Abstract
To detect pre-patent parasitemia, we developed a real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for the asexual 18S ribosomal RNA (rRNAs) of Plasmodium falciparum. Total nucleic acids extracted from whole blood were combined with control RNA and tested by qRT-PCR. The assay quantified > 98.7% of parasite-containing samples to ±0.5 log10 parasites/mL of the nominal value without false positives. The analytical sensitivity was ≥ 20 parasites/mL. The coefficient of variation was 0.6% and 1.8% within runs and 1.6% and 4.0% between runs for high and low parasitemia specimens, respectively. Using this assay, we determined that A-type 18S rRNAs are stably expressed at 1×104 copies per ring-stage parasite. When used to monitor experimental P. falciparum infection of human volunteers, the assay detected blood-stage infections 3.7 days earlier on average than thick blood smears. This validated, internally controlled qRT-PCR method also uses a small (50 μL) sample volume requiring minimal pre-analytical handling, making it useful for clinical trials.
Introduction
Infection by Plasmodium falciparum results in nearly one million deaths annually, mostly in children in sub-Saharan Africa. The effort to eradicate this parasite is based on interrupting its lifecycle using vaccines, anti-malarial drugs, bed nets, and insecticides. Currently, microscopic examination of thick blood smears is the gold standard for detection of parasites because of its availability, low cost, and ability to provide species identification and rudimentary quantification. With expert microscopists, thick blood smears detect 2–20 asexual parasites/μL of whole blood (2,000–20,000/mL). However, even at this parasite density, infected patients with sub-patent parasitemia may receive false negative diagnoses. 1,2 Rapid diagnostic tests for malarial antigens can sometimes outperform blood smears in developed world settings, 3 but they are less specific in regions of high prevalence and following drug treatment. 4
A variety of molecular methods of malaria parasite detection were developed in the past 20 years and include single-step and nested polymerase chain reaction (PCR) with gel detection, RNA-specific nucleic acid-based sequence amplification (NASBA), and probe-based real-time PCR and reverse transcription (RT)-PCR (using various combinations of genus- or species-specific primers and TaqMan probes or molecular beacons). 5–23 Species identification can be achieved by SYBR Green melting curve analysis, 20 by real-time PCR with species-specific probes, 5,6 or most sensitively using labor-intensive species-specific nested PCR. 24,25 In addition to diagnosing clinical cases with low parasite densities, some species-specific methods can detect pre-patent parasitemia in samples from field studies and from malaria human challenge trials. 26 Although the most common primary endpoint for blood-stage infection in human challenge trials is detection of ring-stage parasites on thick blood smears, both PCR- and nucleic acid sequence-based amplification (NASBA)-based methods have been used in concert with blood smears in several studies to detect sub-patent parasitemia, confirm diagnosis, and model parasite dynamics. 27–29 With test sensitivities approaching 20 parasites/mL, P. falciparum parasites have been detected by molecular methods in the blood of human volunteers 2–4 days earlier than corresponding thick blood smears. 30–32 Quantification by molecular methods allows the determination of asexual parasite growth kinetics before drug treatment, which may help to prioritize partially effective vaccine candidates.
Most molecular approaches are based on detection of conserved regions of parasite DNA encoding the 18S ribosomal RNA (rDNA) or the 18S rRNA itself. In P. falciparum, there are four 18S (or small subunit) rRNA genes differentially expressed during the lifecycle. 33,34 Asexual or “A”-type rRNAs encoded on chromosomes 5 (MAL5_18S) and 7 (MAL7_18Sa) are found in the blood-stage of infection and persist into the mosquito blood meal, whereas “S” type rRNAs encoded on chromosomes 1 (MAL1_18S) and 11 (MAL11_rRNA) are found during oocyst maturation through sporozoite formation; in some non-P. falciparum species, a second sexual-type 18S rRNA (type “O”) is present as well. 34
We sought to develop a highly sensitive, precise, and reproducible internally controlled P. falciparum-specific quantitative assay on a small volume of whole blood with minimal pre-analytical processing to support clinical care and malaria vaccine research. Here, we describe the development and evaluation of a real-time qRT-PCR assay that fulfills these criteria and its application to monitor infections in a recent human challenge trial.
Methods
Reagents
Unless otherwise specified, all chemicals were reagent-grade from Fisher Scientific (Pittsburgh, PA).
Target sequence selection
The PL1473/PL1679 primers were reported to detect all Plasmodium spp. known to infect humans and primer sequence numbering was originally based on GenBank accession no. M19173 for 18S rRNAs not present in asexual stage parasites. Thus, the primer names are listed here as initially reported. 20 The primers match the asexual-stage 18S rRNA P. falciparum genes (MAL5_18S, MAL7_18Sa) and the P1679 primer lacks the 2-bp insertion present in both sexual-type 18S rRNA transcripts (Figure 1).
Figure 1.
Alignment of Plasmodium falciparum 18S sexual- and asexual-type rRNAs and assay reagents. CLUSTAL-W alignments of the assay reagents to the four 18S ribosomal RNA (rRNA) genes of P. falciparum 3D7. Asterisks denote homology between assay reagents and all S- and A-type genes. The assay reagents are completely matched to the A-type 18S rRNAs (MAL5_18S and MAL7_18Sa). PF1473F18 and PL1679R18 (Rev. Comp.) refer to the forward primer and the reverse complement of the reverse primer; PFALF and PFALR refer to the donor and acceptor probes, respectively.
Primers and probes
Primers (PL1473F18: 5′-TAACGAACGAGATCTTAA-3′; PL1679R18: 5′-GTTCCTCTAAGAAGCTTT-3′) were stored in sterile RNase-/DNase-free water as 10 μM stock solutions at −20°C. Control primers were human β-globin (βg1: 5′-GGGCTGGGCATAAAAGTCA-3′; βg2: 5′-AATAGACCAATAGGCAGA-3′) and human acidic ribosomal protein RPLP2 (NM_001004; forward primer: TCTTGCGTCGGCGCCTTC; reverse primer: CACGCTGTCCAAGATCTTC). All primers were from Invitrogen (Carlsbad, CA).
HPLC-grade dual hybridization fluorescence resonance energy transfer (FRET) probes from TIB Molbiol (Adelphia, NJ) were diluted in HPLC-grade water and stored as 10 μM stock solutions at −80°C protected from light. The P. falciparum-specific probes (donor probe PFALF: 5′-TTGAAATTGAACATAGGTAACTATA-[3′ fluorescein]; acceptor probe PFALR: [5′ LightCycler Red 610]-ATTTATTCAGTAATCAAATTAGGAT-[3′ phosphate]) match the P. falciparum A-type 18S rRNAs, but have multiple mismatches to S-type transcripts (Figure 1). A competitor RNA acceptor probe was also obtained ([5′ LightCycler Red 705]-TAGTGTTATCAGTATAAGAACTCAT-[3′ phosphate]); the competitor RNA is described below. The 18S rRNA target and the competitive control RNA were differentiated on the basis of different acceptor hybridization probes (Figure 2).
Figure 2.
Plasmodium falciparum real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) reagents. The full-length P. falciparum A-type 18S rRNA and in vitro transcribed (IVT) synthetic RNAs corresponding to the 18S ribosomal RNA (rRNA) amplicon and a competitor RNA with a mutated internal region can all be amplified with the same common PCR primers and all hybridize to the same FITC-labeled donor probe. An LC610-labeled acceptor probe binds to the native 18S rRNA sequence present in the full-length sequence and IVT RNA sequence, whereas a unique LC705-labeled acceptor probe for the competitor RNA is used to bind the mutated region. In this way, competitive amplification is achieved with minimal multiplexing.
DNA-based PCR
Extractions for species-identification PCR on whole blood were performed as described 20 on archival de-identified blood samples collected from 2004 to 2009 and previously speciated by the Clinical Microbiology service of the Department of Laboratory Medicine at the University of Washington Medical Center and Harborview Medical Center (Seattle, WA). Samples consisted of either the original EDTA-anticoagulated whole blood frozen at −20°C or Giemsa-stained thin blood smears stored at room temperature in segregated slide trays. Some EDTA-anticoagulated blood samples collected within 2 weeks of testing were stored at 4°C without freezing. Blood smears were scraped into a microfuge tube with a clean razor blade before extraction. De-identified clinical histories and results of thick and thin blood smears and/or BinaxNOW point-of-care malaria immunoassays were evaluated, if available. Samples with an initial microscopic diagnosis of “unspecified Plasmodium” were not included in the overall P. falciparum sensitivity and specificity analyses. The SYBR Green PCR was performed using the FastStart DNA Master SYBR Green I kit (Roche, Indianapolis, IN); probe-based PCR was performed using a LightCycler FastStart DNA Master Hybridization Probe kit (Roche). Both assays used 2 μL of extracted DNA template. Thermal cycling on a LightCycler 2.0 consisted of 95°C (10 min) followed by 45 PCR cycles (95°C [20 sec], 50°C [20 sec], 72°C [20 sec]) with melting curve analysis (35–95°C). Control β-globin PCR consisted of 95°C (10 min) followed by 35 PCR cycles (95°C [10 sec], 55°C [15 sec], 72°C [15 sec]) and melting curve analysis (40–95°C).
RNA calibration standards and competitive RNA control
Plasmodium falciparum RNA standards and competitive control RNA were prepared using a modification of published methods. 35 The P. falciparum A-type 18S rRNA sequence (nucleotides 1392–1722 of MAL5_18S:rRNA) and a corresponding competitor RNA differing only at the acceptor probe binding site were selected and the upstream (nucleotides 1392–1421) and downstream (nucleotides 1636–1772) sequences were modified to encode > 35% overall G/C content to optimize synthesis (See Supplementary Data for sequences and competitor RNA design). The DNA was synthesized and cloned into pIDT-Blue (Integrated DNA Technologies, Coralville, IA), transformed into Escherichia coli, purified, and sequenced. Positive-sense RNA transcripts were produced using the T7 MEGAshortscript High Yield Transcription kit (Ambion, Austin, TX) from linearized, RNase-treated (Ambion) plasmid DNA. The RNA was treated with two rounds of DNA-free DNase (Ambion) in the presence of RNase inhibitors, purified, analyzed for size and purity (Bioanalyzer 2100, Agilent, Santa Clara, CA), and quantified (260 nm). Transcripts showing no contaminating DNA by real-time PCR were diluted to 1×108 copies/μL in 10 mM Tris (pH 7) containing 10 μg/mL carrier tRNA (Sigma, St. Louis, MO), aliquoted, frozen on dry ice, and stored at −80°C. The RNA from the plasmid encoding the 18S rRNA fragment were used as RNA calibration standards, whereas RNA from the competitor RNA plasmid was used as a competitive control, as described below.
Parasite culture
Plasmodium falciparum 3D7 strain was cultured in type O human erythrocytes using a modification of the Trager and Jensen method. 36 The RPMI 1640 containing 2 mM L-glutamine and 25 mM HEPES was supplemented with 0.5% Albumax II (GIBCO, San Diego, CA), 26.8 mM NaHCO3, 11 mM dextrose, 360 μM hypoxanthine, 5% human type AB sera, and 11 μg/L gentamicin. Cultures were placed in a Modular Incubator Chamber (Billups-Rothenberg, Inc., Del Mar, CA), gassed with 5% O2 and 5% CO2 (in N2) and incubated at 37°C. Late-stage trophozoites and schizonts were enriched using LS columns (Miltenyi Biotech, Bergisch Gladbach, Germany) with a MidiMACS magnet to exclude remaining ring stages and enrich for a synchronous population of late-stage trophozoites and schizonts. 37 Resulting mature parasites were washed in serum-free medium, resuspended in growth medium, and allowed to invade fresh erythrocytes at 1–5% hematocrit.
For time course studies, MACS-purified mature parasites were cultured with fresh erythrocytes for 1 hr before cultures were treated with 5% D-sorbitol for 15 min at 37°C to destroy remaining mature stages. 38 Sorbitol-treated samples were washed three times in serum-free media, resuspended to 0.5% Hct in growth medium, and incubated as 1 mL cultures in 24-well plates under standard conditions. Wells were harvested in triplicate at various times post-invasion by centrifugation (600×g for 1 min). For some samples, media was removed and the resulting 5 μL cell pellet was lysed in 1 mL EasyMAG lysis buffer (hereafter referred to simply as “lysis buffer,” bioMérieux, Marcy l'Etoile, France) and frozen at −80°C; some samples were used to make a Giemsa-stained thin blood smears at each time point to confirm culture synchrony. Before extraction, time course samples were combined with 1.05 mL lysis buffer containing 50 μL malaria-free whole blood to ensure a comparable whole blood matrix.
Parasite standards
To produce large-scale cultures for controls and standard curves, synchronized, slide-confirmed ring-stage parasites were harvested at 12 hrs post-invasion. Parasites were serially diluted in freshly collected EDTA-treated whole blood (4×109 red blood cell/mL determined by Coulter counting [Beckman Coulter, Inc., Miami, FL]) from 10% to 0.000001% parasitemia; additional standards were prepared at 0.0002% and 0.000002% parasitemia. The parasitemia of diluted samples was reconfirmed by Giemsa staining blood smears from dilutions 10% through 0.01% parasitemia.
Stabilization of whole blood samples
Fifty μL of freshly-collected whole blood (with or without parasites) were added to 2 mL of room temperature lysis buffer, vortexed, and stored at −80°C until processing. Sometimes, samples were stored in 1 mL lysis buffer, with the addition of an additional 1 mL lysis buffer immediately before extraction. Lysed frozen samples were always transported to the central processing laboratory on dry ice.
qRT-PCR
Before extraction, a single-use aliquot of competitor RNA was defrosted and diluted to 1×104 copies/μL in lysis buffer. Competitor RNA (5×104 copies) was added to every sample and samples were extracted for total nucleic acids using the Specific Protocol B (off-board lysis) using 70 μL silica beads and 40 μL elution volume on a silica-based EasyMAG (bioMérieux). All instrument runs included a high parasitemia control (4×105 parasites/mL) and a malaria-negative control. Eluates were processed by qRT-PCR on the same day as extraction. Fifty μL qRT-PCR reactions were performed using 5 μL eluate with the QuantiTect Multiplex RT-PCR NR Kit (Qiagen, Valencia, CA) using a LightCycler 2.0; cycling conditions consisted of 50°C (20 min), 95°C (15 min), 45 cycles of PCR (95°C [20 sec], 50°C [20 sec], 72°C [20 sec]; ramp speed 20°C/sec) with a final melting curve (37–80°C at 0.1°C/sec). After color compensation, data were analyzed at 610/530 nm and 705/530 nm. Quantification was reported for clinical samples with cycle thresholds (CT) ≤ 36.8 cycles (determined to be equivalent to one parasite in the starting 50 μL extraction volume; see Results). In the presence of a large amount of P. falciparum target, the competitor RNA does not amplify, whereas in the presence of little or no P. falciparum target, the competitor reproducibly amplifies within a controlled range.
An RNA standard curve was made by diluting the in vitro transcribed (IVT) RNA calibrators (1×103–1×108 RNA copies/reaction) into total nucleic acids previously extracted from uninfected whole blood (containing the usual amount of competitor RNA). This RNA standard curve was used to determine the log10 RNA copies per RT-PCR reaction. To calculate log10 RNA copies/mL of whole blood, the result is multiplied by 160 to account for both the 5 μL RT-PCR template (12.5% of the extraction eluate) and the initial 50 μL blood sampling volume (160 = [100%/12.5%] × [1000 uL/50 uL]). The absolute parasite density (parasites/mL) was further calculated by dividing the log10 RNA copies/mL by the log10 copy number of RNAs per parasite. This conversion factor (4 log10 copies/ring-stage parasite) was experimentally determined in this study by comparing the RNA standards against whole blood samples with known parasite density; see Results).
For some control reactions, human RPLP2 mRNA was amplified using the iScript One-Step RT-PCR Kit with SYBR Green kit (Biorad, Hercules, CA) with cycling conditions of 50°C (10 min), 95°C (5 min), 45 cycles of PCR (95°C [10 sec], 55°C [30 sec], 72°C [20 sec]; ramp speed 20°C/sec), and melting curve analysis (50–80°C at 0.5°C/sec).
Clinical trial
Six adult human volunteers were experimentally infected with wild-type strain NF54 P. falciparum sporozoites by the bite of five infected Anopheles stephensi mosquitoes under controlled containment conditions; after feeding, mosquitoes were dissected to confirm that salivary glands were sufficiently infected according to standard procedures. 39 Informed consent was obtained under a clinical protocol (study no. NCT01058226) approved by the Western Institutional Review Board (WIRB). Subjects were treated upon first evidence of microscopic parasitemia or at Day 18 if they remained blood smear negative; treatment consisted of a standard oral regimen of chloroquine under direct observation. Blood samples for qRT-PCR were collected twice daily from Day 5 post-challenge until the day of positive blood smear diagnosis and treatment, and then daily until blood smears were negative and once again on Day 28 post-challenge. Whole blood was collected in spray-dried K2-EDTA Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ) by clinical staff, mixed, and transported to the laboratory within 4 hrs of collection. Fifty μL of blood were added to 2 mL lysis buffer, and samples were frozen at −80°C.
The qRT-PCR processing technologists were blinded to the volunteer number, collection date, or visit number. In parallel with qRT-PCR, thick blood smears were prepared and interpreted using standardized methods. Volunteers were treated when two unequivocal parasites were observed by an expert microscopist on a thick blood smear. The qRT-PCR was performed in batches at the conclusion of the trial and was not used to make treatment decisions.
Results
Assay development
A number of factors were evaluated before validating an optimized qRT-PCR assay. The reproducibility of extraction of 50 μL of whole blood in 2 mL lysis buffer was assessed by testing 11 individually extracted samples of whole human blood from the same individual by SYBR Green real-time RT-PCR for the human acidic ribosomal protein RPLP2 mRNA. The RPLP2 CT of all extractions was 19.6 cycles (95% confidence interval [CI] = 18.5–20.8, %CV 3.0%). Thus, extraction was reproducible.
Singleplex reactions using the malaria reagents only (no competitor RNA) were optimized by varying primer and probe concentrations and annealing/extension times and temperatures. Singleplex qRT-PCR on the malaria RNA standards alone or the competitor RNA alone showed linear kinetics (malaria RNA standards: r2 = 0.993, slope = −3.32, efficiency = 100%; Competitor RNA: r2 = 0.990, slope = −3.38, efficiency 98%) with analytical sensitivities of 500 copies per reaction by CT analysis and 100 copies by melting curve analysis (data not shown).
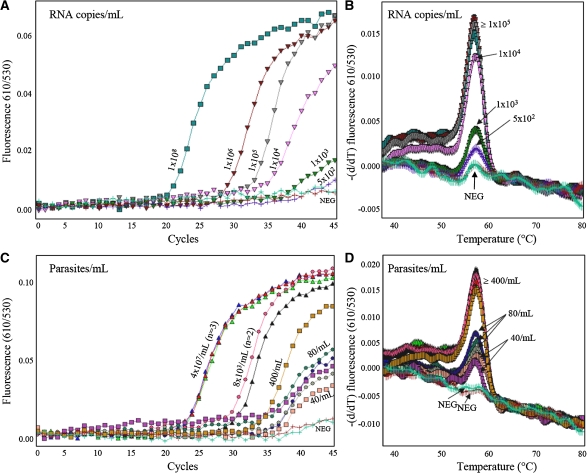
Multiplexed qRT-PCR was performed for the 18S rRNA and the competitor RNA in a single tube. We tested a range of competitor RNA inputs (2.5×104–1×107 copies per extraction). Above ≥ 1×105 copies per extraction, detection of low parasitemia samples was reduced (data not shown). With 5×104 copies of competitor RNA per extraction, the malaria RNA standards were detectable to 1×103 RNA copies per reaction (Figure 3A and B) and parasite-containing malaria standards to 40 parasites/mL (0.000001% parasitemia) (Figure 3C and D). The competitor RNA was reliably extracted from negative controls that were supplemented with 5×104 competitor RNA copies before extraction (mean CT 35.4 cycles: SD 1.4 cycles; %CV 4.0%).
Figure 3.
Sensitivity of multiplexed quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for Plasmodium falciparum A-type 18S ribosomal RNA (rRNA). Dilution series of samples containing a known concentration of IVT RNA in nucleic acids from whole blood (A and B) or a known concentration of whole parasites in whole blood (C and D). Analytical sensitivity was 1×103 copies (dark green curves) of IVT target RNA per reaction (A, representative CT analysis of IVT standards; B, representative melting curve analysis of IVT standards). Lower concentrations of IVT standards were detectable by melting curve analysis (purple curve, 5×102 copies/reaction), but are not quantitatively reportable. Using parasite-containing standards, samples containing 40 parasites/mL (pink/purple curve) were routinely detected (C, representative CT analysis of parasite-containing standards; C, representative melting curve analysis of parasite-containing standards). Numbers refer to RNA copies/reaction (A and B) or parasites/mL (C and D).
The malaria RNA standard curve described in the Methods (1×103–1×108 copies per reaction) was tested daily for 4 days. All standards ≥ 1×103 RNA copies per reaction were detected, whereas samples at 5×102 RNA copies per reaction were positive in only 25% of assays. Samples containing 1×103 RNA copies per reaction were repeatedly positive with ≥ 95% confidence (19 of 19 samples). Thus, the limit of quantification for malaria RNA was 1×103 copies per reaction and the standard curve was only interpreted at ≥ 1×103 RNA copies per reaction. The malaria RNA standard curve in the presence of competitor RNA showed linear and sensitive characteristics: r2 = 0.987, slope = −3.44 (95% CI = −3.33 to −3.54), y-intercept = 47.44 (95% CI = 46.4–48.5), overall efficiency 95%.
Plasmodium falciparum specificity
The species specificity of the qRT-PCR assay could not be directly determined from archival samples because the RNA in the samples was likely degraded. Instead, PCR using the same primers, probes, and reaction conditions without RT was performed, along with a SYBR Green-based pan-Plasmodium species identification assay, on 67 archival EDTA-treated whole blood clinical specimens, including 41 samples with malaria-positive blood smears (Table 1 ). All samples previously reported as blood smear positive were positive by pan-Plasmodium SYBR Green-based PCR (41/41) and 25/26 slide-negative samples were pan-Plasmodium PCR-negative. Melting curve analyses of the SYBR Green pan-Plasmodium PCR products speciated all archival specimens in accordance with their original microscopic diagnoses (data not shown), consistent with the experience of other centers. 20 Our probe-based P. falciparum PCR was positive in 13 of 13 smear-positive P. falciparum cases, negative in 25 of 25 smear-positive non-P. falciparum cases, and negative in 25 of 26 smear-negative cases. One slide-negative, PCR-positive sample tested positive by pan-Plasmodium PCR and P. falciparum-specific PCR; limited information indicated that this case was clinically consistent with malaria and that the patient had received anti-malaria therapy despite the negative microscopic findings. Thus, using microscopy as the gold standard, the P. falciparum-specific probe-based PCR was 100% sensitive and at least 98% specific.
Table 1.
Plasmodium genus and P. falciparum-specific polymerase chain reaction (PCR) on clinical specimens
| Microscopic diagnosis | Pan-Plasmodium PCR | P. falciparum PCR |
|---|---|---|
| P. falciparum (N = 12) | 12/12 | 12/12 |
| P. falciparum/P. ovale (N = 1) | 1/1 | 1/1 |
| P. ovale (N = 11) | 11/11 | 0/11 |
| P. vivax (N = 14) | 14/14 | 0/14 |
| Unspecified Plasmodium (N = 3) | 3/3 | 2/3 |
| No parasites (N = 26) | 1/26* | 1/26* |
One case was compatible with clinical malaria, and the patient was treated with anti-malarial drugs.
18S rRNA copy number per 12-hr ring-stage parasite
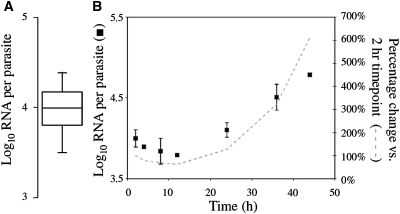
To convert 18S RNA copies/mL to parasites/mL, a conversion factor (18S RNA copies per parasite) was required. The qRT-PCR was performed on synchronized 12-hr blood-stage P. falciparum parasites diluted from 1% to 0.000002% parasitemia in whole human blood (N = 51 samples). Parasite samples were compared with the RNA standard curve, and 3.98 log10 copies (∼1×104) of the 18S rRNA were measured per 12-hr ring-stage parasite (Figure 4A). This conversion factor was also confirmed for two smaller sets of P. falciparum parasite samples obtained from separate in vitro cultures (data not shown).
Figure 4.
Copy number and expression of the A-type 18S ribosomal RNA (rRNA) in cultured Plasmodium falciparum 3D7. (A) Box plot showing an average of 10,000 copies of the A-type 18S rRNA per 12-hr ring-stage parasite (mean 3.98 log10 copies each). Parasites were cultured and synchronized by magnetic activated cell sorting (MACS) and sorbitol treatments, added to whole blood and quantified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (n = 51 samples ranging from 1% to 0.000002% parasitemia). (B) Expression of the A-type 18S rRNA by qRT-PCR in highly synchronized P. falciparum cultures sampled at the indicated time points after the invasion time point. RNA copies per parasite were normalized to the starting parasitemia determined at the 2-hr time point. Black boxes show mean log10 RNA copies/parasite (n = 2, error bars 95% CI; representative experiment shown) and the dashed line shows the percentage change relative to the 2-hr time point throughout the lifecycle.
Target expression throughout intraerythrocytic lifecycle
To determine if the 18S rRNA copy number in 12-hr ring-stage parasites varied during development, cultured parasites were synchronized to within 1 hr using MACS and sorbitol treatments, sampled at 2, 4, 8, 12, 24, 36, and 46 hrs of the lifecycle and tested by qRT-PCR (Figure 4B). The A-type 18S rRNA copy number was stable during the ring stage of the lifecycle (variation < 0.5 log10 parasites/mL during 2–24 hrs), whereas the copy number increased during 24–48 hrs. Thus, the 12-hr 18S rRNA copy number was used as a conversion factor to calculate parasite/mL from RNA copies/mL for 0–24-hr ring-stage P. falciparum parasites.
qRT-PCR assay performance
The assay was evaluated for accuracy, correlation, agreement, precision, analytical sensitivity, analytical specificity (interferences), reportable range, and carryover. 40,41 Sixty-eight parasite-containing whole blood samples prepared by dilution from a 10% parasitemia culture (parasitemia independently confirmed by three trained microscopists) were used to evaluate accuracy, correlation, agreement, precision, analytical sensitivity, analytical specificity (interferences), reportable range, and carryover. The malaria RNA standards (5×102–1×1011 copies per reaction) diluted in extracted whole blood with competitor RNA were additionally used to evaluate the reportable range and carryover.
To assess accuracy, high, medium, and low parasite-containing specimens (4×105, 8×103 and 80 nominal parasites/mL, respectively) and negative control specimens were tested in duplicate over 4–5 days. Some additional samples with higher or lower parasitemias were also tested. All parasite-containing specimens were qRT-PCR-positive across a wide range of parasitemias, and all negative controls were negative (100% sensitive, 100% specific). Nominal and observed data showed excellent correlation across a wide range of parasitemia (Figure 5; r2 = 0.9826, N = 63).
Figure 5.
Assay correlation and difference assessment. Correlation (left) of nominal (expected) and observed (measured) values (log10 RNA copies/mL) showed strong correlation across samples with wide ranging parasitemias (r2 = 0.9826, n = 63); the line of identity is also shown. A Bland-Altman difference plot (right) showed strong agreement between nominal and observed measurements [95% limits of agreement (UL, upper limit; LL, lower limit; dashed lines): + 0.50075 to −0.43184 log10 RNA copies/mL, n = 63], with only one outlier. The overall assay bias was −0.0344 log10 RNA copies/mL. Recovery was not concentration dependent (r2 = 0.0165; p = 0.2548).
Agreement between observed and nominal results (i.e., recovery) was evaluated by comparing average log10 results using a Bland-Altman difference plot (Figure 5; 95% limits of agreement: +0.50 to −0.43 log10 RNA copies/mL, N = 63). Of 63 samples tested, only one showed a difference in recovery > 0.5 log10 RNA copies/mL from the nominal value; this single outlier occurred at the very lowest tested parasite density. Differences in recovery were not concentration dependent (r2 = 0.0165; P = 0.2548). The average log10 difference (i.e., bias) noted in all samples was 0.034 log10 RNA copies/mL.
Within-run (repeatability) and within-laboratory (between-run) precision was determined by testing 2–3 replicates of high, medium, low, and negative samples daily for 3–5 days. Intra- and inter-assay percent coefficients of variation (imprecision = SD/mean) were calculated (Table 2 ). For different parasitemia samples, the within- and between-run assay imprecision was 0.60% and 1.63% (high parasitemia), 1.11% and 1.88% (medium parasitemia), and 1.79% and 4.05% (low parasitemia), respectively. Thus, the assay was more precise than quantitative NASBA, especially at low parasitemias. 22 Because the assay controls are monitored using Levey-Jennings plots, we also evaluated 20 batched runs (performed over several months) and determined that the %CV for the high malaria control and low competitor RNA control were 2.4% and 4.0%, respectively. There were also no false positives for the malaria-negative control during this time.
Table 2.
Precision characteristics of assay
| Control | Samples per run | Total runs | Nominal parasites/mL | Nominal RNA log10 copies/mL | Intra-assay (within run) | Inter-assay (within laboratory) | ||
|---|---|---|---|---|---|---|---|---|
| SD | %CV | SD | %CV | |||||
| High | 3 | 5 | 4 × 105 | 9.60 | 0.057 | 0.60 | 0.154 | 1.63 |
| Mid | 2 | 3 | 8 × 103 | 7.90 | 0.084 | 1.11 | 0.142 | 1.88 |
| Low | 3 | 5 | 80 | 5.90 | 0.106 | 1.79 | 0.241 | 4.05 |
| Negative | 2 | 5 | 0 | None detected | – | – | – | – |
When we tested whole blood samples containing 40 (N = 8), 80 (N = 15), 100 (N = 2), and 400 (N = 3) parasites/mL, all extracted samples were positive by qRT-PCR. The analytical sensitivity of the qRT-PCR reaction was previously determined to be 1×103 RNA copies per reaction, sufficient to detect a single parasite in the 50 μL starting volume of whole blood.
Analytical specificity caused by common laboratory interferences was evaluated by adding the standard quantity of competitor RNA to malaria-negative clinical samples that showed lipemia (triglycerides ∼800 mg/dL; N = 3 samples), hemolysis (N = 3 samples), hyperbilirubinemia (total bilirubin > 15 mg/dL; N = 2 samples), or leukocytosis (> 30,000/μL, N = 1 sample). The competitor RNA CT was < 2 SD different from the normal control for all samples, except for the leukocytosis sample and one hyperbilirubinemic sample, both of which differed by < 3 SD. All samples were within the acceptable Levey-Jennings range for the competitor RNA control.
The reportable range of the assay was determined by testing samples over a wide range of parasitemia. Testing low- and mid-range parasitemia specimens was described previously. Extremely high parasitemia specimens (1% parasitemia or ∼2.5×109 RNA copies per reaction) and high concentration RNA standards (1×109 and 1×1011 RNA copies per reaction, equivalent to 0.4% and 40% parasitemia, respectively) were tested to evaluate assay linearity. Difference of log10 transformed values from nominal values were calculated and compared with log10 nominal value (data not shown). The average difference for all samples (0.000001–40% parasitemia) was −0.38 log10 RNA copies/mL. Aside from the one low parasitemia outlier described previously, only the very highest samples (1×1011 copies per reaction, equivalent to 40% parasitemia) showed a log10 difference > 0.5 log10 units from the nominal values. On the basis of this information, we decided to report CT values from 12.0 to 36.8 cycles, whereas samples with CT < 12 cycles needed to be diluted and re-tested for accurate quantification. In practice, this would be an extremely rare occurrence because even florid hyperparasitemia usually does not reach > 10% in humans.
Carryover was assessed by placing the high parasitemia samples (4×107 parasites/mL) adjacent to negative samples within all runs. There were no false positive samples in any of these experiments.
Sample stability was tested by delaying the addition of lysis buffer to freshly collected EDTA-treated whole blood obtained from known positive samples. There was no significant change in the qRT-PCR results when processing of clinical samples was delayed up to 12 hr (data not shown). However, if aliquots of whole blood were directly frozen before storage and thawed without lysis buffer, the RNA was degraded as indicated by a CT difference in the presence or absence of RT of only 5–7 cycles as compared with a CT difference of > 12.5 cycles with and without RT when using guanidinium-preserved samples (data not shown).
Experimental P. falciparum infection of human volunteers
Six human volunteers were challenged with P. falciparum sporozoites by the bites of five infected mosquitoes in an IRB-approved clinical trial conducted at the Seattle BioMed Malaria Clinical Trials Center (MCTC). One-hundred four EDTA-treated whole blood samples were processed into lysis buffer, frozen, and later extracted and tested by qRT-PCR. In 44 samples, no malaria target was detected. Fifty-four samples were qRT-PCR-positive at or above the limit of quantification (≥ 20 parasites/mL), and six additional samples were positive by melting curve analysis (below quantifiable limits).
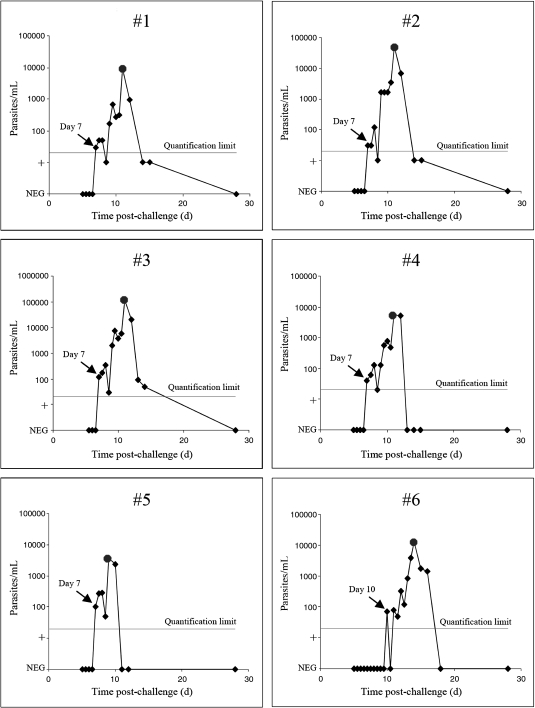
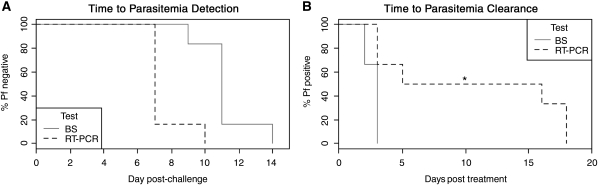
In quantifiable samples, parasitemias ranged from 20 to 120,000 parasites/mL, consistent with ranges observed at other human challenge centers. 30,32,42 The parasite burden rose in ∼44–46 hr increments, with decreased parasitemia in the latter half of this period, consistent with sequestration by P. falciparum. Participants 1–5 were qRT-PCR positive on Day 7 post-challenge (Figure 6). Participant 5 was thick blood smear-positive at Day 9, whereas participants 1–4 were positive on Day 11 (Table 3 ). Parasites were detected in volunteer 6 by qRT-PCR on Day 10 and by blood smear on Day 14. Thus, qRT-PCR detected parasites an average of 3.7 days earlier than thick blood smears (Figure 7A, 95% CI 2.8–4.5 days) and 6 days before the onset of fever (range 3–9 days; only three volunteers developed fevers). Symptomatic volunteers were treated with standard treatment doses of chloroquine when a positive thick smear was identified. Within 3 days of treatment, all blood smears were negative, and 18S rRNA was undetectable by qRT-PCR in two participants (Figure 7B). By 5 days post-treatment, the qRT-PCR target was undetectable in three participants and reduced by > 2 log10 parasites/mL in the remaining three participants. At the follow-up visit 28 days post-challenge, parasites were not detected in any participant by thick blood smear or qRT-PCR. Samples were not obtained in the intervening period, however, two participants who were positive for parasite 18S rRNA after 5 days of therapy had a low positive result (< 20 parasites/mL) and the third participant had only 50 parasites/mL. This result was not surprising because highly sensitive molecular tests for other organisms like pathogenic Neisseria and Chlamydia can sometimes detect nucleic acids from nonviable organisms for days to weeks after effective treatment. 43,44
Figure 6.
Early detection of pre-patent parasitemia in six volunteers experimentally infected with Plasmodium falciparum. Sporozoites kinetics of parasite load determined by real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) during challenge of six human volunteers with P. falciparum 3D7 sporozoites. For qRT-PCR, venous blood was collected twice daily starting on Day 5. The first qRT-PCR positive sample in each participant is denoted with a black arrow listing the day post-challenge. Thick blood smears were positive starting on Day 9 (participant 5), 11 (participants 1–4) and 14 (participant 6), as denoted with circles. Chloroquine treatment was initiated on the first detection of a positive thick smear. The limit of quantification (20 parasites/mL) is indicated with a horizontal line. Samples from subjects 1 and 2 that were positive by melting curve analysis but were below the limit of quantification were plotted at the “+” level on the parasitemia axis.
Table 3.
Human volunteer infection data*
| Parameter | Volunteer | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| First positive qRT-PCR (day†) | 7 | 7 | 7 | 7 | 7 | 10 |
| Parasites/mL by qRT-PCR | 30 | 30 | 120 | 40 | 100 | 70 |
| Patency period - 1st positive BS (day†)‡ | 11 | 11 | 11 | 11 | 9 | 14 |
| Parasites/mL by qRT-PCR at patency | 9,460 | 50,170 | 120,680 | 5,350 | 3,730 | 12,880 |
| Fever > 38.0°C (day) | N/A | 13 | N/A | N/A | 10 | 15 |
qRT-PCR = quantitative reverse transcription-polymerase chain reaction.
Days are days post-challenge.
Blood smears (BS) were evaluated for 360 high-powered fields (hpf) if asymptomatic or treated, or, 1080 hpf if symptomatic.
Figure 7.
Kaplan-Meier curves for time to detection and clearance of parasites by thick blood smears and quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Kaplan-Meier curves for the time to detection (A) and post-treatment clearance (B) of parasites for thick blood smears (BS) and qRT-PCR. *No qRT-PCR testing was performed during this interval 6–15 days following treatment.
We analyzed the clinical trial data using two published models to estimate the number of infected hepatocytes and average parasite burden in challenged non-immune volunteers. 42,45 These models are applicable to our trial because our study and the initial modeling studies used infection by five mosquitoes and achieved comparable times to both 18S rRNA and thick blood smear positivity. The Hermsen model used a cycle-to-cycle multiplication factor of 7.5, whereas our present study showed a mean multiplication factor of 16.2 (median 13.9), closer to that used in the Bejon models. 42 Five of six individuals experienced three cycles of parasite growth before drug treatment in our trial, and the cycle-to-cycle multiplication factor was more conserved within individuals than between individuals, as observed previously. 42 From our data, the Hermsen and Bejon models predicted an average of 28 (range 8–58) and 60 (range 27–111) infected hepatocytes per volunteer, respectively. Both models closely mimicked the rising parasitemia in participants 1–5, but did not adequately model participant 6, in whom the onset of detectable parasitemia was delayed by ∼3 days.
Discussion
Giemsa-stained thick and thin blood smears are the gold standard diagnostic method for malaria. Where microscopy may miss very low density parasitemia infections, molecular assays with sensitivities of 20–50 parasites/mL allow detection of pre-patent infections in humans experimentally infected with malaria parasites. 29,32,42,46 Detection of pre-patent parasitemia and determination of the parasite growth rate can help to determine whether vaccine candidates affect liver- and/or erythrocyte-stage parasite growth. 42 There are many factors to consider when choosing from the diverse molecular methods reported for malaria. 5–23 First, pre-analytical handling steps (such as leukocyte filtering reported in some assays) should be minimized or highly controlled to reduce the risk of contamination. Second, as the sample volume decreases, the limit of detection increases. For instance, if 200 μL of blood is extracted, the lowest possible limit of detection is 5 parasites/mL because there can be at a minimum only 1 parasite/200 μL, whereas for 50 μL samples, the lowest limit of detection is 20 parasites/mL (or 1 parasite/50 μL). The actual limit of detection is further affected by the blood sample volume, the extracted nucleic acid volume, the volume added to the (RT)-PCR assay and the Poisson distribution. Low parasitemia samples can be difficult to detect using single-step DNA PCR approaches, 14 although reports vary. 8 Some RNA assays have improved sensitivities at low parasite densities because of the increased copy number relative to coding DNA. 22 Third, many assays do not include a multiplexed internal control, which is critical for ruling out PCR inhibitors in every sample.
To overcome these limitations, we developed a high sensitivity, internally controlled real-time RT-PCR assay for absolute P. falciparum quantification and demonstrated its use in a clinical trial. The high sensitivity of qRT-PCR from a 50 μL blood sample is possible because 18S rRNAs are abundant within individual parasites (1×104 copies/ring form). Thus, we obtain three pieces of evidence to support the test result: the malaria CT, the malaria PCR product Tm, and a reciprocal decrease in total competitor amplification. We include a competitive internal control RNA in every multiplexed reaction to detect target extraction/amplification failures caused by inhibitors, laboratory errors, and/or other Plasmodium infections.
Using a combination of purified RNA calibrators, synchronized (12-hr) ring-stage parasite standards in whole blood, and synchronous parasite cultures sampled throughout the 48-hr lifecycle, we measured 10,000 copies of the 18S rRNA per ring-stage parasite, in agreement with the conversion factor used for NASBA assays. 7 Although RT-PCR on aliquots of frozen whole blood were recently reported to yield ≤ 1-log increase in parasite detection compared with PCR alone, 23 we find a nearly 4-log increase in detection, probably because 18S rRNA are better preserved when lysed in guanidinium-based buffer before and stored at −80°C. To generate a standard curve for absolute quantification, we used RNA standards of known quantity added to extracted nucleic acids from whole blood of known erythrocyte density. As noted previously, we can convert log10 RNA copies to parasite density, allowing for absolute quantification of P. falciparum from whole blood.
With this system, we validated the qRT-PCR for clinical trial use and used it in a demonstration human challenge trial of six volunteers. The assay was highly sensitive and specific across a wide range of parasitemia and performed favorably in accuracy, correlation, precision, analytical sensitivity, analytical specificity, and carryover evaluations. This assay detected sub-patent parasitemia 3.7 days on average before blood smears were positive, similar to that seen in other trials using other techniques. 42,45 Although it must be remembered that Poisson statistics affect the confidence limits for positive detection at parasite densities < 60 parasites/mL, we routinely detect samples containing as few as one parasite per 50 μL of whole blood (i.e., 20 parasites/mL). However, we did not test samples at a nominal concentration of 20 parasites/mL because parasites would be statistically absent from 37% of such samples. Because our RNA standards can be detected > 95% of the time at copy numbers below that expected for a single parasite in 50 μL of blood, we can quantitatively report parasite densities down to 20 parasites/mL, recognizing again that the Poisson distribution influences the actual outcome.
In P. falciparum, the expression of the A-type 18S rRNA target during ring-stage development was critical for the interpretation of our assay. Here, we report that the A-type 18S rRNA is stably expressed throughout the ring stage of the P. falciparum lifecycle. The 18S rRNA has not been previously profiled by quantitative molecular methods in P. falciparum, but our data is generally consistent with earlier Northern blotting of murine P. berghei, which showed that A-type 18S rRNA expression was maximal in the growing trophozoite stage and lower in the transcriptionally quiescent ring stage. 47 In P. falciparum infections, only ring-stage parasites are detected in peripheral blood during the first 24 hrs after erythrocyte invasion. Although mature trophozoite and schizont forms are easily cultured in vitro, such mature forms are not usually present in peripheral blood in vivo because of cytoadherence during the latter half of the lifecycle. 48 Mature forms are only observed in the peripheral blood in cases of severe hyperparasitemia. For non-P. falciparum species, sequestration does not occur and all lifecycle forms are present in peripheral blood, making quantification of other species more complicated. Thus, for the relevant ring-stage P. falciparum parasites, the 18S rRNA signal is stably expressed in vitro. Indirect evidence for stable ring-stage expression of the target in vivo comes from the clinical trial data, which showed rising cyclical parasitemia in participants by qRT-PCR that was consistent with data from other centers using DNA-based quantification. Such agreement would be unlikely if the RNA target varied dramatically during in vivo infections. Whether stable expression typifies in vivo infections in the presence of sub-therapeutic anti-malarial drugs or in infections with mixed strains is unknown.
RNA-based assays for malaria allow for high sensitivity detection and quantification of parasites from a small sample. To reap the benefits of the high copy number 18S rRNA as a target, samples must be adequately preserved at the time of collection, which is easily accomplished using guanidinium-based lysis buffer and, possibly, even dried blood spots. 49–51
Given the need for reliable, controlled assays to support malaria clinical trials and eradication efforts, we recommend the use of multiplexed internal control targets and in vitro transcribed RNA standards for absolute quantification in all RNA- or total nucleic acid-based assays. Furthermore, a network for inter-laboratory exchange of well-characterized malaria comparator samples would be useful to help laboratories establish and maintain assay performance. Such quality control programs for malaria molecular diagnostics would benefit research and clinical efforts alike.
Supplementary Data
ACKNOWLEDGMENTS
We thank Robert Coombs and Linda Cook for advice and feedback on assay development and validation, and Jane Kuypers and Nancy Wright for valuable advice regarding production of RNA standards.
Disclaimer: The content of the information presented in this paper does not necessarily reflect the position or the policy of the U.S.Government, and no official endorsement should be inferred.
Footnotes
Financial support: Funding in support of the human challenge trial was provided by the PATH Malaria Vaccine Initiative and the U.S. Department of Defense (award no. W81XWH-09-1-0531; the U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014 is the awarding and administering acquisition office.)
Authors' addresses: Sean C. Murphy and Jennifer L. Prentice, Department of Laboratory Medicine, University of Washington Medical Center, Seattle, WA, E-mails: murphysc@u.washington.edu and jprentic@u.washington.edu. Kathryn Williamson and Michal Fried, Seattle Biomedical Research Institute, Seattle, WA, E-mails: kathryn.williamson@seattlebiomed.org and michal.fried@seattlebiomed.org. Carolyn K. Wallis, Department of Laboratory Medicine, Harborview Medical Center, Seattle, WA, E-mail: ckw@u.washington.edu. Ferric C. Fang and Brad T. Cookson, Departments of Laboratory Medicine and Microbiology, University of Washington Medical Center, Seattle, WA, E-mails: fcfang@u.washington.edu and cookson@u.washington.edu. Cris Pinzon and Ruobing Wang, Malaria Clinical Trials Center, Seattle Biomedical Research Institute, Seattle, WA, E-mails: cris.pinzon@seattlebiomed.org and ruobing.wang@seattlebiomed.org. Angela K. Talley, Malaria Clinical Trials Center, Seattle Biomedical Research Institute, Seattle, WA, E-mail: angela.talley@seattlebiomed.org (also affiliated with the Department of Medicine [Allergy and Infectious Diseases], University of Washington Medical Center]. Stefan H. I. Kappe, Seattle Biomedical Research Institute, Seattle, WA, E-mail: stefan.kappe@seattlebiomed.org (also affiliated with the Department of Global Health, University of Washington, Seattle, WA). Patrick E. Duffy, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, E-mail: duffype@niaid.nih.gov (also affiliated with Seattle Biomedical Research Institute, Seattle, WA).
Reprints requests: Sean C. Murphy, Department of Laboratory Medicine, University of Washington, 1959 NE Pacific St., Box 357110, Seattle, WA 98195-7110; Tel: 206-598-6131; Fax: 206-598-6189; E-mail: murphysc@u.washington.edu.
References
- 1.Makler MT, Palmer CJ, Ager AL. A review of practical techniques for the diagnosis of malaria. Ann Trop Med Parasitol. 1998;92:419–433. doi: 10.1080/00034989859401. [DOI] [PubMed] [Google Scholar]
- 2.Bejon P, Andrews L, Hunt-Cooke A, Sanderson F, Gilbert SC, Hill AV. Thick blood film examination for Plasmodium falciparum malaria has reduced sensitivity and underestimates parasite density. Malar J. 2006;5:104. doi: 10.1186/1475-2875-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stauffer WM, Cartwright CP, Olson DA, Juni BA, Taylor CM, Bowers SH, Hanson KL, Rosenblatt JE, Boulware DR. Diagnostic performance of rapid diagnostic tests versus blood smears for malaria in US clinical practice. Clin Infect Dis. 2009;49:908–913. doi: 10.1086/605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinkhumba J, Skarbinski J, Chilima B, Campbell C, Ewing V, San Joaquin M, Sande J, Ali D, Mathanga D. Comparative field performance and adherence to test results of four malaria rapid diagnostic tests among febrile patients more than five years of age in Blantyre, Malawi. Malar J. 2010;9:209. doi: 10.1186/1475-2875-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swan H, Sloan L, Muyombwe A, Chavalitshewinkoon-Petmitr P, Krudsood S, Leowattana W, Wilairatana P, Looareesuwan S, Rosenblatt J. Evaluation of a real-time polymerase chain reaction assay for the diagnosis of malaria in patients from Thailand. Am J Trop Med Hyg. 2005;73:850–854. [PubMed] [Google Scholar]
- 7.Mens PF, Schoone GJ, Kager PA, Schallig HD. Detection and identification of human Plasmodium species with real-time quantitative nucleic acid sequence-based amplification. Malar J. 2006;5:80. doi: 10.1186/1475-2875-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safeukui I, Millet P, Boucher S, Melinard L, Fregeville F, Receveur MC, Pistone T, Fialon P, Vincendeau P, Fleury H, Malvy D. Evaluation of FRET real-time PCR assay for rapid detection and differentiation of Plasmodium species in returning travelers and migrants. Malar J. 2008;7:70. doi: 10.1186/1475-2875-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tangin A, Komichi Y, Wagatsuma Y, Rashidul H, Wataya Y, Kim HS. Detection of malaria parasites in mosquitoes from the malaria-endemic area of Chakaria, Bangladesh. Biol Pharm Bull. 2008;31:703–708. doi: 10.1248/bpb.31.703. [DOI] [PubMed] [Google Scholar]
- 10.Das A, Holloway B, Collins WE, Shama VP, Ghosh SK, Sinha S, Hasnain SE, Talwar GP, Lal AA. Species-specific 18S rRNA gene amplification for the detection of P. falciparum and P. vivax malaria parasites. Mol Cell Probes. 1995;9:161–165. doi: 10.1006/mcpr.1995.0025. [DOI] [PubMed] [Google Scholar]
- 11.Ciceron L, Jaureguiberry G, Gay F, Danis M. Development of a Plasmodium PCR for monitoring efficacy of antimalarial treatment. J Clin Microbiol. 1999;37:35–38. doi: 10.1128/jcm.37.1.35-38.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kho WG, Chung JY, Sim EJ, Kim MY, Kim DW, Jongwutiwes S, Tanabe K. A multiplex polymerase chain reaction for a differential diagnosis of Plasmodium falciparum and Plasmodium vivax. Parasitol Int. 2003;52:229–236. doi: 10.1016/s1383-5769(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 13.Vo TK, Bigot P, Gazin P, Sinou V, De Pina JJ, Huynh DC, Fumoux F, Parzy D. Evaluation of a real-time PCR assay for malaria diagnosis in patients from Vietnam and in returned travelers. Trans R Soc Trop Med Hyg. 2007;101:422–428. doi: 10.1016/j.trstmh.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 15.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 16.Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. doi: 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- 17.Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, Ricci L, Medici MC, Arcangeletti MC, Snounou G, Dettori G, Chezzi C. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol. 2004;42:1214–1219. doi: 10.1128/JCM.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman RE, Sattabongkot J, Promstaporm S, Maneechai N, Tippayachai B, Kengluecha A, Rachapaew N, Zollner G, Miller RS, Vaughan JA, Thimasarn K, Khuntirat B. Comparison of PCR and microscopy for the detection of asymptomatic malaria in a Plasmodium falciparum/vivax endemic area in Thailand. Malar J. 2006;5:121. doi: 10.1186/1475-2875-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, Peterson LR, Kaul KL. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005;43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43:402–405. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoone GJ, Oskam L, Kroon NC, Schallig HD, Omar SA. Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J Clin Microbiol. 2000;38:4072–4075. doi: 10.1128/jcm.38.11.4072-4075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF. Development of a highly sensitive genus-specific qRT-PCR assay for detection and quantitation of Plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J Clin Microbiol. 2011;49:2946–2953. doi: 10.1128/JCM.00276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mixson-Hayden T, Lucchi NW, Udhayakumar V. Evaluation of three PCR-based diagnostic assays for detecting mixed Plasmodium infection. BMC Res Notes. 2010;3:88. doi: 10.1186/1756-0500-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snounou G. Genotyping of Plasmodium spp. Nested PCR. Methods Mol Med. 2002;72:103–116. doi: 10.1385/1-59259-271-6:103. [DOI] [PubMed] [Google Scholar]
- 26.Epstein JE, Rao S, Williams F, Freilich D, Luke T, Sedegah M, de la Vega P, Sacci J, Richie TL, Hoffman SL. Safety and clinical outcome of experimental challenge of human volunteers with Plasmodium falciparum-infected mosquitoes: an update. J Infect Dis. 2007;196:145–154. doi: 10.1086/518510. [DOI] [PubMed] [Google Scholar]
- 27.Webster DP, Dunachie S, Vuola JM, Berthoud T, Keating S, Laidlaw SM, McConkey SJ, Poulton I, Andrews L, Andersen RF, Bejon P, Butcher G, Sinden R, Skinner MA, Gilbert SC, Hill AV. Enhanced T cell-mediated protection against malaria in human challenges by using the recombinant poxviruses FP9 and modified vaccinia virus Ankara. Proc Natl Acad Sci USA. 2005;102:4836–4841. doi: 10.1073/pnas.0406381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walther M, Thompson FM, Dunachie S, Keating S, Todryk S, Berthoud T, Andrews L, Andersen RF, Moore A, Gilbert SC, Poulton I, Dubovsky F, Tierney E, Correa S, Huntcooke A, Butcher G, Williams J, Sinden RE, Hill AV. Safety, immunogenicity, and efficacy of prime-boost immunization with recombinant poxvirus FP9 and modified vaccinia virus Ankara encoding the full-length Plasmodium falciparum circumsporozoite protein. Infect Immun. 2006;74:2706–2716. doi: 10.1128/IAI.74.5.2706-2716.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walther M, Dunachie S, Keating S, Vuola JM, Berthoud T, Schmidt A, Maier C, Andrews L, Andersen RF, Gilbert S, Poulton I, Webster D, Dubovsky F, Tierney E, Sarpotdar P, Correa S, Huntcooke A, Butcher G, Williams J, Sinden RE, Thornton GB, Hill AV. Safety, immunogenicity and efficacy of a pre-erythrocytic malaria candidate vaccine, ICC-1132 formulated in Seppic ISA 720. Vaccine. 2005;23:857–864. doi: 10.1016/j.vaccine.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Hermsen CC, Telgt DS, Linders EH, van de Locht LA, Eling WM, Mensink EJ, Sauerwein RW. Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol Biochem Parasitol. 2001;118:247–251. doi: 10.1016/s0166-6851(01)00379-6. [DOI] [PubMed] [Google Scholar]
- 31.Schneider P, Schoone G, Schallig H, Verhage D, Telgt D, Eling W, Sauerwein R. Quantification of Plasmodium falciparum gametocytes in differential stages of development by quantitative nucleic acid sequence-based amplification. Mol Biochem Parasitol. 2004;137:35–41. doi: 10.1016/j.molbiopara.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Andrews L, Andersen RF, Webster D, Dunachie S, Walther RM, Bejon P, Hunt-Cooke A, Bergson G, Sanderson F, Hill AV, Gilbert SC. Quantitative real-time polymerase chain reaction for malaria diagnosis and its use in malaria vaccine clinical trials. Am J Trop Med Hyg. 2005;73:191–198. [PubMed] [Google Scholar]
- 33.McCutchan TF, de la Cruz VF, Lal AA, Gunderson JH, Elwood HJ, Sogin ML. Primary sequences of two small subunit ribosomal RNA genes from Plasmodium falciparum. Mol Biochem Parasitol. 1988;28:63–68. doi: 10.1016/0166-6851(88)90181-8. [DOI] [PubMed] [Google Scholar]
- 34.McCutchan TF, Li J, McConkey GA, Rogers MJ, Waters AP. The cytoplasmic ribosomal RNAs of Plasmodium spp. Parasitol Today. 1995;11:134–138. doi: 10.1016/0169-4758(95)80132-4. [DOI] [PubMed] [Google Scholar]
- 35.Kuypers J, Wright N, Morrow R. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J Clin Virol. 2004;31:123–129. doi: 10.1016/j.jcv.2004.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 37.Ribaut C, Berry A, Chevalley S, Reybier K, Morlais I, Parzy D, Nepveu F, Benoit-Vical F, Valentin A. Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar J. 2008;7:45. doi: 10.1186/1475-2875-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambros C, Vanderberg JP, Ribaut C, Berry A, Chevalley S, Reybier K, Morlais I, Parzy D, Nepveu F, Benoit-Vical F, Valentin A. Synchronization of Plasmodium falciparum erythrocytic stages in culture Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 39.Chulay JD, Schneider I, Cosgriff TM, Hoffman SL, Ballou WR, Quakyi IA, Carter R, Trosper JH, Hockmeyer WT. Malaria transmitted to humans by mosquitoes infected from cultured Plasmodium falciparum. Am J Trop Med Hyg. 1986;35:66–68. doi: 10.4269/ajtmh.1986.35.66. [DOI] [PubMed] [Google Scholar]
- 40.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 41.Chesher D. Evaluating assay precision. Clin Biochem Rev. 2008;29((Suppl 1)):S23–S26. [PMC free article] [PubMed] [Google Scholar]
- 42.Bejon P, Andrews L, Andersen RF, Dunachie S, Webster D, Walther M, Gilbert SC, Peto T, Hill AV. Calculation of liver-to-blood inocula, parasite growth rates, and preerythrocytic vaccine efficacy, from serial quantitative polymerase chain reaction studies of volunteers challenged with malaria sporozoites. J Infect Dis. 2005;191:619–626. doi: 10.1086/427243. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann LH, Desmond RA, Stephens J, Hughes A, Hook EWI. Duration of persistence of gonococcal DNA detected by ligase chain reaction in men and women following recommended therapy for uncomplicated gonorrhea. J Clin Microbiol. 2002;40:3596–3601. doi: 10.1128/JCM.40.10.3596-3601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RE, Newhall WJ, Papp JR, Knapp JS, Black CM, Gift TL, Steece R, Markowitz LE, Devine OJ, Walsh CM, Wang S, Gunter DC, Irwin KL, DeLisle S, Berman SM. Screening tests to detect Chlamydia trachomatis and Neisseria gonorrhoeae infections. MMWR Recomm Rep. 2002;18:1–38. [PubMed] [Google Scholar]
- 45.Hermsen CC, de Vlas SJ, van Gemert GJ, Telgt DS, Verhage DF, Sauerwein RW. Testing vaccines in human experimental malaria: statistical analysis of parasitemia measured by a quantitative real-time polymerase chain reaction. Am J Trop Med Hyg. 2004;71:196–201. [PubMed] [Google Scholar]
- 46.McConkey SJ, Reece WH, Moorthy VS, Webster D, Dunachie S, Butcher G, Vuola JM, Blanchard TJ, Gothard P, Watkins K, Hannan CM, Everaere S, Brown K, Kester KE, Cummings J, Williams J, Heppner DG, Pathan A, Flanagan K, Arulanantham N, Roberts MT, Roy M, Smith GL, Schneider J, Peto T, Sinden RE, Gilbert SC, Hill AV. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat Med. 2003;9:729–735. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 47.Waters AP, van Spaendonk RM, Ramesar J, Vervenne RA, Dirks RW, Thompson J, Janse CJ. Species-specific regulation and switching of transcription between stage-specific ribosomal RNA genes in Plasmodium berghei. J Biol Chem. 1997;272:3583–3589. doi: 10.1074/jbc.272.6.3583. [DOI] [PubMed] [Google Scholar]
- 48.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 49.Waters AP, McCutchan TF. Rapid, sensitive diagnosis of malaria based on ribosomal RNA. Lancet. 1989;333:1343–1346. doi: 10.1016/s0140-6736(89)92800-6. [DOI] [PubMed] [Google Scholar]
- 50.Gauffin F, Nordgren A, Barbany G, Gustafsson B, Karlsson H. Quantitation of RNA decay in dried blood spots during 20 years of storage. Clin Chem Lab Med. 2009;47:1467–1469. doi: 10.1515/CCLM.2009.351. [DOI] [PubMed] [Google Scholar]
- 51.Karlsson H, Guthenberg C, von Dobeln U, Kristenssson K. Extraction of RNA from dried blood on filter papers after long-term storage. Clin Chem. 2003;46:979–981. doi: 10.1373/49.6.979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.