Abstract
To address the problem of the health impacts of unsafe drinking water, methods are needed to assess microbiologic contamination in water. However, indicators of water quality have provided mixed results. We evaluate five assays (three for Escherichia coli and one each for enterococci and somatic coliphage) of microbial contamination in villages in rural Ecuador that rely mostly on untreated drinking water. Only membrane filtration for E. coli using mI agar detected a significant association with household diarrheal disease outcome (odds ratio = 1.29, 95% confidence interval = 1.02–1.65 in household containers and odds ratio = 1.18, 95% confidence interval = 1.02–1.37) in source samples. Our analysis and other published research points to the need for further consideration of study design factors, such as sample size and variability in measurements, when using indicator organisms, especially when relating water quality exposure to health outcomes. Although indicator organisms are used extensively in health studies, we argue that their use requires a full understanding of their purposes and limitations.
Introduction
Contaminated drinking water is a major contributor to the problem of diarrheal disease, which continues to plague children worldwide. Despite improving trends, diarrhea still accounts for 2.5 million annual deaths of children worldwide.1 To address the problem of unsafe drinking water, methods are needed to assess what constitutes good quality versus contaminated drinking water. In this report, we compare water quality data collected from communities in rural northern coastal Ecuador by using a variety of standard techniques and indicator organisms to provide recommendations for the use of indicator data as a measure of infectious disease health risk in untreated drinking water.
Rather than directly assessing presence of pathogens in water, indicator organisms characteristic of fecal contamination are used as a proxy measure of a recent fecal contamination event. To date, epidemiologic studies have found weak relationships between drinking water quality at the point of use and incidence of diarrheal disease.2,3 This lack of evidence to support a clear relationship between microbial contamination of household drinking water and diarrheal disease may be caused by the importance of other transmission pathways in spreading pathogenic organisms, or may be caused by difficulty in measuring microbial contamination of water by using indicator organisms. Interventions to improve water quality have been associated with reductions in diarrheal illness,2,4,5 suggesting that measurement issues may be a contributing factor.
The difficulty in finding associations between indicator organisms and health outcomes highlights the challenges in water quality research. Methods for assessing the microbiologic quality of water were developed in temperate areas of the world, and have subsequently been expanded geographically, to developing country settings that are often located in tropical areas, and to evaluate other endpoints. This expansion has been carried out despite differences in physio-chemical and microbiologic characteristics of source waters, and social and economic differences in tropical and temperate locations.6 In tropical regions, where diarrheal disease poses the biggest health burden, the organisms that practitioners have traditionally used to assess fecal contamination of waterways (total and fecal coliforms, Escherichia coli, enterococci) have been found to naturally occur in tropical areas, bringing into question their utility as indicators of fecal contamination.6–10 In addition, the application of indicator organisms to evaluate microbiologic contamination of water as a proxy for health outcomes is not warranted, given the lack of evidence to support these associations.
We report on a study that we carried out in rural coastal Ecuador to evaluate a suite of indicators of water quality for health research in tropical developing country settings. We present data on associations between water quality and diarrheal disease to illustrate the utility and limitations of water quality indicators in this and other tropical developing country settings. We discuss some of the reasons why it has been difficult to establish relationships between concentrations of indicator organisms in untreated drinking water and health outcomes. We evaluate the utility of indicators in field situations, where ideal laboratory conditions may not exist. Lastly, we point to applications where indicator organisms can provide useful insights, despite their limitations, such as assessing treatment efficacy, household recontamination, or comparative studies in water bodies with similar background levels of contamination.
The results of our study point to the importance of careful consideration of the research questions and appropriate sample size when designing a study to evaluate water quality, especially when relating water quality exposure to health outcomes. Although indicator organisms are used extensively in health studies, we argue that their use requires a full understanding of their purposes and limitations.
Materials and Methods
Study site
The study area was in northern coastal Ecuador in the province of Esmeraldas, Canton Eloy Alfaro. The 125 villages in this region are situated along the Santiago, Cayapas, and Onzole Rivers, all of which drain into the major population center of Borbón. Little sanitation infrastructure exists in these communities. Although some persons use private or community latrines, in household surveys that we have carried out, 60% of persons report that they dispose of human waste out in the open by digging a hole or directly into the rivers. These same rivers serve as the primary water source for 68% of households in the region, and 60% of households report drinking their water without treating it. High rates of diarrheal disease have been observed in this study area.11–13
Sample collection
Water samples were collected in six villages: two with a simple piped water system that transports untreated surface water, two that rely on unimproved surface water from fast-flowing rivers, one that relies on unimproved surface water from a small stream, and one with a more extensive water distribution system. In this last town (Borbón), a more advanced water treatment plant was installed between the first and the second sampling visits. In addition to their primary water source (tap or surface water), some villagers also use simple wells and/or collect rainwater as source waters for drinking. Samples were collected from sources identified as source waters by household members and from storage containers within the house. The number of samples collected from each type of source (both directly from the source and from storage containers in the household) is shown in Table 1. Sample collection and processing took place during March 2005–March 2006.
Table 1.
Sources of water sampled from containers stored in the household and directly from the source, Ecuador
| Water source | Household containers, no. samples | Source, no. samples |
|---|---|---|
| Harvested rainwater | 104 | 6 |
| Unprotected well | 25 | 14 |
| Piped (untreated) | 259 | 95 |
| Surface (river) | 122 | 49 |
| Surface (small stream) | 117 | 16 |
| Total | 627 | 180 |
Village and household water samples were collected in conjunction with a case–control study of diarrhea incidence in each village (for details on the study, see Eisenberg and others11). Each village was visited for 15 days and during this period all cases of diarrhea (defined as three loose stools in a 24-hour period) were identified through daily visits to the households in the community. For every household with a case of diarrhea, a control household without a case of diarrhea was randomly selected from a subset of the households with non-symptomatic persons enrolled in the case–control study. Household drinking water samples and source water samples were collected for as many case and control households as possible (Table 2). The Institutional Review Boards of the University of California at Berkeley and Universidad San Francisco de Quito approved all contact with human subjects.
Table 2.
Characteristics of study villages at the time of each visit*
| Village | River basin | Water system | No. households | Population | Month/year of visit | No. cases | Two-week prevalence per household | No. case households sampled | No. control households sampled |
|---|---|---|---|---|---|---|---|---|---|
| Borbón | Santiago-Cayapas-Onzole | Piped (pre-water treatment plant) | 202† | 877† | 7/2005 | 40 | 0.20 | 8 | 8 |
| Borbón | Santiago-Cayapas-Onzole | Piped (post-water treatment plant) | 202† | 862† | 3/2006 | 25 | 0.12 | 23 | 24 |
| Colon Eloy | Estero Maria | Surface | 170 | 709 | 8/2005 | 16 | 0.09 | 12 | 18 |
| Playa de Oro | Santiago | Piped surface | 56 | 235 | 9/2005 | 3 | 0.05 | 3 | 15 |
| Guayabal | Santiago | Surface | 27 | 107 | 9/2005 | 3 | 0.11 | 3 | 7 |
| Telembí | Cayapas | Piped surface | 46 | 268 | 3/2005 | 7 | 0.39 | 6 | 15 |
| Telembí | Cayapas | Piped surface | 58 | 290 | 1/2006 | 12 | 0.26 | 11 | 12 |
| Trinidad | Cayapas | Surface | 15 | 79 | 3/2005 | 1 | 0.07 | 1 | 12 |
| Trinidad | Cayapas | Surface | 18 | 89 | 1/2006 | 3 | 0.05 | 3 | 3 |
A water treatment plant was installed in Borbón between the first and the second visits.
These numbers represent a sample of the population of approximately 5,000 persons living in approximately 1,000 houses.
Sample processing
Samples were collected in a manner consistent with the way users collect and serve their drinking water. All samples were collected in Whirl-Pak bags (Nasco, Fort Atkinson, WI) and kept on ice until processed within 24 hours. A field laboratory was set up in a house or health dispensary in the villages in which samples were collected by using a bunsen burner inside a modular field hood made from plexiglass and metal to avoid contamination. Enterococci plates were incubated at 41 ± 2°C by using an egg incubator and generator where electricity was not available, and E. coli plates were incubated at ambient temperatures (30 ± 2°C). Agar plates were poured at a microbiology laboratory in Quito, wrapped individually in Parafilm (Pechiney Plastic Packaging Co., Chicago, IL), packaged in plastic bags, and then transferred to the field site in coolers. Five assays of water quality were used: Petrifilms (3M, St. Paul, MN) for E. coli, membrane filtration for E. coli using m-Coliblue medium (Millipore, Billerica, MA) and mI agar (BD Difco, Sparks, MD), membrane filtration for enterococci by using membrane filtration with membrane Enterococcus indoxyl-β-D-glucoside agar (mEI agar; BD Difco), and double-layer somatic coliphage using CN-13 E. coli as the host strain (ATCC no. 700609).
It should be emphasized that testing was optimized for limitations on space during transport by bus, car, and dugout canoe. This feature is not a trivial concern when considering the types of conditions involved in setting up a field laboratory. Financial considerations may also be a concern in a developing country context. Thus, the indicator assays chosen were the best suite of indicators given the constraints of our study, which may be representative of constraints of other studies carried out in similar contexts. Below we further describe the assay techniques used.
Petrifilms
Petrifilm E. coli/total coliform plates consist of plastic films with grids that are coated with gelling agents and violet red bile nutrients, an indicator of glucuronidase activity.14 Petrifilms were inoculated with 1 mL of water spread over the gel and incubated for 24 hours at 30 ± 2°C. Blue colonies were counted as E. coli.
Membrane filtration
A sample of water was passed through a 47-mm diameter 0.45-μm cellulose filter (Millipore) and then rinsed with a phosphate-buffered saline solution (pH 7.4 ± 0.2) before transferring to a growth medium plate. The stainless steel membrane filtration apparatus (Millipore) was dipped in alcohol, flame-sterilized, and cooled between each sample. Growth medium used included mEI agar (BD Difco; prepared according to Environmental ProtectionAgency (EPA) Method 1600),15 mI agar (BD Difco; prepared according to EPA Method 1604),16 and m-Coliblue (mCB) ampules (Millipore).17 Results were read after 24 hours of incubation at 30 ± 2°C (E. coli) and 41 ± 2°C (Enterococcus spp.).
Coliphage
A 2–3-mL sample of water was mixed in a sterile tube with three drops of CN-13 E. coli (ATTC no. 700609), which was grown overnight in tryptic soy broth with 0.01% naladixic acid (Acros). Glass tubes containing 7 mL of 0.7% tryptic soy agar plus 0.2% MgSO4 were heated in a water bath and allowed to cool to approximately 50°C before adding 0.07 mL of a 0.01% solution of naladixic acid. The water and E. coli mixture was then added to the agar, agitated, and poured over 100×15-mm plates of 1.5% tryptic soy agar containing 0.2% MgSO4 and 0.01% naladixic acid. Plates were sealed with Parafilm, inverted, and incubated at air temperature (30 ± 2°C). Results were read after 18 and 24 hours of incubation. These methods were based on the enumeration procedure described in EPA Method 1601.18
Volume of water filtered
As noted above, logistical limitations of the research context limited the amount of supplies that could be transported to the field, requiring optimization of protocols for testing the water samples. Thus, serial dilutions were not possible because of the multiple tubes that would have been required to transport, and also because of considerations of tradeoffs between total numbers of samples versus the need for duplication of assays. Thus, in most cases, only one volume was processed for each sample.
For most (62–76%, depending on the culture media) sample, a volume of 10 mL was filtered through the membrane filtration unit. To minimize non-detectable results, if a sample was suspected of being particularly clean (rain water, treated drinking water), a volume of 50 mL was filtered; this occurred in 15–25% of samples (depending on the culture media). To minimize the number of results too numerous to count (TNTC), if a sample was suspected of being particularly contaminated (based on previous samples from the same source), a smaller volume (usually 5 mL but in a few cases 1 mL) was filtered; this occurred in 8–22% of samples. If the level of contamination was uncertain, more than one volume was filtered for the same sample; this only occurred in only 1% of samples. In these cases, if one of the volumes tested had a result of zero or TNTC, only the sample in the countable range was included in the analysis. If both results were in a countable range, or if both resulted in a count of zero, the result for both volumes were summed and divided by the total volume sampled. If both were TNTC, then the smaller volume was used in the denominator. In the case of petrifilms, if a sample was expected of being particularly clean, the test was carried out in triplicate; this occurred in 24% of samples. The results of the three tests were summed and divided by 3 mL. In the case of coliphage, a volume of 2 mL (23% of samples) or 3 mL (77% of samples) was used.
In all cases the number of colony-forming units (CFU) or plaque-forming units was normalized by the volume of water processed, and multiplied by 100 to obtain a standardized total count per 100 mL. Non-detected results were included in the analysis as half of the lower detection limit. A value of 450/plate was assigned to the TNTC results because the highest reported count was 400/plate. Note that these values for the lower and upper detection limits varied depending on the volume of water processed. For petrifilms, possible results therefore ranged from 16.7 CFU/100 mL (halfway between zero and the lower detection limit of 1 CFU/3 mL×100 mL) to 45,000 CFU/100 mL (the upper limit of 450 CFU/1 mL×100 mL), whereas for the membrane filtration techniques, possible results ranged from 1 CFU/100 mL (halfway between zero and the lower detection limit of 1 CFU/50 mL) and 9,000 (the upper limit of 450 CFU/5 mL×100 mL). The choice of volume to process was based on knowledge of the sample to limit the number of non-detected or TNTC results. Of the samples processed with volumes > 10 mL, only 1–3% (depending on the culture media) reached the upper detection limit, and only 0–2% of the samples processed with volumes < 10 mL reached the lower detection limit. Thus, the decision to sample a smaller or larger volume and the resulting change in the range of detectable values did not affect a large number of samples. The effect of the variability in volume of water filtered in the different assays is further discussed below.
Quality control
Some basic quality control measures were used. Daily negative controls were used with each test, and if contamination was detected the results for that assay were not used for that day; this occurred only for the coliphage assays. A duplicate of one randomly selected water sample was carried out for all tests for half of the days when laboratory work was carried out. To test the specificity of the assay for the organism being tested, several isolates were subjected to further testing. Of 117 E. coli isolates, 95% (111) were also positive for E. coli by using the Colilert (IDEXX, Westbrook, ME) presence/absence test, which is based on activity of β-glucuronidase. Using Kovac’s reagent, which tests for the ability to cleave indole from tryptophan, 91% (49 of 54 isolates) were confirmed as E. coli. Following the procedures outlined in EPA Method 1600,15 we determined that 76% (28 of 37 isolates grown on mEI agar) were enterococci.
Evaluation of results
Indicators were evaluated for performance on the basis of their association with diarrheal disease and other considerations, as outlined below. All analysis was carried out using STATA 9.0 (Stat Corp., College Station, TX).
Associations with disease
Household exposure was examined with logistic regression at several levels: 1) for samples of source water only; 2) for samples of water from the source and from household storage containers combined; 3) for only samples from household storage containers; and 4) for only samples from household storage containers identified by household members as drinking water. If exposure to indicator organisms at the household level were associated with increased diarrheal disease, we would expect the logistic regression to result in an odds ratio above unity. We would also expect an increase in this association from levels 1 to 4 as water type becomes more specific to drinking water consumed in the home.
Because more than one sample was often collected for the same household and case was defined at the household level, we pooled results for all drinking water samples for a given household and used this number in the logistic regression analysis. The log of the median CFU value was used in logistic models, and binary case status was the outcome variable. The median household value was used because it is a more robust measure of central tendency than the mean. Data were log-transformed to evaluate the effect of an order of magnitude, rather than a unit change in microbial contamination. The logistic model determines the odds ratio, which is a measure of the power of each indicator to predict diarrheal disease outcome. An odds ratio of 1 indicates that diarrheal disease is equally likely regardless of exposure to indicator organisms in the water. Any value above unity suggests that increasing exposure to indicator organisms increases the odds of a household of one of its members having a case of diarrhea.
Using the procedure of Moe and others,19 we carried out a stratified analysis to determine the risk ratio of a household becoming a case depending on five categories of exposure (0, 1–10, 11–100, 101–1,000, and > 1,000 CFUs/100 mL), and on high versus low exposure (≥ 1,000 versus < 1,000 CFUs/100 mL).
Reliability
We also compared the different indicators according to other criteria to assess the reliability of the assays. To test the ability of the indicators to detect contamination as a result of human activities above the background levels of indicator organisms, samples were taken from waters at two sites considered to be free of human contamination: 1) a fast-flowing river upstream of any human settlement; and 2) surface water flowing over a vertical rock face upstream of human traffic. The distributions of 36 samples from these relatively undisturbed sites were compared with those of surface water samples downstream of human habitation by using the Wilcoxon rank-sum (Mann-Whitney) test.
To test for reproducibility of indicator test results, repeated laboratory assays of the same sample (laboratory duplicates) and repeated samples of the same water source (field duplicates) were compared. Agreement of paired samples of 41 field duplicates and 49 laboratory duplicates were assessed by using Spearman’s rank correlation and percentage agreement of presence-absence results. Additionally, we report cross-comparisons of results of the different tests for all samples tested.
Results
A total of 1,405 samples were collected and processed, although not all samples were processed with each assay. A summary of results by water type and indicator assay is shown in Table 3. All of the sample types are poor quality water according to the World Health Organization drinking water quality guidelines of ≤ 60% of samples negative for E. coli as characteristic of a poor quality of water supply.20
Table 3.
Levels of microbial contamination in water samples, Ecuador*
| Indicator organism | Assay method | Result | All samples | Source samples | All household container samples | Household drinking water samples only |
|---|---|---|---|---|---|---|
| Escherichia coli | Membrane filtration (mI agar) | Geometric mean | 129.2 (112.7–148.2) | 409.3 (331.8–504.8) | 60.2 (49.5–73.3) | 41.9 (33.0–53.3) |
| % E. coli (–) | 15.2 | 0.07 | 22.2 | 25.1 | ||
| No. | 1,276 | 455 | 618 | 407 | ||
| Membrane filtration (mCB agar) | Geometric mean | 43.4 (35.6–52.8) | 138.3 (91.6–208.8) | 24.7 (19.4–31.5) | 18.8 (14.1–25.1) | |
| % E. coli (–) | 22.9 | 18.4 | 27.1 | 28.8 | ||
| No. | 585 | 174 | 321 | 217 | ||
| Petrifilms | Geometric mean | 201.2 (182.9–221.3) | 498.0 (422.2–587.4) | 123.5 (109.4–139.4) | 94.8 (82.0–109.6) | |
| % E. coli (–) | 36.0 | 19.2 | 48.4 | 51.4 | ||
| No. | 1,404 | 494 | 705 | 407 | ||
| Enterococci | Membrane filtration (mEI agar) | Geometric mean | 146.4 (130.2–164.6) | 327.4 (273.4–391.9) | 88.1 (74.5–104.2) | 63.9 (50.8–80.4) |
| % Enterococci (–) | 12.6 | 0.05 | 17.0 | 20.1 | ||
| No. | 1,401 | 494 | 701 | 407 | ||
| Somatic coliphage | Double-layer method | Geometric mean | 97.5 (87.7–108.4) | 198.2 (164.2–239.3) | 62.1 (54.3–71.0) | 55.9 (47.8–65.5) |
| % E. coli (–) | 38.6 | 28.8 | 47.9 | 51.0 | ||
| No. | 1,117 | 416 | 536 | 365 |
Overall geometric mean and 95% confidence intervals (colony-forming units/100mL) and proportion (%) of samples with negative results is shown for each indicator assay.
Associations with disease
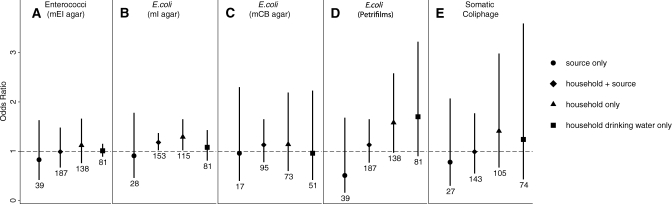
The two-week period prevalence and characteristics of each of the villages visited is summarized in Table 2. The results of the logistic analysis suggest that none of the indicator assays was a good predictor of diarrheal disease outcome in this setting. Almost all of the 95% confidence intervals (CIs) of the odds ratios cross unity (Figure 1), suggesting that a one order of magnitude increase in exposure to indicator organisms did not have a significant effect on disease outcome. The only exception was for E. coli measured by mI medium, for which the entire 95% CI was > 1.0 for samples from household containers and for samples from household containers and source waters for a particular house. Using this assay, we found that a one unit increase in log10 household median counts of E. coli in household containers resulted in a 1.29 (95% CI = 1.02–1.65) increased odds of a household having a case of diarrhea; a one unit increase in log10 household median counts of E. coli in household containers combined with samples collected at the source resulted in a 1.18 (95% CI = 1.02–1.37) increased odds of a case of diarrhea in the household. For all of the estimates of association between exposure to indicator organisms in source water and diarrheal disease outcome, the point estimate of the odds ratio is < 1.0, but the 95% CI crosses unity. Stratified analyses did not show a dose-response effect with different categories or a threshold effect with two categories of exposure (≤1,000 versus > 1,000 CFUs/100 mL).
Figure 1.
Comparison of indicators ability to predict disease outcome, Ecuador. Odds ratios and 95% confidence intervals (error bars) are shown on the basis of values calculated with logistic regression of household case status versus the log10 of the median number of indicator organisms/100 mL for samples collected in a particular household. Results are shown for regressions using four categories of water associated with a particular household: samples collected from the original water source (circles); samples collected at the source and from containers within the household (diamonds); samples collected only from containers within the house (triangles); and samples collected from within the house that specifically were identified as drinking water (squares). Sample sizes are shown below each line.
Reliability
The assays that best distinguished between water samples from sites upstream of human contamination versus samples from sites downstream of human contamination were the three tests for E. coli: membrane filtration with mCB medium (P = 0.0001), petrifilms (P = 0.01), and mI medium (P = 0.05). A high number of enterococci was cultured from the samples taken from the vertical rock face. No significant difference was seen between upstream and downstream sites when assayed for enterococci (using mEI medium) or somatic coliphage (Table 4).
Table 4.
Ability to detect difference between samples from sites upstream versus downstream of human contamination, Ecuador*
| Organism | Assay | P | Upstream sites | Downstream sites | ||||
|---|---|---|---|---|---|---|---|---|
| No. samples | Geometric mean CFU | Median CFU (95% CI) | No. samples | Geometric mean CFU | Median CFU (95% CI) | |||
| Escherichia coli | mCB | 0.0001 | 25 | 39 | 30 (13–77) | 13 | 415 | 400 (190–72) |
| E. coli | Petrifilms | 0.01 | 36 | 176 | 217 (119–381) | 81 | 546 | 400 (300–83) |
| E. coli | mI | 0.05 | 36 | 300 | 260 (123–473) | 60 | 507 | 525 (259–74) |
| Enterococci | mEI | 0.44 | 36 | 567 | 650 (391–908) | 81 | 435 | 580 (372–20) |
| Somatic coliphage | Phage | 0.60 | 32 | 125 | 117 (33–300) | 51 | 91 | 67 (33–33) |
P value tests difference between samples by using a two-sample Wilcoxin rank-sum (Mann Whitney) test. CFU = colony-forming units; CI = confidence intervals.
Agreement of test results for all techniques assessed is shown in Table 5. The E. coli tests have the highest agreement with one another, and the coliphage has the lowest agreement with any of the other tests because it is the only non-bacterial indicator organism. These values are similar to correlations between indicators found by Moe and others,19 which ranged from 0.58 to 0.85.
Table 5.
Agreement among test results using the five assays, Ecuador
| Organism | Assay | Escherichia coli | Enterococci | Coliphage | ||
|---|---|---|---|---|---|---|
| mI medium | mCB medium | Petrifilms | mEI medium | phage | ||
| E. coli | mI medium | 1.00 (n = 1,276) | ||||
| mCB medium | ρ = 0.86, P/A = 0.87 (n = 585) | 1.00 (n = 585) | ||||
| Petrifilms | ρ = 0.83, P/A = 0.76 (n = 1,272) | ρ = 0.78, P/A = 0.76 (n = 583) | 1.00 (n = 1,404) | |||
| Enterococci | mEI medium | ρ = 0.72, P/A = 0.88 (n = 1,274) | ρ = 0.74, P/A = 0.82 (n = 585) | ρ = 0.65, P/A = 0.73 (n = 1,397) | 1.00 (n = 1,401) | |
| Coliphage | Phage | ρ = 0.60, P/A = 0.69 (n = 1,113) | ρ = 0.62, P/A = 0.70 (n = 536) | ρ = 0.57, P/A = 0.70 (n = 1,115) | ρ = 0.51, P/A = 0.66 (n = 1,112) | 1.00 (n = 1,117) |
ρ = Spearman’s rank correlation coefficient; P/A = % presence absence agreement.
The duplicate test results for both field and laboratory duplicates are shown in Table 6. Surprisingly, across all indicator assays, laboratory duplicates showed less consistency than field duplicates. Petrifilms showed the poorest consistency for laboratory duplicates. The results for mCB medium must be interpreted with caution because of the small sample sizes.
Table 6.
Consistency of test results for duplicate tests, Ecuador*
| Assay | Organism | Field duplicates | Laboratory duplicates |
|---|---|---|---|
| Membrane filtration | Enterococci (mEI agar) | ρ = 0.72, P/A = 0.88 (n = 41) | ρ = 0.68, P/A = 0.90 (n = 41) |
| Escherichia coli (mI agar) | ρ = 0.78, P/A = 0.95 (n = 41) | ρ = 0.64, P/A = 0.96 (n = 49) | |
| E. coli (mCB medium) | ρ = 0.72, P/A = 1.00 (n = 7) | ρ = 0.25, P/A = 1.00 (n = 9) | |
| Other technique | E. coli (petrifilms) | ρ = 0.78, P/A = 0.78 (n = 40) | ρ = 0.49, P/A = 0.65 (n = 49) |
| Somatic coliphage | ρ = 0.78, P/A = 0.83 (n = 36) | ρ = 0.72, P/A = 0.77 (n = 44) |
Field duplicates are duplicate samples from the same source and laboratory duplicates are duplicate assays of the same sample.
Because they were not limited by sample volume, the three membrane filtration techniques had fewer non-detectable results and therefore a higher percentage of results in a countable range compared with petrifilms and coliphage, which were limited by low sample volumes (Table 7).
Table 7.
Number and percentage of test results falling within a countable range, Ecuador
| Assay | Organism | Total | Below detection limit, No. (%) | Above detection limit, No. (%) | In countable range, No. (%) |
|---|---|---|---|---|---|
| Membrane filtration | |||||
| mEI medium | Enterococci | 1,401 | 176 (12.6) | 74 (5.3) | 1,151 (82.2) |
| mI medium | Escherichia coli | 1,276 | 194 (15.2) | 131 (10.3) | 951 (74.5) |
| mCB medium | E. coli | 585 | 134 (22.9) | 39 (6.7) | 412 (70.4) |
| Other technique | |||||
| Petrifilm | E. coli | 1,404 | 506 (36.0) | 36 (2.1) | 868 (61.8) |
| Coliphage | Somatic coliphage | 1,117 | 431 (38.6) | 10 (0.9) | 676 (60.5) |
Discussion
In this study, we evaluate a group of microbiologic indicators of water quality to assess infectious disease health risk in six villages in rural Ecuador that rely mostly on untreated drinking water. In this context, E. coli outperform the other indicator organisms tested, yet our results point to the importance of careful consideration of sample size and other study design issues when using any indicator of water quality for purposes other than evaluation of the effectiveness of water treatment processes.
Of the five assays tested, only E. coli as measured with mI agar detected a significant association with diarrheal disease outcome. The association was fairly small and inconsistent between categories of samples analyzed. Petrifilms exhibited an even greater association with disease outcome (higher point estimate for the odds ratio), but the 95% CI crossed 1.0; the CIs for this indicator assay were greater because of the larger range of values over which petrifilms were estimated (detection limit ranged from 16.7 to 45,000 CFUs/100 mL) (Figure 1).
In this study, the number of cases of diarrhea and number of household water samples might not have been high enough to detect associations between indicator organisms and diarrheal disease. This observation can be observed in the differing results for the E. coli assays using mCB medium compared with those using mI agar. The point estimates are similar, yet the CIs are greater for the mCB assays because of lower sample sizes. The estimates for petrifilms had higher point values for household samples and household drinking water samples than any other OR estimate, and the CIs were almost all > 1.0, despite the high uncertainty in the estimates (Figure 1). With even higher sample sizes, all of the assays for E. coli might show a significant relationship with disease. Conversely, the assay for enterococci had higher sample sizes than the mI assay for E. coli and still showed no association with disease outcome. The coliphage assay also had similar sample sizes to the mI assay, and had a similar sample volume compared with petrifilms, yet showed weak associations with disease. Thus, of the indicator organisms tested, E. coli appears to be the best predictor of diarrheal disease.
A similar study comparing several different indicators of fecal contamination in a tropical environment (in Cebu, The Philippines) found a significant association with disease for enterococci by using mE and testing for esculin hydrolysis on esculin-iron agar, and for E. coli by using mTEC agar.20 This study used a serial cross-sectional case study design, and had the advantage of a higher total number of cases of diarrhea (277) and number of water samples tested (> 2,000), which may have led to the increased significance of results. Using the same analysis techniques as used here (logistic regression of case status versus log10 of the indicator density per 100 mL), the authors found an odds ratio of 1.18 (95% CI = 1.05–1.31) and 1.14 (95% CI = 0.99–1.31) for case status as predicted by E. coli and enterococci, respectively. The results also showed an exposure threshold effect of water quality on disease rates, with exposure ≥ 1,000 CFUs/100 mL associated with an increased risk of disease. A similar categorical threshold effect was not observed here, but this might have been caused by the lower number of samples in the highest exposure category.
In addition to a larger sample size, another difference from the study of Moe and others19 is the types of water samples tested. In that study, the water sources included protected springs, wells, boreholes, and a piped water supply, as opposed to the unprotected surface waters in our Ecuador site. Because the initial source waters were cleaner, Moe and others19 may have had increased power to detect a differential effect of clean versus contaminated waters.
All of the indicator organisms used in this study are excreted by warm-blooded animals,21 and also have the potential to grow in the environment, especially in tropical climates.6–10 However, indicator organisms may still be able to identify recent contamination from human feces if the relative level of contamination is higher from humans compared with the other sources. In our field site, enterococci or somatic coliphage could not distinguish between contamination levels in samples taken from sites upstream of human settlement versus sites downstream or in the midst of human settlements that were subject to ample sources of human fecal contamination given the sanitation conditions in the villages. The three E. coli tests distinguished between the two sites, but geometric mean counts of 39–300 CFUs/100 mL were still observed in the upstream sites.
The assay for enterococci was less successful than that for E. coli in detecting a contamination gradient,22 suggesting that it might be more naturally prevalent in surface waters of the study region. The ratio of fecal streptococci to fecal coliform is known to be higher in most animal feces than in human feces,23 which might explain the higher enterococci (a subset of fecal streptococci) concentrations that we found in upstream waters, which were likely contaminated by animal feces. Thus, enterococci may not be a good indicator of drinking water quality for untreated surface water sources compared with the protected sources studied by Moe and others.19 In addition, in our own laboratory spiking experiments, enterococci were consistently absent from the feces of one person,22 and in other studies of human populations enterococci were absent from both neonates and adults,24 which is disconcerting, given that presence in human feces is a key requirement of indicator organisms. In a review of pathogen-indicator correlations in water samples, enterococci were the only indicators that were not significantly correlated with pathogens.25 For these reasons, we do not recommend enterococci as an indicator of household or source drinking water quality for untreated surface waters in tropical regions.
There was little basis for differentiating the assays by consistency of results (Tables 5 and 6). None of the assays were perfect, and the E. coli results for petrifilms had low consistency for laboratory duplicates. Not enough samples were duplicated for the samples tested using mCB agar to evaluate this assay for consistency.
The three membrane filtration techniques had a higher percentage of results in a countable range as compared with petrifilms and coliphage because the latter two techniques were limited by low sample volumes (Table 7). In situations with high levels of contamination, such as found in our study area, these low volume techniques can be useful for water quality testing, but in sites with improved water supplies their utility would be limited. A previous study comparing petrifilms to mCB, mTEC, and the IDEXX Quanti-Tray system, which all assay E. coli, found comparable results when 1-mL samples were consistently used.26
As discussed above, ease of use in a field situation is of paramount importance for research in remote regions or locations without a developed infrastructure, and cost may also be a factor. Because of the amount of material required and the complexity of the protocols involved (maintaining a host organism, warming up agar tubes), coliphage was the most difficult test to carry out in the field. In situations where better laboratory facilities are available, coliphage (specifically F-specific RNA coliphages) might prove more useful.27 Of the three membrane filtration techniques, mCB was the easiest to use because no medium had to be prepared ahead of time, as with the mI and mEI agars. However, other researchers have found problems with this culture medium.28,29 Petrifilms were the easiest test to carry out and could be handled by someone with little microbiology experience. Petrifilms had the lowest cost ($1.00/test), followed by membrane filtration with mI agar ($1.40/test), coliphage ($1.80/test), membrane filtration with mCB ($2.20/test), and mEI ($2.40/test).
Our study was limited by logistical constraints of our field setting. Thus, some of the variability may have been contributed by field laboratory conditions. For example, our E. coli assays were incubated at ambient air temperatures, whereas the commercial tests and media used have been optimized for use at 35 ± 2°C. However, other assessments suggest that reliable results are found when incubating E. coli at these or similar temperatures with the same culture media.30,31
Petrifilms can also be useful as a pedagologic tool for community educational seminars about drinking water quality. We used petrifilms to teach community health workers with no microbiology training about water contamination and the effects of water treatment. They proved to be an effective teaching tool because the health workers could inoculate the plates themselves and observe bacterial growth in their water supply.
This is not the first study in which weak or no relationships have been found between indicators of water quality and incidence of diarrheal disease, and we believe this finding warrants extensive consideration of the use of indicators in studies of waterborne disease. In a systematic review of water quality and diarrhea, no clear relationship was found between diarrhea and microbial contamination of drinking water at the point of use.2 Jensen and others28 also found no association between bacterial drinking water quality and incidence of diarrhea in a one-year prospective study in rural Pakistan. In rural India, a recent study identified positive but non-statistically significant associations between diarrhea in children less than five years old and H2S-producing bacteria and E. coli indicators (Colford JM Jr., unpublished data). Even in temperate areas, there is poor evidence relating indicators of drinking water quality to health outcomes.3
There are several possible explanations for the lack of association observed between the indicators assessed and diarrheal disease. First, not all diarrheal disease pathogens are transmitted exclusively via water. The bacterial, viral, and protozoan agents of diarrheal illness can also be transmitted by food, fomites, personal contact, and in some cases via droplets in air. By measuring relative levels of contamination of water, we are therefore focusing only on one of several possible routes of exposure to these enteric pathogens. Thus, we should expect water contamination to explain only a portion of disease incidence.
Second, measuring the fraction of diarrheal disease attributable to waterborne transmission suffers from a lack of specificity in measures of outcome and exposure. Diarrhea, the outcome of interest, is a non-specific symptom, rather than a disease caused by one particular etiologic agent. Only a fraction of diarrheal illness can be traced to a particular infectious agent. Some diarrhea is known to be non-infectious in nature and rather caused by lactose intolerance, irritable bowel syndrome, and various other conditions. Nutritional status, immunologic status, and genetic factors of a person also play a large role in determining disease outcome.
Third and less often recognized is the inherent difficulty in measuring exposure. By definition indicator organisms, the current most often used measures of exposure to microbiologic quality of water are only a proxy for pathogens. In a recent review of the literature analyzing a dataset of 540 pathogen-indicator relationships published during 1970–2009, Wu and others25 found that only 41% showed correlations between indicators and pathogens. A comprehensive report on indicators for waterborne pathogens by the U.S. National Research Council also noted the lack of evidence correlating indicators of waterborne pathogens, the pathogens themselves, and adverse human health effects.3
Measurement variability is also a serious issue in assessing water quality, the exposure of interest. This finding is especially relevant when assessing surface water supplies but also applies to household water samples. Household water quality may be influenced by original source water quality, turbidity, matrix effects, periodic contamination events, die-off, and other factors. High variability in measurements of indicator concentrations have been observed in this study region32 and in temperate coastal waters33 from short to long timescales. Wu and others34 found that the density of E. coli in an urban watershed in Massachusetts displayed a high degree of spatial and temporal variability, and was highly influenced by both weather and surrounding land use. Wide variability has been observed between the ten U.S. EPA–approved tests to detect and quantify total coliforms and E. coli.29
Because of the inherent noise in the system caused by such extreme variability, sample size is a critical consideration when carrying out studies of water quality and health. In the review by Wu and others,25 the most important factor in determining correlations between indicator-pathogen pairs were the sample size and the number of samples positive for pathogens. Pathogen sources, detection methods, and other variables had little influence.
Most epidemiologic studies have been based on a low water quality sampling frequency, and sampling is not carried out over a long period of time. To gain more accurate results, researchers should take measurement variability into account when designing and powering studies that aim to detect an association between water quality indicators and health outcomes, and adjusting the sampling frequency as needed to properly characterize the microbial contamination of water samples according to the noise inherent in the water quality samples in their system.
The practice of using indicator organisms to detect fecal contamination was developed, mostly in temperate areas, to suggest the presence of pathogens.21 Indicators have subsequently been applied in other contexts, such as studies using indicators as a proxy measurement to assess the relative risk of disease under differing conditions, which may not be appropriate given the lack of evidence supporting strong associations between indicator organisms and pathogens that represent true disease risk.
Given the problems with measuring indicators of fecal contamination as a proxy for disease risk, indicator organisms can still be useful as a comparative tool when examining environmental contamination. For example, these organisms can be used to test relative contamination levels of different water sources or containers from source waters with similar background levels of indicators to assess recontamination in the household and to assess seasonal changes in source waters. Indicator organisms can also be quite useful when assessing the effectiveness or efficacy of in-home treatment devices or source water supply improvements, as process indicators about whether a device works, and also for monitoring for intrusion of sewage into water delivery systems.
In this study location, we have carried out several studies using indicator organisms to understand patterns in-home and source water quality and treatment efficacy and effectiveness. In one study, we observed a decrease in indicator bacteria after water was transferred from the source to household storage containers, but then an increase caused by recontamination of water in the home, providing insights into the phenomenon of in-home recontamination.35 In another, despite extensive variability in indicator counts in source waters observed on an hourly and daily basis (> 2 log difference), we were able to observe seasonal differences in counts and relate these differences to environmental and anthropogenic drivers of contamination, and the different time scales at which these factors act.32
In this study, E. coli indicators performed better overall than the other indicator organisms with respect to associations with disease outcomes, reliability, and field utility. Given the extensive variability inherent in indicator measurements, the fact that we were able to detect some associations between household drinking water and diarrheal disease outcome suggests that the associations may exist. However, higher sample sizes are needed to account for the inherent variability in water quality exposure measurements using indicator organisms. Future studies should take this variability into account when designing studies relating water quality to health outcomes. Researchers must recognize that indicator organisms are proxy measures that may be too blunt a tool for the purposes of water and health research.
Review of the literature provides weak evidence to support the idea that microbial indicators and health risk are associated. On the basis of this finding and consistent with the results of this case study, researchers should exercise caution in using indicators as direct proxies for evidence of health risk. Further work is necessary to create more precise ways of studying the role of water in the transmission of waterborne disease, including development of time and cost-effective methods for detecting pathogens directly, microbial source tracking, and other methods, especially in resource-limited settings.
ACKNOWLEDGMENTS
We thank Khalid Kadir and Sarah Brownell (University of California, Berkeley, CA), Rick Danielson (BioVir Laboratories, Inc., Benicia, CA), and Scott Meschke (University of Washington, Seattle, WA) for help with elaboration of field methods and research ideas; Patricio Bueno, Junior Mina Estupiñan, Sangam Tiwari, and Owen Solberg for assisting with processing water samples; Geovanny Hurtado, Maritza Renteria, Deni David Tenorio, Chelo Ortiz, and Melecio Quintero for helping to collect water samples; and Karina Ponce, William Cevallos, Sarah Bates, and Laura McLaughlin for providing considerable logistical and technical support in the field.
Footnotes
Financial support: This study was supported by grants from the National Institute of Allergy and Infectious Diseases (RO1AI050038), the University of California Pacific Rim Research Program, and the University of California Center for Occupational and Environmental Health.
Authors’ addresses: Karen Levy, Department of Environmental Health, Emory University Rollins School of Public Health, Atlanta, GA, E-mail: karen.levy@emory.edu. Kara L. Nelson, Department of Civil and Environmental Engineering, University of California, Berkeley, CA, E-mail: nelson@ce.berkeley.edu. Alan Hubbard, Division of Biostatistics, University of California Berkeley School of Public Health, Berkeley, CA, E-mail: hubbard@stat.berkeley.edu. Joseph N. S. Eisenberg, Department of Epidemiology, University of Michigan School of Public Health, Ann Arbor, MI, E-mail: jnse@umich.edu.
References
- 1.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- 2.Gundry S, Wright J, Conroy R. A systematic review of health outcomes related to household water quality in developing countries. J Water Health. 2004;2:1–13. [PubMed] [Google Scholar]
- 3.National Research Council . Health Effects Assessment. Committee on Indicators for Waterborne Pathogens; Board on Life Sciences; Water Science and Technology Board, ed. Indicators for Waterborne Pathogens. Washington, DC: The National Academies Press; 2005. pp. 53–108. [Google Scholar]
- 4.Clasen T, Schmidt WP, Rabie T, Roberts I, Cairncross S. Interventions to improve water quality for preventing diarrhoea: systematic review and meta-analysis. BMJ. 2007;334:782–791. doi: 10.1136/bmj.39118.489931.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fewtrell L, Kaufmann RB, Kay D, Enanoria W, Haller L, Colford JM., Jr Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect Dis. 2005;5:42–52. doi: 10.1016/S1473-3099(04)01253-8. [DOI] [PubMed] [Google Scholar]
- 6.Hazen TC, Toraznos GA. In: Drinking Water Microbiology: Progress and Recent Developments. GA McFeters., editor. New York: 1990. pp. 32–53. (Tropical source water). [Google Scholar]
- 7.Rivera S, Hazen TC, Toranzos GA. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl Environ Microbiol. 1988;54:513–517. doi: 10.1128/aem.54.2.513-517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. Sources of Escherichia coli in a coastal subtropical environment. Appl Environ Microbiol. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toranzos GA. Current and possible alternate indicators of fecal contamination in tropical waters: a short review. Environ Toxicol Water Qual. 1991;6:121–130. [Google Scholar]
- 10.Fujioka RS, Tenno K, Kansako S. Naturally occurring fecal coliforms and fecal streptococci in Hawaii's freshwater streams. Toxicity Assessment. 1988;3:613–630. [Google Scholar]
- 11.Eisenberg JN, Cevallos W, Ponce K, Levy K, Bates SJ, Scott JC, Hubbard A, Vieira N, Endara P, Espinel M, Trueba G, Riley LW, Trostle J. Environmental change and infectious disease: how new roads affect the transmission of diarrheal pathogens in rural Ecuador. Proc Natl Acad Sci USA. 2006;103:19460–19465. doi: 10.1073/pnas.0609431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endara P, Trueba G, Solberg OD, Bates SJ, Ponce K, Cevallos W, Matthijnssens J, Eisenberg JNS. Symptomatic and subclinical infection with rotavirus P[8]G9, rural Ecuador. Emerg Infect Dis. 2007;13:574–580. doi: 10.3201/eid1304.061285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieira N, Bates SJ, Solberg OD, Ponce K, Howsmon R, Cevallos W, Trueba G, Riley L, Eisenberg JNS. High prevalence of enteroinvasive Escherichia coli isolated in a remote region of northern coastal Ecuador. Am J Trop Med Hyg. 2007;76:528–533. [PMC free article] [PubMed] [Google Scholar]
- 14.3M Petrifilm E. coli/Coliform Count Plate Interpretation Guide. 2007. http://www.3m.com/microbiology Available at. Accessed May 1, 2007.
- 15.US EPA . Method 1600: Enterococci in Water by Membrane Filtration Using Membrane-Enterococcus Indoxyl-β-d-Glucoside Agar (mEI) Washington, DC: U.S. Environmental Protection Agency Office of Water; 2002. [Google Scholar]
- 16.US EPA . Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium) Washington, DC: U.S. Environmental Protection Agency Office of Water; 2002. [Google Scholar]
- 17.US EPA . National Primary and Secondary Drinking Water Regulations: Analytical Methods for Chemical and Microbiological Contaminants and Revisions to Laboratory Certification Requirements; Final Rule. Washington, DC: U.S. Environmental Protection Agency, Federal Register; 1999. pp. 67449–67467. [Google Scholar]
- 18.US EPA . Method 1601: Male-specific (F+) and Somatic Coliphage in Water by Two-Step Enrichment Procedure. Washington, DC: U.S. Environmental Protection Agency Office of Water.; 2001. [Google Scholar]
- 19.Moe C, Sobsey M, Samsa G, Mesolo V. Bacterial indicators of risk of diarrhoeal disease from drinking-water in the Philippines. Bull World Health Organ. 1991;69:305–317. [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization . Chapter 5: Surveillance. Guidelines for Drinking-Water Quality. Geneva: World Health Organization; 2008. [Google Scholar]
- 21.Ashbolt NJ, Grabow WOK, Snozzi M. Fewtrell L, Bartram J. Water Quality: Guidelines, Standards and Health. London: World Health Organization/IWA Publishing; 2001. Indicators of microbial water quality; pp. 289–314. [Google Scholar]
- 22.Levy K. Environmental Determinants of Water Quality and Waterborne Disease in the Tropics with a Particular Focus on Northern Coastal Ecuador. Environmental Science, Policy, and Management. Berkeley, CA: University of California, Berkeley; 2007. [Google Scholar]
- 23.Geldreich EE, Kenner BA. Concepts of faecal streptococci in stream pollution. J Water Pollut Control Fed. 1969;41:R336–R352. [PubMed] [Google Scholar]
- 24.Chenoweth C, Schaberg D. The epidemiology of enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:80–89. doi: 10.1007/BF01963631. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Long SC, Das D, Dorner SM. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J Water Health. 2011;9:265–278. doi: 10.2166/wh.2011.117. [DOI] [PubMed] [Google Scholar]
- 26.Vail JH, Morgan R, Merino CR, Gonzales F, Miller R, Ram JL. Enumeration of waterborne Escherichia coli with petrifilm plates: comparison to standard methods. J Environ Qual. 2003;32:368–373. doi: 10.2134/jeq2003.3680. [DOI] [PubMed] [Google Scholar]
- 27.Luther K, Fujioka R. Usefulness of monitoring tropical streams for male-specific RNA coliphages. J Water Health. 2004;2:171–181. [PubMed] [Google Scholar]
- 28.Jensen PK, Jayasinghe G, van der Hoek W, Cairncross S, Dalsgaard A. Is there an association between bacteriological drinking water quality and childhood diarrhoea in developing countries? Trop Med Int Health. 2004;9:1210–1215. doi: 10.1111/j.1365-3156.2004.01329.x. [DOI] [PubMed] [Google Scholar]
- 29.Olstadt J, Schauer JJ, Standridge J, Kluender S. A comparison of ten USEPA approved total coliform/E. coli tests. J Water Health. 2007;5:267–282. [PubMed] [Google Scholar]
- 30.Allen E, Dillman L, Weigl B. Efficacy of Standard Methods for Detection of Fecal Contamination at a Range of Nonstandard Temperatures. Seattle, WA: USAID and PATH; 2007. [Google Scholar]
- 31.Brown J, Stauber CE, Murphy JL, Khan A, Mu T, Elliott M, Sobsey MD. Ambient-temperature incubation for the field detection of Escherichia coli in drinking water. J Appl Microbiol. 2011;110:915–923. doi: 10.1111/j.1365-2672.2011.04940.x. [DOI] [PubMed] [Google Scholar]
- 32.Levy K, Hubbard A, Nelson KL, Eisenberg JNS. Drivers of water quality variability in northern coastal Ecuador. Environ Sci Technol. 2009;43:1788–1797. doi: 10.1021/es8022545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehm AB, Grant SB, Kim JH, Mowbray SL, McGee CD, Clark CD, Foley DM, Wellman DE. Decadal and shorter period variability of surf zone water quality at Huntington Beach, California. Environ Sci Technol. 2002;36:3885–3892. doi: 10.1021/es020524u. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Rees P, Dorner SM. Variability of E. coli density and sources in an urban watershed. J Water Health. 2011;9:94–106. doi: 10.2166/wh.2010.063. [DOI] [PubMed] [Google Scholar]
- 35.Levy K, Nelson KL, Hubbard A, Eisenberg JN. Following the water: a controlled study of drinking water storage in northern coastal Ecuador. Environ Health Perspect. 2008;116:1533–1540. doi: 10.1289/ehp.11296. [DOI] [PMC free article] [PubMed] [Google Scholar]