Abstract
Mass spectrometry-based proteomics has greatly benefitted from enormous advances in high resolution instrumentation in recent years. In particular, the combination of a linear ion trap with the Orbitrap analyzer has proven to be a popular instrument configuration. Complementing this hybrid trap-trap instrument, as well as the standalone Orbitrap analyzer termed Exactive, we here present coupling of a quadrupole mass filter to an Orbitrap analyzer. This “Q Exactive” instrument features high ion currents because of an S-lens, and fast high-energy collision-induced dissociation peptide fragmentation because of parallel filling and detection modes. The image current from the detector is processed by an “enhanced Fourier Transformation” algorithm, doubling mass spectrometric resolution. Together with almost instantaneous isolation and fragmentation, the instrument achieves overall cycle times of 1 s for a top10 higher energy collisional dissociation method. More than 2500 proteins can be identified in standard 90-min gradients of tryptic digests of mammalian cell lysate— a significant improvement over previous Orbitrap mass spectrometers. Furthermore, the quadrupole Orbitrap analyzer combination enables multiplexed operation at the MS and tandem MS levels. This is demonstrated in a multiplexed single ion monitoring mode, in which the quadrupole rapidly switches among different narrow mass ranges that are analyzed in a single composite MS spectrum. Similarly, the quadrupole allows fragmentation of different precursor masses in rapid succession, followed by joint analysis of the higher energy collisional dissociation fragment ions in the Orbitrap analyzer. High performance in a robust benchtop format together with the ability to perform complex multiplexed scan modes make the Q Exactive an exciting new instrument for the proteomics and general analytical communities.
Mass spectrometry-based proteomics often involves the analysis of complex mixtures of proteins derived from cell or tissue lysates or from body fluids, posing tremendous analytical challenges (1–3). After proteolytic digestion, the resulting peptide mixtures are separated by liquid chromatography and online electrosprayed for mass spectrometric (MS) and tandem mass spectrometric (MS/MS) analysis. Because tens of thousands of peptides elute over a relatively short time and with ion signals different by many orders of magnitude (4, 5), mass spectrometers have been pushed to even higher sensitivity, sequencing speed, and resolution (6, 7). In current shotgun proteomics there are mainly four mass spectrometric separation principles: quadrupole mass filters, time of flight (TOF)1 mass analyzers, linear ion traps, and Orbitrap™ analyzers. These are typically combined in hybrid configurations. Quadrupole TOF instruments use a quadrupole mass filter to either transmit the entire mass range produced by the ion source (for analysis of all ions in MS mode) or to transmit only a defined mass window around a precursor ion of choice (MS/MS mode). In the latter case ions are activated in a collision cell and resulting fragments are analyzed in the TOF part of the instrument with very high repetition rate. This TOF part of quadrupole TOF instruments replaces the final quadrupole section of triple quadrupole instruments, which are today mainly used for targeted proteomics (8–10).
The quadrupole TOF instruments achieve peptide separation “in space”, meaning the ions are separated nearly instantaneously by passing through either the quadrupole section, in which only a chosen small mass range has stable trajectories, or by traversing the TOF section. In contrast, trapping instruments such as linear ion traps separate ions “in time” by applying external RF-DC fields to a stationary ion population that allow only a certain ion population to stably remain in the trap (see ref (11)) for the concept of separation and fragmentation in time versus in space).
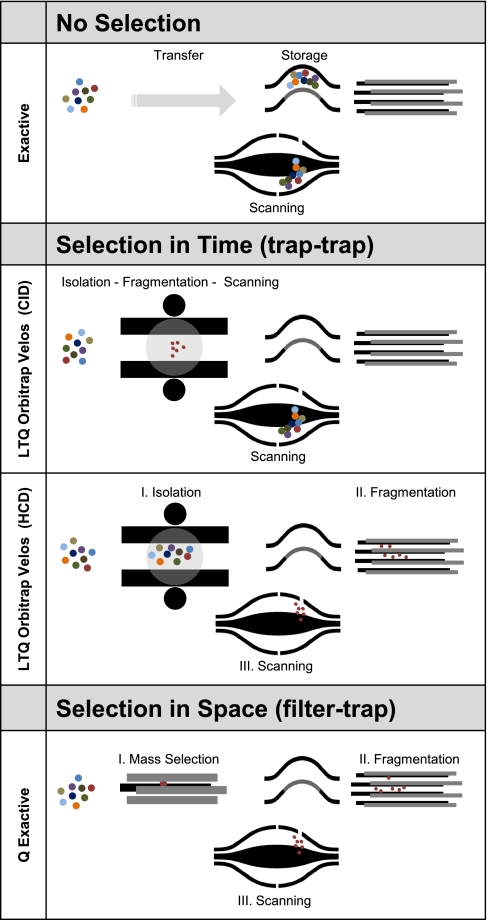
The Orbitrap mass analyzer was developed about ten years ago by Makarov. It consists of a small electrostatic device into which ion packets are injected at high energies to orbit around a central, spindle-shaped electrode (12–14). The image current of the axial motion of the ions is picked up by the detector and this signal is Fourier transformed (FT) to yield high resolution mass spectra. Commercially, the Orbitrap analyzer was first introduced in 2005 in a hybrid instrument (15). In proteomics and related fields, this combination of a low resolution linear ion trap with the high resolution Orbitrap analyzer—termed “LTQ Orbitrap”—has now become widespread (16, 17). The LTQ Orbitrap instruments represent a multistage trap combination (Fig. 1). In MS mode the linear trap performs the function of collecting the ion population, passing them on to an intermediate C-trap for injection and analysis in the Orbitrap analyzer at high resolution. In MS/MS mode the linear ion trap only retains a chosen mass window, which is activated by a supplemental RF field leading to fragmentation of the trapped precursor ions, and records the signal of a mass dependent scan at low resolution. Note that the high resolution MS scan can be performed at the same time as the low resolution MS/MS scans in the linear ion trap. Recently, an improved linear ion trap Orbitrap analyzer combination termed “LTQ Orbitrap Velos” has been introduced (18). It features an S-lens with up to 10-fold improved ion transmission from the atmosphere, a dual linear ion trap, and a more efficient Higher energy Collisional Dissociation (HCD) cell interfaced directly to the C-trap (18). HCD fragmentation is similar to the fragmentation in triple quadrupole or quadrupole TOF instruments and its products are analyzed with high mass accuracy in the Orbitrap analyzer (19). Thus, the LTQ Orbitrap or LTQ Orbitrap Velos instruments offer versatile fragmentation modes depending on the analytical problem (20–22).
Fig. 1.
Mass spectrometers incorporating an Orbitrap analyzer. The Exactive is a standalone instrument without mass selection. The total ion population is collected in the C-trap and injected into the Orbitrap analyzer (see text and Fig. 2 for details on detector components). In the LTQ Orbitrap Velos combination, ions can be selected “in time” by mass selective scans in the linear ion trap. In CID mode, the LTQ and Orbitrap operate as separate mass spectrometers. In HCD mode its function is to isolate a particular precursor, which is then fragmented in the HCD cell. In contrast, in the Q Exactive mass selection is “in space” as ions of only a specified m/z range have stable trajectories and are transferred to the storage or fragmentation devices before Orbitrap analysis.
Taking advantage of the small size of the Orbitrap analyzer a standalone benchtop instrument termed “Exactive” has been introduced mainly for small molecule applications. However, because of the absence of mass selection, its use in proteomics is limited to non-mass selective fragmentation of the entire mass range (called “All Ion Fragmentation” (AIF) on this instrument (23)).
The combination of a quadrupole mass filter with an Orbitrap analyzer has not yet been reported. We reasoned that such a quadrupole trap combination might offer unique and complementary advantages to the hybrid mass spectrometers described above. In particular, a quadrupole Exactive instrument or “Q Exactive” would be able to select ions virtually instantaneously because of the fast switching times of quadrupoles and it would be able to fragment peptides in HCD mode on a similarly fast time scale. Furthermore, because of the small size and mature technology used in current quadrupole mass filters, this analyzer combination should have a small footprint and be particularly robust. Finally, the ability to separate “in space” and analyze MS and MS/MS ranges at high resolution in the Orbitrap analyzer offers the promise of enabling efficient multiplexed scan modes not currently applied in proteomics research using trapping instruments.
EXPERIMENTAL PROCEDURES
Construction of a Quadrupole Orbitrap Instrument
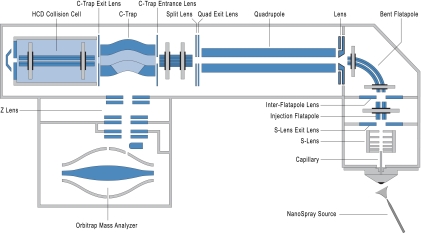
The Q Exactive instrument includes an atmospheric pressure ion source (API), a stacked-ring ion guide (S-lens) in the source region, a quadrupole mass filter, a C-trap, an HCD cell, and an Orbitrap mass analyzer as shown in Fig. 2. Ions are formed at atmospheric pressure (in this work in a nanoelectrospray ion source), pass through a transfer tube to an S-lens described in (18) and then via an injection multipole into a bent flatapole. The bent flatapole has 2-mm gaps between its rods, oriented in such a way that the line of sight from the S-lens is open for clusters and droplets to fly unimpeded out of the flatapole.
Fig. 2.
Construction details of the Q Exactive. This instrument is based on the Exactive platform but incorporates an S-lens, a mass selective quadrupole, and an HCD collision cell directly interfaced to the C-trap. Note that the drawing is not to scale.
After collisional cooling in the bent flatapole, ions are transmitted via a lens into a hyperbolic quadrupole (r0 = 4 mm), capable of isolating ions down to an isolation width of 0.4 Th at m/z 400. The quadrupole is followed by its exit lens combined with a split lens used to gate the incoming ion beam. A short octapole then brings ions into the C-trap interfaced to an HCD cell with axial field (18). The gas-filled HCD cell is separated from the C-trap only by a single diaphragm, allowing easy HCD tuning. Fragmentation of ions in the HCD cell is achieved by adjusting the offset of the RF rods and the axial field to provide the required collision energy. As long as this offset remains negative relative to the C-trap and the HCD exit lenses, all fragments remain trapped inside the HCD cell, even if the offset of the RF rods is varied. This allows to introduce multiple precursor ions and to fragment them at their optimum collision energy without compromising the storage of preceding injections. The summed ion population can then be transferred back into the C-trap, ejected into the Orbitrap analyzer and analyzed in a single Orbitrap detection cycle. This opens the possibility of fundamentally new, “multiplexing” modes of operation. In practice, the useful number of ion injections for a single Orbitrap detection is limited by the sum of the individual inject times being lower than the time for the Orbitrap scan.
A new challenge posed by interfacing the Orbitrap analyzer to a quadrupole is the automatic gain control (AGC) of weak ion signals. This problem was addressed by using an AGC pre-scan for a full MS spectrum with subsequent prediction of the ion currents for the weak signals on the basis of their share in total ion current (predictive AGC).
The mass range covered by the instrument is m/z 50–4000, with the range of mass selection reaching m/z 2500. Acquisition speed ranges from 12 Hz for resolving power 17,500 at m/z 200 (corresponding to 12,500 at m/z 400) to 1.5 Hz for resolving power 140,000 at m/z 200 (corresponding to 100,000 at m/z 400). Vacuum in the Orbitrap compartment is typically below 7 × 10−10 mBar, which makes the analyzer adequate for high resolution analysis of most analytes, including large peptides and small proteins.
The ability to fill the HCD cell or the C-trap with ions while a previous Orbitrap detection cycle is still ongoing is another important innovation that allows to significantly reduce the influence of low ion currents on acquisition speed and quality of spectra.
Processing of Transients Using Magnitude and Phase Information (eFT)
Transients detected in the Orbitrap mass analyzer are processed using an enhanced version of Fourier Transformation (eFT™) for conversion of transients into frequency and then m/z. Details of the technique can be found in (24). Both eFT and conventional FT make use of complex numbers, which can be represented by magnitude and phase, or by real and imaginary components. As the initial phase of the ion package appears to be dependent on initial parameters of the ions in a very complex way (25), FT spectra have to be presented in the so-called magnitude mode, which amounts to disregarding the phase information. However, in Orbitrap mass spectrometers the built-in excitation-by-injection mechanism (26) provides an initial phase of ion oscillations that is almost m/z independent. This synchronization allows converting spectra in such a way that the real component of data can be utilized, which results in narrower peaks. In practice, eFT uses a combination of the magnitude and the real component of the signal to improve mass accuracy and peak shape.
Better accuracy of synchronization is achieved, if detection starts as early as possible after ion injection. For this reason, modifications of preamplifier and Orbitrap analyzer were introduced to reduce the delay between ion injection and start of transient detection from almost 10 ms to a fraction of a millisecond.
Practical implementation of eFT achieves between 1.8- and 2-fold increase of resolving power for the same transient (except for rapidly decaying signals, for example from proteins, where the gain is reduced to about 1.4-fold because of “hard sphere” collisions with background gas). The dual-spectrum online processing is computationally demanding but still fast enough to be completed in the LC MS time scale. Thus cycle time is still determined by transient acquisition and ion injection times and not by processing of the data. The eFT method is sensitive to precise synchronization of the instrument electronics and remaining shot-to-shot jitter, so that final mass accuracy is comparable to that of traditional magnitude mode FT spectra. Side-lobes in eFT spectra are comparable to those in conventional FT spectra.
Preparation of HeLa Lysates
HeLa cells were lysed and the pellet was dissolved in a urea (6 m) and thiourea (2 m) solution. Proteins were reduced with dithiotreitol (1 mm) for 30 min at room temperature followed by alkylation with iodoacetamide (55 mm) for 20 min in the dark. The mixture was incubated with LysC (1 μg/50 μg protein) (Wako, Richmond, VA) at room temperature for 3 h before 1:4 dilution with water. Incubation with trypsin (1 μg/50 μg protein) (Promega, Madison, WI) was carried out for 12 h at room temperature. The digestion was stopped by addition of formic acid (3%). Organic solvent was removed in a SpeedVac concentrator. The peptide mixture was desalted on reversed phase C18 StageTips (27). Directly before analysis, peptides were eluted into 8 well autosampler vials with 60 μl buffer B (80% acetonitrile in 0.5% acetic acid). Organic solvent was removed in a SpeedVac concentrator and the final sample volume was adjusted with buffer A* (2% acetonitrile in 0.1% trifluoroacetic acid) to 12 μl.
LC MS/MS Analysis for Q Exactive and LTQ Orbitrap Velos
A nanoflow HPLC instrument (Easy nLC, Proxeon Biosystems, now Thermo Fisher Scientific) was coupled on-line to a Q Exactive or an LTQ Orbitrap Velos mass spectrometer (both from Thermo Fisher Scientific) with a nanoelectrospray ion source (Proxeon). Chromatography columns were packed in-house with ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH) in buffer A (0.5% acetic acid). The peptide mixture (5 μg) was loaded onto a C18-reversed phase column (15 cm long, 75 μm inner diameter) and separated with a linear gradient of 5–60% buffer B (80% acetonitrile and 0.5% acetic acid) at a flow rate of 250 nL/min controlled by IntelliFlow technology over 90 min. Because of loading and washing steps, the total time for an LC MS/MS run was about 40–50 min longer.
MS data was acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1650 Th) for HCD fragmentation. Target values on Q Exactive were similar to those typically used on an LTQ Orbitrap Velos. Determination of the target value is based on predictive Automatic Gain Control (pAGC) in both instruments. However, the LTQ Orbitrap Velos is equipped with electron multipliers, which allows scaling of the number of ions in a direct manner. In contrast, scaling of the number of ions is more indirect on the Q Exactive accounting for the difference in target values for the same S/N. Dynamic exclusion duration was 60 s with early expiration disabled on the LTQ Orbitrap Velos. Isolation of precursors was performed with a 4-Th window and MS/MS scans were acquired with a starting mass of 100 Th. Survey scans were acquired at a resolution of 70,000 at m/z 200 on the Q Exactive and 30,000 at m/z 400 on the LTQ Orbitrap Velos (see Results and Discussion and Table I for conversion of resolution values to different m/z values). Resolution for HCD spectra was set to 17,500 at m/z 200 on the Q Exactive and 7500 at m/z 400 on the LTQ Orbitrap Velos. Normalized collision energy was 30 eV for the Q Exactive and 35 eV for the LTQ Orbitrap Velos—they are not identical because of different scaling functions in the instrument software. The underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1% on the Q Exactive. For the LTQ Orbitrap Velos the lower threshold for targeting a precursor ion in the MS scans was 5,000 counts. Both instruments were run with peptide recognition mode enabled, but exclusion of singly charged and unassigned precursor ions was only enabled on the LTQ Orbitrap Velos. This was because of the higher sequencing speed of the Q Exactive and a slightly different precursor selection algorithm for the data-dependent scans. However, in practice there was not much difference between the settings with regard to the number of identified unique peptides and proteins.
Table I. Four Q Exactive resolution settings and transient times.
| Resolution @ m/z = 200 Th | Resolution @ m/z = 400 Th | Transient length |
|---|---|---|
| 17,500 | 12,500 | 64 ms |
| 35,000 | 25,000 | 128 ms |
| 70,000 | 50,000 | 256 ms |
| 140,000 | 100,000 | 512 ms |
To demonstrate multiplexing of selected ion monitoring (SIM) scans, a method alternating full scans and SIM scans over the entire gradient was set up on the Q Exactive. The 92 min range in which peptides eluted was divided into 23 segments of 4 min duration. For each of these segments, three SIM windows of 2 Th width were defined, centered around 69 randomly chosen, low abundance precursor ions observed in these elution time windows in a previous top10 run. Pre-selection of these low abundance peptides was carried out manually based on the msms.txt file resulting from MaxQuant analysis. The method for multiplexed SIM scans was specified using the “Targeted SIM” template in the Q Exactive method editor. Resolution was set to 140,000 at m/z 200 and a target value of 1e6 ions for both scan types was chosen. The maximum ion injection time was set to 10 ms for the full scan and to 100 ms for each of the multiplexed SIMs. The inclusion list was saved in the global list features and in the data-dependent settings page “inclusion” was set to “on.” Multiplexing of MS/MS spectra was done in exactly the same format as the standard top10 method, except that “msx” in the method setup of the data-dependent scans was set to 2 for multiplexing the fragment ions of two consecutively selected precursors.
Analysis of Proteomic Data
The mass spectrometric raw data from top10 methods were analyzed with the MaxQuant software (developmental version 1.1.1.32) (28). The false discovery rate (FDR) was set to 0.01 for proteins and peptides, which had to have a minimum length of 6 amino acids. MaxQuant was used to score peptides for identification based on a search with an initial allowed mass deviation of the precursor ion of up to 7 ppm. The allowed fragment mass deviation was 20 ppm. Search of the MS/MS spectra against the International Protein Index human data base (version 3.68, 87,061 entries) combined with 262 common contaminants was performed using the Andromeda search engine (29). Enzyme specificity was set as C-terminal to Arg and Lys, also allowing cleavage at proline bonds and a maximum of two missed cleavages. Carbamidomethylation of cysteine was set as fixed modification and N-terminal protein acetylation and methionine oxidation as variable modifications. MaxQuant applied time-dependent recalibration to the precursor masses for improved mass accuracy. Further analysis of the data provided by MaxQuant was performed in the R scripting and statistical environment (30). The data sets used for analysis are deposited at Tranche (www.proteomecommons.org).
RESULTS AND DISCUSSION
Our goal was to construct a high performance quadrupole Orbitrap mass spectrometer in a compact format. Details of the hardware are in Experimental Procedures but here we give a brief overview. We started by building on the Exactive platform. The Exactive does not have mass selection capability and was developed mainly for small molecule applications (31). However, it can be equipped with a higher energy collisional dissociation cell (HCD) at the far side of the C-trap. Thus the detection system of the Exactive already allows HCD fragmentation (19) albeit without mass selection. This mode is called “AIF” for All Ion Fragmentation on this instrument and can also be used in proteomics (23). To support mass selective MS/MS scans in the Q Exactive, the transmission from electrospray source to vacuum was increased up to 10-fold, for which we used the S-lens employed in the LTQ Orbitrap Velos (18). New, rapidly switching electronics systems controlling the instrument were incorporated. Apart from some inlet ion optics changes the Orbitrap analyzer is the same as in previous Orbitrap analyzers. The Orbitrap voltage is 5 kV as it is on the Exactive and therefore higher than the 3.5 kV on LTQ Orbitrap instruments. The Q Exactive also employs a 90° bent ion path from the source toward the mass analyzer in common with the Exactive and in contrast to the LTQ Orbitrap instruments. The defining difference of the Q Exactive compared with the Exactive is the presence of a mass selective quadrupole analyzer between the ion source and the C-trap (Fig. 2). This quadrupole is the same as that used in triple quadrupole Access instruments, however, it features a modified RF-generator capable of driving selection of wide mass selection windows.
From a practical point of view, maintenance of the Q Exactive is similar to that of the Exactive. The quadrupole mass filter has very few tunable parameters and the instrument is automatically calibrated in a few minutes.
Mass Spectrometric Resolution
In the analysis of complex mixtures, peptides of similar mass often co-elute and therefore resolution is a key parameter of a mass spectrometer in these applications (7). Shotgun proteomics on the LTQ Orbitrap instruments is usually performed with 30,000 or 60,000 resolution at m/z 400. (Note that resolution decreases with the square root of the m/z value in Orbitrap analyzers.) High intrinsic resolution of an instrument allows short transients and hence short cycle times in topN methods—facilitating deep coverage of the proteome.
Because of the higher voltage of the Q Exactive, resolution at the same transient length is 20% higher. More importantly, we here employ eFT of the transients, which boosts resolution by a factor 1.8 to 2.0 (for further explanation see Experimental Methods). A similar principle has recently been described by Marshall and coworkers for FT ICR (32).
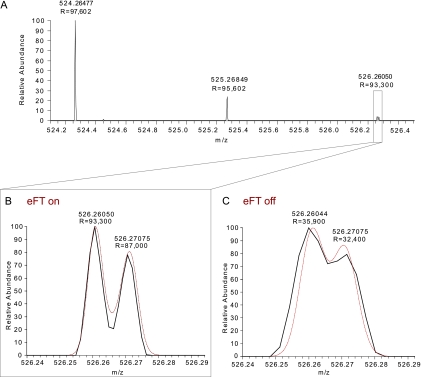
In Fig. 3, the resolution of the Q Exactive is demonstrated for the tetra peptide MRFA. As apparent from the widths of the isotope peaks and by their spacing, a resolution of more than 90,000 was achieved at m/z 524. The figure also illustrates the effect of turning the eFT algorithm on and off (although in normal operation of the Q Exactive, eFT is always on). With eFT enabled, the instrument clearly resolves the two isotopes of the same nominal mass that are because of two 13C carbon atoms or the sulfur atom contained in methionine (13C2 versus 34S).
Fig. 3.
Resolution of the Q Exactive using eFT. A, Isotope cluster of the MRFA peptide from a mass scan with a 512 ms transient employing eFT. B, Zoom into A demonstrating resolution of the 13C2 isotope from the 34S isotope. The red curve is the simulated signal for MRFA. C, The same isotopes as in B measured with the same transient but without enabling eFT.
In the Exactive instrument, resolution is specified at m/z 200 because of its small molecule applications and this convention is kept in the Q Exactive. We provide a table to aid comparison between resolution values at m/z 200 and m/z 400 used with the LTQ Orbitrap instruments for the four possible transient lengths on the Q Exactive (Table I). The standard 60,000 resolution scan on the LTQ Orbitrap instruments uses a 768 ms transient. On the Q Exactive, a resolution of 100,000 (at m/z 400) is reached with a 512 ms transient. We use a resolution of 50,000 (at m/z 400), corresponding to a 256 ms transient length as a standard in proteomics experiments. This resolution is only slightly lower than that normally used on LTQ Orbitrap instruments, but takes less than half of the time. For MS/MS experiments we employ a resolution of 17,500 at m/z 200 (12,500 at m/z 400), which is achieved with a transient length of 64 ms. This value is substantially higher than the 7,500 resolution (at m/z 400) typical for HCD experiments on the LTQ Orbitrap Velos. The higher resolution in MS/MS spectra helps in assigning fragments of large precursors, however, the 64 ms transient was mainly chosen because even shorter transients would decrease the signal to noise in the MS/MS spectra.
Cycle Times for MS and MS/MS Analysis
As the quadrupole only serves as a selection device, the Q Exactive cannot perform MS and MS/MS operations in parallel (Fig. 1). On the other hand, the Q Exactive—unlike the LTQ Orbitrap Velos—fills ions in parallel to Orbitrap transient acquisition. Therefore, we next tested its overall cycle times and compared them to other Orbitrap instruments.
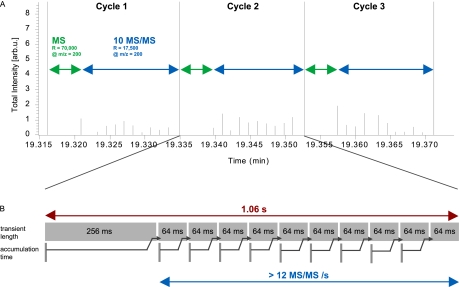
For the analysis of complex peptide mixtures, topN experiments consisting of a survey scan followed by N MS/MS scans are typically performed. Depending on the complexity of the mixture, N is usually between three and 20, and top10 is a widely used standard method. As explained above, a transient of 256 ms results in a resolution of 50,000 (at m/z 400), which is appropriate for proteomic applications. We combined this survey scan with ten 64 ms MS/MS scans (resolution 12,500 at m/z 400). If accumulation of the ions to the desired target count happened entirely in parallel with transient detection and if there was no overhead, this method would take 896 ms. The actually measured time for this sequence was 1.06 s, indicating that all overhead times together amounted only to about 160 ms (Fig. 4). This figure even included the automatic gain control scan performed before each full scan (whereas fill times for MS/MS scans are determined by “predictive AGC”).
Fig. 4.
Cycle times for a top10 method on the Q Exactive. A, Large ticks represent the total cycle consisting of MS and MS/MS scans. Duration for the MS survey scans is indicated by the green arrows (resolution 70,000 at m/z 200 or 50,000 at m/z 400) and for the MS/MS scans by the blue arrows (resolution 17,500 at m/z 200 or 12,500 at m/z 200). The x axis indicates chromatographic elution time and the y axis the total spectral intensity. B, Total cycle time for a top10 method is about 1 s and fragmentation frequency is more than 12 Hz. The lower trace indicates parallel ion accumulation for the following scan. Note that peptide ion accumulation times for typical LC column loads are generally shorter than transient times (as indicated in this example) and that they therefore do not add to cycle times.
Completion of a full top10 method in about 1 s is exceedingly fast and compares favorably with top10 HCD methods on a Velos instrument. For example, the number of HCD spectra in such a Velos based method in a recent study was 3.3 MS/MS/s over the entire gradient (4). Even compared with top10 CID methods on the Velos instrument, which have the advantage of parallel acquisition, the Q Exactive proved to be faster in our hands (18). The reasons for the very fast cycle times are fivefold: (1) eFT allows using short transient times (2) ion filling is done in parallel with detection (3) overhead times of electronics components have been minimized (4) precursor selection is done “in space” in a few ms and (5) HCD peptide fragmentation is nearly instantaneous.
Because of the parallel ion accumulation in the Q Exactive, fill times shorter than the transient length do not affect the overall cycle time. In our experiments with complex mixtures (see below), fill times for full scans were in the range of 1 to 10 ms, and for MS/MS scans they were generally between 5 and 50 ms leading to completely parallel acquisition and detection in almost all cases. The fill times observed here are similar to those of the LTQ Orbitrap Velos (18), indicating comparable sensitivity of both instrument types in full scan and HCD MS/MS mode.
Q Exactive Performance for Proteome Analysis
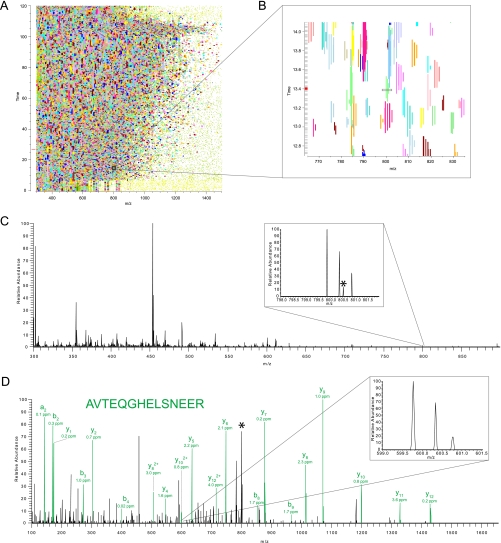
To characterize the performance of the Q Exactive for shotgun proteomics, we prepared a digest of a mammalian cell line (Experimental Methods). The peptide mixture was separated by on-line HPLC in a 90 min gradient by standard methods used in our laboratories. The entire analysis was done in triplicate and for comparison it was also performed on an LTQ Orbitrap Velos. Fig. 5A shows a heat map of the MS signals generated by peptides eluting from the column over the 90 min. The inset in the heat map is a zoom of a typical region (Fig. 5B), showing the complexity of eluting isotope patterns in this peptide mixture derived from whole cell lysate. As can be seen on the left hand scale, MS scans occurred every second and consequently eluting peptide peaks were well sampled. The MS spectrum in Fig. 5C depicts a single MS scan intersecting the zoomed region and indicates a triply charged precursor that was selected for fragmentation. Note that on the Q Exactive all fragmentation is performed by HCD and MS/MS spectra are always acquired with high resolution. This enables unambiguous recognition of charge states as illustrated in Fig. 5D and high fragment mass accuracy.
Fig. 5.
Proteome analysis with the Q Exactive. A, Heat map of an LC MS/MS run of a peptide mixture resulting from proteolytic digestion of a HeLa lysate. B, Zoom of a typical part of the heat map. Marks on the left hand side represent the MS survey scans of each MS and MS/MS cycle and are separated by 1 s. C, Survey spectrum showing 50,000 resolution (at m/z 400) and the isotope pattern of a triply charged precursor in green; the asterisk indicates a co-eluting precursor ion. D, MS/MS spectrum of the precursor shown in C with 12,500 resolution (at m/z 400) and zoom of a doubly charged fragment ion.
The data were analyzed in MaxQuant with the integrated Andromeda search engine (28, 29). Table IIA lists the results of the database search of the Q Exactive data. The total number of MS scans was in excess of 5,000 and the total number of MS/MS scans in excess of 35,000. (Note that top10 sequencing is only performed when there are sufficient peptide candidates in the MS scan that meet selection criteria for fragmentation.) The number of isotope patterns detected was close to 150,000, a very high number considering that the gradient was not particularly long (4, 5), presumably because of the short MS and MS/MS cycle time of 1 s. On average 12,563 unique peptides were identified in each run, for a total of 16,255 peptides in the triplicate analysis. These peptides mapped to an average of 2,557 proteins per run, and a total of 2,864 proteins of the HeLa proteome with the three 90 min gradients (supplemental Tables S1–S5).
Table II. A, Peptide identification from HeLa lysate triplicate analysis on a Q Exactive (90 min gradient).
| MS spectra | MSMS spectra | Identifications [%] | Unique peptides | Proteins | Isotope clusters | |
|---|---|---|---|---|---|---|
| HeLa (1) | 5427 | 35203 | 37.23 | 12298 | 2513 | 146138 |
| HeLa (2) | 5098 | 35911 | 38.35 | 12830 | 2601 | 143556 |
| HeLa (3) | 5274 | 35348 | 38.23 | 12560 | 2557 | 144336 |
| Σ Triplicates | 37.94 | 16255 | 2864 |
| B, Peptide identification from HeLa lysate triplicate analysis on an LTQ Orbitrap Velos (90 min gradient) | ||||||
|---|---|---|---|---|---|---|
| MS spectra | MSMS spectra | Identifications [%] | Unique peptides | Proteins | Isotope clusters | |
| HeLa (1) | 2012 | 19818 | 55.64 | 10420 | 1895 | 125738 |
| HeLa (2) | 2102 | 19103 | 56.64 | 9855 | 1843 | 120553 |
| HeLa (3) | 2005 | 19634 | 56.37 | 10347 | 1906 | 126717 |
| Σ Triplicates | 56.21 | 14401 | 2242 | |||
For comparison, we performed the same analysis on an LTQ Orbitrap Velos. As expected, significantly more unique peptides were identified by the Q Exactive in the single LC runs (12,253 versus 10,207 on average; increase of 23%). Thus the Q Exactive, despite its compact format, represents an advance in the analysis of complex peptide mixtures as typically analyzed in shotgun proteomics. Note, however, that the above comparison only considers relatively short analysis of very complex mixtures and compares HCD fragmentation on both instruments. A detailed comparison of the two instrument types would require additional experiments and should also take into account that the Velos instrument can perform high or low resolution CID, in addition to HCD. Furthermore, the LTQ Orbitrap Velos, unlike the Q Exactive, is available with an ETD unit thereby providing a complementary fragmentation approach.
Multiplexing at the MS and MS/MS Levels
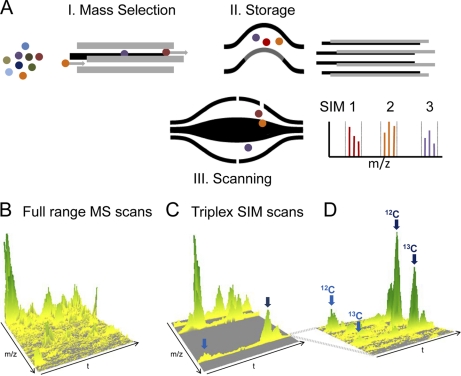
The linear ion trap Orbitrap analyzer combination is very versatile because it is comprised of two fully functional mass spectrometers. This allows isolation and fragmentation in different parts of the instruments and offers considerable flexibility in combining isolation and fragmentation events. Although many of these operation modes are not possible on the Q Exactive, it turns out that this novel combination of a mass filter and an Orbitrap analyzer also enables unique scan events. The principle feature making these scan modes possible is the fact that mass selection occurs “in space” which is extremely fast (Fig. 1). This should allow almost arbitrarily complex “mixing and matching” of MS and MS/MS mass ranges followed by high resolution analysis in the Orbitrap analyzer.
Fig. 6 illustrates two such multiplexed scan modes, one at the MS level and one at the MS/MS level. In selected ion monitoring (SIM) scans, a narrow mass range is accumulated providing increased signal to noise for particular ions of interest. SIM scans are useful in many applications but they are not often performed on Orbitrap instruments. This is because (1) the isolation of the SIM mass range in the linear ion trap is relatively time consuming, (2) there is a space charge limit on the number of ions that can be cleanly isolated, and (3) the analysis of even a single SIM scan takes considerable time. With few exceptions such as the lock mass injection to correct the mass scale (33), multiple mass range filling of the Orbitrap has not been implemented. The Q Exactive does not have the above limitations and, for example, allows selecting several SIM mass ranges of interest (Fig. 6A). In this mode, the C-trap is used as a storage device, which is filled with the desired number of ions from up to ten different SIM windows. These ions are together injected into the Orbitrap analyzer and measured in the same way as full mass ranges. Because fill times are typically much shorter than MS transient times, multiplexed SIM scans use Orbitrap instrument time much more efficiently. We demonstrate this concept in Fig. 6B–6D where a complete HeLa cell lysate was run in a 90 min gradient. (Note that such a gain does not occur on TOF instruments because they are not limited by scan times.) Three peptides known to be of low abundance from a previous top10 run were selected to define mass ranges (LTGMAFRVPTANVSVVDLTCR, DMIILPEMVGSMVGVYNGK, DAATIMQPYFTSNGLVTK). Fig. 6B represents the full range MS scans. In Fig. 6C and 6D, depicting a zoom into one of the SIM windows, the peptides are clearly visible with very good signal to noise. The ion injection time of most full scan was less than 1 ms, whereas ions for each of the SIM scans were accumulated for 100 ms accounting for the drastically improved signal-to-noise. At this high sensitivity, other peptides emerged from the background but were clearly resolved from the targeted peptide. These three multiplexed SIM windows were analyzed together in 140,000 resolution scans (0.5 s), adding little to the overall cycle time. Switching time to position the quadrupole at each mass window was 6 ms. Clearly such multiplexed SIMs could play an important role in targeted peptide analysis and peptide quantification.
Fig. 6.
Multiplexing at the MS level. A, The quadrupole mass filter is set to transmit a specific SIM mass range. After accumulation of the desired number of ions, the quadrupole is rapidly switched to the next SIM window up to the total number of SIM windows to be monitored. The combined SIM ranges are analyzed together in one high-resolution scan as shown in cartoon form in the inset. B, Three-dimensional representation of a 4-min segment of full range MS scans from a 90 min LC run of HeLa peptides. C, Visualization of the data from triplex scans acquired directly after each full scan in the same segment. Signals of the targeted low abundant peptides are clearly visible in the three SIM scans in C but virtually absent in the full scans in B. D, Zoom of one of the SIM ranges in C show that the SIM range contains peptides of even lower abundance than the targeted one marked with the dark blue arrow.
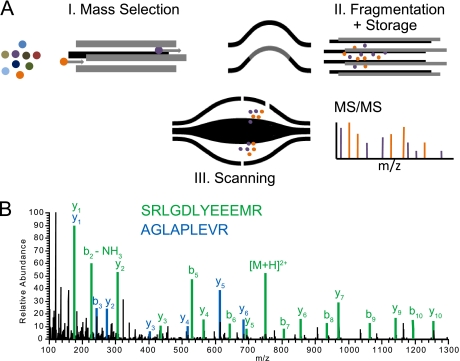
In complex mixture analysis, sequencing speed can be a limiting factor. In principle, fragmentation of several precursors with simultaneous recording of the fragments can further boost the number of analyzable MS/MS events per unit time. Although in principle possible with a linear ion trap, in practice the necessary multiple rounds of peptide isolation and fragmentation preclude such an option on the LTQ Orbitrap. The Q Exactive, however, can successively isolate different precursor ions and fragment them in the HCD cell each at an individual normalized collision energy suitable for its properties. As each population of precursor ions is only fragmented once during injection into the HCD cell, all fragment ions are stored successively in the HCD cell and then they undergo joint analysis in the Orbitrap analyzer (see Fig. 7A). To demonstrate this, we specified a top10 method with multiplex degree of two. Fig. 7B shows a representative example of a multiplexed MS/MS spectrum analyzed at the normal MS/MS resolution setting of 12,500 (at m/z 400). Visual inspection of the spectrum clearly reveals extensive sequence information from both sequences. Interpretation of these deliberately multiplexed spectra, as opposed to cofragmented precursors, is aided by the fact that the same number of ions can be fragmented for each targeted precursor. Although it is clear that multiplexed MS/MS scans are easily possible on the Q Exactive platform, further investigations will be necessary to determine any resulting gain in peptide identifications. However, increased sequencing speed is not the only application of multiplexed MS/MS. As an example, the fragmentation of different charge states of multiply charged proteins could yield interesting structural information.
Fig. 7.
Multiplexing at the MS/MS level. A, Different precursor ions are mass selected in the quadrupole, fragmented in turn by HCD and stored in the HCD cell. The combined fragment populations are measured together in the Orbitrap analyzer (depicted in cartoon form in the inset) B, Duplexed MS/MS spectrum.
Conclusions and Outlook
We have described the construction and initial performance evaluation of a new type of mass spectrometer, the combination of a quadrupole mass filter with the Orbitrap analyzer. The quadrupole is one of the most robust and mature mass filters. Its combination with the relatively recently introduced Orbitrap mass spectrometer allowed realization of a high performance instrument with a small footprint and straightforward operation. These characteristics make the instrument an interesting addition to the proteomics toolbox, especially as proteomics is performed more and more by nonspecialist groups with biological or biomedical background.
Performance of the Q Exactive for complex peptide mixtures compares well with current LTQ Orbitrap instruments such as the LTQ Orbitrap Velos. Although the Q Exactive only offers the HCD fragmentation mode, we have shown here that HCD speed and sensitivity are not limiting. In fact, parallel filling of the ions combined with nearly instantaneous ion selection and fragmentation allowed implementation of a top10 method with 1-s cycle times. In comparison, the MS/MS scan rate of quadrupole TOF instrumentation could reach nominal speed up to 50 MS/MS per second, but, because of the lower transmission of TOF, signal to noise in each scan will be severely compromised unless high sample loads are used. An interesting novel feature of the Q Exactive is its ability to multiplex MS and MS/MS mass ranges, almost without limitations, which we have demonstrated here with two examples: multiplexed SIM mass ranges and multiplexed MS/MS spectra. In contrast to quadrupole TOF instrumentation, where scan speed in SIM mode is limited mainly by the time needed to reach acceptable signal to noise ratio, multiplexed SIM scans allow decoupling spectral acquisition speed from the speed of acquiring SIM scans and thus to utilize the full high transmission to the Orbitrap analyzer. We anticipate that the Q Exactive will enable additional interesting multiplexing capabilities in the future.
Acknowledgments
We thank our colleagues at Thermo Fisher Scientific, especially Ulf Froehlich, Andreas Kuehn, and Florian Grosse-Coosmann, and at the Max Planck Institute, especially Nadin Neuhauser, Tami Geiger, and Richard Scheltema, for help and fruitful discussions.
Footnotes
* This project was supported by the European Commission's 7th Framework Program PROteomics SPECificat ion in Time and Space (PROSPECTS, HEALTH-F4-2008-021,648).
 This article contains supplemental Tables S1 to S4.
This article contains supplemental Tables S1 to S4.
Data availability: Supplementary data is available with this publication at the MCP web site. Raw MS files are uploaded to Tranche (www.proteomecommons.org) as “Michalski et al. Q Exactive” Hash code to access 8 RAW files: Nbh0v8NbSxDGuN/qunMhsCz2z+rNP6YtKM1/uW2r2a2FEt9fUWESQH5XE1mGzU1BsPxpVWSlHtfTeeufl2hkW54eL54AAAAAAAAIDQ ==.
1 The abbreviations used are:
- TOF
- time-of-flight
- AIF
- all ion fragmentation
- CID
- collision induced dissociation
- ETD
- electron transfer dissociation
- FDR
- false discovery rate
- FT
- Fourier transform
- HCD
- higher energy collisional dissociation
- HPLC
- high performance liquid chromatography
- LTQ
- linear trap quadrupole
- MS/MS
- tandem mass spectrometry
- pAGC
- predictive automatic gain control
- RF
- radio frequency
- SIM
- selected ion monitoring.
REFERENCES
- 1. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 3. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michalski A., Cox J., Mann M. (2011) More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 5. Köcher T., Swart R., Mechtler K. (2011) Ultra-High-Pressure RPLC Hyphenated to an LTQ-Orbitrap Velos Reveals a Linear Relation between Peak Capacity and Number of Identified Peptides. Anal. Chem. 83, 2699–2704 [DOI] [PubMed] [Google Scholar]
- 6. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 7. Mann M., Kelleher N. L. (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. U. S. A. 105, 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U. S. A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louris J. N., Cooks R. G., Syka J. E. P., Kelley P. E., Stafford G. C., Todd J. F. J. (1987) Instrumentation, applications, and energy deposition in quadrupole ion-trap tandem mass-spectrometry. Anal. Chem. 59, 1677–1685 [Google Scholar]
- 12. Hardman M., Makarov A. A. (2003) Interfacing the orbitrap mass analyzer to an electrospray ion source. Anal. Chem. 75, 1699–1705 [DOI] [PubMed] [Google Scholar]
- 13. Makarov A. (2000) Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal. Chem. 72, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 14. Scigelova M., Makarov A. (2006) Orbitrap mass analyzer–overview and applications in proteomics. Proteomics 6, 16–21 [DOI] [PubMed] [Google Scholar]
- 15. Syka J. E., Marto J. A., Bai D. L., Horning S., Senko M. W., Schwartz J. C., Ueberheide B., Garcia B., Busby S., Muratore T., Shabanowitz J., Hunt D. F. (2004) Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 [DOI] [PubMed] [Google Scholar]
- 16. Makarov A., Denisov E., Lange O., Horning S. (2006) Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 17, 977–982 [DOI] [PubMed] [Google Scholar]
- 17. Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., Horning S. (2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 18. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 20. Macek B., Waanders L. F., Olsen J. V., Mann M. (2006) Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol. Cell Proteomics 5, 949–958 [DOI] [PubMed] [Google Scholar]
- 21. McAlister G. C., Berggren W. T., Griep-Raming J., Horning S., Makarov A., Phanstiel D., Stafford G., Swaney D. L., Syka J. E., Zabrouskov V., Coon J. J. (2008) A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J. Proteome Res. 7, 3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McAlister G. C., Phanstiel D. H., Westphall M. S., Coon J. J. (2011) Higher-energy collision-activated dissociation without a dedicated collision cell. Mol. Cellular Proteomics, 10(5):O111.009456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geiger T., Cox J., Mann M. (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell Proteomics 9, 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lange O., Makarov A., Denisov E., Balschun W. (2010) Accelerating spectral acquisition rate of Orbitrap mass spectrometry. Proc. 58th Conf. Amer. Soc. Mass Spectrom [Google Scholar]
- 25. Vining B. A., Bossio R. E., Marshall A. G. (1999) Phase correction for collision model analysis and enhanced resolving power of fourier transform ion cyclotron resonance mass spectra. Anal. Chem. 71, 460–467 [DOI] [PubMed] [Google Scholar]
- 26. Makarov A. (2009) In: March R. E., Todd J. F. J., eds. Practical Aspects of Trapped Ion Mass Spectrometry. Vol 4: Theory and Instrumentation, CRC Press; (Taylor & Francis) [Google Scholar]
- 27. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 11, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 30. Ihaka R., Gentleman R. (1996) R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314 [Google Scholar]
- 31. Bateman K. P., Kellmann M., Muenster H., Papp R., Taylor L. (2009) Quantitative-qualitative data acquisition using a benchtop Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 20, 1441–1450 [DOI] [PubMed] [Google Scholar]
- 32. Beu S. C., Blakney G. T., Quinn J. P., Hendrickson C. L., Marshall A. G. (2004) Broadband phase correction of FT-ICR mass spectra via simultaneous excitation and detection. Anal. Chem. 76, 5756–5761 [DOI] [PubMed] [Google Scholar]
- 33. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]