
| Lipids | WT | Dgat1 | Dgat1 | WT | Dgat1 | Dgat1 |
| TG (nmol/mg) | 20.3 ± 2.1 | 19.3 ± 1.6 | 18 ± 2.5 | 23.3 ± 1.6a | 21.9 ± 2.3 | 21.1 ± 1.8 |
| FFA (nmol/mg) | 7.4 ± 0.9 | 7 ± 0.9 | 8.3 ± 0.8 | 10.2 ± 1.3b | 9.7 ± 1.7 | 9.3 ± 2.4 |
| DAG (pmol/mg) | 293 ± 42 | 250 ± 38 | 224 ± 58a | 371 ± 57a | 342 ± 78 | 310 ± 66 |
| Ceramid (pmol/mg) | 287 ± 53 | 244 ± 32 | 215 ± 49a | 345 ± 70 | 323 ± 44 | 267 ± 41c |
DAG, diacylglycerol; DGAT, diacylglycerol acyl transferase; TG, triglyceride; WT, wild type.
P < 0.05 versus chow diet WT.
P < 0.01 versus chow diet WT.
P < 0.05 versus high fat diet WT
Heart and muscle weights were increased in Dgat1−/− mice
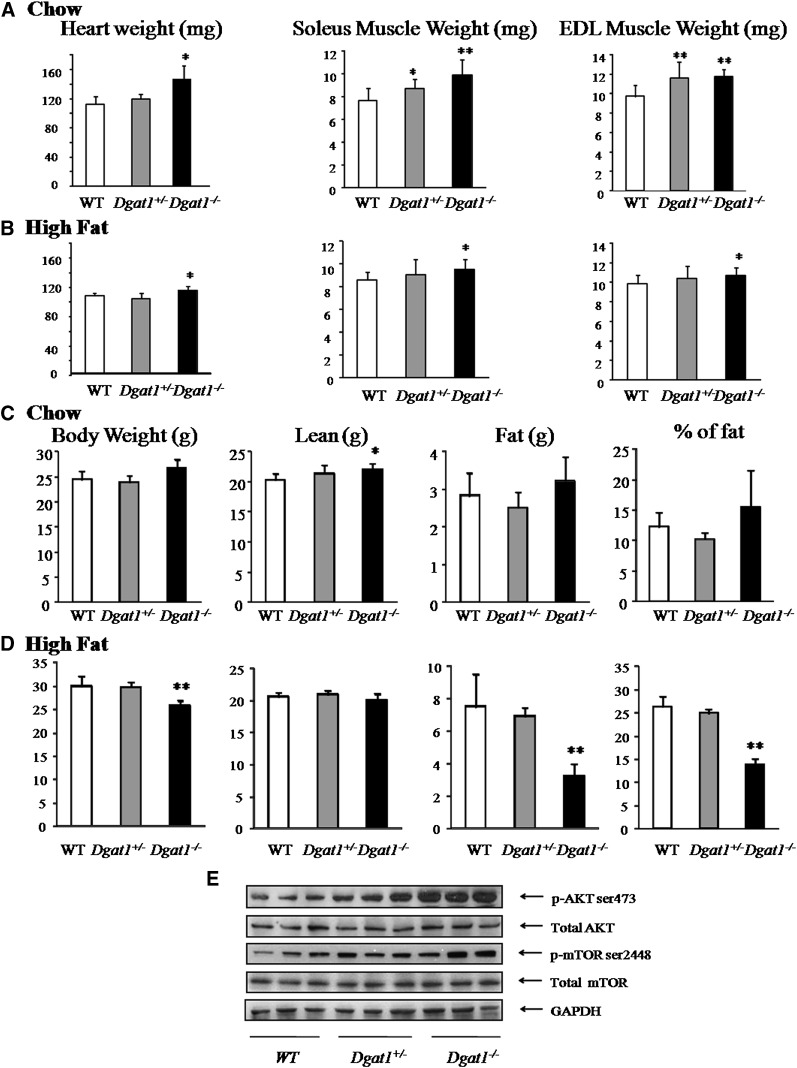
Heart weight was increased 29% (P < 0.01) in chow diet and 7% (P < 0.05) in high-fat diet Dgat1−/− mice. Soleus and extensor digitorum longus (EDL) muscle weight increased 28% and 21%, respectively, in chow diet Dgat1−/− mice (P < 0.05) and 10% and 9%, respectively, in high-fat diet Dgat1−/− mice (P < 0.05) (Fig. 2A, B). Soleus and EDL muscle weights also were increased 13% (P < 0.05) and 20% (P < 0.01) in chow diet Dgat1+/− mice (Fig. 2A). Thus, Dgat1 deficiency increased muscle weight, especially on a chow diet.
TABLE 2.
Heart gene expression in Dgat1 and Dgat1 mice (n = 5–6)
| Chow Diet |
High-Fat Diet |
|||||
|---|---|---|---|---|---|---|
| Gene | WT | Dgat1/− | Dgat1−/− | WT | Dgat1 | Dgat1−/− |
| % | % | % | % | % | % | |
| Ppara | 100 ± 37 | 89 ± 35 | 38 ± 17a | 175 ± 41a | 122 ± 41 | 111 ± 22c |
| Ppard | 100 ± 20 | 76 ± 26 | 44 ± 21a | 124 ± 47 | 131 ± 58 | 69 ± 13c |
| Pparg | 100 ± 31 | 88 ± 17 | 28 ± 11b | 244 ± 69b | 147 ± 40 | 124 ± 38c |
| Cd36 | 100 ± 28 | 82 ± 49 | 43 ± 21a | 307 ± 113b | 153 ± 46a | 12 ± 33d |
| Lpl | 100 ± 55 | 47 ± 15 | 34 ± 14a | 78 ± 32 | 118 ± 41 | 52 ± 30 |
| Atgl | 100 ± 47 | 66 ± 26 | 34 ± 17a | 185 ± 54a | 135 ± 38 | 71 ± 21c |
| Pdk4 | 100 ± 53 | 62 ± 34 | 30 ± 12a | 345 ± 127b | 242 ± 84a | 88 ± 35d |
| Aox | 100 ± 17 | 86 ± 31 | 67 ± 27a | 211 ± 77b | 155 ± 57 | 97 ± 33c |
| Cptlb | 100 ± 25 | 88 ± 23 | 63 ± 19a | 241 ± 65b | 147 ± 26a | 105 ± 42c |
| PGCla | 100 ± 46 | 127 ± 26 | 148 ± 39 | 57 ± 11a | 73 ± 31 | 114 ± 35c |
| Glut1 | 100 ± 34 | 151 ± 56 | 215 ± 45a | 123 ± 28 | 159 ± 47 | 187 ± 39c |
| Glut4 | 100 ± 30 | 274 ± 53b | 479 ± 133b | 83 ± 23 | 129 ± 42 | 260 ± 58d |
AOX, acylCoA oxidase; ATGL, adipose TG lipase; CPT, carnitine palmitoyl transferase; DGAT, diacylglycerol acyl transferase; GLUT, glucose transporter; PDK, pyruvate dehydrogenase kinase; PGC, proliferator-activated receptor gamma coactivator; PPAR, peroxisome proliferator-activated receptor; WT, wild type.
P < 0.05 versus chow diet WT.
P < 0.01 versus chow diet WT.
P < 0.05 versus high-fat diet WT.
P < 0.01 versus high-fat diet WT.
Fig. 2.
Heart and skeletal muscle hypertrophy in Dgat1−/− mice. Heart and skeletal muscle (soleus and EDL) weight in WT, Dgat1+/−, and Dgat1−/− mice (n = 5, *P < 0.05 vs. WT, **P < 0.01 vs. WT) fed chow (A) and high fat (n = 5, *P < 0.05 vs. WT) (B). C and D: Lean and fat weight in chow- and high-fat diet WT, Dgat1+/−, and Dgat1−/− mice (n = 5, *P < 0.05 vs. chow diet WT, **P < 0.01 vs. high-fat diet WT). E: Western blots for p-AKT (ser473), total AKT, p-mTOR (ser2448), and total mTOR in chow diet hearts from WT, Dgat1+/−, and Dgat1−/− mice. DGAT, diacylglycerol acyl transferase; EDL, digitorum longus; WT, wild type.
DEXA showed that lean tissue weight was increased 8% in chow diet Dgat1−/− mice (Fig. 2C). No significant change was observed for lean weight in high-fat diet mice with DEXA. However, as has been reported (10), fat tissue was reduced 56% in high-fat diet Dgat1−/− mice (Fig. 2D). Photographs of DEXA are shown in supplementary Fig. I.
There are several signaling pathways that can mediate cardiac hypertrophy (18, 19). We assessed markers for both pathological and physiologic hypertrophy. mRNA levels of heart failure markers brain-type natriuretic peptide (BNP) and atrial natriuretic factor (ANF) were not increased in the heart (supplementary Fig. IC, D). However, protein levels of p-AKT, a mediator of physiologic hypertrophy (18, 19), and its downstream target p-mTOR were increased in heart (Fig. 2E); p-AKT increased 183%, and p-mTOR increased 126%. These increases were also found in high-fat diet mice (supplementary Fig. IIA). Soleus muscle from chow diet Dgat1−/− mice also had increased p-AKT and p-mTOR (supplementary Fig. IIB). Muscle total AKT and mTOR protein did not differ among tissues of Dgat1−/−, Dagt1+/−, and WT mice (Fig. 2E and supplementary Fig. IIA, B).
mRNA levels of Ppar genes and their downstream targets were reduced in heart and skeletal muscle in Dgat1−/− mice
Heart muscle from chow diet Dgat1−/− mice had striking reductions in mRNA levels of genes involved in lipid metabolism and Ppara, Ppard, and Pparg (all reduced 56-72%, Table 2). The protein levels of PPARs were also reduced (supplementary Fig. III). Cd36 mRNA level was decreased ∼60-80%, and adipose TG lipase (Atgl) and lipoprotein lipase (Lpl) mRNA levels were decreased ∼65%. mRNA levels of genes coding for pyruvate dehydrogenase kinase (Pdk) 4, a regulator of glucose oxidation, acylCoA oxidase (Aox), and carnitine palmitoyl transferase (Cpt)1b decreased ∼60-80% (Table 2). Proliferator-activated receptor γ coactivator (Pgc)1a mRNA level, a coactivator of PPARs involved in fatty acid oxidation and mitochondria genesis, was not changed in chow fed Dgat1−/− cardiac muscle. It was decreased in cardiac muscle from high-fat-diet mice, but in Dgat1−/− hearts, mRNA levels were the same as those found with chow (Table 2). Glucose transporter (Glut)1 and 4 mRNA levels were upregulated 115% and 379% (P < 0.05) in Dgat1−/− hearts; this implies an energy supply shift from fatty acid consumption to glucose use. High-fat diets increased mRNA levels of a number of genes, but loss of Dgat1 significantly reduced these genes (Table 2). Skeletal muscle showed similar results to heart (supplementary Table III).
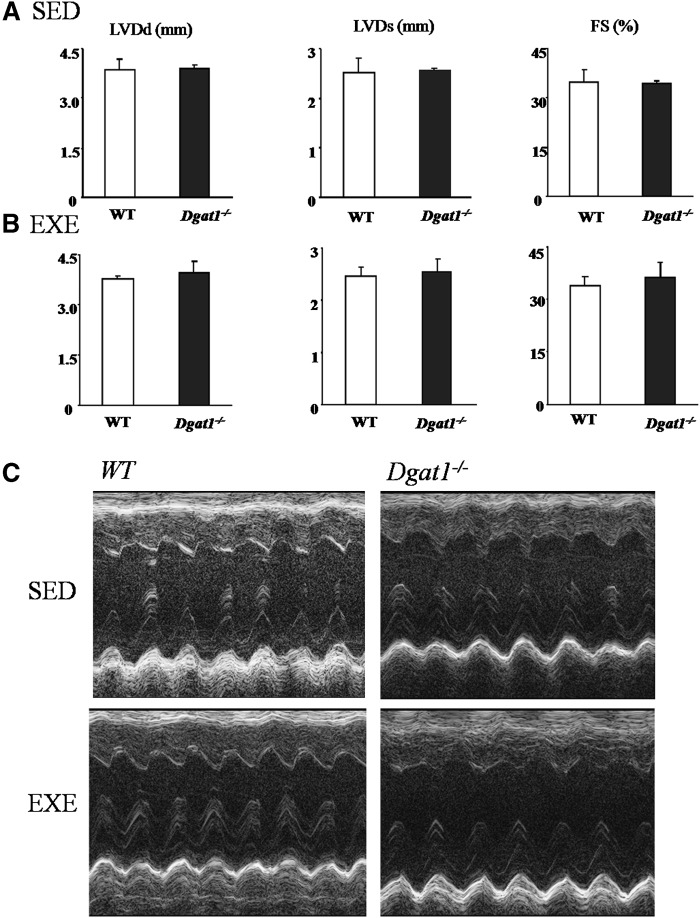
Dgat1−/− mice have normal heart function
Cardiac function of Dgat1−/− mice evaluated by echocardiography was normal. We also exercised the Dgat1−/− mice for two weeks to see if there was any change in heart function after exercise challenge. LVDd, LVDs, and fractional shortening were not significantly altered in Dgat1−/− mouse hearts before and after exercise (Fig. 3A, B). Representative echocardiograms are shown in Fig. 3C.
Fig. 3.
Effects of exercise on cardiac function in WT, Dgat1+/−, and Dgat1−/− mice. Echocardiography showing left ventricular systolic dimension and fractional shortening in WT, Dgat1+/−, and Dgat1−/− mice before exercise (A) (n = 5-6) and after exercise (B) (n = 4-5). C: Representative echocardiograms. DGAT, diacylglycerol acyl transferase; EXE, exercise; LVDd, left ventricular end-diastolic dimension; LVDs, left ventricular end-systolic dimension; SED, sedimentary; WT, wild type.
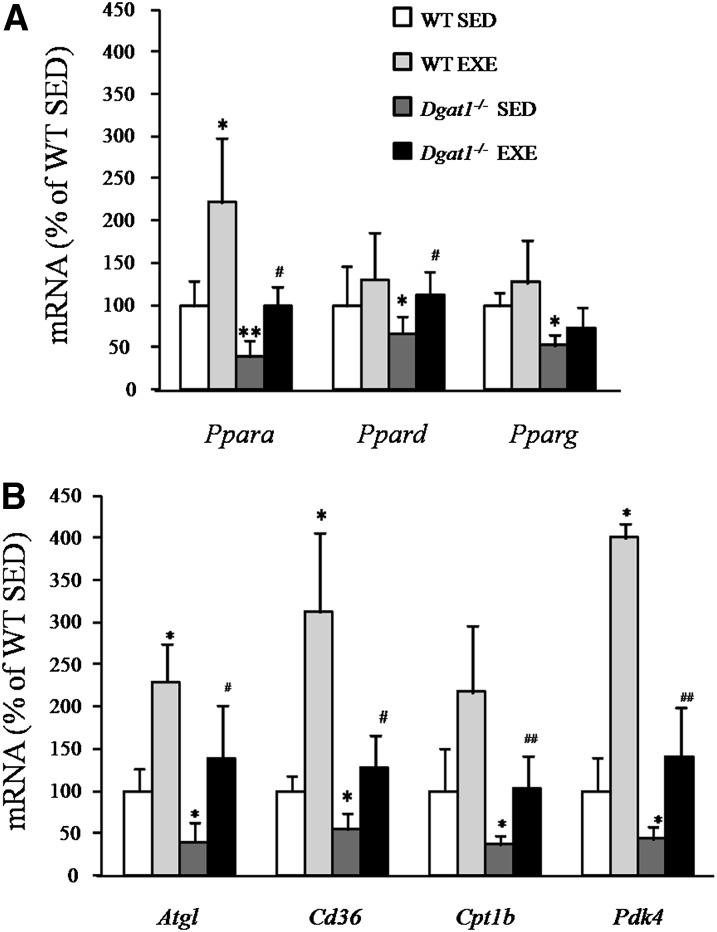
Exercise alters mRNA levels of genes involved in fatty acid oxidation in Dgat1−/− cardiac muscle
Heart Ppara mRNA expression was doubled by exercise (Fig. 4A). This finding was associated with greater mRNA levels of Atgl, Cd36, and Pdk4 (Fig. 4B); these changes are consistent with greater lipid uptake and oxidation as reported by others (20–23). Dgat1−/− mice also had exercise-induced increases in mRNA levels of Ppara and Ppard and their downstream target genes (Fig. 4A, B). Thus, although the exercised Dgat1−/− hearts still had lower mRNA levels of many of these genes compared with exercised WT mice, these genes were increased in the exercised Dgat1−/− hearts, leading to similar mRNA levels as were found in the unexercised WT mouse hearts. These results indicated that exercise was able to induce metabolic changes independent of DGAT1 actions.
Fig. 4.
Gene expression in exercised WT and Dgat1−/− mice. A: Ppara, Ppard, and Pparg mRNA expression in SED WT and Dgat1−/− mice compared with EXE WT and Dgat1−/− mice. B: Atgl, Cd36, Cpt1, and Pdk4 mRNA levels in SED WT and Dgat1−/− mice compared with EXE WT and Dgat1−/− mice. *P < 0.05, **P < 0.01 vs. SED, #P < 0.05, ##P < 0.01 vs. Dgat1−/− SED, n = 4-5. ATGL, adipose TG lipase; CPT1, carnitine palmitoyl transferase–1; DGAT, diacylglycerol acyl transferase; EXE, exercise; PDK, pyruvate dehydrogenase kinase; PPAR, peroxisome proliferator-activated receptor; SED, sedimentary; WT, wild type.
DGAT1 deficiency reduces cardiac fatty acid oxidation and alters VLDL and glucose uptake
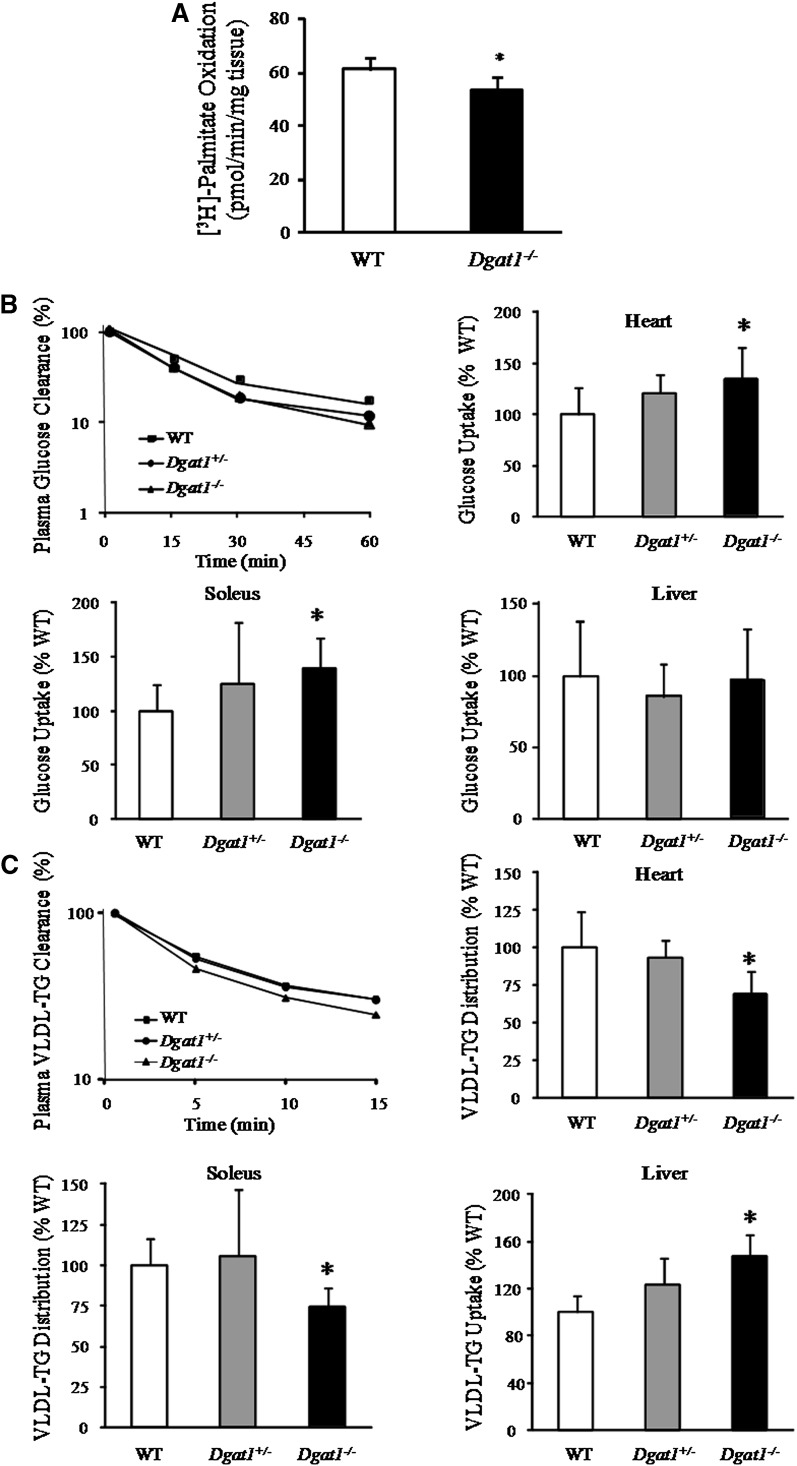
Previous studies showed that Dgat1−/− skeletal muscle is more insulin-sensitive when exposed to hyperinsulinemic conditions (24), but the reasons were not revealed. Because our study demonstrated that mRNA levels of genes involved in lipid metabolism were reduced in Dgat1−/− mice, we hypothesized that under basal conditions (without addition of insulin) Dgat1−/− mice switch from lipid to more glucose usage. First, we tested fatty acid oxidation in isolated heart slices. Fatty acid oxidation rates were reduced in Dgat1−/− mice (Fig. 5A).
Fig. 5.
DGAT1 deficiency reduced fatty acid oxidation, diminished VLDL uptake, and increased glucose uptake into the hearts. A: Palmitic acid oxidation in cardiac muscle slices (P < 0.05, n = 5). B: Plasma glucose clearance, and liver, heart, and soleus muscle glucose uptake in WT, Dgat1+/−, and Dgat1−/− mice. C: Plasma VLDL-TG clearance, liver VLDL-TG uptake, and VLDL-TG uptake distribution in heart and soleus muscle from WT, Dgat1+/−, and Dgat1−/− mice (n = 5-6, *P < 0.05 vs. WT). DGAT, diacylglycerol acyl transferase; TG, triglyceride; WT, wild type.
To assess the metabolic effects of DGAT1 deficiency in vivo, plasma turnover and tissue distribution of tracer 2-deoxy-glucose and VLDL-triglyceride were determined. Despite normal plasma decay (Fig. 5B), 2-deoxy-glucose uptake was increased in Dgat1−/− heart (34.4%, P < 0.05) and skeletal muscle (soleus: 39%, P < 0.05). Glucose uptake by the liver did not change. VLDL-TG uptake into muscle was compared with that of liver to assess distribution of this tracer among organs. Plasma decay of VLDL is shown in Fig. 5C. VLDL-TG uptake was 47% greater in Dgat1−/− liver. Although the amount of recovered tracer in heart and soleus was unchanged, when compared with liver uptake, relatively less VLDL-TG was distributed to muscles; 31% less TG was recovered in hearts, and 26% less in soleus (Fig. 5B). Thus, Dgat1 deficiency led to greater uptake of lipid into the liver, while muscles switched to more glucose use.
Effect of acute DGAT1 inhibition on lipid contents and gene expression
The metabolic changes found in Dgat1−/− mice could reflect developmental compensation. For this reason, we tested whether pharmacological inhibition of DGAT1 also leads to reduced mRNA levels of genes mediating lipid uptake and oxidation. First, we measured the lipid content of DGAT1i-treated muscles. DGAT1i reduced the content of TG ∼16% (P < 0.05) and ceramides ∼18% (P < 0.05) in chow diet WT mouse hearts; DAG levels were not changed (Table 3). DGAT1i-treated mice had reduced mRNA levels of all three forms of Ppar, ranging from 47 to 63% of control (Table 4). mRNA levels of genes coding for proteins involved in lipid uptake were reduced [Cd36 to ∼50%, Atgl to ∼50%, Pdk4 to ∼50%, and Cpt1b to ∼20% of WT (Table 4)]. mRNA levels of genes coding for heart failure markers (Anf and Bnp) were not changed (data not shown).
TABLE 3.
Effect of DGAT1i on heart lipids (n = 5)
| Chow Diet |
High-Fat Diet |
|||
|---|---|---|---|---|
| Lipids | WT | DGAT1i | WT | DGAT1i |
| TG (nmol/mg) | 19.3 ± 2.5 | 16.1 ± 2.2 | 25.6 ± 3.7a | 18.9 ± 2.5b |
| FFA (nmol/mg) | 7.8 ± 0.9 | 9.6 ± 1.6 | 10.2 ± 1.7a | 9.1 ± 1.8 |
| DAG (pmol/mg) | 248 ± 51 | 215 ± 37 | 319 ± 39 | 284 ± 66 |
| Ceramide (pmol/mg) | 219 ± 32 | 179 ± 26a | 307 ± 50a | 277 ± 55 |
DAG, diacylglycerol; DGAT, diacylglycerol acyl transferase; DGATli, DGAT1 inhibitor; TG, triglyceride; WT, wild type.
P < 0.05 versus chow diet WT.
P < 0.05 versus high fat diet WT.
TABLE 4.
Effect of DGATli on heart gene expression (n = 5)
| Chow Diet |
High-Fat Diet |
|||
|---|---|---|---|---|
| Gene | Control | DGAT1i | Control | DGAT1i |
| % | % | % | % | |
| Ppara | 100 ± 41 | 46 ± 28a | 189 ± 51a | 107 ± 26c |
| Ppard | 100 ± 22 | 63 ± 11a | 128 ± 47 | 56 ± 29c |
| Pparg | 100 ± 15 | 39 ± 8b | 190 ± 85a | 51 ± 10d |
| Cd36 | 100 ± 33 | 53 ± 23a | 276 ± 121a | 86 ± 49d |
| Lpl | 100 ± 33 | 62 ± 22a | 137 ± 45 | 76 ± 30c |
| Atgl | 100 ± 26 | 51 ± 27b | 183 ± 100a | 20 ± 7d |
| Pdk4 | 100 ± 23 | 56 ± 34a | 258 ± 118a | 121 ± 38c |
| Aox | 100 ± 41 | 77 ± 11a | 149 ± 65 | 84 ± 33c |
| Cpt1b | 100 ± 63 | 26 ± 15a | 159 ± 68 | 28 ± 17d |
AOX, acylCoA oxidase; ATGL, adipose TG lipase; CPT, carnitine palmitoyl transferase; DGAT, diacylglycerol acyl transferase; DGATli, DGAT1 inhibitor; PDK, pyruvate dehydrogenase kinase; PGC, proliferator-activated receptor gamma coactivator; PPAR, peroxisome proliferator-activated receptor; WT, wild type.
P < 0.05 versus chow diet WT.
P < 0.01 versus chow diet WT.
P < 0.05 versus high fat diet WT.
P < 0.01 versus high fat diet WT.
We next tested whether the DGAT1i had similar effects in mice fed a high-fat diet. As reported by others (25, 26), the levels of mRNA expression of genes involved in fatty acid oxidation were increased in hearts of mice on a high-fat diet. However, many of these increases were totally blocked by the acute administration of DGAT1i; increases in mRNA levels corresponding to Atgl and Cpt1b were totally blocked by the DGAT1i (Table 4). The mRNA levels for genes encoding Cd36 and Pdk4 were increased by high fat, but they remained much lower than was seen in untreated control hearts from high-fat diet mice.
Expression of genes involved in fatty acid oxidation in liver was increased in Dgat1−/− mice and in DGAT1i-treated WT mice
Although our results demonstrated reduced mRNA levels of genes involved in fatty acid oxidation in heart and skeletal muscle in Dgat1−/− mice, other studies reported that Dgat1−/− mice on high-fat diets had increased fatty acid oxidation in whole body (10) and liver (27). However, hepatic mRNA levels of many metabolic genes were not reported. Unlike in muscle, some mRNA levels of genes involved in hepatic fatty acid oxidation were increased in Dgat1−/− mice (Table 5). mRNA levels of Ppard, Cpt1a, and Pdk4 were increased to 159 ± 49, 137 ± 18, and 151 ± 29% of WT (P < 0.05). mRNA levels of other Ppar genes that mediate fatty acid uptake and metabolism and hepatocyte nuclear factor (Hnf)4a did not change in Dgat1−/− livers. Similar results were found in liver from DGAT1i-treated mice; mRNA levels of genes coding for Cpt1a and Pdk4 increased to 189 ± 58 and 146 ± 16% of control, respectively (P < 0.05) (Table 5).
TABLE 5.
Gene expression in DGAT1 genetic and inhibitor knockdown mouse liver (high-fat diet) (n = 4–6)
| Gene | WT | Dgat1+/− | Dgat1−/− | DGAT1i |
|---|---|---|---|---|
| % | % | % | % | |
| Ppara | 100 ± 48 | 79 ± 26 | 72 ± 31 | 87 ± 13 |
| Ppard | 100 ± 13 | 110 ± 34 | 159 ± 49a | 85 ± 28 |
| Pparg | 100 ± 56 | 67 ± 12 | 62 ± 18 | 117 ± 45 |
| Cpt1a | 100 ± 17 | 109 ± 23 | 137 ± 18a | 189 ± 58a |
| Pdk4 | 100 ± 25 | 96 ± 32 | 151 ± 29a | 146 ± 16a |
| Hnf4a | 100 ± 28 | 122 ± 29 | 135 ± 34 | 142 ± 51 |
CPT, carnitine palmitoyl transferase–1; DGAT, diacylglycerol acyl transferase; DGATli, DGAT1 inhibitor; HNF, hepatocyte nuclear factor; PDK, pyruvate dehydrogenase kinase; PPAR, peroxisome proliferator-activated receptor; WT, wild type.
P < 0.05 versus WT.
Inhibition of DGAT1 reduces mRNA levels of genes involved in lipid metabolism in AC16 cardiomyocytes and C2C12 myocytes
To determine if the effects of DGAT1i treatment were direct or secondary to some other in vivo metabolic effect of this drug, AC16 and C2C12 cells were treated with DGAT1i in the presence or absence of palmitic acid. DGAT1i markedly reduced expression of Ppar and genes involved in fatty acid oxidation in both cell types (Table 6). In addition, the increased mRNA levels due to 0.5 mM palmitic acid (Ppara, Ppard, Pparg, Cd36, Atgl, and Pdk4 increased from 1- to 4-fold) were totally prevented by DGAT1i treatment. DGAT1i attenuated, but did not completely block, the palmitic acid-mediated increase of mRNA levels of the gene coding for Cpt1b. Treating AC16 cardiomyocytes with DGAT1i prior to addition of labeled palmitic acid reduced palmitate uptake (supplementary Fig. IV). These results are similar to what we observed in vivo with the effects of high-fat diets and indicate that DGAT1 deficiency in muscle cells leads to marked reduction in mRNA levels of genes involved in lipid metabolism, an effect that is only partially compensated by addition of exogenous fatty acids.
TABLE 6.
DGAT1i effect on PPARs and their downstream genes in control and palmitic acid-treated myocytes
| Gene | Control | DGAT1i | Palmitic acid | DGAT1i + Palmitic acid | |
|---|---|---|---|---|---|
| % | % | % | % | % | |
| AC16 cells | Ppara | 100 ± 37 | 49 ± 21a | 210 ± 54a | 98 ± 31d |
| Ppard | 100 ± 26 | 53 ± 24a | 278 ± 60b | 82 ± 29d | |
| Pparg | 100 ± 43 | 41 ± 17a | 389 ± 91b | 71 ± 24d | |
| Cd36 | 100 ± 15 | 66 ± 28a | 191 ± 55a | 84 ± 19c | |
| Atgl | 100 ± 27 | 55 ± 19a | 197 ± 35a | 52 ± 18d | |
| Cpt1b | 100 ± 38 | 61 ± 17a | 148 ± 49a | 74 ± 18d | |
| Pdk4 | 100 ± 22 | 38 ± 10b | 333 ± 82b | 80 ± 26d | |
| C2C12 cells | Ppara | 100 ± 20 | 50 ± 12a | 266 ± 76b | 101 ± 22d |
| Ppard | 100 ± 35 | 49 ± 23a | 501 ± 123b | 88 ± 38d | |
| Pparg | 100 ± 27 | 58 ± 22a | 452 ± 89b | 68 ± 15d | |
| Cd36 | 100 ± 21 | 53 ± 14a | 219 ± 65b | 116 ± 27c | |
| Atgl | 100 ± 33 | 21 ± 6b | 238 ± 61b | 65 ± 28d | |
| Cpt1b | 100 ± 38 | 75 ± 11 | 1520 ± 355b | 312 ± 148d | |
| Pdk4 | 100 ± 22 | 44 ± 27a | 328 ± 103b | 74 ± 17d |
Data are from three separate experiments performed in duplicate. ATGL, adipose TG lipase; CPT, carnitine palmitoyl transferase; DGAT, diacylglycerol acyl transferase; DGATli, DGAT1 inhibitor; PDK, pyruvate dehydrogenase kinase; PPAR, peroxisome proliferator-activated receptor; WT, wild type.
P < 0.05 versus control.
P < 0.01 versus control.
P < 0.05 versus palmitic acid treatment.
P < 0.01 versus palmitic acid treatment.
DISCUSSION
Lipid uptake and oxidation regulate cellular lipid stores. When this regulation is dysfunctional due to greater lipid accumulation than usage, it leads to a group of diseases classified as lipotoxicities, including nonalcoholic fatty liver disease, type 2 diabetes, and metabolic cardiomyopathy. Data from our lab (5, 6) and others (28) have shown that TG accumulation itself is not toxic. In contrast, it is believed that several lipid intermediates, especially ceramides and DAGs, are harmful (29–34). Ceramides are thought to directly cause cellular apoptosis (35), and DAGs via activation of PKCs lead to insulin resistance (33, 34). For this reason, we initially hypothesized that loss of DGAT1 activity would prevent conversion of DAG to TG and lead to cellular toxicity, if not on chow then on a high-fat diet. We were wrong! But, in the process of our studies on Dgat1 deficiency, we obtained information that explains the increased insulin sensitivity in Dgat1−/− muscles and the reason that Dgat1−/− mice have greater fatty acid oxidation (lower respiratory quotient). We show that buildup of toxic lipids does not occur because expression of genes involved in lipid uptake (Cd36 and Lpl) is reduced. This finding is a direct effect of loss of DGAT1 activity in myocytes, not a secondary phenomenon. Finally, we show that deficiency of DGAT1 has a different effect on the liver.
Much of the interest in DGAT1 resulted from studies in mice with a genetic deletion of this enzyme. Dgat1−/− mice have less diet-induced insulin resistance and obesity (10). Although older Dgat1−/− mice have alopecia (36), these abnormalities were not observed in the young mice in our studies. In addition, this skin phenotype is not seen with partially reduced Dgat1 expression. Thus, inhibition of DGAT1 appeared to be a reasonable target for treatment of metabolic disorders.
In some situations, defective lipid storage in adipose leads to greater muscle lipid content and insulin resistance (37, 38). The reason this does not occur in Dgat1−/− mice or with acute pharmacologic DGAT1 inhibition became clear when we studied muscles from these mice. Chronic Dgat1 deficiency did not grossly alter heart content of TG, although acute use of DGAT1i led to reduced heart TG content. In both situations, we had expected that reduced conversion of DAG to TG would elevate tissue levels of DAG and perhaps ceramide (i.e., we should find the opposite of the effects of genetic overexpression of DGAT1 in muscles (5)). This did not occur. We previously incubated isolated Dgat1−/− skeletal muscle in high concentrations of fatty acids (a combination of oleic and palmitic acids) to test whether DGAT1 actions mediated detoxification of fatty acids by their conversion to TG, resulting in less DAG and ceramide (5). This acute intervention and its effect on muscle insulin resistance did not use any conditions that were comparable to those found in vivo or in our in vitro studies of gene expression in cultured cells. In contrast, other studies have shown that Dgat1−/− mice have improved insulin sensitivity (10), and our data suggest that this is due, at least in part, to changes in skeletal muscle metabolism.
The reasons that Dgat1 deficiency did not lead to lipotoxicity were shown. Levels of mRNA of genes involved in lipid uptake were markedly reduced. Although not specifically studied, a reduction in cluster of differentiation 36 (CD36) is likely to have led to reduced uptake of free fatty acids (39), while a decrease in LpL would decrease muscle uptake of lipoprotein-derived fatty acids (14). These changes are also likely to have caused the redistribution of VLDL-TG with relatively more lipid directed to the liver and less to muscle. Although both overexpression and knockout of DGAT1 in cardiac muscle resulted in the downregulation of DAG and ceramide, the mechanisms are different. Overexpression of DGAT1 decreases DAG as more DAG is used for TG synthesis and fatty acid oxidation in the muscle. Fatty acid levels in cardiac muscle of DGAT1 overexpression were decreased, whereas CD36 was increased as compensation for the outcome of this result (6). In contrast, the reduction in ceramide and DAG with DGAT1 deficiency is, we assume, due to a decrease in lipid uptake and the availability of precursors.
Neither heart nor skeletal muscle appeared to have pathological changes due to reduced lipid uptake. In part, these tissues compensated by increasing glucose uptake. In many isolated perfused heart models of ischemia/reperfusion, reduced fatty acid and increased glucose oxidation leads to less cardiac damage (40). Dgat1−/− hearts appeared to retain their metabolic flexibility with changes in metabolic genes, allowing greater fatty acid oxidation with exercise and high-fat diets. The greater heart and skeletal glucose uptake in the presence of nonelevated insulin is due to greater insulin sensitivity, as shown previously in clamp studies (41). In our study, greater glucose uptake was associated with a marked reduction in mRNA levels of the Ppar target genes regulating fatty acid oxidation. A reduction in PDK4 would be expected to lead to greater glucose oxidation.
Chronic Dgat1 deficiency was also associated with increased p-AKT and p-mTOR, well-established causes of muscle hypertrophy (18, 19). Ceramide content may reduce AKT phosphorylation (30, 42). In our study, ceramide levels were significantly decreased in hearts of Dgat1−/− and DGAT1i-treated mice. This change might have accounted for the increase of p-AKT and p-mTOR. In the heart, muscle hypertrophy occurs with both physiologic compensation and in response to pathologic stimuli (43). Hearts of Dgat1−/− mice functioned normally under control and exercise training. In addition, the increase in the p-AKT/p-mTOR pathway without increased expression of genes coding for the natiuretic proteins ANF and BNP is consistent with physiologic hypertrophy.
We performed two experiments to determine if mRNA levels of proteins mediating fatty acid uptake and oxidation would increase with physiologic interventions. Although both exercise and high-fat diets increased gene expression, Dgat1−/− mouse hearts were not normalized. Nonetheless, it was apparent that some activation of fatty acid oxidation pathways was via processes exclusive of those blocked by Dgat1 deficiency.
As for DGAT1, studies of overexpression and deletion of PPAR in the heart have not led to the expected opposite phenotypes. Three PPAR genes have been upregulated in the heart. PPARa and PPARg lead to lipotoxic cardiomyopathy (44, 45). PPARd, which does not cause increased lipid accumulation, does not cause cardiomyopathy (46). Reduced lipid use would be expected to occur with deletion of these transcription factors. Presumably that is why PPARa, PPARd, and PPARg cardiomyocyte deletion all lead to toxicity (47–49). We should note that unlike the situation in Dgat1−/− mice, these deletions of Ppar are total and do not cause partial loss of the transcription factor.
Why does loss of DGAT1 activity lead to such marked changes in Ppar mRNA levels and expression of their downstream target genes? By performing studies in cultured cells, we showed that these changes are not secondary to other metabolic changes occurring in vivo. We postulate that Dgat1 deficiency affects a pathway needed for uptake of lipids into the cells and movement of endogenous PPAR agonists to the nucleus. This might occur because Dgat1 deficiency reduces input into the lipid droplet and creation of PPAR ligands via the activity of ATGL. LpL also generates PPAR ligands and is downregulated in Dgat1−/− mice. How other ligands, such as the phospholipid ligand described by the Semenkovich Lab (50), get into tissues is unclear. It is also not clear whether the reduction in lipid uptake or Ppar gene downregulation is primary; this is the direction of future investigation. Note that overexpression of DGAT1 in heart did not lead to as marked an alteration in PPAR downstream gene expression or regulation of PPAR mRNA levels as was found with knockout. Of all tissues, we suspect that the heart is bathed in PPAR agonists and any DGAT1-mediated increase in ligands leads to a relatively less dramatic effect than is seen with the knockout.
Dgat1−/− mice on high-fat diets had increased fatty acid oxidation in whole body (10) and liver (27). Some mRNA levels of genes Ppard, Cpt1a, and Pdk4 involved in hepatic fatty acid oxidation were increased in Dgat1−/− and DGAT1i-treated mice. It is likely that the high level of expression of DGAT2 in the liver is sufficient to obviate effects of DGAT1 loss in TG production (2, 4).
In summary, studies of muscle effects of Dgat1 deficiency uncovered a new PPAR regulatory pathway. Downregulation of genes involved in lipid uptake and oxidation is likely responsible for improved insulin sensitivity and lack of toxic lipid accumulation with Dgat1 deficiency. Muscles from chow diet Dgat1−/− mice are hypertrophied but do not have reduced function. Thus, as a therapeutic target, DGAT1 inhibition appears to be relatively free of cardiac and skeletal muscle toxicity.
Supplementary Material
Footnotes
Abbreviations:
- ACO
- acyl-CoA oxidase
- ANF
- atrial natriuretic factor
- AOX
- acylCoA oxidase
- ATGL
- adipose TG lipase
- BNP
- brain-type natriuretic peptide
- CD36
- cluster of differentiation 36
- CPT
- carnitine palmitoyl transferase
- DAG
- diacylglycerol
- DEXA
- dual energy X-ray absorptiometry
- DGAT
- diacylglycerol acyl transferase
- DGATli
- DGAT1 inhibitor
- EDL
- digitorum longus
- EXE
- exercise
- FS
- fraction shorting
- GLUT
- glucose transporter
- HNF
- hepatocyte nuclear factor
- LVDd
- left ventricular end-diastolic dimension
- LVDs
- left ventricular end-systolic dimension
- p-AKT
- phospho-AKT
- p-mTOR
- phospho-mTOR
- PDK
- pyruvate dehydrogenase kinase
- PGC
- proliferator-activated receptor gamma coactivator
- PPAR
- peroxisome proliferator-activated receptor
- SED
- sedimentary
- TC
- total cholesterol
- TG
- triglyceride
- WT
- wild type
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figures and three tables.
This work was supported by National Institutes of Health Grants HL-73029 and HL-45095 (I.J.G.); DK-79221 and AA-10914 (W.S.B.); HL-62583 (L.S.H.); and T32-07343 (R.K. and L.L). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. K.G.B. was supported by a mentored postdoctoral fellowship from the American Diabetes Association. This work was also supported by the Astra-Zeneca Company.
REFERENCES
- 1.Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 95: 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen C. L., Monetti M., Burri B. J., Farese R. V., Jr 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 1502–1511. [DOI] [PubMed] [Google Scholar]
- 3.Oelkers P., Behari A., Cromley D., Billheimer J. T., Sturley S. L. 1998. Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes. J. Biol. Chem. 273: 26765–26771. [DOI] [PubMed] [Google Scholar]
- 4.Choi C. S., Savage D. B., Kulkarni A., Yu X. X., Liu Z. X., Morino K., Kim S., Distefano A., Samuel V. T., Neschen S., et al. 2007. Suppression of diacylglycerol acyltransferase-2 (DGAT2), but not DGAT1, with antisense oligonucleotides reverses diet-induced hepatic steatosis and insulin resistance. J. Biol. Chem. 282: 22678–22688. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Zhang Y., Chen N., Shi X., Tsang B., Yu Y. H. 2007. Upregulation of myocellular DGAT1 augments triglyceride synthesis in skeletal muscle and protects against fat-induced insulin resistance. J. Clin. Invest. 117: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L., Shi X., Bharadwaj K. G., Ikeda S., Yamashita H., Yagyu H., Schaffer J. E., Yu Y. H., Goldberg I. J. 2009. DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J. Biol. Chem. 284: 36312–36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodpaster B. H., He J., Watkins S., Kelley D. E. 2001. Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J. Clin. Endocrinol. Metab. 86: 5755–5761. [DOI] [PubMed] [Google Scholar]
- 8.Bruce C. R., Thrush A. B., Mertz V. A., Bezaire V., Chabowski A., Heigenhauser G. J., Dyck D. J. 2006. Endurance training in obese humans improves glucose tolerance and mitochondrial fatty acid oxidation and alters muscle lipid content. Am. J. Physiol. Endocrinol. Metab. 291: E99–E107. [DOI] [PubMed] [Google Scholar]
- 9.Koliwad S. K., Streeper R. S., Monetti M., Cornelissen I., Chan L., Terayama K., Naylor S., Rao M., Hubbard B., Farese R. V., Jr 2010. DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. J. Clin. Invest. 120: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr 2000. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat. Genet. 25: 87–90. [DOI] [PubMed] [Google Scholar]
- 11.Chen H. C., Jensen D. R., Myers H. M., Eckel R. H., Farese R. V., Jr 2003. Obesity resistance and enhanced glucose metabolism in mice transplanted with white adipose tissue lacking acyl CoA:diacylglycerol acyltransferase 1. J. Clin. Invest. 111: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H. C., Smith S. J., Ladha Z., Jensen D. R., Ferreira L. D., Pulawa L. K., McGuire J. G., Pitas R. E., Eckel R. H., Farese R. V., Jr 2002. Increased insulin and leptin sensitivity in mice lacking acyl CoA:diacylglycerol acyltransferase 1. J. Clin. Invest. 109: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryder J. W., Kawano Y., Galuska D., Fahlman R., Wallberg-Henriksson H., Charron M. J., Zierath J. R. 1999. Postexercise glucose uptake and glycogen synthesis in skeletal muscle from GLUT4-deficient mice. FASEB J. 13: 2246–2256. [DOI] [PubMed] [Google Scholar]
- 14.Augustus A. S., Buchanan J., Park T. S., Hirata K., Noh H. L., Sun J., Homma S., D'Armiento J., Abel E. D., Goldberg I. J. 2006. Loss of lipoprotein lipase-derived fatty acids leads to increased cardiac glucose metabolism and heart dysfunction. J. Biol. Chem. 281: 8716–8723. [DOI] [PubMed] [Google Scholar]
- 15.Pillutla P., Hwang Y. C., Augustus A., Yokoyama M., Yagyu H., Johnston T. P., Kaneko M., Ramasamy R., Goldberg I. J. 2005. Perfusion of hearts with triglyceride-rich particles reproduces the metabolic abnormalities in lipotoxic cardiomyopathy. Am. J. Physiol. Endocrinol. Metab. 288: E1229–E1235. [DOI] [PubMed] [Google Scholar]
- 16.Birch A. M., Birtles S., Buckett L. K., Kemmitt P. D., Smith G. J., Smith T. J., Turnbull A. V., Wang S. J. 2009. Discovery of a potent, selective, and orally efficacious pyrimidinooxazinyl bicyclooctaneacetic acid diacylglycerol acyltransferase-1 inhibitor. J. Med. Chem. 52: 1558–1568. [DOI] [PubMed] [Google Scholar]
- 17.Birch A. M., Buckett L. K., Turnbull A. V. 2010. DGAT1 inhibitors as anti-obesity and anti-diabetic agents. Curr. Opin. Drug Discov. Devel. 13: 489–496. [PubMed] [Google Scholar]
- 18.Dorn G. W., 2nd 2007. The fuzzy logic of physiological cardiac hypertrophy. Hypertension. 49: 962–970. [DOI] [PubMed] [Google Scholar]
- 19.Spangenburg E. E. 2009. Changes in muscle mass with mechanical load: possible cellular mechanisms. Appl. Physiol. Nutr. Metab. 34: 328–335. [DOI] [PubMed] [Google Scholar]
- 20.Iemitsu M., Miyauchi T., Maeda S., Tanabe T., Takanashi M., Irukayama-Tomobe Y., Sakai S., Ohmori H., Matsuda M., Yamaguchi I. 2002. Aging-induced decrease in the PPAR-alpha level in hearts is improved by exercise training. Am. J. Physiol. Heart Circ. Physiol. 283: H1750–H1760. [DOI] [PubMed] [Google Scholar]
- 21.Alsted T. J., Nybo L., Schweiger M., Fledelius C., Jacobsen P., Zimmermann R., Zechner R., Kiens B. 2009. Adipose triglyceride lipase in human skeletal muscle is upregulated by exercise training. Am. J. Physiol. Endocrinol. Metab. 296: E445–E453. [DOI] [PubMed] [Google Scholar]
- 22.Barger P. M., Kelly D. P. 2000. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc. Med. 10: 238–245. [DOI] [PubMed] [Google Scholar]
- 23.Tunstall R. J., Cameron-Smith D. 2005. Effect of elevated lipid concentrations on human skeletal muscle gene expression. Metabolism. 54: 952–959. [DOI] [PubMed] [Google Scholar]
- 24.Chen H. C., Rao M., Sajan M. P., Standaert M., Kanoh Y., Miura A., Farese R. V., Jr, Farese R. V. 2004. Role of adipocyte-derived factors in enhancing insulin signaling in skeletal muscle and white adipose tissue of mice lacking Acyl CoA:diacylglycerol acyltransferase 1. Diabetes. 53: 1445–1451. [DOI] [PubMed] [Google Scholar]
- 25.Peters S. J., Harris R. A., Wu P., Pehleman T. L., Heigenhauser G. J., Spriet L. L. 2001. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am. J. Physiol. Endocrinol. Metab. 281: E1151–E1158. [DOI] [PubMed] [Google Scholar]
- 26.Pilegaard H., Saltin B., Neufer P. D. 2003. Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes. 52: 657–662. [DOI] [PubMed] [Google Scholar]
- 27.Villanueva C. J., Monetti M., Shih M., Zhou P., Watkins S. M., Bhanot S., Farese R. V., Jr 2009. Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 50: 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H. C., Stone S. J., Zhou P., Buhman K. K., Farese R. V., Jr 2002. Dissociation of obesity and impaired glucose disposal in mice overexpressing acyl coenzyme a:diacylglycerol acyltransferase 1 in white adipose tissue. Diabetes. 51: 3189–3195. [DOI] [PubMed] [Google Scholar]
- 29.Holland W. L., Brozinick J. T., Wang L. P., Hawkins E. D., Sargent K. M., Liu Y., Narra K., Hoehn K. L., Knotts T. A., Siesky A., et al. 2007. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 5: 167–179. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz-Peiffer C., Craig D. L., Biden T. J. 1999. Ceramide generation is sufficient to account for the inhibition of the insulin-stimulated PKB pathway in C2C12 skeletal muscle cells pretreated with palmitate. J. Biol. Chem. 274: 24202–24210. [DOI] [PubMed] [Google Scholar]
- 31.Avignon A., Yamada K., Zhou X., Spencer B., Cardona O., Saba-Siddique S., Galloway L., Standaert M. L., Farese R. V. 1996. Chronic activation of protein kinase C in soleus muscles and other tissues of insulin-resistant type II diabetic Goto-Kakizaki (GK), obese/aged, and obese/Zucker rats. A mechanism for inhibiting glycogen synthesis. Diabetes. 45: 1396–1404. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz-Peiffer C., Browne C. L., Oakes N. D., Watkinson A., Chisholm D. J., Kraegen E. W., Biden T. J. 1997. Alterations in the expression and cellular localization of protein kinase C isozymes epsilon and theta are associated with insulin resistance in skeletal muscle of the high-fat diet rat. Diabetes. 46: 169–178. [DOI] [PubMed] [Google Scholar]
- 33.Itani S. I., Ruderman N. B., Schmieder F., Boden G. 2002. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. 51: 2005–2011. [DOI] [PubMed] [Google Scholar]
- 34.Kim J. K., Fillmore J. J., Sunshine M. J., Albrecht B., Higashimori T., Kim D. W., Liu Z. X., Soos T. J., Cline G. W., O'Brien W. R., et al. 2004. PKC-theta knockout mice are protected from fat-induced insulin resistance. J. Clin. Invest. 114: 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turpin S. M., Lancaster G. I., Darby I., Febbraio M. A., Watt M. J. 2006. Apoptosis in skeletal muscle myotubes is induced by ceramides and is positively related to insulin resistance. Am. J. Physiol. Endocrinol. Metab. 291: E1341–E1350. [DOI] [PubMed] [Google Scholar]
- 36.Chen H. C., Farese R. V., Jr 2005. Inhibition of triglyceride synthesis as a treatment strategy for obesity: lessons from DGAT1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 25: 482–486. [DOI] [PubMed] [Google Scholar]
- 37.Kirk E. P., Klein S. 2009. Pathogenesis and pathophysiology of the cardiometabolic syndrome. J. Clin. Hypertens. (Greenwich). 11: 761–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y. H., Ginsberg H. N. 2004. The role of acyl-CoA:diacylglycerol acyltransferase (DGAT) in energy metabolism. Ann. Med. 36: 252–261. [DOI] [PubMed] [Google Scholar]
- 39.Coburn C. T., Knapp F. F., Jr, Febbraio M., Beets A. L., Silverstein R. L., Abumrad N. A. 2000. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275: 32523–32529. [DOI] [PubMed] [Google Scholar]
- 40.Luptak I., Yan J., Cui L., Jain M., Liao R., Tian R. 2007. Long-term effects of increased glucose entry on mouse hearts during normal aging and ischemic stress. Circulation. 116: 901–909. [DOI] [PubMed] [Google Scholar]
- 41.Baron A. D., Brechtel G., Wallace P., Edelman S. V. 1988. Rates and tissue sites of non-insulin- and insulin-mediated glucose uptake in humans. Am. J. Physiol. 255: E769–E774. [DOI] [PubMed] [Google Scholar]
- 42.Summers S. A., Garza L. A., Zhou H., Birnbaum M. J. 1998. Regulation of insulin-stimulated glucose transporter GLUT4 translocation and Akt kinase activity by ceramide. Mol. Cell. Biol. 18: 5457–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heineke J., Molkentin J. D. 2006. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7: 589–600. [DOI] [PubMed] [Google Scholar]
- 44.Park S. Y., Cho Y. R., Finck B. N., Kim H. J., Higashimori T., Hong E. G., Lee M. K., Danton C., Deshmukh S., Cline G. W., et al. 2005. Cardiac-specific overexpression of peroxisome proliferator-activated receptor-alpha causes insulin resistance in heart and liver. Diabetes. 54: 2514–2524. [DOI] [PubMed] [Google Scholar]
- 45.Son N. H., Park T. S., Yamashita H., Yokoyama M., Huggins L. A., Okajima K., Homma S., Szabolcs M. J., Huang L. S., Goldberg I. J. 2007. Cardiomyocyte expression of PPARgamma leads to cardiac dysfunction in mice. J. Clin. Invest. 117: 2791–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkart E. M., Sambandam N., Han X., Gross R. W., Courtois M., Gierasch C. M., Shoghi K., Welch M. J., Kelly D. P. 2007. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J. Clin. Invest. 117: 3930–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Djouadi F., Weinheimer C. J., Saffitz J. E., Pitchford C., Bastin J., Gonzalez F. J., Kelly D. P. 1998. A gender-related defect in lipid metabolism and glucose homeostasis in peroxisome proliferator- activated receptor alpha- deficient mice. J. Clin. Invest. 102: 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng L., Ding G., Qin Q., Huang Y., Lewis W., He N., Evans R. M., Schneider M. D., Brako F. A., Xiao Y., et al. 2004. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat. Med. 10: 1245–1250. [DOI] [PubMed] [Google Scholar]
- 49.Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., Koder A., Evans R. M. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 4: 585–595. [DOI] [PubMed] [Google Scholar]
- 50.Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. 2009. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 138: 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.