Abstract
Serial homologs are similar structures that develop at different positions within a body plan. These structures share some, but not all, aspects of developmental patterning, and their evolution is thought to be constrained by shared, pleiotropic gene functions. Here we describe the functions of 17 developmental genes during metamorphic development of the legs in the red flour beetle, Tribolium castaneum. This study provides informative comparisons between appendage development in Drosophila melanogaster and T. castaneum, between embryonic and adult development in T. castaneum, and between the development of serially homologous appendages. The leg gap genes Distal-less and dachshund are conserved in function. Notch signaling, the zinc-finger transcription factors related to odd-skipped, and bric-à-brac have conserved functions in promoting joint development. homothorax knockdown alters the identity of proximal leg segments but does not reduce growth. Lim1 is required for intermediate leg development but not distal tarsus and pretarsus development as in D. melanogaster. Development of the tarsus requires decapentaplegic, rotund, spineless, abrupt, and bric-à-brac and the EGF ligand encoded by Keren. Metathoracic legs of T. castaneum have four tarsomeres, whereas other legs have five. Patterns of gene activity in the tarsus suggest that patterning in the middle of the tarsal region, not the proximal- or distal-most areas, is responsible for this difference in segment number. Through comparisons with other recent studies of T. castaneum appendage development, we test hypotheses for the modularity or interdependence of development during evolution of serial homologs.
Keywords: appendage patterning, adult development, metamorphosis, developmental constraint, serial homology
Goethe (1790) and Owen (1848) recognized the importance of serial homology in plants and vertebrates, respectively, long before Darwin (1859) highlighted this phenomenon in support of evolution by natural selection. Anatomical duplication results in serial homologs, similar morphological structures that are repeated at different positions within an organism’s body plan, such as the appendages of arthropods. Serial homologs have widely divergent forms. Owen saw these “teleological modifications” as deviations (in his words, “adaptive masks”) of a group’s archetype, but Darwin claimed differences in serial homologs as evidence of adaptation through descent with modification. Among arthropods, modifications of the appendages, which form as extensions from the body, have generated structures adapted for sensation, feeding, and locomotion. More recently, duplication and diversification have emerged as crucial processes in evolution at the level of genes (Lynch and Force 2000) and genomes (Dehal and Boore 2005; Ohno 1970) as well as anatomy (Boxshall 2004; Snodgrass 1935).
Arthropod ventral appendages are organized with a proximal-to-distal (PD) axis with jointed segments. Among arthropods, ventral appendages share many aspects of developmental patterning (Abzhanov and Kaufman 2000; Angelini and Kaufman 2005b; Beermann et al. 2001; Inoue et al. 2002; Jockusch et al. 2000; Palopoli and Patel 1998; Panganiban et al. 1994; Prpic and Damen 2009; Rogers et al. 2002; Schoppmeier and Damen 2001). Nevertheless, appendage morphologies have diversified across body segments and species, and even in cases of conserved anatomy, divergent developmental genetic processes have been found (Angelini and Kaufman 2005a; Jockusch et al. 2000, 2004; Ronco et al. 2008; see also True and Haag 2001).
Here we evaluate two models for the evolution of developmental mechanisms controlling serially homologous appendages. The first model assumes that pleiotropic functions act as a strong constraint because mutations result in concerted changes in gene function that affect all appendage types (dependent model). The alternative model posits that pleiotropy is easily broken and that gene functions may evolve independently between appendages. These models represent ends of a spectrum and are not necessarily exclusive. A general understanding of the prevalence of dependent or independent evolution in the development of serial homologs is possible through a survey of developmental mechanisms in the appendages of multiple species. To test these hypotheses, we have examined the function of 17 candidate regulatory genes during metamorphosis in the legs of the red flour beetle, Tribolium castaneum (Table 1), and we consider these results in comparison with studies of other appendages in T. castaneum and the fruit fly Drosophila melanogaster.
Table 1 . Candidate genes.
| Gene Name | Symbol | Protein Class | LG | GenBank | Clone Source |
|---|---|---|---|---|---|
| decapentaplegic | dpp | TGFβ ligand | 4 | NM_001039451 | Sanchez-Salazar et al. 1996 |
| Distal-less | Dll | homeobox TXF | 7 | NM_001039439 | Jockusch et al. 2004 |
| dachshund | dac | Ski/Sno-related TXF | 4 | XM_964678 | Prpic et al. 2001 |
| homothorax | hth | homeobox TXF | 7 | NM_001039400 | Angelini and Kaufman 2004 |
| Lim1 | homeobox TXF | 6 | XM_964391 | Angelini et al. 2009 | |
| Notch | N | Notch receptor | 10 | NM_001114381 | " |
| Serrate | Ser | Delta/Serrate-type EGF | 7 | XM_964393 | " |
| Delta | Dl | Delta/Serrate-type EGF | X | XM_964994 | Current study |
| odd-skipped | odd | Zn-finger TXF | 8 | XM_966993 | Angelini et al. 2009 |
| brother of odd with entrails limited | bowl | Zn-finger TXF | 8 | XM_967045 | " |
| sister of odd and bowl | sob | Zn-finger TXF | 8 | XM_966942 | " |
| drumstick | drm | Zn-finger TXF | 8 | XM_966887 | " |
| Keren | Krn | EGF ligand | 3 | XM_001813564 | " |
| bric-a-brac | bab | BTB/Psq TXF | 3 | XM_001812888 | " |
| abrupt | ab | BTB/Zn-finger TXF | 5 | XM_969854 | " |
| rotund | rn | Zn-finger TXF | ? | XM_966094 | Current study |
| spineless | ss | bHLH/PAS TXF | 10 | XM_962783 | Angelini et al. 2009 |
The chromosomal linkage group (LG) is listed, as well as the GenBank accession number of the known or predicted transcript. bHLH, basic helix-loop-helix; BTB, Bric-a-brac/Tramtrak/Broad complex domain; EGF, epidermal growth factor; PAS, Per/Arnt/Sim domain; Psq, pipsqueak; TXF, transcription factor.
Studies of leg development in the fruit fly D. melanogaster (reviewed by Angelini and Kaufman 2005b; Kojima 2004) provide a useful model for considering appendage development in other insect species and appendage types. In D. melanogaster, broad domains along the PD axis of the leg imaginal disc are established by gradients of secreted signaling molecules encoded by wingless and decapentaplegic (dpp) (Diaz-Benjumea et al. 1994; Lecuit and Cohen 1997; Wu and Cohen 1999). The distal “gap gene” in the leg disc is Distal-less (Dll) (Cohen and Jürgens 1989); dachshund (dac) is required for development of intermediate structures (Mardon et al. 1994); and proximal appendage development depends on the function of homothorax (hth) (Casares and Mann 2001). The transcription factors bric-à-brac 1 and bric-à-brac 2 (collectively bab) are encoded by paralogous, functionally redundant genes (Couderc et al. 2002) and are required for distal segmentation (Chu et al. 2002; Godt et al. 1993). The position of bab expression in the distal leg is influenced by repression from odd-skipped (odd) and its paralog brother of odd with entrails limited (bowl), which are expressed in the regions of the first (t1) and fifth tarsomeres (t5) (de Celis Ibeas and Bray 2003). Meanwhile, during the mid-third instar, other genes specific to distinct, presumptive PD regions become expressed in the leg disc, such as spineless (ss) and rotund (rn) from t2 to t4 (Duncan et al. 1998; St. Pierre et al. 2002). Epidermal growth factor (EGF) signaling is activated by Dll in cells giving rise to t5 and the pretarsus (Galindo et al. 2002). EGF activity is also key to activating genes responsible for pretarsus development, such as the homeobox transcription factor Lim1 (Campbell 2005; Galindo et al. 2002; Kojima et al. 2005; Tsuji et al. 2000). The formation of joints is directed by Notch signaling and expression of the Notch ligands encoded by Serrate (Ser) and Delta results from the interaction of many of these genes (Bishop et al. 1999; de Celis et al. 1998; Greenberg and Hatini 2009; Rauskolb 2001; Rauskolb and Irvine 1999). These events pattern a leg with a single PD axis comprised of six primary segments (podomeres) separated by joints (the coxa, trochanter, femur, tibia, tarsus—which is subdivided into annuli called tarsomeres—and the pretarsus). This leg structure is conserved across insects.
In most Holometabola, adult appendages develop from fully functional external larval appendages, which are morphologically similar to the adult appendages (Svacha 1992; Tanaka and Truman 2005). Larval development is highly modified in Brachycera (Daly et al. 1998), such as D. melanogaster, meaning that some aspects of appendage morphogenesis in flies may not be representative of most insects. The appendages of D. melanogaster are suppressed in larval instars (Keilin 1915), and adult appendages develop from discs of presumptive imaginal tissue. Growth and patterning of these appendage primordia occurs internally during larval stages, and at metamorphosis the discs evert to form the adult appendages. Unlike the legless larvae of Brachycera, larvae of the tenebrionid beetle Tribolium castaneum have well-developed legs divided into five segments: coxa, trochanter, femur, tibiotarsus, and pretarsus (Figure 1B′; Sokoloff 1972). Metamorphosis involves an increase in leg size, segmentation within the tibiotarsus, and major changes in the shape and sensillae of appendages. Work in Tenebrio molitor, another tenebrionid beetle, has shown that the entire larval leg epidermis contributes to the adult leg epidermis. Each larval leg segment gives rise to the corresponding adult segment, with the exception that the larval trochanter gives rise to the proximal femur as well as the adult trochanter (Huet and Lenoir-Rousseaux 1976).
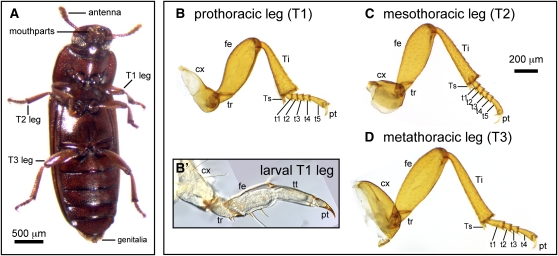
Figure 1 .
The ventral appendages of Tribolium castaneum include the antennae, mouthparts (mandibles, maxillae, and labium), thoracic legs, and genitalia. (A) Ventral view of an adult female. The thoracic legs of T. castaneum are roughly similar, with six true adult podomeres: coxa, trochanter, femur, tibia, tarsus, and pretarsus. The tarsus is subdivided by joints into tarsomeres, which lack independent muscles. The prothoracic leg (B) and mesothoracic leg (C) have five tarsomeres, whereas the metathoracic leg has four tarsomeres (D). The coxae of each type of leg have distinct shapes and sizes. (B′) Larval legs consist of five main segments: the coxa, trochanter, femur, tibiotarsus, and pretarsus. Abbreviations: cx, coxa; fe, femur; pt, pretarsus; t1-5, tarsomeres 1-5; T1–T3, thoracic segments 1-3; Ti, tibia; Ts, tibial spur; tt, tibiotarsus; tr, trochanter.
Comparative studies have found evidence for both conservation and divergence in aspects of appendage patterning across species and appendage types. For example, the “leg gap genes” have strongly conserved functions in embryonic and later development in many arthropods (e.g., Angelini and Kaufman 2004; Moczek and Rose 2009; Schoppmeier and Damen 2001; Suzuki et al. 2009). However, gene functions in appendage allocation and PD axis specification have been found to vary (e.g., Angelini and Kaufman 2005a; Jockusch et al. 2000; Ober and Jockusch 2006; Ronco et al. 2008).
Tribolium castaneum is a useful species for comparison with D. melanogaster because it is a holometabolous insect, like Drosophila, but the adult legs undergo development from larval legs. Variation in the number of tarsomeres in T. castaneum also makes it an attractive system. Pro- and mesothoracic legs have five tarsal elements (Figure 1, B and C), as do all legs of D. melanogaster, whereas the hindlegs of T. castaneum have four tarsomeres (Figure 1D). Compared with other leg traits, tarsomere number varies greatly among insects, but this heteromerous “5-5-4” tarsal pattern is characteristic of tenebrionoid beetles. A focus on metamorphic leg development also allows comparisons with previous studies of embryonic leg development in T. castaneum and other insects.
Our results illustrate a number of similarities between the development of the legs in D. melanogaster and T. castaneum. However, several important differences were found, including changes in the PD level of function for several genes as well as distinct gene functions in T. castaneum. We use these data, in combination with data on the metamorphic patterning of the antennae (Angelini et al. 2009) and mouthparts (Angelini et al. 2012) in T. castaneum, to examine the degree of modularity in the development of serial homologs and the implications this may have for evolution of appendage diversity.
Materials and Methods
General methods
Wild-type cultures of Tribolium castaneum were obtained from Carolina Biological Supply Company and reared using the supplier’s recommendations. The Fused tarsi and antennae (Fta) mutation of Dll was obtained from Susan J. Brown. The 17 candidate genes selected for study include transcription factors and components of conserved signaling networks (Table 1). Each gene has known roles in leg or antennal patterning in D. melanogaster. Cloned gene fragments were provided by colleagues as indicated by the references in Table 1, or cloned using standard methods that have been detailed elsewhere (Angelini et al. 2009). Additional genes were also cloned and tested functionally using RNAi; however, phenotypes in the legs were absent or not penetrant enough to be informative. These genes included apterous and apterous-related, aristaless, clawless/C15, pdm/nubbin, and spalt, which will not be described further in this study.
Relative real-time polymerase chain reaction (PCR) determination of expression
The relative expression levels of genes in the tarsus and proximal leg were determined using real-time PCR. The pro- and mesothoracic legs of approximately 20 pupae were bisected at the tibia−tarsus joint to generate separate pools of proximal and distal tissue for RNA extraction using Trizol (Invitrogen/Life Technologies) followed by DNase I treatment to remove genomic DNA. The iScript cDNA Synthesis Kit (BioRad) was used for first-strand cDNA synthesis. Amplification of target cDNAs used a SYBR Green realtime PCR kit (Absolute Scientific) on a BioRad iCycler instrument. Calculations of relative target sequence abundance were normalized with 18S ribosomal RNA and adjusted for measured primer efficiency (Pfaffl 2001). All primer sequences are available from the authors upon request.
RNA interference
Knockdown phenotypes were generated in adult beetles using RNA interference (RNAi). Prepupal larvae were injected with double-stranded (ds) RNA following the methods of previous studies (Angelini et al. 2009, 2012; Tomoyasu and Denell 2004). GFP dsRNA was used as a control to determine the rates of spontaneous or injection-induced malformations. Because of the potential for functional redundancy of some genes studied here, we also included some treatments in which multiple genes were targeted by injecting multiple dsRNAs (Table 2). Validation of RNAi knockdown was performed by real-time PCR comparison of target gene expression in control (GFP) dsRNA and gene-specific dsRNA treatments (Table 2), as described in an accompanying article in Genetics, Angelini et al. (2012; see also Aspiras et al. 2011).
Table 2 . Summary of RNA inference effects.
| Phenotypic Penetrance | ||||||||
|---|---|---|---|---|---|---|---|---|
| dsRNA Sequence | dsRNA Size, bp | Target Gene Knockdowna | Number Scored | Legs | Defects in Other Appendages | |||
| Unaffected | Mild | Moderate | Severe | |||||
| GFP | 600 | 83 | 96.4% | 2.4%b | 0 | 1.2%b | 1.2%b | |
| dpp | 812 | 66% ± 13%* | 35 | 37% | 17% | 17% | 29% | 40% |
| Dll | 462 | 75% ± 4.9%* | 42 | 17% | 4.8% | 17% | 62% | 91% |
| dac | 359 | 30% ± 16% | 66 | 38% | 11% | 23% | 29% | 76% |
| hth | 335 | 44% ± 7.5%* | 205 | 34% | 39% | 22% | 6% | 90% |
| Lim1 | 388 | 56% ± 31% | 16 | 31% | 6.3% | 25% | 38% | 75% |
| Notch | 409 | 24% ± 9.0%* | 31 | 23% | 6.5% | 19% | 52% | 94% |
| Ser | 329 | n/ac | 151 | 14% | 0.7% | 2.6% | 83% | 97% |
| Delta | 180 | 71% ± 14%* | 75 | 85% | 6.7% | 6.7% | 1.3% | 38% |
| Ser, Delta | — | 23 | 44% | 0 | 4.3% | 52% | 78% | |
| odd | 211 | 65% ± 7.3%* | 50 | 36% | 14% | 8.0% | 42% | 86% |
| bowl | 342 | 63% ± 5.2%* | 57 | 46% | 8.8% | 8.8% | 37% | 79% |
| sob | 321 | 61% ± 8.0%* | 16 | 6.3% | 0 | 56% | 38% | 94% |
| drm | 225 | n/ac | 30 | 3.3% | 0 | 3.3% | 93% | 100% |
| odd, bowl, sob | — | 16 | 50% | 6.3% | 19% | 25% | 81% | |
| odd, bowl, sob, drm | — | 37 | 5.4% | 5.4% | 0 | 89% | 100% | |
| Krn | 188 | 67% ± 9.2%* | 19 | 11% | 5.3% | 21% | 63% | 90% |
| bab | 774 | n/ac | 22 | 18% | 4.5% | 9.1% | 68% | 68% |
| ab | 198 | 42% ± 6.3%* | 31 | 26% | 6.5% | 0 | 68% | 81% |
| rn | 127 | 78% ± 9.0%* | 52 | 75% | 2% | 14% | 10% | 21% |
| ss | 355 | n/ac | 19 | 58% | 16% | 5.3% | 21% | 100% |
| total | 1076 | 33% | 76% | |||||
The level of target gene knockdown was determined by real-time PCR comparisons of pooled pupae to nonspecific GFP dsRNA controls. The phenotypic effects on legs and other appendages were scored after metamorphosis.
* Significant difference from gene expression in GFP control specimens (Welch’s t-test, P < 0.05).
0% represents no reduction in activity, while 100% is complete suppression.
One control specimen eclosed with truncated T1 legs (scored as severe). Two specimens were missing a single tarsal joint. One specimen had a fusion of the distal- most segments of one antenna.
Suitable primers for this gene were unavailable (i.e., n/a).
Adult morphology was examined after clearing overnight in a solution of 20% glycerol in glacial acetic acid at 50° (modified from van der Meer 1977). Pharate or eclosed adults (n = 1076) were scored for up to 64 anatomical characters related to the legs, including the presence or absence of each appendage segment, degree of fusion between segments, and changes in size, shape, and characteristic bristle patterns. This data matrix (supporting information, File S1) was used to quantify the penetrance of RNAi for each gene in each leg structure and to assess the severity of phenotypes.
Microscopy and imaging
Photomicrographs of dissected appendages were obtained with an Olympus digital camera on a Zeiss Axioskop compound microscope. For electron microscopy, specimens were prepared by overnight dehydration in ethanol, followed by a 15-min immersion in hexamethyldisilazane. Specimens were then sputter coated in gold palladium and imaged with a Zeiss DSM982 Gemini field emission scanning electron microscope.
Results
On the basis of the development of D. melanogaster, candidate genes were identified for study in T. castaneum (Table 1). Expression and functional studies were conducted with the resulting group of 17 genes in T. castaneum. Real-time PCR confirmed that RNAi resulted in a reduction in the expression of target genes, with knockdown levels ranging from 24% to 78% (mean of 57% reduction) in pooled pupal samples (Table 2). RNAi phenotypes were qualitatively consistent within dsRNA treatments, and for many genes, these phenotypes could be ordered into a series from mild to severe, resembling a hypomorphic mutant series. RNAi was highly penetrant, with an average of 84% of individuals having defects in at least one appendage. Penetrance in the legs averaged 67% (Table 2) and did not vary significantly between thoracic segments; in general, affected individuals showed similar defects in multiple legs. Sample sizes of individuals scored for leg defects ranged among treatments from 16 to 205 (mean = 48; Table 2). The RNAi phenotypes are described under the subheadings to follow, arranged by the developmental processes affected.
Proximal-to-distal gene expression bias in the leg
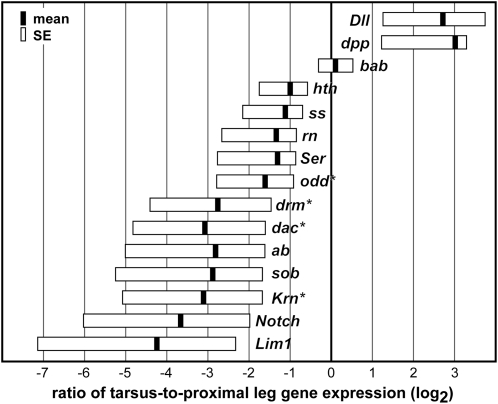
The relative PD expression bias for genes was determined in the pupal leg using relative real-time PCR (Figure 2). Expression of several genes was significantly enriched in either the tarsus or proximal leg at the pupal stage. For most genes, regions of expression-bias correlated well with regions affected by RNAi. Two genes, Dll and dpp, whose depletion produced phenotypes in the tarsus, showed significantly elevated expression in that tissue. Notch signaling components (Notch and Ser), which affected the entire leg, and genes with RNAi phenotypes in the tibia (dac, Lim1, and Krn) had a significant proximal bias in expression. We detected an unexpected proximal bias in expression for three genes with depletion phenotypes in the tarsus (rn, ss, and abrupt); however, the expression ratio for these genes was not significantly different from 1 (Welch’s t-test, P > 0.05).
Figure 2 .
Relative expression of candidate genes is compared in the tarsus and proximal leg of pupae. Expression ratios are given in log2 scale. Zero represents equal expression in each region; genes appearing to the right are enriched in the tarsus, relative to the rest of the leg. Black bars indicate the mean expression ratio, whereas boxes indicate standard error. Genes with expression ratios significantly different from 1 (equal expression in both regions) are denoted with an asterisk (Welch’s t-test, P < 0.05).
Genes required for growth of large leg regions
Two of the classic leg “gap genes,” Distal-less (Dll) and dachshund (dac), were required for the specification and growth of large regions of the developing metamorphic leg in T. castaneum. Similar RNA interference results for these genes have been reported by Suzuki et al. (2009), and we expand the anatomical description of phenotypes here. RNAi targeting Dll produced a range of phenotypes. In the mildest phenotypes, the tarsus lacked joints and the tarsus, tibia, and femur were shorter than in wild type (Figures 3C and 4C). Hypomorphic alleles of Dll have been isolated in T. castaneum (Beermann et al. 2001); their phenotypes closely resemble this mild Dll RNAi effect (Figure 3, B and C). More severely affected Dll RNAi specimens had legs truncated at the level of the tibia (Figure 3D) or femur (Figure 3E). In the most severely affected individuals, the joints between proximal segments were also incomplete (Figure 3E). Depletion of dac resulted in loss of an intermediate portion of the legs, extending from the middle of the femur to the proximal tarsus (Figure 3F). The joint at the distal end of the femur was abnormal and did not resemble either the wild-type femur−tibia or tibia−tarsus joint. The proximal tarsal elements were also affected, with tarsomere 1 (t1) being deleted or fused to t2 (Figure 4D). RNAi depletion of each of these genes closely paralleled mutant phenotypes from D. melanogaster (Cohen and Jürgens 1989; Mardon et al. 1994).
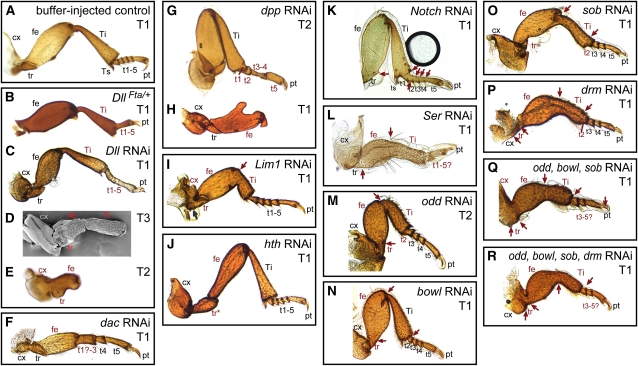
Figure 3 .
RNA interference effects on adult legs. (A) Control treatments were indistinguishable from unmanipulated beetles. (B) DllFta heterozygotes develop with reduced tarsi, lacking joints. (C) This phenotype is similar to mild Dll RNAi specimens. (D) Dll RNAi also resulted in stronger phenotypes, in which structures distal to the tibia are deleted. (E) In the most severe Dll-depleted individuals, the legs are truncated within the femur, which is reduced. Joints are also absent from the remaining segments. (F) Severe dac RNAi specimens had deletions of the distal femur, tibia, and proximal tarsomeres. (G) RNAi targeting dpp caused alterations of the proximal tarsomeres in mildly affected specimens. (H) In more severe dpp RNAi specimens, the legs are truncated in the mid-femur. (I) Lim1 RNAi caused the loss of proximal leg joints (red arrow) and a reduction of the distal femur and proximal tibia. Although the femur−tibia joint did not form normally, an anatomical boundary was present at this position. (J) Knockdown of hth caused a homeotic transformation of the coxa, trochanter and proximal femur toward more distal morphologies. (K) Notch RNAi eliminated most joints from the leg (red arrows). (L) Ser RNAi eliminated joints (red arrows) and reduced the overall length of the leg. (M-R) Depletion of odd-related genes caused reduction of the leg and loss of proximal joints (red arrows), as well as deletion of proximal tarsomeres. Abbreviations: cx, coxa; fe, femur; pt, pretarsus; t1-5, tarsomeres 1-5; T1, prothoracic leg; T2, mesothoracic leg; T3, metathoracic leg; Ti, tibia; tr, trochanter; Ts, tibial spur. Structures with a defect are labeled in red.
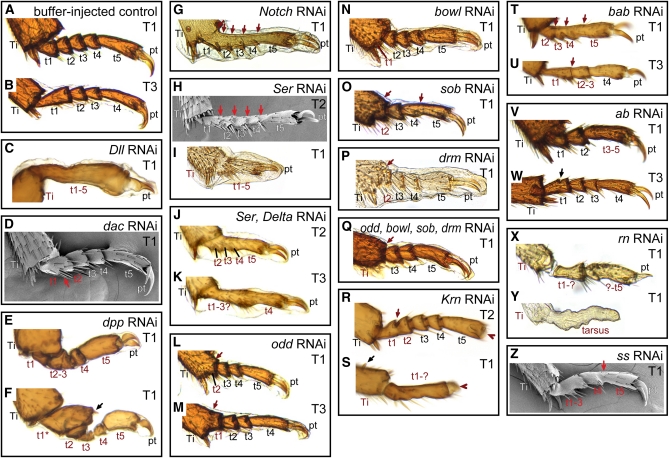
Figure 4 .
Phenotypes in the tarsus produced through RNAi. (A) Normal tarsus anatomy in the pro- and mesothoracic legs includes five tarsomeres. The distal tibia is also visible, showing the distal tibial spurs. (B) The metathoracic legs normally have four tarsomeres. (C) Dll RNAi produces mild specimens in which the tarsomeres lack joints and the region is reduced. (D) Depletion of dac produced fusions in the proximal tarsus. (E) Knockdown of dpp reduced t1-t4. (F) Rarely dpp RNAi caused a transformation of the first tarsomere (t1*) toward a distal tibia identity, as indicated by an ectopic tibial spur (black arrow). (G) Notch RNAi caused the loss of joints in the tarsus (red arrows). (H) Mild Ser-depleted specimens also lacked joints between tarsomeres (red arrows). (I) Moderate Ser knockdown phenotypes included fusion and reduction of the tarsus. (J–K) Simultaneous depletion of Ser and Delta produced phenotypes similar to Ser RNAi alone. (L—Q) RNAi targeting the odd-related genes caused fusion of proximal tarsomeres with the tibia (L, M, O), or adjacent tarsomeres (O). The first tarsomere (L, N, P) and sometimes also the second (Q) were deleted in moderate and severe specimens. (R) Krn RNAi eliminated the pretarsus (red arrowhead), and caused occasional loss of joints (red arrow) in mild specimens. (S) More severely affected Krn RNAi specimens had a reduction of the tarsus with complete loss of joints. Tibial spurs were also deleted, although ectopic tibial spurs could sometimes be found (black arrow). (T–U) Knockdown of bab caused loss of joints in the tarsus (red arrows) and fusion of tarsomeres. (V) Depletion of ab caused fusions of tarsomeres. (W) Rarely partial ectopic joint formation was observed in ab RNAi (black arrow). (X) RNAi targeting rn caused a reduction of the tarsus and failure of joint formation. (Y) In severely affected rn RNAi specimens the pretarsus also failed to form. (Z) Knockdown of ss also caused fusions of tarsomeres (red arrow). Abbreviations are as in Figure 3.
RNAi disruption of dpp activity in T. castaneum prepupae also produced some adults (5 of 35 scored) with large deletion phenotypes (Figure 3H), although most specimens had defects restricted to the tarsus (Figures 3G and 4E). Truncations occurred in the middle of the femur, with loss of more distal podomeres. The coxa and trochanter of these legs appeared normal; however, the femur was malformed and its distal end was often bifurcated into two prongs (Figure 3H). This dpp RNAi phenotype was dramatically asymmetrical (which was unusual for the RNAi phenotypes of most genes we investigated) and all individuals with one or more truncated legs also had multiple anatomically normal legs. This asymmetry was also observed with milder dpp RNAi effects in the tarsus. Truncation or bifurcation of the leg was also found after clonal disruption of Dpp signaling in the fly leg (Theisen et al. 1996). In the tarsus, dpp depletion caused reduction and malformation of t1–t4 (Figures 3G and 4, E and F). The pretarsus was not affected.
RNAi targeting Lim1 in T. castaneum prepupae caused a reduction of the intermediate region of the leg (Figure 3I), which we interpret as a partial deletion of the distal femur and proximal tibia. Shape and bristle patterns indicate that the proximal femur and distal tibia developed normally in Lim1 RNAi. The region between these podomeres, where the presumptive femur-tibia joint normally forms, was sharply angled and constricted ventrally, suggesting that some differentiation of separate podomeres still occurred. The intervening joint was lost or malformed (8 of 16 specimens scored). Loss of the coxa-trochanter joint was also observed in severely affected specimens (3 of 16 scored). In D. melanogaster, Lim1 is expressed in four discrete domains along the PD axis (Campbell 2005; Lilly et al. 1999; Tsuji et al. 2000), and RNAi phenotypes in T. castaneum closely parallel those in the proximal leg of D. melanogaster Lim1 mutants, in which the coxa is deformed or absent and the femur is greatly reduced and fused to an abnormally bent tibia (Pueyo et al. 2000; Tsuji et al. 2000). The pretarsus in the legs of D. melanogaster mutants is also absent (Pueyo et al. 2000; Tsuji et al. 2000), whereas in all T. castaneum Lim1 RNAi specimens the tarsus and pretarsus were unaffected.
Overall, these phenotypes reveal that appendage regions which are already present in the T. castaneum larva can be deleted at metamorphosis. These results indicate an ongoing requirement for Dll, dac, dpp, and Lim1 in maintenance and adult development of specific leg regions.
Homeotic transformation of the proximal leg as a result of homothorax RNAi
Among the genes examined in this study, only homothorax depletion produced a homeotic phenotype. The proximal region of the leg imaginal disc of D. melanogaster is specified in part by homothorax (hth), which encodes a homeobox transcription factor (Wu and Cohen 1999). Leg imaginal discs lacking hth develop a normal tarsus but have a single reduced, proximal segment with identity from more than one of the missing podomeres (Casares and Mann 2001). Depletion of hth in T. castaneum prepupae also caused defects in the proximal adult legs (Figure 3J). Unlike the reduced phenotype of the D. melanogaster hth null legs, hth-depleted T. castaneum had fully elongated legs with the normal number of leg segments. However, the morphology of the three proximal podomeres was altered. The most obvious effects were in the coxa and trochanter, which were enlarged and had bristle patterns more like those of more distal leg segments. The presumptive femur was narrower but similar in length to the wild-type femur. Joints between the coxa, trochanter, and femur were present, but resembled the wild-type femur–tibia joint in morphology. The prothoracic coxa was more rounded in shape than in wild type. This may represent either a transformation of the coxa toward distal identity, or it may represent a transformation of the prothoracic coxa toward the identity of a mesothroracic coxa (compare Figures 1, B and C, and 3J). However, the placement of the coxa-trochanter joint differed between the transformed prothoracic and normal mesothoracic coxae. The distal leg was unaffected. Taken together, these phenotypes suggest that depletion of hth removes the normal identity cues in the proximal leg, but does not affect growth along the PD axis or the location and formation of joints. The resulting podomeres have the shape and bristle pattern suggestive of the femur, but their identity remains ambiguous. One hypothesis is that these segments have a mixed femur/tibia identity, as in D. melanogaster hth loss-of-function (Casares and Mann 2001).
The Notch signaling pathway and odd-skipped paralogs are require for both leg growth and joint formation
Notch signaling is required early for elongation of the PD axis in D. melanogaster leg discs, as well as for joint formation later in development (Bishop et al. 1999; de Celis et al. 1998; Rauskolb and Irvine 1999). Notch-mediated joint formation has been proposed as a defining characteristic of arthropods (Prpic and Damen 2009). In T. castaneum, we examined depletion phenotypes for Notch as well as its ligands, encoded by Serrate (Ser) and Delta, and found evidence that both the axis elongation and joint formation roles are conserved.
Knockdown of Notch or Ser blocked joint formation in the legs (Figure 3, K−L). Notch RNAi individuals had legs that were relatively normal in size and shape but that lacked most joints (Figure 3K). Joint loss phenotypes were much less penetrant in the femur–tibia (26 of 186) and tarsus–pretarsus (10 of 186) joints than they were in the other joints between the primary segments (18%–60% joint loss). Joints between the tarsomeres were especially sensitive to depletion of Notch (Figure 4G), and in the mildest phenotypes, joint loss only occurred between tarsomeres. Despite the absence of joints, differentiation of individual tarsomeres was suggested by the retention of the distal bristles present on t1–t4 (on pro- and mesothoracic legs—or t1–t3 on metathoracic legs) which were spaced as in wild-type individuals. Delta RNAi was far less penetrant (11 of 75 Delta-depleted specimens had leg defects vs. 28 of 31 Notch-depleted specimens) despite significant reduction in Delta transcripts after RNAi (71% reduction ±14%). Delta-depleted specimens with phenotypes in the legs had a failure of joint formation. These individuals lacked all joints in the tarsus (8 of 443 legs) or joints between coxa, trochanter, and femur (10 of 444 legs) or both of these states (1 of 443 legs). Ser RNAi phenotypes were highly penetrant and typically severe (Table 2), including loss of joints and reduction of the legs overall. The pretarsus was only rarely affected (5% of specimens). RNAi targeting Ser (but not Notch or Delta) resulted in drastic reduction in the length of legs (Figure 3L), with the tarsus and tibia more severely reduced compared with more proximal segments. The structures most affected by Ser RNAi are also those that undergo the greatest increase in size during normal metamorphosis (compare Figure 1, B and B′). Mild Ser RNAi phenotypes lacked tarsal joints and showed fusions of tarsomeres (Figure 4, H and I). Despite penetrant phenotypes in the tibia and tarsus, the pretarsus and tibial spurs were not strongly affected by depletion of any Notch signaling component, having been deleted in only 3.6% (61/1679) or 4.8% (76/1572) of legs scored, respectively, in all Notch signaling RNAi experiments.
Depletion of Notch, Ser, and Delta had different effects on bristle development. In Notch RNAi individuals, most bristles were absent, although the large pegs at the end of each tarsomere remained (Figure 3K). In contrast, many bristles remained after Ser RNAi, including the regularly arranged pattern of bristles on the femur and tibia (Figure 3L). With the exception of this effect on bristles, depletion of Notch had less severe effects on leg development than depletion of Ser. The Ser ligand acts redundantly with Delta in some contexts in D. melanogaster (Zeng et al. 1998). Therefore, it would be predicted that Notch RNAi phenotypes should be more severe than those of Ser. However, it is possible that severe Notch depletion is lethal and that only relatively mild phenotypes were recovered here. Simultaneous depletion of Ser and Delta produced phenotypes similar to Ser RNAi alone, with reduction of leg length, loss of joints, and fusions of tarsomeres (Figure 4, J and K).
Genes related to D. melanogaster odd-skipped (odd) also affected large regions of the legs in T. castaneum. This gene family includes four linked zinc-finger transcription factors: odd, brother of odd with entrails limited (bowl), sister of odd and bowl (sob), and drumstick (drm). The similarity of odd-paralog and Ser RNAi phenotypes suggests that these genes lie in the same pathway in T. castaneum as they do during leg development in D. melanogaster. In the leg imaginal disc of D. melanogaster, odd-related genes are required for proper patterning of the tarsus (de Celis Ibeas and Bray 2003; Hao et al. 2003), where they may act to stabilize Notch signaling (Greenberg and Hatini 2009). Three of these genes (odd, bowl, sob) share substantial sequence similarity in the regions we targeted for knockdown in T. castaneum; therefore, we do not distinguish specific roles for each of these genes. Phenotypes for these single-gene knockdowns were similar to one another and to phenotypes resulting from simultaneous targeting of all four paralogs.
The legs of T. castaneum were reduced in length after knockdown of odd paralogs, with most of the reduction occurring in the femur and tibia (Figure 3, M−2R). The shape changes in these segments also resembled the shape changes observed in response to Ser RNAi (Figure 3L). In the most extreme phenotypes, the tibia was broadest centrally, with rounded lateral edges. Joints between the trochanter, femur, tibia, and tarsus also frequently failed to form. Unlike the joint loss found with RNAi targeting Notch signaling, which affected all tarsomeres equally, proximal tarsal elements (t1–t2) were far more sensitive to odd-paralog depletion than were more distal tarsal elements. Fusion occurred in 39% of t1–t2 joints vs. 10% to 13% of more distal tarsomere joints (for scored T1 and T2 legs; e.g., Figure 4O). In mildly affected RNAi specimens, the only defects we observed were in the tarsus, where t1 was deleted or fused to the distal tibia (Figure 4, M, N, and P). Occasionally the second tarsomere was also deleted or fused to the distal tibia (Figure 4, L and O−Q). Often, loss of the proximal tarsomere was accompanied by deletion of the spurs at the distal end of the tibia (333 of 874 legs scored, e.g., Figure 4, O and Q).
Genes required primarily in the tarsus and pretarsus
Depletion of several genes yielded phenotypes that were restricted to the tarsus or pretarsus. In D. melanogaster, several EGF ligands are secreted from the distal appendage tip and loss of EGF signaling leads to the loss of t5 and the pretarsus (Clifford and Schupbach 1989; Galindo et al. 2002) with rare reduction of the entire tarsus and pretarsus to a single tarsomere-like structure (Campbell 2002). The T. castaneum genome has only one activating EGF ligand, with greatest sequence similarity to Krn (Tribolium Genome Sequencing Consortium 2008). Knockdown of Krn in T. castaneum led to dramatic defects in the antennae and mouthparts (Angelini et al. 2009; see also accompanying article in Genetics, Angelini et al. 2012). However, in the legs, Krn RNAi phenotypes were restricted to the distal leg (Figure 4, R and S). The most common phenotype was loss of the pretarsus (17 of 19 scored). In more strongly affected individuals, tarsomeres were also fused to one another (Figure 4R), and in the most severely affected individuals, the entire tarsal region was reduced and lacked joints (Figure 4S). In moderately and severely affected specimens the distal tibia, near the tibia−tarsus joint, was also abnormally shaped, with absent or misplaced tibial spurs (Figure 4S, red arrow).
Downstream of EGF signaling in D. melanogaster the pretarsus and distal tarsomeres are patterned through the activity of a feed-forward gene circuit that includes Lim1. Loss of Lim1 function results in deletion of the claw and reduction or fusion of t4–t5 (Campbell 2005; Pueyo et al. 2000; Tsuji et al. 2000). As described previously, Lim1 RNAi caused deletions spanning the femur-tibia joint and failure of joint formation in the proximal leg in T. castaneum. Defects in the tarsus were rare and consisted of the incomplete fusion of adjacent tarsomeres (2 of 16 specimens; this rate is not significantly different from that seen in GFP RNAi, Fisher’s exact test, P = 0.18).
Depletion of dpp also produced tarsal defects that were pronounced in the proximal tarsus (Figure 4, E and F), in addition to defects in the more proximal leg segments (Figure 3H). Defects included fusions and reduction of tarsomeres, but these defects were qualitatively different from reduction and fusion occurring with other dsRNA treatments. Tarsomeres were typically reduced in width, not length (Figure 4F, t3-t4). In two specimens, the proximal tarsomere was enlarged and bore the spurs indicative of tibial identity (Figure 4F, t1*).
Depletion of two BTB-class transcription factors, bric-à-brac (bab) and abrupt (ab), produced tarsal defects with high penetrance. In D. melanogaster redundant bab paralogs are required for the proper development of the distal tarsomeres and mutations in bab affect t2-t5, causing distal deletions, fusions, or transformation to more proximal identity (Couderc et al. 2002; Godt et al. 1993). RNA interference targeting the single bab ortholog in T. castaneum caused a similar phenotype wherein tarsomeres were reduced and fused (Figure 4, T and U). In contrast to bab phenotypes in D. melanogaster, T. castaneum bab RNAi phenotypes often included t1 in fusions (91 of 104 legs scored). Depletion of ab also caused the loss of joints, fusion of adjacent segments, and/or a small deletion in the tarsal region (Figure 4, V and W), but in a different pattern than bab depletion. Loss of joints or fusions of tarsomeres 1-2 and 3-5 (or 3-4 in the metathoracic tarsi) occurred in 37 of 42 ab RNAi specimen legs, with the remainder being wild type or missing only a single joint. Rarely, ab knockdown individuals were recovered with a partial ectopic joint and accompanying ventral macrochetes in the metathoracic tarsus (2 of 57 T3 legs scored; Figure 4W, arrowhead). In D. melanogaster, abrupt mutants have altered legs, with distal regions more strongly affected or deleted (Hu et al. 1995); the mutant phenotype has not been described in detail.
RNAi targeting two other transcription factors produced leg defects that were limited to the tarsal region with relatively low penetrance: rotund (rn) and spineless (ss). In D. melanogaster, rn and ss are transiently expressed in an intermediate region of the tarsus, and t2-t4 are lost in response to mutations in either gene (Cavener et al. 1986; Duncan et al. 1998; Kozu et al. 2006; St. Pierre et al. 2002). Depletion of each of these genes in T. castaneum also caused the loss of joints, fusion of adjacent tarsomeres, and reduction of the tarsal region (Figure 4, X−Z). Knockdown of rn also frequently produced fusion between the tarsus and pretarsus accompanied by abnormal development of the pretarsus (38 of 77 legs in specimens with rn-depletion defects; Figure 4, X and Y). Shippy et al. (2009) reported similar ss RNAi phenotypes in making the case that the classic T. castaneum antennapedia mutations are allelic to ss. The strongest of these ss alleles cause reductions in the tarsus resembling ss RNAi phenotypes (Shippy et al. 2009).
Discussion
To explore how distinct appendage types are patterned, we have examined the function of 17 genes during metamorphic appendage development of the red flour beetle Tribolium castaneum. The majority of the genes studied here have conserved roles in leg patterning in D. melanogaster and T. castaneum (summarized in Figure 5), but several have differences in their area of functional effect; these differences range from slight to dramatic. After comparing leg patterning in Tribolium and Drosophila and across developmental stages within Tribolium, we conclude by using the genes with divergent functions to test conflicting predictions about how the appendage patterning networks of serial homologs evolve.
Figure 5 .
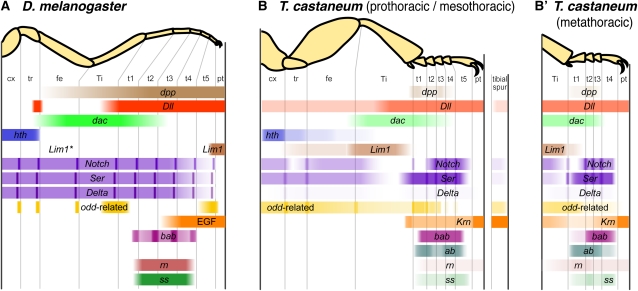
Summary of RNAi phenotypes in the legs. (A) Functional domains of genes in the leg of D. melanogaster. For each gene, colored bars represent the region on the corresponding adult leg in which the gene is expressed and/or functions, focusing on the late third instar larval imaginal leg disc. Dll, EGF, and dpp are also required in the embryo for initiation of imaginal leg disc primordia. odd-related genes are expressed more broadly in the tarsus earlier in development, and are required for proper formation of tarsomere joints. In D. melanogaster, ab has been observed in the leg discs, but its expression pattern has not been described in detail (Hu et al. 1995). *Lim1 expression has also been detected in multiple domains in the proximal imaginal leg disc, including the femur (Tsuji et al. 2000). (B) Patterning of the pro- and mesothoracic legs of T. castaneum. Intensity of expression indicates phenotypic penetrance of RNAi effects. The tibial spur is represented by the column at right. (B′) The metathoracic leg has only four tarsomeres, and its patterning of the intermediate tarsomeres (t2-t3) differs from the more anterior legs.
Patterning of the primary leg segments
All insects share a conserved leg morphology consisting of six primary segments, including an annulated tarsus. For many genes in this study (although not all) gene functions in the legs appear to be very similar in T. castaneum and D. melanogaster, as expected if an ancestral leg patterning network has been conserved in both lineages. Genes involved in the outgrowth of appendages, such as Dll, EGF, and Notch signaling components, are conserved in that role in legs (Figures 3, D, E, and L and 4, I and T), as well as in other appendage types. Similarly, Notch signaling is conserved it its role in joint formation in both insects. Loss of function of Notch or one of its ligands causes a failure of appendage joint development (Figures 3, K and L and 4, G−K). On the basis of studies of Notch signaling in a spider, it has recently been proposed that Notch-mediated cuticular joint formation is a synapomorphy of arthropods, which may have contributed to their success and diversification (Prpic and Damen 2009).
The odd-skipped family of zinc-finger transcription factors is an important regulator of leg development; these proteins interact with Notch signaling to specify joints in the primary leg segments. In D. melanogaster, all four family members are expressed at the joints between primary leg segments, where they function in joint formation. bowl is more broadly expressed in the tarsus, where loss of function leads to loss of joints, as well as reduction and other patterning defects (de Celis Ibeas and Bray 2003; Hao et al. 2003). In T. castaneum, odd-related genes are also required for tarsal development, as well as for elongation and joint formation in more proximal leg segments, as shown by the RNAi phenotypes (Figures 3, M−R and 4, L−Q). Considering the complex patterns of expression and function in these genes, their conservation is noteworthy.
Another group of genes with largely conserved functions is the “limb gap genes.” Distal-less, dac, and hth specify broad domains of identity along the leg PD axis in D. melanogaster, T. castaneum, and other species. The loss of function phenotypes for Dll and dac result in deletion of the distal and intermediate domains, respectively (Cohen and Jürgens 1989; Mardon et al. 1994; Suzuki et al. 2009; Figure 3, B−F). Similar RNAi phenotypes have also been reported for these genes in the beetles Harmonia axyridis (Dll: Niimi et al. 2005) and Onthophagus taurus (Dll, dac, hth: Moczek and Rose 2009), the milkweed bug Oncopeltus fasciatus (Dll, dac, hth: Angelini and Kaufman 2004), and the spider Cupiennius salei (Dll: Schoppmeier and Damen 2001). In both D. melanogaster (Casares and Mann 2001) and T. castaneum, hth is required for proper development of the proximal most leg segments. However, aspects of the hth phenotypes differ in significant ways between these species; in particular, in D. melanogaster, loss of hth expression leads to a reduction in the number of podomeres, while in T. castaneum, all podomeres are retained, although their identities are altered.
The role of dpp in leg development has been an important test case for understanding the conservation of developmental systems. In the leg imaginal disc of D. melanogaster, dpp is required for dorsal-ventral axis formation, and null clones lacking one of the Dpp receptors (encoded by thickveins and punt) lead to truncations and bifurcations of the limb axis (Theisen et al. 1996). This is similar to the severe phenotype obtained here for dpp RNAi in adult legs (Figure 3H). The role of dpp in proximodistal patterning of embryonic appendages in T. castaneum is unclear. In mild embryonic RNAi phenotypes, T. castaneum limb buds develop normally (Ober and Jockusch 2006), while in stronger phenotypes, early embryonic dorsoventral axis patterning is greatly altered, and appendages fail to form (van der Zee et al. 2006).
Tarsal development and evolution
It is interesting that the basic developmental mechanisms responsible for overall leg patterning (i.e., differentiation of the PD axis into the six true segments) and for tarsal patterning (subdivision of the tarsus into jointed tarsomeres) are fundamentally similar, whereas the evolutionary diversity of these traits is very different. The pattern of subdivision of the PD axis of the leg differs between arthropod classes, but it has remained fixed within insects for 396 million years (Engel and Grimaldi 2004). By contrast, the number of tarsomeres never exceeds five, but otherwise varies among insect groups. In addition, in some hemimetabolous insects, tarsomere number varies between instars (e.g., Heteroptera), whereas in other groups, different legs may have different numbers of tarsomeres, such as the 5-5-4 pattern of tenebrionoid beetles. In T. castaneum, the pretarsus forms a discrete leg segment in the larva, but a single more proximal segment (the tibiotarsus) gives rise to both the tibia and the tarsus of the adult (Figure 1). Thus, tarsal subdivision occurs during appendage metamorphosis. In D. melanogaster, the identity of tarsomeres is intercalated between already established pretarsal and tibial identities during mid-to-late third instar imaginal disc development (Kojima et al. 2000).
Our data suggest that most genes are conserved in their tarsal patterning roles between D. melanogaster and T. castaneum, but that several have undergone changes in the PD extent of activity. Genes with highly conserved functions in the tarsus include dac, Dll, and several genes regulated by Dll. Mutations in dac in D. melanogaster affect tarsomeres 1-3 (t1-t3; Mardon et al. 1994), although dac is only expressed in t1 (Abu-Shaar and Mann 1998; Lecuit and Cohen 1997), suggesting that the more distal phenotypes result from indirect effects. Similarly, T. castaneum dac had a significant bias toward proximal expression in the leg (Figure 2), and dac RNAi strongly affected t1-t2 with rarer defects in t3 and t4. The DllFta mutation (Figure 3B) and mildly affected specimens recovered from Dll RNAi in T. castaneum (Figures 3C and 4C; Suzuki et al. 2009) closely resemble weak hypomorphic combinations of Dll alleles in D. melanogaster (Panganiban 2000). These similar phenotypes suggest that the role of Dll as an early activator of tarsus-specific genes is conserved in T. castaneum. Dll initially activates several genes in a broad tarsal domain, including bab and ss (reviewed by Kojima 2004). bab is required for joint formation within the tarsus in D. melanogaster (Godt et al. 1993). Similarly, RNAi targeting bab during T. castaneum metamorphosis caused reduction of the tarsus and loss of joints (Figure 4, T and U). Depletion of odd-related genes in D. melanogaster and T. castaneum also had similar phenotypes in the tarsus. Expression of ss is transiently activated in the central tarsus, and D. melanogaster ss mutants develop without t2-t4 (Duncan et al. 1998; Kozu et al. 2006). Knockdown of ss in T. castaneum produced phenotypes with fusions in the central tarsus, resembling D. melanogaster phenotypes (Figure 5).
High levels of EGF signaling at the distal region of the D. melanogaster leg disc are also initiated by Dll and are required for the development of the pretarsus and distal tarsus; more proximal regions may require a low level of EGF signaling (Campbell 2002; Galindo et al. 2002, 2005). Krn was also required for development of these structures in T. castaneum, where depletion caused deletion of the pretarsus, and reduction and joint loss throughout the tarsus. Frequent defects in the proximal tarsus, deletion of the tibial spurs, and malformation of the distal tibia (Figure 4, R and S) may indicate a more extensive proximal requirement for EGF in the legs of T. castaneum compared with D. melanogaster. Alternatively, because the T. castaneum genome has only one activating EGF ligand (Krn), we may detect functions that are redundantly covered by multiple ligands in D. melanogaster. In D. melanogaster, ultimately, tarsomeres are distinguished by the expression of different combinations of transcription factors, which activate the Notch pathway at each of the intratarsal joints (reviewed by Kojima 2004). The activity of the Notch pathway is also well conserved as discussed above.
EGF activity is key to activating other genes responsible for pretarsus development in D. melanogaster, including Lim1 (Campbell 2005; Galindo et al. 2002; Kojima et al. 2005; Tsuji et al. 2000). Surprisingly, depletion of Lim1 in T. castaneum did not affect distal leg development, although it led to reduction of the femur and tibia and loss of proximal leg joints (Figure 3I), paralleling other aspects of Lim1 mutant phenotypes in D. melanogaster (Pueyo et al. 2000). The pretarsus remained unaffected in T. castaneum (in 96 of 96 legs scored). One possible explanation for this apparent difference would be that RNAi was performed too late to affect pretarsus development; the claw expression domain is the earliest to appear in D. melanogaster, whereas the femur and tibia rings appear last. However, this explanation is insufficient because Krn RNAi eliminates the claw and Krn is expected to act earlier than Lim1 if claw development is conserved. Moreover, Lim1 expression in the leg may be biased proximally (Figure 2; two-tailed t-test, P = 0.0759).
Another component of the D. melanogaster tarsal patterning pathway that appears to have a different domain of effect in T. castaneum is rn. In D. melanogaster, Dll activates rn in a broad tarsal domain spanning t2-t4 (reviewed by Kojima 2004). Expression of rn is transient, but the rn null tarsus consists of a single fused segment (Cavener et al. 1986; St. Pierre et al. 2002). Knockdown of rn in T. castaneum produced phenotypes that extended more distally than the D. melanogaster phenotypes (Figure 5), as loss of the pretarsus was also observed (15 of 95 legs scored). Whether this represents a direct or indirect effect of loss of rn expression is unclear.
An interesting aspect of anatomy in T. castaneum is the divergence in tarsus segmentation across the legs. Tribolium and other tenebrionoid beetles are characterized by having five tarsomeres in the pro- and mesothoracic legs but only four tarsomeres in the metathoracic legs (Figure 1, B−D). This heteromery can be analyzed in the context of our functional genetic data (Figure 5, B and B′). The proximal- and distal-most tarsomeres of the metathoracic tarsus are elongated relative to other tarsomeres. Comparison with serial homologs suggests two alternative hypotheses for how the metathoracic tarsus develops: by the suppression of either the t1/t2 or t4/t5 joint present in the pro- and mesothoracic tarsi. In all three legs, several genes have limits of activity that demarcate the basitarsus (t1). The depletion of odd-related paralogs has a high penetrance in t1. Furthermore, Notch and bab RNAi cause joint loss but not reduction in the proximal tarsomere of all legs. Therefore, it does not appear that the metathoracic tarsus develops by a suppression of the most proximal joint within the tarsus. Serial homology of the distitarsus (t5 in T1-T2; t4 in T3) is supported by the boundaries of function for both dac and dpp, which are required for the adjacent proximal segments in all legs. These results lead us to reject the hypothesis that the metathoracic tarsus develops by suppression of a presumptive distal joint. The remaining possibility is that the intermediate tarsal region is differently segmented in pro- and mesothoracic vs. metathoracic legs. In particular, our data suggest that the presumptive t2/t3 joint is unique to pro- and mesothoracic legs. This hypothesis is supported by the fact that odd-related gene knockdown has a relatively high penetrance in the second tarsal joint in pro- and mesothoracic legs but not in metathoracic legs. Conversely, ab RNAi causes fusion of tarsal joints with high penetrance, except in the t2/t3 joint of the pro- and mesothoracic legs (e.g., Figure 4V). This hypothesis is also consistent with a model of tarsus segmentation in which intermediate tarsal identities are intercalated between proximal and distal tarsal identities.
Ontogeny of appendage patterning
The functions of two genes studied here in metamorphic leg patterning have also been examined during embryonic patterning of the larval leg in T. castaneum, allowing for comparisons of leg development over ontogeny. The proximal-to-distal extent of Dll function is conserved between embryonic and metamorphic patterning. Dll mutants (Beermann et al. 2001; Figure 3B) and Dll RNAi specimens (Suzuki et al. 2009; Figure 3, C−E) lack the distal femur and all more distal appendages regions. While Dll mutants always have some distal leg identity as adults (Beermann et al. 2001), the distal leg can be completely deleted in response to RNAi targeting Dll (Figure 3E; Suzuki et al. 2009). Presumably, the retention of distal leg identity in Dll mutants reflects lethality of stronger Dll alleles before the adult stage, whereas RNAi provides a targeted way to reduce its expression at metamorphosis. The deletion of the tibia and tarsus (and the distal femur) in response to Dll RNAi also shows that leg segments present in the larval leg can be completely lost at metamorphosis, a result also found in response to dac RNAi, in which distal femur, tibia and proximal tarsus are deleted, (Suzuki et al. 2009; Figure 3F).
Although Dll and dac function to pattern and maintain large domains of the leg that persist through all life stages, what about genes involved in uniquely adult features of the leg? Depletion of ss during metamorphosis caused fusions or deletions of tarsomeres, but not elsewhere in the leg (Shippy et al. 2009; Figure 4Z). In parental ss RNAi, larval leg defects were not observed (Shippy et al. 2009; Toegel et al. 2009). These differences are consistent with the absence of discrete tarsomeres in the larval leg and with their development during metamorphosis.
The evolution of serially homologous appendages
We have proposed two models representing extreme scenarios for the evolution of developmental mechanisms controlling serially homologous appendages. One model assumes that developmental processes may evolve independently in different serial homologs. However, serial homologs share the same genome making serial homology fundamentally different from special homology (Owen’s terminology, e.g., the leg of T. castaneum and the leg of D. melanogaster). The evolution of serial homologs may be influenced by the sharing of developmental genes and fitness trade-offs for the organism (Boyden 1947; Wagner 2007), such that serial homologs instead evolve in a constrained, dependent manner.
The independent model predicts that gene functions may differ in the development of only a single appendage type between species. In contrast, the dependent model predicts that changes to the leg-patterning network in a species would also cause changes in the patterning network of other appendages in that same species, leading to greater similarity in patterning of serial homologs within species than special homologs between species. The data presented in this study are considered with results from D. melanogaster to test these predictions. Legs are a convenient starting point for such an analysis because their conserved anatomy allows unambiguous determination of whether the PD level of effect has changed across species. For genes with functions that differ in the legs of two species, we then examine whether the gene has a parallel functional difference in any other appendage type. To the extent that serial homologs are developmentally independent within a species, there should be no such parallel changes. To the extent that serial homology stems from shared gene functions, parallel changes are expected.
Several genes function at different PD levels in the legs of T. castaneum and D. melanogaster (see Figure 5, A and C). Lim1 RNAi produced defects only in the proximal to intermediate leg in T. castaneum (Figure 3I), without the pretarsus defects found in D. melanogaster Lim1 mutants (Pueyo et al. 2000; Tsuji et al. 2000). In the adult antennae of T. castaneum, Lim1 was also required for development of the proximal-most segments (Angelini et al. 2009), whereas D. melanogaster Lim1 mutant antennae are either absent, deformed overall, or deformed distally (Pueyo et al. 2000). The high sensitivity of distal appendage regions to loss of Lim1 expression in D. melanogaster compared with the insensitivity of these regions to Lim1 depletion in T. castaneum supports the dependent model. A second example comes from Delta. In D. melanogaster both Ser and Delta are necessary for joint formation (de Celis et al. 1998; Rauskolb and Irvine 1999). However, in T. castaneum Ser RNAi caused widespread joint loss, but only 15% of Delta RNAi specimens had any joint defects despite 71% reduction in pupal transcript numbers as a result of RNAi (Table 2). Low penetrance for Delta RNAi was also seen in the antenna (data not shown) and mouthparts (Angelini et al. 2012). Thus, the features of these two genes that are divergent between species are consistent across appendage types. These results underscore the influence of pleiotropy in serial homologs.
However, other genes provide evidence that serial homologs may evolve in a more independent way. hth functions as a regulator of proximal identity in both T. castaneum and D. melanogaster legs, but the extent of proximal reduction differs between these species. When lacking hth activity, the leg discs of D. melanogaster develop a normal tarsus, but they have a single fused proximal segment (Casares and Mann 2001). Thus, hth is required for cell growth and maintenance in the proximal leg of D. melanogaster. A similar role for hth in leg development is known for a more distantly related species, the cricket Gryllus bimaculatus (Ronco et al. 2008). By contrast, all of the primary leg segments are retained in response to hth depletion in T. castaneum, although the identity of the coxa, trochanter and femur is affected (Figure 3J). However in other T. castaneum appendage types, hth has a role in both growth and patterning. In the maxillae and labium hth knockdown causes proximal transformations, but in severely affected individuals a proximal segment of the palps was deleted (Angelini et al. 2012). Moreover, hth RNAi produced strong reductions in antenna length, through fusions or loss of the intermediate segments (Angelini et al. 2009). In addition, RNAi targeting rn in T. castaneum produced defects in the pretarsus, which is not seen in D. melanogaster rn mutants (Cavener et al. 1986; St. Pierre et al. 2002). However rn loss-of-function in the antenna results in defects in the intermediate funicle region in T. castaneum (Angelini et al. 2009) and the intermediate (a3-a4) segments in D. melanogaster (Cavener et al. 1986). No defects are seen in the distal-most antenna structures of either species. Therefore, the novel aspects of hth and rn function are unique to specific appendage types.
In the case of EGF signaling, differences were observed across species, but in a pattern that is potentially consistent with either hypothesis, depending on the ancestral state. Krn was required in a broader domain including more proximal structures in the legs of T. castaneum compared to the relatively distal requirement for EGF signaling in D. melanogaster legs (Campbell 2002). Krn was also required throughout the antenna (Angelini et al. 2009), and maxillary and labial palps of T. castaneum (Angelini et al. 2012) at metamorphosis. In contrast, in D. melanogaster, loss of EGF receptor function has not produced described defects in antennal PD patterning (Amin and Finkelstein 1999; Clifford and Schupbach 1989).
These functional comparisons among serial homologs reveal some support for each hypothesis. Although negative results, such as the absence of phenotypes in the pretarsus for Lim1 RNAi, low penetrance for Delta knockdown, and the lack of leg reduction in hth RNAi, are relatively weak support for each hypothesis, positive results, such as the novel function for rn in T. castaneum, provide stronger support. Complete dependence or independence in the evolution of serial homologs is not expected. Instead our data suggest a complex mix of divergence and constraint among appendages. Despite the breadth of this study, we still have only a limited number of comparisons. Data from other genes and taxa also suggest mixed support for the two hypotheses. For example, pdm/nubbbin has exceptionally labile expression (Li and Popadić 2004) and functional domains (Hrycaj et al. 2008; Turchyn et al. 2011) among insects, but there is some consistency across special homologs, as predicted by the independent model. More extensive comparative functional studies will be necessary in the future to evaluate the extent of developmental dependence among serial homologs, and to examine whether genes involved in certain developmental processes are more prone to divergence in limited or universal ways.
Conclusion
Serial homology has been considered in biology for more than two centuries, and it is characterized by the influence of shared developmental processes or pleiotropy (Boyden 1947; Owen 1848; Roth 1984; Wagner 2007). On one hand, shared development indicates that serial homology presents an accessible pathway through which novel structures may originate. On the other hand, shared development may constrain the subsequent evolution of serial homologs. To the extent that selection favors divergence in the function of serial homologs, any genes deployed in these structures may be subject to antagonistic pleiotropy. Developmental changes favored by selection on a structure may result in developmental changes of the serially homologous structures at other positions in the body. Thus, it is likely that trade-offs resulting from pleiotropy bias the mutations that are ultimately fixed by selection during evolution (Stern and Orgogozo 2009). The prevalence of this influence remains an important gap in our knowledge of evolution. This study has made a small-scale test of the question here, finding that changes in leg patterning between groups are often accompanied by parallel changes in other appendages. Comparative studies in diverse species and other types of serially homologous structures will help to resolve the issue.
Supplementary Material
Acknowledgments
We thank Martin Klingler for generously sharing cDNA clones of T. castaneum dachshund and Sue Brown for providing the Dll mutant stock. An anonymous reviewer provided helpful comments on the manuscript. We thank Moto Kikuchi for assistance with microinjections and phenotype scoring and Ariel Aspiras for assistance with real-time PCR. Assistance with microinjections was also provided by Sean D. Burn, Derek Cornetta, Matthew Gaudio, Ashley Johnson, Jessica Klein, Abigail Labella, Avis Thompson, and Chelsea Willet. Electron microscopy training and assistance were provided by James Romanow. This work was supported through an National Institutes of Health National Research Service Award Kirschstein postdoctoral fellowship to D.R.A. (5F32GM074365-02) and a U.S.D.A. Cooperative State Research, Education, and Extension Service seed grant (2006-35604-16746) to E.L.J.
Footnotes
Communicating editor: R. Kulathinal
Literature Cited
- Abu-Shaar M., Mann R. S., 1998. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development 125: 3821–3830 [DOI] [PubMed] [Google Scholar]
- Abzhanov A., Kaufman T. C., 2000. Homologs of Drosophila appendage genes in the patterning of arthropod limbs. Dev. Biol. 227: 673–689 [DOI] [PubMed] [Google Scholar]
- Amin A., Li Y., Finkelstein R., 1999. Hedgehog activates the EGF receptor pathway during Drosophila head development. Development 126: 2623–2630 [DOI] [PubMed] [Google Scholar]
- Angelini D. R., Kaufman T. C., 2004. Functional analyses in the hemipteran Oncopeltus fasciatus reveal conserved and derived aspects of appendage patterning in insects. Dev. Biol. 271: 306–321 [DOI] [PubMed] [Google Scholar]
- Angelini D. R., Kaufman T. C., 2005a Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev. Biol. 283: 409–423 [DOI] [PubMed] [Google Scholar]
- Angelini D. R., Kaufman T. C., 2005b Insect appendages and comparative ontogenetics. Dev. Biol. 286: 57–77 [DOI] [PubMed] [Google Scholar]
- Angelini D. R., Kikuchi M., Jockusch E. L., 2009. Genetic patterning in the adult capitate antenna of the beetle Tribolium castaneum. Dev. Biol. 327: 240–251 [DOI] [PubMed] [Google Scholar]
- Angelini D. R., Smith F. W., Aspiras A. C., Kikuchi M., Jockusch E. L., 2012. Patterning of adult mandibulate mouthparts in the red flour beetle, Tribolium castaneum. Genetics 190: 639–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras A. C., Smith F. W., Angelini D. R., 2011. Sex-specific gene interactions in the patterning of insect genitalia. Dev. Biol. 360: 369–380 [DOI] [PubMed] [Google Scholar]
- Beermann A., Jay D. G., Beeman R. W., Hulskamp M., Tautz D., et al. , 2001. The Short antennae gene of Tribolium is required for limb development and encodes the orthologue of the Drosophila Distal-less protein. Development 128: 287–297 [DOI] [PubMed] [Google Scholar]
- Bishop S. A., Klein T., Arias A. M., Couso J. P., 1999. Composite signalling from Serrate and Delta establishes leg segments in Drosophila through Notch. Development 126: 2993–3003 [DOI] [PubMed] [Google Scholar]
- Boyden A., 1947. Homology and analogy. A critical review of the meanings and implications of these concepts in biology. Am. Midl. Nat. 37: 648–669 [Google Scholar]
- Boxshall G. A., 2004. The evolution of arthropod limbs. Biol. Rev. Camb. Philos. Soc. 79: 253–300 [DOI] [PubMed] [Google Scholar]
- Campbell G., 2002. Distalization of the Drosophila leg by graded EGF-receptor activity. Nature 418: 781–785 [DOI] [PubMed] [Google Scholar]
- Campbell G., 2005. Regulation of gene expression in the distal region of the Drosophila leg by the Hox11 homolog, C15. Dev. Biol. 278: 607–618 [DOI] [PubMed] [Google Scholar]
- Casares F., Mann R. S., 2001. The ground state of the ventral appendage in Drosophila. Science 293: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Otteson D. C., Kaufman T. C., 1986. A rehabilitation of the genetic map of the 84B-D region in Drosophila melanogaster. Genetics 114: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Dong P. D. S., Panganiban G., 2002. Limb type-specific regulation of bric a brac contributes to morphological diversity. Development 129: 695–704 [DOI] [PubMed] [Google Scholar]
- Clifford R. J., Schupbach T., 1989. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics 123: 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. M., Jürgens G., 1989. Proximal-distal pattern formation in Drosophila: graded requirement for Distal-less gene activity during limb development. Roux’s Arch. Dev. Biol. 198: 157–169 [DOI] [PubMed] [Google Scholar]
- Couderc J.-L., Godt D., Zollman S., Chen J., Li M., et al. , 2002. The bric à brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development 129: 2419–2433 [DOI] [PubMed] [Google Scholar]
- Daly H. V., Doyen J. T., Purcell A. H. I., 1998. Introduction to Insect Biology and Diversity. Oxford University Press, Oxford [Google Scholar]
- Darwin C., 1859. On The Origin of Species by Means of Natural Selection. Murray, London [Google Scholar]
- De Celis Ibeas J. M., Bray S. J., 2003. Bowl is required downstream of Notch for elaboration of distal limb patterning. Development 130: 5943–5952 [DOI] [PubMed] [Google Scholar]
- De Celis J. F., Tyler D. M., De Celis J., Bray S. J., 1998. Notch signalling mediates segmentation of the Drosophila leg. Development 125: 4617–4626 [DOI] [PubMed] [Google Scholar]
- Dehal P., Boore J. L., 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Benjumea F. J., Cohen B., Cohen S. M., 1994. Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature 372: 175–179 [DOI] [PubMed] [Google Scholar]
- Duncan D. M., Burgess E. A., Duncan I., 1998. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 12: 1290–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M. S., Grimaldi D. A., 2004. New light shed on the oldest insect. Nature 427: 627–630 [DOI] [PubMed] [Google Scholar]
- Galindo M. I., Bishop S. A., Greig S., Couso J. P., 2002. Leg patterning driven by proximal-distal interactions and EGFR signaling. Science 297: 256–259 [DOI] [PubMed] [Google Scholar]
- Galindo M. I., Bishop S. A., Couso J. P., 2005. Dynamic EGFR-Ras signalling in Drosophila leg development. Dev. Dyn. 233: 1496–1508 [DOI] [PubMed] [Google Scholar]
- Goethe J. W. v., 1790 Versuch die Metamorphose der Pflanzen zu erklaren. Ettingersche Buchhandlung, Gotha. [Google Scholar]
- Godt D., Couderc J. L., Cramton S. E., Laski F. A., 1993. Pattern formation in the limbs of Drosophila: bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development 119: 799–812 [DOI] [PubMed] [Google Scholar]
- Greenberg L., Hatini V., 2009. Essential roles for lines in mediating leg and antennal proximodistal patterning and generating a stable Notch signaling interface at segment borders. Dev. Biol. 330: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao I., Green R. B., Dunaevsky O., Lengyel J. A., Rauskolb C., 2003. The odd-skipped family of zinc finger genes promotes Drosophila leg segmentation. Dev. Biol. 263: 282–295 [DOI] [PubMed] [Google Scholar]
- Hrycaj S., Mihajlovic M., Mahfooz N., Couso J. P., Popadić A., 2008. RNAi analysis of nubbin embryonic functions in a hemimetabolous insect, Oncopeltus fasciatus. Evol. Dev. 10: 705–716 [DOI] [PubMed] [Google Scholar]
- Hu S., Fambrough D., Atashi J. R., Goodman C. S., Crews S. T., 1995. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 9: 2936–2948 [DOI] [PubMed] [Google Scholar]
- Huet C., Lenoir-Rousseaux J. J., 1976. Etude de la mise en place de la patte imaginable de Tenebrio molitor. 1. Analyse expérimentale des processus de restauration au cours de la morphogenése. J. Embryol. Exp. Morphol. 35: 303–321 [PubMed] [Google Scholar]
- Inoue Y., Mito T., Miyawaki K., Matsushima K., Shinmyo Y., et al. , 2002. Correlation of expression patterns of homothorax, dachshund, and Distal-less with the proximodistal segmentation of the cricket leg bud. Mech. Dev. 113: 141–148 [DOI] [PubMed] [Google Scholar]
- Jockusch E. L., Nulsen C., Newfeld S. J., Nagy L. M., 2000. Leg development in flies vs. grasshoppers: differences in dpp expression do not lead to differences in the expression of downstream components of the leg patterning pathway. Development 127: 1617–1626 [DOI] [PubMed] [Google Scholar]
- Jockusch E. L., Williams T. A., Nagy L. M., 2004. The evolution of patterning of serially homologous appendages in insects. Dev. Genes Evol. 214: 324–338 [DOI] [PubMed] [Google Scholar]
- Keilin D., 1915. Recherches sur les larves de diptères cyclorhaphes. Bull. Sci. Fr. Belg. 49: 15–198 [Google Scholar]
- Kojima T., 2004. The mechanism of Drosophila leg development along the proximodistal axis. Dev. Growth Differ. 46: 115–129 [DOI] [PubMed] [Google Scholar]
- Kojima T., Sato M., Saigo K., 2000. Formation and specification of distal leg segments in Drosophila by dual Bar homeobox genes, BarH1 and BarH2. Development 127: 769–778 [DOI] [PubMed] [Google Scholar]
- Kojima T., Tsuji T., Saigo K., 2005. A concerted action of a paired-type homeobox gene, aristaless, and a homolog of Hox11/tlx homeobox gene, clawless, is essential for the distal tip development of the Drosophila leg. Dev. Biol. 279: 434–445 [DOI] [PubMed] [Google Scholar]
- Kozu S., Tajiri R., Tsuji T., Michiue T., Saigo K., et al. , 2006. Temporal regulation of late expression of Bar homeobox genes during Drosophila leg development by Spineless, a homolog of the mammalian dioxin receptor. Dev. Biol. 294: 497–508 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Cohen S. M., 1997. Proximal-distal axis formation in the Drosophila leg. Nature 388: 139–145 [DOI] [PubMed] [Google Scholar]
- Li H., Popadić A., 2004. Analysis of nubbin expression patterns in insects. Evol. Dev. 6: 310–324 [DOI] [PubMed] [Google Scholar]
- Lilly B., O’Keefe D. D., Thomas J. B., Botas J., 1999. The LIM homeodomain protein dLim1 defines a subclass of neurons within the embryonic ventral nerve cord of Drosophila. Mech. Dev. 88: 195–205 [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G., Solomon N. M., Rubin G. M., 1994. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120: 3473–3486 [DOI] [PubMed] [Google Scholar]
- Moczek A. P., Rose D. J., 2009. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl. Acad. Sci. USA 106: 8992–8997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Kuwayama H., Yaginuma T., 2005. Larval RNAi applied to the analysis of postembryonic development in the ladybird beetle, Harmonia axyridis. J. Insect Biotechnol. Sericology 74: 95–102 [Google Scholar]
- Ober K. A., Jockusch E. L., 2006. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev. Biol. 294: 391–405 [DOI] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin [Google Scholar]
- Owen R., 1848. On the Archetype and Homologies of the Vertebrate Skeleton. Richard and John E. Taylor, London [Google Scholar]
- Palopoli M. F., Patel N. H., 1998. Evolution of the interaction between Hox genes and a downstream target. Curr. Biol. 8: 587–590 [DOI] [PubMed] [Google Scholar]
- Panganiban G., 2000. Distal-less function during Drosophila appendage and sense organ development. Dev. Dyn. 218: 554–562 [DOI] [PubMed] [Google Scholar]
- Panganiban G., Nagy L., Carroll S. B., 1994. The role of the Distal-less gene in the development and evolution of insect limbs. Curr. Biol. 4: 671–675 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpic N.-M., Damen W. G. M., 2009. Notch-mediated segmentation of the appendages is a molecular phylotypic trait of the arthropods. Dev. Biol. 326: 262–271 [DOI] [PubMed] [Google Scholar]
- Prpic N. M., Wigand B., Damen W. G., Klingler M., 2011. Expression of dachshund in wild-type and Distal-less mutant Tribolium corroborates serial homologies in insect appendages. Dev. Genes Evol. 211: 467–477 [DOI] [PubMed] [Google Scholar]
- Pueyo J. I., Galindo M. I., Bishop S. A., Couso J. P., 2000. Proximal-distal leg development in Drosophila requires the apterous gene and the Lim1 homologue dlim1. Development 127: 5391–5402 [DOI] [PubMed] [Google Scholar]
- Rauskolb C., 2001. The establishment of segmentation in the Drosophila leg. Development 128: 4511–4521 [DOI] [PubMed] [Google Scholar]
- Rauskolb C., Irvine K. D., 1999. Notch-mediated segmentation and growth control of the Drosophila leg. Dev. Biol. 210: 339–350 [DOI] [PubMed] [Google Scholar]
- Rogers B. T., Peterson M. D., Kaufman T. C., 2002. The development and evolution of insect mouthparts as revealed by the expression Patterns of Gnathocephalic genes. Evol. Dev. 4: 96–110 [DOI] [PubMed] [Google Scholar]
- Roth V. L., 1984. On homology. Biol. J. Linn. Soc. Lond. 22: 13–29 [Google Scholar]
- Ronco M., Uda T., Mito T., Minelli A., Noji S., et al. , 2008. Antenna and all gnathal appendages are similarly transformed by homothorax knock-down in the cricket Gryllus bimaculatus. Dev. Biol. 313: 80–92 [DOI] [PubMed] [Google Scholar]
- Sanchez-Salazar J., Pletcher M. T., Bennett R. L., Brown S. J., Dandamudi T. J., et al. , 1996. The Tribolium decapentaplegic gene is similar in sequence, structure, and expression to the Drosophila dpp gene. Dev. Genes Evol. 206: 237–246 [DOI] [PubMed] [Google Scholar]
- Schoppmeier M., Damen W. G., 2001. Double-stranded RNA interference in the spider Cupiennius salei: the role of Distal-less is evolutionarily conserved in arthropod appendage formation. Dev. Genes Evol. 211: 76–82 [DOI] [PubMed] [Google Scholar]
- Shippy T., Yeager S., Denell R., 2009. The Tribolium spineless ortholog specifies both larval and adult antennal identity. Dev. Genes Evol. 219: 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass R. E., 1935. Principles of Insect Morphology. McGraw-Hill, New York [Google Scholar]
- Sokoloff A., 1972. The Biology of Tribolium, Vol. 1 Clarendon Press, Oxford [Google Scholar]
- Stern D. L., Orgogozo V., 2009. Is genetic evolution predictable? Science 323: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Pierre S. E., Galindo M. I., Couso J. P., Thor S., 2002. Control of Drosophila imaginal disc development by rotund and roughened eye: differentially expressed transcripts of the same gene encoding functionally distinct zinc finger proteins. Development 129: 1273–1281 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Squires D. C., Riddiford L. M., 2009. Larval leg integrity is maintained by Distal-less and is required for proper timing of metamorphosis in the flour beetle, Tribolium castaneum. Dev. Biol. 326: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svacha P., 1992. What are and what are not imaginal discs: reevaluation of some basic concepts (Insecta, Holometabola). Dev. Biol. 154: 101–117 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Truman J. W., 2005. Development of the adult leg epidermis in Manduca sexta: contribution of different larval cell populations. Dev. Genes Evol. 215: 78–89 [DOI] [PubMed] [Google Scholar]
- Theisen H., Haerry T. E., O'connor M. B., Marsh J. L., 1996. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development 122: 3939–3948 [DOI] [PubMed] [Google Scholar]
- Toegel J., Wimmer E., Prpic N.-M., 2009. Loss of spineless function transforms the Tribolium antenna into a thoracic leg with pretarsal, tibiotarsal, and femoral identity. Dev. Genes Evol. 219: 53–58 [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y., Denell R. E., 2004. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214: 575–578 [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium, 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955 [DOI] [PubMed] [Google Scholar]
- True J. R., Haag E. S., 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119 [DOI] [PubMed] [Google Scholar]
- Tsuji T., Sato A., Hiratani I., Taira M., Saigo K., et al. , 2000. Requirements of Lim1, a Drosophila LIM-homeobox gene, for normal leg and antennal development. Development 127: 4315–4323 [DOI] [PubMed] [Google Scholar]
- Turchyn N., Chesebro J., Hrycaj S., Couso J. P., Popadić A., 2011. Evolution of nubbin function in hemimetabolous and holometabolous insect appendages. Dev. Biol. 357: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. M., 1977. Optical clean and permanent whole mount preparation for phase-contrast microscopy of cuticular structures of insect larvae. Dros. Inf. Serv. 52: 160 [Google Scholar]
- van der Zee M., Stockhammer O., Von Levetzow C., Nunes Da Fonseca R., Roth S., 2006. Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. Proc. Natl. Acad. Sci. USA 103: 16307–16312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., 2007. The developmental genetics of homology. Nat. Rev. Genet. 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Wu J., Cohen S. M., 1999. Proximodistal axis formation in the Drosophila leg: subdivision into proximal and distal domains by homothorax and Distal-less. Development 126: 109–117 [DOI] [PubMed] [Google Scholar]
- Zeng C., Justice N. J., Abdelilah S., Chan Y. M., Jan L. Y., et al. , 1998. The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc. Natl. Acad. Sci. USA 95: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.